Abstract
The genes MPN141 and MPN142 encode the major adhesin P1 and the cytadherence-related B/C proteins (P90/P40), respectively, in Mycoplasma pneumoniae. Using reverse transcriptase PCR we found open reading frames MPN140 to MPN142 constitute a polycistronic transcriptional unit. Cytadherence mutant IV-22 has a frameshift mutation in MPN141 and lacks the P1, B, or C proteins. Recombinant MPN141 and/or MPN142 were introduced into mutant IV-22 by transposon delivery in several configurations, and the levels of the P1, B, and C proteins were assessed by immunoblotting. MPN142 in mutant IV-22 has a wild-type nucleotide sequence, yet the introduction of recombinant MPN141 alone to mutant IV-22, although it restored P1 levels, failed to restore levels of B or C. In contrast, recombinant MPN141 and MPN142 delivered in cis or in trans were sufficient to restore all three proteins. Taken together, our data indicated that some but not all synthesis of B or C is dependent on coupling to the translation of P1 immediately upstream of MPN142 and demonstrated that proteins B and C are not stable in the absence of P1. The linkage of MPN141 and MPN142 at the levels of transcription, translation, and protein stability, in addition to their previously demonstrated colocalization and the requirement of B and/or C for P1 function, reinforces the conclusion that these proteins constitute a multiprotein complex that functions in receptor binding.
The cell wall-less bacterium Mycoplasma pneumoniae is a common agent of respiratory infections, including tracheobronchitis, pharyngitis, and atypical or “walking” pneumonia in humans (29). A crucial event in the initiation of infection is the intimate attachment of the bacteria to the respiratory epithelium (cytadherence). This process is essential to pathogenesis, and cytadherence mutants are avirulent (17). M. pneumoniae possesses a differentiated polar extension of the cell body called the attachment organelle, which also plays a role in cell division and gliding motility (25). Many of the proteins required for cytadherence are located at the attachment organelle, and their proper localization is important for attachment organelle function (14). P1 is a large integral membrane protein found clustered primarily, but not exclusively, at the attachment organelle of M. pneumoniae and is a major cytadhesin (3, 5, 11). Localization of P1 to the attachment organelle is necessary but not sufficient for function, and the generation of properly localized and functional P1 appears to be a complex process requiring many accessory proteins (14).
The gene encoding P1 (MPN141) is the second of three genes thought to constitute a transcriptional unit expressed from a single promoter upstream of the first gene and having a predicted stem-loop terminator following the third (Fig. 1A) (12). MPN140, the first gene of the three and also known as ORF4, encodes a predicted phosphoesterase (1). The presence but not the predicted biochemical activity of the MPN140 gene product has been demonstrated (13). Immediately downstream of MPN141 is MPN142, also known as ORF6. The MPN142 gene product has a predicted size of 130 kDa but is cleaved to yield two polypeptides of sizes 90 and 40 kDa (18) also known as P90 and P40 (26) or B and C (9), respectively. Interestingly, the gene products of MPN142 homologs in the closely related Mycoplasma genitalium and Mycoplasma gallisepticum are present as single polypeptides (20, 22), indicating that the roles of B and C are highly interrelated. Proteins B and C are integral membrane proteins located at the attachment organelle in proximity to P1 (6, 19), are required for P1 function, and with P1 likely constitute a major adhesin complex.
FIG. 1.
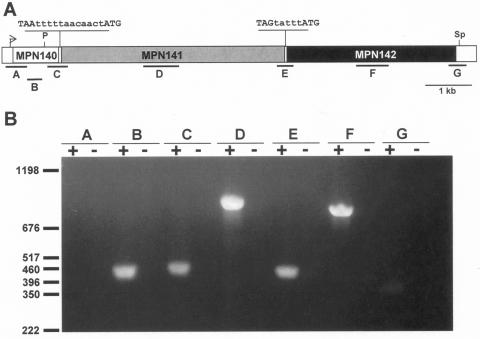
RT-PCR analysis of the MPN140 to MPN142 locus of M. pneumoniae. (A) Map of the locus in M. pneumoniae. The arrow indicates transcriptional promoter upstream of MPN140. The intergenic sequences are shown above the map, with 12 and 5 nt (in lowercase) separating MPN140 and MPN141, and MPN141 and MPN142, respectively. A predicted stem-loop terminator (not indicated) begins 17 nt downstream of MPN142. Locations corresponding to annealing sites of oligonucleotide primer sets A-G (Table 2) used for RT-PCR are shown below the map. Restriction enzyme sites for PmeI (P) and SphI (Sp) referenced in Fig. 2 are shown. Scale bar is 1 kb. (B) Agarose gel electrophoresis of RT-PCR products. RT-PCRs for primer pairs A-G with wild-type M. pneumoniae RNA template are shown. RT was included (+) or omitted (−). Locations of DNA markers and their sizes in base pairs are indicated.
Ribosome-binding sites in Mycoplasma are poorly defined. Only five nucleotides separate the stop codon of MPN141 and the start of MPN142 (12), raising the possibility that the two are translationally linked. This scenario is supported by both the nature of the P1/B/C complex, which implies a role for stoichiometric protein levels, and the observation that a noncytadherent M. pneumoniae mutant designated IV-22, which does not make P1 due to a frameshift in MPN141, also lacks proteins B and C. Noncytadherent mutant III-4 has a frameshift in MPN142 and lacks proteins B and C but produces wild-type levels of P1 (Table 1). Mutants III-4 and IV-22 also each lack the 70-kDa protein A (9, 17), for which the gene is unknown. However, the noncytadherent phenotype of mutant III-4 is not due to the loss of protein A (30), and the same is probably true for mutant IV-22.
TABLE 1.
Strains examined in this study
| Strain | Relevant genotypea | Relevant phenotypeb
|
Source or reference | |
|---|---|---|---|---|
| P1 (MPN141) | B/C (MPN142) | |||
| M129 | Wild type | ++ | ++ | 17 |
| III-4 | MPN142 frameshift | ++ | − | 17, 30 |
| IV-22 | MPN141 frameshift | − | − | 17, 27 |
| III-4 + pKV258 | III-4 + MPN141 and MPN142* | ++ | ++ | 30 |
| III-4 + pKV264 | III-4 + MPN142 | ++ | ++ | 30 |
| III-4 + pKV265 | III-4 + del MPN141† and MPN142 | ++ | + | This study |
| IV-22 + pKV258 | IV-22 + MPN141 and MPN142 | ++ | ++ | This study |
| IV-22 + pKV264 | IV-22 + MPN142 | − | +/− | This study |
| IV-22 + pKV265 | IV-22 + delMPN141† and MPN142 | − | − | This study |
| IV-22 + pKV299 | IV-22 + MPN141 | ++ | +/− | This study |
| IV-22 + pKV299 and pKV264 | IV-22 + MPN141 and MPN142‡ | ++ | ++ | This study |
*, All restored genes are recombinant and delivered by transposons borne by the indicated plasmids; †, the 5′ end of MPN141 is removed, rendering it untranslatable; ‡, MPN141 and MPN142 were delivered by different transposons.
++, Protein present at or near wild-type levels; +, protein present at reduced levels; +/−, protein present at trace levels; −, protein undetected.
We examined here in greater detail the relationship of MPN141 and MPN142. Analysis by reverse transcriptase PCR (RT-PCR) demonstrated that genes MPN140 to MPN142 are transcribed as a unit. Furthermore, using a series of genetic studies with recombinant MPN141 and MPN142 in mutants IV-22 and III-4, we examined the factors involved in their translation initiation and the interrelated fates of the P1, B, and C proteins. The results strongly suggested that some but not all translation of MPN142 is coupled to translation of MPN141 and that proteins B and C are unstable and quickly degraded in the absence of P1.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The M. pneumoniae strains used in the present study included wild-type M129 (broth passage 18) and noncytadherent mutants III-4 and IV-22 derived from M129 (17). Cultures of mutant IV-22 were initially found to possess trace levels of the B and C proteins (data not shown). Upon filter cloning (28), this phenotype disappeared and was likely due to a subpopulation of spontaneous revertants (16). The filter-cloned strain was used for all experiments. Cultures were grown and harvested in Hayflick broth and PPLO agar, as described previously (7). Transformations with Tn4001mod-bearing suicide vectors were performed as previously described (10). Gentamicin (Gm) was included at 18 μg/ml for selection and maintenance of Tn4001mod (10), while chloramphenicol (Cm) was included at 12 or 24 μg/ml for selection and maintenance, respectively, of Tn4001cat (8). To generate strains carrying both transposons, M. pneumoniae cells were electroporated and transformants isolated sequentially, first with Tn4001mod and then Tn4001cat constructs. In all cases multiple transformants from independent transformations were isolated and characterized, with representative results shown here. The Escherichia coli strains used to construct plasmids were Sure (Stratagene, La Jolla, Calif.) and JM109 (Promega, Madison Wis.), and ampicillin resistance was selected on Luria media with 50 μg of antibiotic/ml.
RNA preparation and RT-PCR.
All RNA experiments used RNase-free reagents, and labware was treated to eliminate RNase (24). RNA was purified from 10-ml mid-logarithmic-phase cultures by using the RNAqueous-4PCR kit (Ambion, Austin, Tex.) according to the instructions, except that the DNase treatment required 10% (vol/vol) enzyme incubated for 2 h. The boundaries and gene linkage for the transcriptional unit encompassing MPN142 were analyzed by RT-PCR using the Access RT-PCR kit (Promega), with amplicons visualized by using agarose gel electrophoresis and ethidium bromide staining. PCR primers were designed to amplify regions 407 to 812 nucleotides in length which were internal to each gene of the predicted transcriptional unit, spanned their junctions, or were outside its predicted limits (Table 2) (Fig. 1A). Controls without RT were used to detect residual DNA contamination. RNA yields were below the limit of detection of UV spectroscopy, and the amounts necessary for RT-PCR were determined empirically with equal volumes used in each set of reactions for a given preparation.
TABLE 2.
Summary of RT-PCR analysis of the P1 operon
| Set/location | nta | Primer sequence | Results
|
|||
|---|---|---|---|---|---|---|
| Size (bp) | RT-PCR
|
PCR | ||||
| +RT | −RT | |||||
| A/MPN140 upstream | (+) −73 | 5′-TTTTTGCACCAAAATGGCG | 468 | + | − | + |
| (−) 395 | 5′-CATCAACGAAGTTATGGGTCG | |||||
| B/MPN140 internal | (+) 332 | 5′-ACCAAAACCAGTTTAAGGCAG | 407 | + | − | + |
| (−) 739 | 5′-TGCACATTCTAAGGGTGTTAC | |||||
| C/MPN140-141 junction | (+) 708 | 5′-GGCCGATAAGAAGTTCCAAAAG | 420 | + | − | + |
| (−) 1128 | 5′-GGGTGTAGCTAAATTGCTGG | |||||
| D/MPN141 internal | (+) 2876 | 5′-TGCAACTCTTCACACCCTAC | 812 | + | − | + |
| (−) 3688 | 5′-GCCTTATCATTCCTTCACCC | |||||
| E/MPN141-142 junction | (+) 5642 | 5′-CAATGCACAAGAACAAACAGG | 414 | + | − | + |
| (−) 6055 | 5′-CGTATGAccccTGTAAATGGGb | |||||
| F/MPN142 internal | (+) 7424 | 5′-GACCTGATGAGCGAAAACC | 794 | + | − | + |
| (−) 8218 | 5′-GCTAATCTTCACCCCATTCC | |||||
| G/MPN142 downstream | (+) 9478 | 5′-AAGCACCAGTTAAACCAGC | 360 | +/− | − | + |
| (−) 9838 | 5′-ACCTTCTTTTTCCAGTAAGACC | |||||
Nucleotide (nt) position of 5′ end of primer in relation to the first +1 transcript position.
Due to an oversight, the minus-strand primer was synthesized with the incorrect sequence. Each lowercase nucleotide (c) should be G. However, the amplification worked, and the results are reported.
Sequencing of MPN142.
The MPN142 gene of mutant IV-22 was sequenced after PCR amplification using genomic DNA from the mutant as a template. PCR primer pairs were designed to amplify the gene and flanking sequence in five overlapping segments. These PCR products were then either sequenced directly by the same primers or cloned into pCRII by using the TA Cloning Kit (Invitrogen, Carlsbad, Calif.) and sequenced with the T7 or SP6 primers of pCRII (sequencing by IBL, University of Georgia).
Plasmid construction.
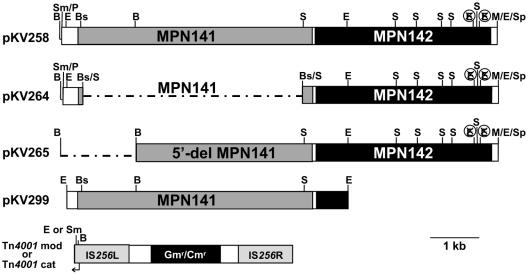
Recombinant genes can be introduced into M. pneumoniae using a modified Tn4001 (7). The plasmids used in the present study are shown in Fig. 2. Plasmids pKV258 and pKV264, whose construction is described elsewhere (30), were used here extensively. In pKV258, MPN141 and MPN142 are expressed from the POUT promoter of Tn4001. In pKV264, MPN142 is likewise expressed from this promoter. The ability to couple translation of MPN142 to MPN141 is preserved, but 95% of the MPN141 gene is missing due to an internal in-frame deletion. For pKV265, the 1.5-kb BamHI fragment of pKV258 was removed, and the remainder of the plasmid was religated, leaving MPN142 and a large region of the upstream DNA intact but disrupting the 5′ end of MPN141, along with possible translation initiation signals. Plasmid pKV104 carries a modified version of Tn4001 (Tn4001cat) for which the original gentamicin resistance gene was replaced with a chloramphenicol resistance gene (8). A 5.7-kb EcoRI fragment bearing MPN141 in pUC19 (a gift from J. Baseman) was excised and subjected to a Klenow fill-in reaction and then ligated into the SmaI site of pKV104 oriented such that MPN141 is under the control of the POUT promoter of Tn4001cat to generate pKV299.
FIG. 2.
Map of the DNA constructs cloned into IS256L of Tn4001mod of pKV74 or Tn4001cat of pKV104 to create pKV258, pKV264, pKV265, and pKV299. Except for the IS elements, all open reading frames are shown with the 5′ end on the left. The region encoding the 3′ end of MPN140 is not indicated. B, BamHI; Bs, BsaBI; E, EcoRI; S, StuI; Sm/P, SmaI/PmeI junction; M/E/Sp, former SphI site of the P1 operon replaced by MfeI linker and present as an MfeI/EcoRI junction; E or Sm, EcoRI in pKV74 or SmaI in pKV104. The barred symbol indicates a silent change to destroy the site. Gentamicin resistance (Gmr) is found in pKV74, and chloramphenicol resistance (Cmr) is found in pKV104. The arrow from Tn4001mod/cat shows the location and direction of the POUT promoter. Note that the orientation of the inserts is inverted when cloned into Tn4001mod/cat.
Analysis of cytadherence proteins and cytadherence-related phenotypes.
Wild-type, mutant, and transformant cells were analyzed for the restoration of several phenotypes related to cytadherence which were lacking in the mutants. Western blotting was performed as described previously (7) with polyclonal rabbit anti-P1 (1:1,000), anti-protein B (1:2,000), and anti-protein C (1:1,000). Anti-P1 serum was raised as described previously (15). Qualitative hemadsorption assays were performed as previously described (7), except that sheep blood was used.
RESULTS
Transcriptional analysis of the MPN140 to MPN142 locus.
RT-PCR analysis of wild-type M. pneumoniae RNA using primer pairs internal to each gene, spanning the junctions between genes, or extending upstream or downstream beyond the MPN140 to MPN142 locus (Fig. 1A and Table 2) demonstrated that the three genes are linked and likely present as a single transcriptional unit. No product was detected with primer pair A, which flanks the promoter, and a weakly positive RT-PCR was seen with primer pair G, which flanks the predicted terminator; hence, termination was not complete. Comparably abundant products were seen with all other primer pairs (Fig. 1B). Similar results were seen using RNA from mutant IV-22 (data not shown); however, quantitative comparisons were not pursued.
Restoration of proteins P1/B/C in mutant IV-22.
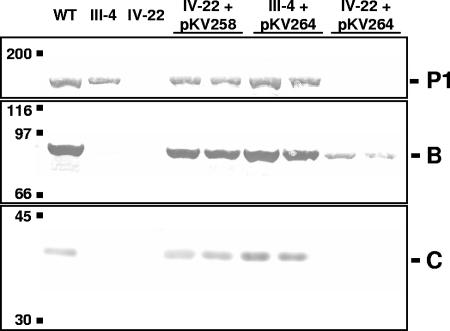
When mutant IV-22, which has a frameshift mutation in MPN141, was transformed with pKV258 (wild-type recombinant MPN141 and MPN142; Fig. 2), recombinant proteins P1, B, and C were each observed at approximately wild-type levels (Fig. 3). However, when mutant IV-22 was transformed with pKV264 (near total in-frame deletion in MPN141 and wild-type MPN142; Fig. 2), protein B was present at greatly reduced levels, while protein C was barely detectable in immunoblots and P1, as expected, remained absent (Fig. 3). In contrast, mutant III-4 transformed with pKV264 produced wild-type levels of B and C (30) (Fig. 3). Thus, the recombinant MPN142 in pKV264 is functional and expressed at approximately wild-type levels, but the fate of B and C differed between the III-4 and IV-22 mutant backgrounds.
FIG. 3.
Western blot analysis of lysates of wild-type (WT) or mutant (IV-22 or III-4) M. pneumoniae untransformed or transformed with pKV258 or pKV264. Two independent transformants are shown for each plasmid-background combination. A total of 60 μg of protein was used per lane. Blots were probed with antiserum to P1, B, or C as indicated.
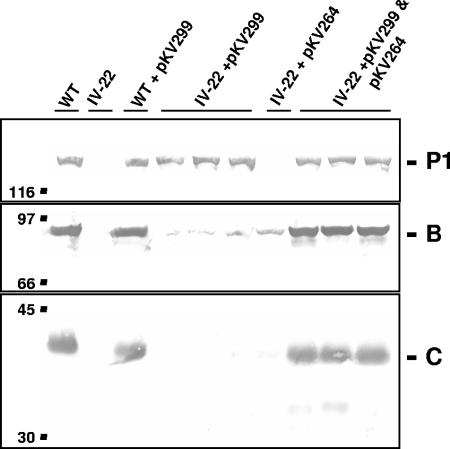
Because mutant IV-22 lacks proteins B and C while the gene for them, MPN142, is apparently wild type (data not shown), we explored whether production of recombinant P1 alone in trans was sufficient to restore B and C to wild-type levels. When mutant IV-22 was transformed with pKV299 (wild-type MPN141; Fig. 2), production of P1 was restored, trace amounts of protein B were observed, and protein C was barely detectable (Fig. 4). When mutant IV-22 was transformed with both pKV264 and pKV299 (recombinant MPN142 and MPN141, respectively), proteins P1, B, and C were fully restored (Fig. 4). An additional minor band was detected in some transformants with the C-specific antiserum, presumably representing a derivative of C. The significance of this minor band is not known.
FIG. 4.
Western blot analysis of lysates of wild-type (WT) or mutant (IV-22) M. pneumoniae untransformed or transformed with pKV264, pKV299, or pKV264 and pKV299. Three independent transformants are shown for each construct in the mutant IV-22 background except IV-22+pKV264 (see Fig. 3). A total of 60 μg of protein was used per lane. Blots were probed with antiserum to P1, B, or C as indicated.
Expression of MPN142 without translation of MPN141.
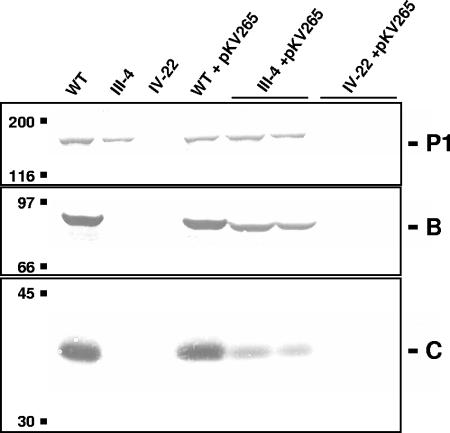
The observation that low levels of B were seen in mutant IV-22 transformed with pKV299 alone suggested that some translation of B or C was possible independent of translational coupling. This possibility was examined further using pKV265 (wild-type MPN142 but untranslatable MPN141; Fig. 2). Low levels of B or C were detected in mutant III-4 transformed with pKV265, while no B or C was detected in mutant IV-22 transformants (Fig. 5).
FIG. 5.
Western blot analysis of lysates of wild-type (WT) or mutant (IV-22 or III-4) M. pneumoniae untransformed or transformed with pKV265. Two independent transformants are shown for each construct-background combination. A total of 60 μg of protein was used per lane. Blots were probed with antiserum to P1, B, or C as indicated.
DISCUSSION
Cytadherence in M. pneumoniae is a complex process requiring many proteins that function in roles beyond simple receptor binding. The major adhesin P1 is necessary but not sufficient for attachment. A series of events involving cytadherence-accessory proteins is required for P1 function (14); these include, not necessarily in order, insertion into the cytoplasmic membrane, trafficking to the attachment organelle, folding in the correct conformation, and interacting (stably, transiently, or both) with likely partner proteins. The interrelationships between cytadherence-accessory proteins in processes leading to the formation of a functional attachment organelle and establishment of proper cellular morphology and functional P1 have been examined in detail (14). However, little is known about the relationship between P1 and the proteins B and C except that the spatial arrangement of the genes (12) and gene products (19) suggest that these proteins are linked functionally. Here we show that proteins P1, B, and C are related at the levels of transcription, translation, and protein stability.
Analysis of MPN140 to MPN142 in M. pneumoniae by RT-PCR demonstrated that these three genes constitute a transcriptional unit, as suggested by others previously (12). However, the nonquantitative nature of simple RT-PCR limits the conclusions that might be drawn about transcription and prompted our genetic approach. Proteins P1, B, and C were restored to wild-type levels in mutant IV-22 by the introduction of recombinant MPN141 and MPN142 by transposon delivery in cis (pKV258; Fig. 3) or in trans (pKV264 and pKV299; Fig. 4). When wild-type MPN142 was present downstream of translatable but nonfunctional MPN141 having a major in-frame deletion (pKV264), recombinant proteins B and C were observed at wild-type levels in the mutant III-4 background (30) (Fig. 3). However, when introduced into a P1-less background (mutant IV-22), the same construct yielded only low levels of proteins B and C. We conclude that recombinant MPN142 was expressed at normal levels in both backgrounds, but proteins B and C were unstable in the absence of P1. The converse is not seen, however, as P1 is stable in the absence of proteins B and C, as with mutant III-4 (17). We were unable to track the fate of newly synthesized proteins B and C by pulse-chase radioimmunoprecipitation analysis (data not shown). However, transposon delivery of wild-type MPN141 in trans (pKV299) into mutant IV-22 transformed with pKV264 yielded wild-type levels of B and C (Fig. 4), supporting the interpretation that recombinant MPN142 is indeed expressed normally from pKV264 in mutant IV-22 and that the stability of proteins B and C requires the presence of P1. This nonreciprocal requirement for stability is similar to that seen in M. pneumoniae with HMW3 and the two proteins HMW1 and HMW2. In that case, HMW3 is unstable if HMW1 or HMW2 levels are reduced (23, 31), but HMW1 and HMW2 are present at wild-type levels in the absence of HMW3 (32).
The MPN142 gene in mutant IV-22 has a wild-type nucleotide sequence (data not shown), but the restoration of just recombinant MPN141 in trans using pKV299 was not sufficient to restore levels of B and C fully (Fig. 4), with only trace amounts of B and little or no C detected. This is similar to what is seen in the closely related M. gallisepticum, where coexpression of the recombinant homologs for MPN141 and MPN142 is necessary (21). The MPN141 gene in mutant IV-22 encodes a drastically truncated protein (46 versus 1,627 residues) due to a frameshift (27). Once the translation machinery has encountered the premature stop codon in the corresponding MPN141 transcript the ribosome likely disassociates from the mRNA. In contrast, the translation of MPN142 in wild-type M. pneumoniae probably begins before the ribosome can disassociate. This coupling of translation between MPN141 and MPN142 would likely ensure near-equimolar amounts of the two gene products. However, when wild-type MPN141 was delivered to mutant IV-22 in trans, small amounts of proteins B and C were detected and could only have come from the native allele, suggesting one or more of the following: (i) translation of MPN142 can proceed at low levels despite premature termination of MPN141 translation, (ii) limited translation reinitiation occurs independent of coupling, or (iii) the untranslated message is less stable, resulting in less protein than would otherwise be produced. Regardless, the products (B and C) could only be seen in the presence of P1, indicating that their stability requires the presence of the adhesin.
In order to distinguish the factors driving the limited translation of MPN142 seen in mutant IV-22, we produced a construct possessing wild-type MPN142 and a large portion of upstream sequence but lacking the 5′ end of MPN141, including any likely translation initiation signals (pKV265). When pKV265 was introduced into mutant IV-22, no MPN142 products were detected (Fig. 5), as expected in the absence of P1. Mutant III-4 transformed with pKV265 produced less than wild-type amounts of proteins B and C, probably the result of translation initiation independent of coupling to MPN141, with the balance of protein B or C synthesis due to coupling. Significantly, the M. pneumoniae gene cluster encoding the cell division protein FtsZ exhibits a decrease in mRNA levels with distance from the promoter (4) and, if the same holds for the MPN140 to MPN142 gene cluster, this might account for the need for coupling-independent translation initiation.
When P1, B, and C were restored in mutant IV-22 by transformation with pKV258 or both pKV264 and pKV299, the transformants exhibited a phenotype that was clearly distinct from both wild-type and mutant IV-22 M. pneumoniae, with an intermediate morphology and attachment to plastic but no adherence to erythrocytes (data not shown). The essential role of proteins B and C in hemadsorption has recently been confirmed (30). Furthermore, cytadherence and virulence in an avirulent high-passage M. gallisepticum strain can be restored by the introduction of recombinant homologs of MPN141 and MPN142 (21), suggesting that restoration of each should be sufficient to confer an otherwise fully wild-type phenotype to the mutant IV-22 transformants. There are at least three possible explanations for the failure to restore mutant IV-22 transformants fully to a wild-type phenotype. First, transposon insertion could have affected a gene whose product was associated with cytadherence, but this seems unlikely given that multiple transformants with pKV258 were examined. Alternatively, a secondary mutation may have occurred during filter cloning of mutant IV-22. However, this strain has wild-type levels of all other known cytadherence accessory proteins for which antibody is available (2), and spontaneous hemadsorption-positive revertants of mutant IV-22 were obtained from the original non-filter-cloned population (16), suggesting a single defect is responsible for the mutant phenotype. The possibility of a secondary mutation might be reconciled by assessing lower-passage non-filter-cloned stocks of mutant IV-22 to isolate what we believe to be the original mutant IV-22. A third possibility is that MPN140 is required in cis to MPN141 in order for the P1/B/C complex to be fully functional. Regardless, the current study significantly expands our understanding of the relationship between MPN141 and MPN142 and their products.
A model of the protein interactions necessary for the formation of the M. pneumoniae attachment organelle and cytadherence competence was recently described (14). In that model, several protein interactions are shown to be important for the proper localization of functional P1, B, and C, which are treated as a single unit. Here evidence is presented supporting the hypothesis that P1, B, and C are closely tied in function and possibly act in concert to confer cytadherence, because MPN141 and MPN142 are connected at the levels of transcription and translation. Furthermore, it is shown that there are distinct, measurable, and nonreciprocal characteristics of these proteins, namely, that B and C require the presence of P1, but the converse does not hold, although B and C are required for P1 function. It is possible that interaction with P1 promotes the proper folding of B and C. This finding and the genetic tools used here now allow an exploration of the regions of P1 necessary for the stabilization and thus likely interaction and binding with B and C.
Acknowledgments
We thank Kyung Lee and Ed Sheppard for technical assistance and Richard Herrmann for kindly providing protein B- and C-specific antisera. The plasmid containing MPN141 was a gift from Joel Baseman.
This study was supported by the Public Health Service research grant AI23362 from the National Institute of Allergy and Infectious Diseases (D.C.K.) and a National Science Foundation Research Training Grant in Prokaryotic Diversity (NSF BIR9413235) (R.H.W.).
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1998. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 23:17-19. [DOI] [PubMed] [Google Scholar]
- 2.Balish, M. F., and D. C. Krause. 2002. Cytadherence and the cytoskeleton, p. 491-518. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum, New York, N.Y.
- 3.Baseman, J. B., R. M. Cole, D. C. Krause, and D. K. Leith. 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 151:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benders, G. A., B. C. Powell, and C. A. Hutchison. 2005. Transcriptional analysis of the conserved ftsZ gene cluster in Mycoplasma genitalium and Mycoplasma pneumoniae. J. Bacteriol. 187:4542-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldner, J., U. Gobel, and W. Bredt. 1982. Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature 298:765-767. [DOI] [PubMed] [Google Scholar]
- 6.Franzoso, G., P. C. Hu, G. A. Meloni, and M. F. Barile. 1993. The immunodominant 90-kilodalton protein is localized on the terminal tip structure of Mycoplasma pneumoniae. Infect. Immun. 61:1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn, T. W., K. A. Krebes, and D. C. Krause. 1996. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol. Microbiol. 19:1085-1093. [DOI] [PubMed] [Google Scholar]
- 8.Hahn, T. W., E. A. Mothershed, R. H. Waldo, and D. C. Krause. 1999. Construction and analysis of a modified Tn4001 conferring chloramphenicol resistance in Mycoplasma pneumoniae. Plasmid 41:120-124. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, E. J., R. M. Wilson, and J. B. Baseman. 1979. Two-dimensional gel electrophoretic comparison of proteins from virulent and avirulent strains of Mycoplasma pneumoniae. Infect. Immun. 24:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170-175. [DOI] [PubMed] [Google Scholar]
- 11.Hu, P. C., R. M. Cole, Y. S. Huang, J. A. Graham, D. E. Gardner, A. M. Collier, and W. A. J. Clyde. 1982. Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216:313-315. [DOI] [PubMed] [Google Scholar]
- 12.Inamine, J. M., S. Loechel, and P. C. Hu. 1988. Analysis of the nucleotide sequence of the P1 operon of Mycoplasma pneumoniae. Gene 73:175-183. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe, J. D., H. C. Berg, and G. M. Church. 2004. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4:59-77. [DOI] [PubMed] [Google Scholar]
- 14.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 15.Krause, D. C., and J. B. Baseman. 1983. Inhibition of Mycoplasma pneumoniae hemadsorption and adherence to respiratory epithelium by antibodies to a membrane protein. Infect. Immun. 39:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause, D. C., D. K. Leith, and J. B. Baseman. 1983. Reacquisition of specific proteins confers virulence in Mycoplasma pneumoniae. Infect. Immun. 39:830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Layh-Schmitt, G., and R. Herrmann. 1992. Localization and biochemical characterization of the ORF6 gene product of the Mycoplasma pneumoniae P1 operon. Infect. Immun. 60:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Layh-Schmitt, G., and R. Herrmann. 1994. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect. Immun. 62:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mernaugh, G. R., S. F. Dallo, S. C. Holt, and J. B. Baseman. 1993. Properties of adhering and nonadhering populations of Mycoplasma genitalium. Clin. Infect. Dis. 17(Suppl. 1):S69-S78. [DOI] [PubMed] [Google Scholar]
- 21.Papazisi, L., S. J. Frasca, M. Gladd, X. Liao, D. Yogev, and S. J. Geary. 2002. GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence. Infect. Immun. 70:6839-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papazisi, L., K. E. Troy, T. S. Gorton, X. Liao, and S. J. Geary. 2000. Analysis of cytadherence-deficient, GapA-negative Mycoplasma gallisepticum strain R. Infect. Immun. 68:6643-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popham, P. L., T. W. Hahn, K. A. Krebes, and D. C. Krause. 1997. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs posttranslationally. Proc. Natl. Acad. Sci. USA 94:13979-13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sperker, B., P. Hu, and R. Herrmann. 1991. Identification of gene products of the P1 operon of Mycoplasma pneumoniae. Mol. Microbiol. 5:299-306. [DOI] [PubMed] [Google Scholar]
- 27.Su, C. J., A. Chavoya, and J. B. Baseman. 1989. Spontaneous mutation results in loss of the cytadhesin (P1) of Mycoplasma pneumoniae. Infect. Immun. 57:3237-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tully, J. G. 1983. New laboratory techniques for isolation of Mycoplasma pneumoniae. Yale J. Biol. Med. 56:511-515. [PMC free article] [PubMed] [Google Scholar]
- 29.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldo, R. H., J. L. Jordan, and D. C. Krause. 2005. Identification and complementation of a mutation associated with loss of Mycoplasma pneumoniae virulence-specific proteins B and C. J. Bacteriol. 187:747-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willby, M. J., M. F. Balish, S. M. Ross, K. K. Lee, J. L. Jordan, and D. C. Krause. 2004. HMW1 is required for stability and localization of HMW2 to the attachment organelle of Mycoplasma pneumoniae. J. Bacteriol. 186:8221-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willby, M. J., and D. C. Krause. 2002. Characterization of a Mycoplasma pneumoniae hmw3 mutant: implications for attachment organelle assembly. J. Bacteriol. 184:3061-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]