Abstract
In Saccharomyces cerevisiae, the WASP (Wiskott-Aldrich syndrome protein) homologue Las17p (also called Bee1p) is an important component of cortical actin patches. Las17p is part of a high-molecular-weight protein complex that regulates Arp2/3 complex-dependent actin polymerization at the cell cortex and that includes the type I myosins Myo3p and Myo5p and verprolin (Vrp1p). To identify other factors implicated with this complex in actin regulation, we isolated proteins that bind to Las17p by two-hybrid screening and affinity chromatography. Here, we report the characterization of Lsb7/Bzz1p (for Las seventeen binding protein 7), an Src homology 3 (SH3) domain protein that interacts directly with Las17p via a polyproline-SH3 interaction. Bzz1p coimmunoprecipitates in a complex with Las17p, Vrp1p, Myo3/5p, Bbc1p, Hsp70p, and actin. It colocalizes with cortical actin patches and with Las17p. This localization is dependent on Las17p, but not on F-actin. Bzz1p interacts physically and genetically with type I myosins. While deletion of BZZ1 shows no obvious phenotype, simultaneous deletion of the BZZ1, MYO3, and MYO5 genes is lethal. Overexpression of Bzz1p inhibits cell growth, and a bzz1Δ myo5Δ double mutant is unable to restore actin polarity after NaCl stress. Finally, Bzz1p in vitro is able to recruit a functional actin polymerization machinery through its SH3 domains. Its interactions with Las17p, Vrp1p, and the type I myosins are essential for this process. This suggests that Bzz1p could be implicated in the regulation of actin polymerization.
Actin polymerization is a crucial event implicated in diverse cellular processes such as cell movement, polarization, endocytosis, cytokinesis, and morphogenesis. Comprehension of how actin polymerization and organization is regulated is of particular interest. One aspect of this regulation has been shown to be mediated by WASP (Wiskott-Aldrich syndrome protein) family proteins. The WASP family proteins are adapters that integrate cellular signals through interaction with such molecules as Cdc42, PIP2, or Nck to activate and regulate the nucleation of actin polymerization by the Arp2/3 complex (39, 42, 61, 67). In Saccharomyces cerevisiae, the WASP family member Las17p is an important component of the cortical actin cytoskeleton involved in actin assembly at the cell cortex, in endocytosis, and in polarity (31, 34, 41, 49). Las17p, like other WASP family proteins, interacts with globular actin and with the Arp2/3 complex (41, 68). The carboxyl-terminal region of Las17p, containing a presumptive actin monomer binding domain (WH2/V) and an acidic motif (A), binds Arp2/3 complex; this interaction stimulates actin nucleation activity of the complex in vitro (68). This is consistent with the mammalian model in which the WASP acidic motif binds and activates the Arp2/3 complex, whereas the WH2/V domain interacts with and presents actin monomer to the complex, helping to nucleate actin. Surprisingly, deletion of the yeast Las17p VA region does not affect actin organization and Arp2p localization (68). This suggests the existence in S. cerevisiae of a functionally redundant activity for the Las17p carboxy-terminal region.
One Las17p partner is the proline-rich protein verprolin (Vrp1p), the yeast homologue of mammalian WASP-interacting protein (WIP) (49). Deletion of Vrp1p, a cortical component, provokes defects in growth, endocytosis, and actin patch polarization (11, 47, 64). The amino-terminal part of Vrp1p contains a WH2/V domain similar to the WH2/V domain in the carboxy-terminal region of Las17p. This is important for verprolin functions and allows interaction with monomeric actin (64). Las17p interacts functionally with Vrp1p, since increased expression of Las17p partially cures the growth and endocytic defects of a vrp1-1 mutant (49). They coimmunoprecipitate from cell extracts and the carboxy-terminal 35 amino acids (aa) of Vrp1p bind the amino-terminal part of Las17p (14, 41, 49). Recently, Lechler et al. have shown that Las17p and Vrp1p are part of a macromolecular complex (∼1,000 kDa) that could mark the sites of cortical actin polymerization in a Cdc42p-dependent way (30).
Two other proteins have been designated as part of the Vrp1p/Las17p complex, the type I myosins Myo3p and Myo5p. These type I myosins are redundant proteins that together ensure essential functions in endocytosis and actin polarization and are required for actin assembly in a permeabilized cell assay (19, 21, 32). Both proteins interact with Las17p and Vrp1p through Src homology 3 (SH3)-polyproline binding (1, 14, 32). They colocalize with Vrp1p in cortical actin patches, and at least Myo5p localization depends on Vrp1p (1, 14, 21). Myo5p is able to interact with F-actin and to recruit the actin polymerization machinery in vitro in a Vrp1p-dependent manner (18). Like other members of the myosin I family, Myo3p and Myo5p are organized in three structural domains: a catalytic motor head domain that binds ATP and F-actin, a tail domain, and a junction allowing interaction with light chains (for a review, see reference 5). Besides the TH1, TH2, and SH3 domains, yeast type I myosin tails contain an acidic carboxyl-terminal motif that is homologous to and functionally redundant with Las17p's acidic domain (14, 32). These acidic domains are the sites of interaction with subunits of the Arp2/3 complex (14, 68). Deletion of the acidic motifs from all three proteins leads to an important synthetic effect on cell growth as well as depolarization and disassembly of cortical actin patches (14, 32). Furthermore, recent studies have shown that the acidic region of Myo3p, in association with the WH2/V G-actin binding domain of Vrp1p, activates the Arp2/3 complex in vitro in a similar way to the Las17p WH2/V and acidic domain (30). This is relevant to the finding that the unique fission yeast type I myosin Myo1p is also able to activate the Arp2/3 complex in vitro (33). A model was proposed in which Las17p, Vrp1p, and Myo3/5p form a complex containing two redundant activators of the Arp2/3 complex: Las17p on the one hand and Vrp1p in association with type I myosins on the other (14, 30).
The WASP family proteins are regulated by several effectors, and in particular by the Rho-type GTPase Cdc42p, which can bind directly with the CRIB domain of certain WASP family proteins (WASP or N-WASP, but not SCAR/WAVE proteins) (61). Although Cdc42p is known to be essential for actin polymerization and organization in S. cerevisiae (25), Las17p does not contain a CRIB motif, and no direct interaction between Cdc42p and Las17p has been reported. Thus, it is important to elucidate the regulation of the Las17p-containing complex. One aspect that appears to be important for the regulation of this complex is the phosphorylation of type I myosins. Myo3p is phosphorylated on its motor domain by two Cdc42 effectors: the PAK kinases Ste20p and Cla4p. This phosphorylation is essential for myosin I functions (30, 32, 69) and for polarized actin polymerization (30, 32, 69). Furthermore, Bni1p, a formin-like effector of Cdc42p, appears to be important for Las17p/Vrp1p function, since deletion of BNI1 precludes the polarized localization of Las17p (30).
In a two-hybrid screen to identify other proteins implicated in the regulation of Las17p, we previously identified several uncharacterized proteins containing SH3 domains, Lsb1p to Lsb4p (for Las seventeen binding protein) (41). Several recent studies in different organisms highlight the roles of SH3 domain-containing proteins in activation of WASP family proteins and the Arp2/3 complex (for reviews, see references 9, 42, and 54). For example, the SH2/SH3 adapter Nck binds WASP and N-WASP, as well as WIP, and is implicated in recruitment of these proteins to sites of tyrosine phosphorylation (2, 15, 35, 55). Nck and PIP2 synergistically activate N-WASP and Arp2/3-dependent actin polymerization in vitro (56). Another adapter, Grb2, directly activates N-WASP and stimulates Arp2/3 complex nucleation activity (8, 43). In S. cerevisiae, other SH3 proteins than type I myosins (described above) have been shown to interact with Las17p (Sla1p, Rvs167p, and Lsb1-4p) (6, 34, 41). Sla1p, a three-SH3 domain-containing protein, is a cortical actin patch protein that interacts with Las17p and might be involved in actin patch assembly (34). Thus, the study of yeast SH3 domain-containing proteins is essential to understand different aspects of Las17p and actin cytoskeleton regulation.
Here, we report the characterization of another SH3 domain-containing Las seventeen binding protein, Lsb7/Bzz1/YHR114Wp (hereafter called “Bzz1p”), identified both in an extension of our previous Las17p two-hybrid screen and by affinity purification of Las17-associated proteins. The BZZ1 gene codes for a 633-aa protein that is a member of the PCH (pombe Cdc15 homology) family, containing proteins such as Cdc15p in Schizosaccharomyces pombe, Hof1p in S. cerevisiae, PSTPIP in Mus musculus, CIP4 (Cdc42-interacting protein) in Homo sapiens, etc. (37). All of the proteins of this family present a highly conserved organization of their predicted structural domains. They contain an N-terminal FER-CIP4 homology (FCH) domain (3), a putative coiled-coil region near their amino terminus, one or two Src homology 3 domains (SH3) at the carboxy terminus, and proline-glutamic acid-serine-threonine-rich (PEST) sequences between the coiled-coil region and the SH3 domain(s) (37). Interestingly, many of the characterized proteins of the PCH family are known to be involved in actin cytoskeleton functions such as cytokinesis, endocytosis, and actin organization, and to interact with WASP family proteins (for a review, see reference 37). We focused our study on Bzz1p and its interactions with Las17p and type I myosins to describe in vivo and in vitro functions.
MATERIALS AND METHODS
Yeast media, strain constructions, and genetic manipulations.
The yeast strains used in this study are listed in Table 1. Yeast manipulations, including cell cultures, sporulation, tetrad dissections, and genetic techniques, were carried out essentially as described by Guthrie and Fink (22). The standard media used were rich YPD (1% yeast extract, 1% peptone, 2% dextrose, plus 2% agar for solid media) for strains that did not bear plasmids and minimal synthetic medium (yeast nitrogen base at 6.7 g/liter, 2% dextrose, CSM [Difco] minus amino acids) used for vector selection, plus 2% agar for solid media. Transformations of S. cerevisiae cells were done by the lithium acetate method with single-strand carrier DNA and dimethyl sulfoxide (DMSO) (23). All deletion strains were constructed by PCR targeting with short flanking homology directly upstream and downstream of the desired open reading frame (ORF) (65). For the deletion of the complete BZZ1 coding region, the oligonucleotides 5′-AATCTATCCTTAAACGCCAACTACTACATTACTTGCAATAAAAATGCTGCATCGACGGATC-3′ and 5′-CGGCCAGGGAAAATATTTAATAGTTTCAGTTCATTCCTTCGTTCATCGATGAATTCGAG-3′ were used to amplify the kanMX4 fragment of pFA6a-kanMX4 (kanMX4 homology underlined). Amplified fragment was precipitated and used to transform the wild-type diploid strain. Recombinants bearing the kanMX4 marker were selected on YPD containing G418 (200 μg/ml). Correct integration of the kanMX4 cassette was checked by PCR analysis of genomic DNA and genetic analysis. Haploid-deleted strains were obtained by sporulation and dissection of heterozygous diploid disruptants. Deletion of LAS17 was previously described by Madania et al. (41). Bzz1p was tagged at its carboxy terminus with three copies of the peptide epitope from the hemagglutinin (HA) protein of human influenza virus by a PCR-based strategy (28). Oligonucleotides 5′-GAATGTGACGGATTGAAAGGTCTATTTCCTACAAGTTACTGTAAACTGCAGGTCGACGGATCCGGA-3′ and 5′-CGGCCAGGGAAAATATTTAATAGTTTCAGTTCATTCCTTCGTTCATCGATGAATTCGAG-3′ were used to PCR amplify from the pYM1 plasmid the cassette coding for three copies of the HA epitope and the kanMX resistance marker. The resulting cassette with short flanking BZZ1 homology was transformed into strain FY1679. Cells bearing the integrated cassette were selected by growth on YPD containing G418 (200 μg/ml). Correct integration of the cassette was checked by PCR analysis of genomic DNA. HA tagging of Bzz1p was confirmed by Western blot analysis with anti-HA monoclonal antibody (clone 12CA5; Boehringer Mannheim). Bzz1-HAp migrated on sodium dodecyl sulfate (SDS) gels as a protein of about 80 kDa, which corresponds to its predicted molecular mass. Other strain constructions were made by mating the appropriate strains (Table 1), sporulation, tetrad dissection, and scoring the segregants for markers.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| FY1679 | MATahis3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 GAL2/ MATα HIS3 LEU2 TRP1 ura3-52 GAL2 | B. Dujon and F. Winston EUROFAN strain |
| FSW700K | MATabzz1Δ::kanMX4 his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 GAL2/ MATα BZZ1 HIS3 LEU2 TRP1 ura3-52 GAL2 | This study |
| FSW701K | MATα bzz1Δ::kanMX4 his3-Δ200 leu2-Δ1 TRP1 ura3-52 GAL2 | This study |
| FSW702K | MATα bzz7Δ::kanMX4 his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 GAL2 | This study |
| FSW711K | MATα bzz1Δ::kanMX4 his3-Δ200 leu2-Δ1 TRP1 ura3-52 GAL2 (+pGFPNfus-BZZ1) | This study |
| FSW71HAK | MATa BZZ1:3HAC:kanMX4 his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 GAL2 | |
| FKW171H | MATalas17Δ::HIS3 his3-Δ200 TRP1 leu2-Δ1 ura3-52 GAL2 | Winsor laboratory |
| FSW717KH | MATα bzz1Δ::kanMX4 las17Δ::HISMX6 his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 GAL2 | This study |
| FSW727KH | MATα bzz1Δ::kanMX4 las17Δ::HISMX6 his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 GAL2 (+pGFPNfus-BZZ1) | This study |
| FSW737KGK | MATα bzz1Δ::kanMX4 LAS17:GFP:kanMX4 HIS3 leu2-Δ1 TRP1 ura3-52 GAL2 | This study |
| LB OR88 | MATavrp1Δ::kanMX4 ura3-52 leu2Δ1 trp1Δ63 (FY1679 derivative) | Crouzet laboratory |
| FSW751KK | MATabzz1Δ::kanMX4 vrp1Δ::kanMX4 leu2-Δ1 trp1-Δ63 ura3-52 GAL2 | This study |
| FKW201 | MATaarp2-2(G19D):URA3 his3-Δ200 TRP1 leu2-Δ1 ura3-52 GAL2 | Winsor laboratory |
| RLY1 | MATahis3-Δ200 leu2-3,112 lys2-801 ura3-52 | Drubin laboratory |
| RLY650 | MATalas17Δ::LEU2 his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 pBEE1-PrA (pTL84) | 68 |
| RLY822 | MATamyo3Δ::HIS3 myo5Δ::TRP1 his3-Δ200 leu2-3 lys2-80 trp1-1 ura3-52 | 30 |
| SLW735HWK | MATaBZZ1 myo3Δ::HIS3 myo5Δ::TRP1 his3-Δ200 leu2-3 trp1-1 ura3-52/ | This study |
| MATα bzz1Δ::kanMX4 MYO3 MYO5 his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 | ||
| SLW001 | MATahis3-Δ200 ura3-52 | This study |
| SLW701K | MATabzz1Δ::kanMX4 his3-Δ200 ura3-52 | This study |
| SLW501W | MATamyo5Δ::TRP1 his3-Δ200 ura3-52 | This study |
| SLW571WK | MATamyo5Δ::TRP1 bzz1Δ::kanMX4 his3-Δ200 ura3-52 | This study |
| SLW572WK | MATamyo5Δ::TRP1 bzz1Δ::kanMX4 his3-Δ200 ura3-52 pRS416-BZZ1 | This study |
| SLW371HK | MATamyo3Δ::HIS3 bzz1Δ::kanMX4 his3-Δ200 ura3-5 | This study |
| Y190 | MATagal4Δ gal80Δ his3 trp-901 ade2-101 leu2-3,112 ura3-52 URA3::GAL1(UAS)-LacZ LYS2::GAL1(UAS)-HIS3 Cyhr | 13 |
| Y187 | MATα gal4Δ gal80Δ his3 trp1-901 ade2-101 leu2-3.112 ura3-52 URA3::GAL1(UAS)-LacZ | 13 |
| CG1945 | MATagal4-542 gal80-538 his3 trp-901 ade2-101 leu2-3,112 ura3-52 URA::GAL4 (3×17-mers)-CYC1 (TATA)-LacZ LYS2::GAL1 (UAS)-GAL1 (TATA)-HIS3 Cyhr | 16 |
DNA techniques, plasmid constructions, and gene banks.
DNA and bacterial manipulations were carried out by standard techniques (57). Restriction enzymes, T4 DNA ligase, and alkaline phosphatase were obtained from Boehringer Mannheim, New England Biolabs, or GibcoBRL. Plasmids were purified with the Bio-Rad plasmid miniprep kit or the Genomed JetStar kit. Transformations of Escherichia coli were performed by electroporation. Eurogentec, GibcoBRL, and MWG-Biotech AG synthesized the oligonucleotides used for PCR. DNA amplifications for cloning were made by using DNA polymerase with proofreading activity (Pwo, Eurogentec; and DyNAzyme Ext, Finzymes) and an Amplitron II thermocycler (Thermolyne). Sequences were determined by the sequencing service of the IBMP (Strasbourg, France).
To construct the BZZ1 rescue plasmid, a 2.5-kb DNA fragment containing the full-length BZZ1 coding region flanked with 400 bp upstream and 250 bp downstream was PCR amplified from yeast genomic DNA with primers 5′-GAATCCGAATTCGAAAACGAGGACGAAGAT-3′ and 5′-AGACAGGTATTAGAACTCTAGACTTGGGTA-3′, which, respectively, incorporate EcoRI and XbaI sites (underlined). This PCR fragment was cloned into the EcoRI and XbaI sites of pRS416 plasmid (60). To create the plasmid encoding an NH2-terminal green fluorescent protein (GFP)-Bzz1p fusion protein, we used a pGFPNfus derivative vector (50), which was mutated to the brighter pGFPNfusL64T65 (P. Dumoulin and B. Winsor, unpublished data). This plasmid contains the GFP coding sequence under the control of the MET25 regulatable promoter with a 3′ multicloning site. Then the full-length BZZ1 coding region was amplified by PCR with the primers 5′-TACTTGCAATAAAGCTTAGTGCAGATTTATCGATTGGTAATG-3′ and 5′-TATATAACCTCGAGTCGTCATACCTCTC-3′, which, respectively, span HindIII and XhoI sites (underlined). After restriction, the PCR fragment was cloned in frame with the GFP gene to create the pGFPNfus-L64T65-BZZ1 vector.
For two-hybrid constructs of BZZ1, the BZZ1 ORF was amplified from pRS416-BZZ1 with oligonucleotides 5′-TACTTGCAATAACCATGGGTGCAGATTTATCGATTGGT-3′ (with an NcoI site at the ATG of BZZ1) and 5′-TATATAACCTCGAGTCGTCATACCTCTC-3′ (with an XhoI site just after the Stop codon). The 1.9-kb NcoI-XhoI PCR fragment was cloned into NcoI-SalI sites of pASΔΔ (13) in fusion with the GAL4 binding domain (GAL4BD) to create pASΔΔ-BZZ1 and into NcoI-XhoI sites of pACTII (13) in fusion with GAL4 activation domain (GAL4AD) to create pACTII-BZZ1. The pACTII construct containing only the two SH3 domains of Bzz1p (aa 494 to 663) was obtained in an extension of the Las17p two-hybrid screen (41; unpublished data). The pACTII-BZZ1-FCH-coiled-coil vector was made by SalI restriction, by eliminating from pACTII-BZZ1 the sequence coding for the two SH3 domains of Bzz1p. Then the DNA fragment containing the FCH-coiled-coil N-terminal coding sequence (aa 1 to 431) of BZZ1 and the vector was purified and self-ligated. All GAL4 fusions constructed were controlled by sequencing the fusion site. Full-length LAS17 two-hybrid constructions in pAS2 and pACTII were described previously (41). To map interactions between Las17p and Bzz1p, different fragments of LAS17 were fused with GAL4 in pASΔΔ in the following way. For the Las17p-WH1-polyproline (Polypro)-WH2 fragment (aa 1 to 571), a 1,714-bp fragment of LAS17 was PCR amplified with primers I-17 (5′-GGCGTGATTTACCATGGGACTCC-3′; NcoI site) and A-17 5′-TGAGGGCTTCTCGAGCTGCGATTTGTCAACTTTT-3′ (XhoI site) with pACTII-LAS17 as a template. The Las17p-WH1-Polypro fragment (aa 1 to 544), a 1,630-bp fragment of LAS17, was PCR amplified with pACTII-LAS17 as a template and oligonucleotides I-17 and B-17 (5′-GACCTGCATCTCGAGTAGTTTCAGCGAATGAACCGCC-3′; XhoI site). For the Las17p-WH1 fragment (aa 1 to 132), a 396-bp fragment of LAS17 was PCR amplified with oligonucleotides I-17 and C-17 (5′-CATTTTTGCTCGAGAAAGTTTTCCTGTTAGCATATCTTTCACGC-3′; XhoI site) with pACTII-LAS17 as a template. The Las17p-polyproline-WH2-Ac fragment (aa 322 to 633) was created by amplification of a 971-bp fragment of LAS17 with primers II-17 (5′-CACAG CAGACCATGGCCCTTCCACAGTTGCCTAAC-3′; NcoI site) and D-17 (5′-CATCTTCTCGAGCATTCCATTACCAA-3′; XhoI site). The Las17p-Polypro-WH2 fragment (aa 322 to 571) was generated by amplification of a 747-bp fragment of LAS17 with primers II-17 and A-17. For the Las17p-Polypro fragment (aa 322 to 544), a 703-bp fragment of LAS17 was PCR amplified with II-17 and B-17. For the Las17p-WH2-Ac fragment (aa 539 to 633), the amplification was made with the primers III-17 (5′-GGCGGTTCCATGGCTGAAACTACTGGAGATGCAGGTC-3′; NcoI site) and D-17. All of these PCR fragments were cloned into the NcoI-SalI sites of pASΔΔ in frame with the GAL4 sequence.
Overexpression constructs were made from the pRS424-derived plasmid p424-GAL1 (46),which allows expression of a gene under the control of the inducible GAL1 promoter. To create p424-GAL1-BZZ1, a 2.1-kb fragment of BZZ1 was PCR amplified with primers 5′-GCGGAAATGGATCCTTAAACGG-3′ (which generates a BamHI site 50 bp upstream of the BZZ1 start codon) and 5′-GAATGGGAATTCGAAAACGAGGACGAAGAT-3′ (which contains an EcoRI site). This fragment was then cloned into the BamHI-EcoRI sites of the p424-GAL1 vector. The sequence of the resulting plasmid, p424-GAL1-BZZ1, was verified.
To construct a plasmid encoding a GST-Bzz1p fusion protein, we first amplified a 2-kb fragment of BZZ1 with oligonucleotides 5′-TACTTGCAATAAGAATTCGTGCAGATTTATCGATTGGT-3′ (with an EcoRI site at the ATG of BZZ1) and 5′-TATATAACCTCGAGTCGTCATACCTCTC-3′ (with an XhoI site just after the Stop codon). Second, this PCR fragment was inserted in frame with the GST sequence into the EcoRI and XhoI sites of the pGEX-5X1 plasmid (Pharmacia). For the plasmid expressing GST-SH3-SH3 (BZZ1), we cloned an 800-bp fragment of BZZ1, PCR amplified with primers 5′-CAACGGAGGATCCATGCATATAACAAGT-3′ (with a BamHI internal site) and 5′-GAATCCGAATTCGAAAACGAGGACGAAGAT-3′ (with an EcoRI internal site) into the BamHI-EcoRI sites of pGEX-2T. A plasmid encoding a six-His-Las17p fusion protein (6His-Las17p) was created by amplification of a 2-kb fragment of LAS17 with oligonucleotides 5′-GGCGTGATTTACCATGGGACTCC-3′ (NcoI site replacing the ATG of LAS17), 5′-CATCTTCTCGAGCATTCCATTACCAA-3′ (with an XhoI internal site), and pRS416-LAS17 as a template (unpublished data). This fragment was then cloned into pET-30a plasmid (Novagen) in frame with the six-His-tagged DNA sequence.
The genomic DNA bank (FRYL) used in two-hybrid screens containing mechanically derived fragments of S. cerevisiae genomic DNA in pACTII vector was a gift from M. Fromont-Racine (16). S. cerevisiae strain Y187 was transformed by standard procedures with DNA amplified from an aliquot of the FRYL library. Ten million yeast transformants were collected and pooled, and aliquots were stored at −80°C.
Two-hybrid experiments with directed two-hybrid assays.
The Y190 yeast strain bearing pAS2-LAS17 (41) or pASΔΔ cloned with different fragments of LAS17 was transformed with pACTII-BZZ1, pACTII-SH3-SH3 (41), pACTII-FCH-coiled coil and empty pACTII. The cells were plated on −Trp, −Leu synthetic medium and allowed to grow at 30°C for 3 days. The resulting transformants were replated on the same medium for 1 day at 30°C, and three of them were tested for β-galactosidase activity.
Genomic DNA screen.
To screen by mating, 2 × 108 FRYL bank cells (described above) were equilibrated in fresh YPD and then mixed with 8 × 108 exponential-phase CG1945 cells previously transformed with plasmid pASΔΔ-BZZ1. The mixture was then incubated at 30°C for 6 h. Cells representing 3.2 × 107 diploids were plated on −His, −Trp, −Leu synthetic medium supplemented with 0.5 mM 3-amino 1,2,4-triazole (3-AT) and incubated for 3 to 4 days at 30°C. The clones thus obtained were assayed for β-galatosidase activity on filters and then recycled. Plasmids were extracted and sequenced.
X-Gal (5-bromo-4-chloro-3-indolyl-β-d-thiogalactopyranoside) colony filter and β-galactosidase activity assays.
Qualitative β-galactosidase activity of strains containing two-hybrid vectors was assessed on nitrocellulose filters according to the method of Breeden and Nasmyth (7). To quantify β-galactosidase activity, freshly streaked Y190 double transformants were inoculated into 2.5 ml of liquid −Trp, −Leu synthetic medium and allowed to grow overnight to an optical density at 600 nm (OD600) of ∼1. Enzymatic activity was measured and expressed in Miller units (27). Values equal to or greater than positive control values were noted by “+,” and values equal or lower than negative control ones were marked by “−.”
Staining and light microscopy.
To observe the actin cytoskeleton, early-exponential-phase cells were fixed and phalloidin stained as previously described (51), except that incubation with phalloidin was for 1 to 2 h with gentle shaking at 0°C in the dark with 1 μM Alexa 594-phalloidin (Molecular Probes, Eugene, Oreg.). Cells were then observed under a fluorescence microscope with a rhodamine filter (excitation, 550/30; dichroic mirror, 575; emission, 615/45). For in vivo observations of GFP fusion proteins, cells were grown in liquid YPD (integrated GFP fusion) or −Ura synthetic medium (GFP plasmid constructs) at 25°C to the early exponential phase and then harvested, washed in phosphate-buffered saline (PBS), and immediately observed with a GFP bandpass filter (excitation, 460 to 500; dichroic mirror, 505; emission, 510 to 560). For simultaneous Alexa-phalloidin and GFP observation, the fixation time was reduced to 15 min in order to minimize GFP destruction and background signal increase caused by formaldehyde. Immunostaining techniques were adapted from “Paula's Immunofluorescence Protocol” web site (http://www.med.unc.edu/∼hdohlman/IF.html). Bzz1p-3HA immunodetection was performed with monoclonal anti-HA primary antibody (clone 12CA5; Boehringer Mannheim) and Alexa-Fluor 568-labeled goat anti-mouse secondary antibody (Molecular Probes). Las17p immunodetection was with a rabbit polyclonal anti-Las17p primary antibody (68) and Alexa-Fluor 568-labeled goat anti-rabbit secondary antibody (Molecular Probes). Observations were made with an Optiphot-2 microscope (Nikon, Melville, N.Y.) equipped with fluorescent optics. Pictures were recorded with a Photonics Science (Miltipas, Calif.) Coolview 10 camera equipped a Gel Grab 2.02 software program.
Latrunculin A treatment of GFP-labeled cells.
Cells were grown to exponential phase at 30°C in −Ura liquid synthetic medium and then concentrated to 108 cells/ml. For each condition tested, latrunculin A (Molecular Probes) in DMSO or an equivalent volume of DMSO was added to 100 μl of cell sample to give a 200 μM final concentration of latrunculin A. Cells were then incubated at 30°C for various periods. For the latrunculin A washout assay, treated cells were extensively washed and incubated with fresh −Ura synthetic medium for 1 generation time. Cells were fixed by addition of 10 volumes of −Ura medium containing 3.7% formaldehyde and incubation with gentle shaking for 15 min.
Purification of GST or six-His recombinant fusion proteins.
The different pGEX vector constructs were transformed into BL21 E. coli (Novagen). Large-scale cultures were grown at 30°C in Luria-Bertani medium (0.5% yeast extract, 1% tryptone, 1% NaCl, 2% agar for solid media) containing 100 μg of ampicillin per ml to an OD600 of 0.6 to 0.8. Then isopropyl-β-d thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM. Cells were further incubated for 1.5 h at 30°C. Whole-cell extracts were made by sonication in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate [DOC], 0.1% SDS, 50 mM Tris [pH 8]), aliquoted, and stocked at −80°C until used. GST fusion proteins were freshly purified with glutathione-Sepharose 4B (Amersham-Pharmacia) to a final concentration of 2 to 3 mg of fusion protein per ml of 50% slurry beads for GST pull-down or actin polymerization assay.
E. coli BL21 cells transformed with pET plasmid encoding 6His-Las17p fusion were grown at 37°C in LB medium containing kanamycin (50 μg/ml) and dextrose (2%), and induced at an OD of 0.6 to 0.7 with 1 mM IPTG for 2.5 h at 30°C. The 6His-Las17p fusion protein was isolated from inclusion bodies under denaturing conditions with isolation buffer (6 M urea, 20 mM Tris-HCl, 0.5 M NaCl, 20 mM imidazole [pH 8]) and purified with a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen). Before elution, recombinant protein was renaturated with a linear 6 to 0 M urea gradient. Protein was eluted, conserved with 50% glycerol and stored at −20°C before use in the pull-down assay.
Yeast extracts.
Depending on the experiment, different methods were used to obtain whole-cell protein extracts from yeast. For Western blot analysis, yeast cells were grown in −Ura synthetic medium at 25°C to a density of 3 × 107 to 4 × 107 cells per ml. Cells were harvested and washed twice with RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS, 50 mM Tris [pH 8]). A 1/4 pellet volume of RIPA containing proteinase inhibitor cocktail (Boehringer-Mannheim, catalog no. 1873580) was added, and the cells were glass bead lysed. The lysate was harvested after centrifugation at 20,000 × g at 4°C. The protein concentration (2 to 5 mg/ml) was measured with the Bio-Rad protein assay. Extract aliquots were frozen in liquid N2 and stored at −80°C. In the actin polymerization assay, protein extracts were prepared with a protocol adapted from Geli et al. (18). Cells were grown in rich medium to ∼4 × 107 cells per ml. Cells were harvested and washed once in PBS and once in XB (100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, 1 mM dithiothreitol [DTT], 1 mM ATP, 10 mM HEPES [pH 7.7]) with 50 mM sucrose. The pellet was resuspended in 1/4 volume of XB-50 mM sucrose containing proteinase inhibitors, and cells were broken by liquid N2 grinding with a mortar and a pestle. Unbroken cells and debris were eliminated by centrifugation at 20,000 × g at 4°C. Extracts were frozen in liquid N2 and stored at −80°C until used in the actin polymerization assay. The protein concentration (20 to 30 mg/ml) was measured with the Bio-Rad protein assay.
Western blotting and antibodies.
Protein samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE [8 to 10% polyacrylamide) according to the method described by Laemmli (29) and blotted onto polyvinylidene difluoride (PVDF) membrane (Hybond-P; Amersham) by semidry transfer. Blots were incubated with the appropriate primary antibody and with secondary antibody conjugated to horseradish peroxidase (HRP). Membranes were revealed with the Pierce Super-signal chemiluminescent kit for HRP, exposed to Kodak BioMax MR-1 film, and developed with a Kodak automatic film developer. Rabbit polyclonal anti-GFP antibody (Molecular Probes) and goat anti-rabbit (immunoglobulin G (IgG [H+L])-HRP-conjugated antibody (Bio-Rad) were employed as primary and secondary antibodies, respectively, to reveal the GFP-Bzz1p fusion protein. Ni-NTA coupled to HRP was used to detect the 6His-Las17p fusion protein. Low-range SDS-PAGE molecular mass standards (Bio-Rad and Pharmacia) were used to determine apparent molecular mass.
GST pull-down assay.
Fifteen microliters of purified recombinant 6His-Las17p fusion protein at 1.7 μg/μl was mixed with 30 μl of 50% glutathione-Sepharose beads bound to 2 to 3 μg of the different GST fusion proteins per μl of matrix. Samples were incubated for 20 min at room temperature and spun at 1,000 × g at 4°C to separate supernatant from beads. Beads were washed three times and resuspended in RIPA buffer. Each fraction was loaded on SDS-PAGE gel. For the actin polymerization assay, beads were washed once more in XB-50 mM sucrose and resuspended in the same buffer to give a 50% slurry bead suspension.
Bead-directed actin polymerization assay.
The bead-directed actin polymerization assay experiment was essentially performed as described by Geli et al. (18), as adapted from Ma et al. (38) with slight modifications. To summarize, 7 μl of the appropriate liquid N2 yeast extracts, adjusted to 20 mg/ml with XB-50 mM sucrose was mixed with 1 μl of ARS (10 mg of creatine kinase per ml, 10 mM ATP, 10 mM MgCl2, 400 mM creatine phosphate) and 1 μl of 10 μM Alexa Fluor 568-labeled actin (Molecular Probes). The actin polymerization reaction was initiated by adding 1 μl of 50% glutathione-Sepharose or Ni-NTA agarose beads bound, respectively, to 2 to 3 μg of GST or six-His fusion proteins or control GST or six-His peptide. After incubation for 20 min at 30°C, samples were visualized with a fluorescence microscope (Nikon Optiphot-2). In latrunculin A experiments, the drug was added to a final concentration of 10 μM prior to addition of the beads. Where necessary, phalloidin was added at 1 μg/ml. For 6His-Las17p add-back assays, glutathione-Sepharose beads coated with the different GST constructs were first incubated with purified 6His-Las17p as described for the GST pull-down assay. After extensive washing, beads were used in the visual actin polymerization assay.
Affinity purification and mass spectrometric identification of Las17p-associated proteins.
Purification of Las17p-associated proteins from yeast extracts prepared from strains containing protein A-tagged Las17p was carried out as described by Lechler et al. (32). Eluted proteins were separated on a one-dimensional polyacrylamide gel and visualized by staining with Coomassie. Specific bands were excised from the gel, in-gel digested with trypsin (unmodified, sequencing grade; Roche Molecular Diagnostics, Mannheim) as described previously(59). Proteins were identified by matrix-assisted laser desorption ionization (MALDI) peptide mass mapping and nanoelectrospray tandem mass spectrometric sequencing as described in reference 58.
RESULTS
Las17p interacts directly with Bzz1p, an uncharacterized PCH family protein.
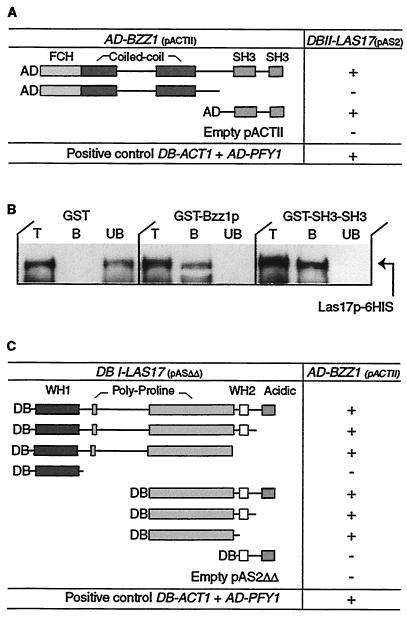
In order to identify proteins that interact with Las17p, we previously screened a yeast two-hybrid genomic DNA library for potential Las17p-interacting proteins (41). Among the different candidates found, four clones represented uncharacterized SH3 domain proteins and were designated LSB1 to LSB4. An extension of this screen revealed several new partners, including a fifth SH3 domain clone. This clone corresponded to the carboxyl-terminal extremity of an uncharacterized protein encoded by the ORF YHR114w, which we called LSB7 (now BZZ1). The plasmid coding for this fusion protein was then purified and transformed into yeast strain Y190 expressing Gal4BD-Las17p (pAS2-LAS17), and the resulting transformants were assayed for β-galactosidase activity. As shown in Fig. 1 A, the recombinant Bzz1p fragment interacts with Las17p compared to known control actin-profilin interaction. However, because we found only the carboxyl-terminal end of BZZ1 in the screen, the question whether full-length Bzz1p would be able to interact with Las17p arose. To answer this question, we constructed a full-length BZZ1 ORF in fusion with Gal4AD coding sequence in pACTII two-hybrid plasmid (see Materials and Methods). This construct was transformed into the Y190 strain expressing Gal4AD-Las17p (pAS2-LAS17), and transformants were assayed for β-galactosidase activity. We determined by Western blotting that each of the fusion proteins was expressed in the two-hybrid receptor strain (data not shown). In view of the varied expression levels, β-galactosidase activity assayed on filters and in cell extracts is reported as positive (+) or undetectable (−). Figure 1A shows that full-length Bzz1p interacts with Las17p. Interestingly, a protein fusion containing the FER-CIP4 homology (FCH) and coiled-coil N-terminal regions of Bzz1p did not interact with Las17p. Therefore, the SH3 domains of Bzz1p are necessary and sufficient for its interaction with Las17p.
FIG. 1.
Las17p interacts with Bzz1p in vivo and in vitro. (A) Two-hybrid interaction between Las17p and portions of Bzz1p. pACTII vectors carrying different GAL4-AD-BZZ1 gene fusions were transformed into Y190 cells expressing GAL4-DB in fusion with full-length LAS17. For each combination, extracts of three transformants were assayed for β-galactosidase activity. Interactions stronger than those of negative controls are shown as “+.” Empty vectors were used as negative controls, and the DB-ACT1 AD-PFY1 vector was used as a positive control. DB, DNA-binding domain; AD, activation domain. (B) Bzz1p interacts directly with Las17p in vitro. GST fused to full-length Bzz1p (GST-Bzz1p), GST fused to the COOH-terminal 188 aa of Bzz1p containing the two SH3 domains (GST-SH3), and six His fused to full-length Las17p (6His-Las17p) were purified from bacteria (see Materials and Methods). The same amount of GST fusion proteins (GST-Bzz1p or GST-SH3SH3) or GST alone bound to glutathione-Sepharose beads was incubated with a fixed amount of purified 6His-Las17p. After washing, total (T), bound (B), and unbound (UB) fractions were analyzed by Western blotting with Ni-NTA-HRP conjugate to reveal the 6His-Las17p fusion protein. (C) Two-hybrid interactions between Bzz1p and fragments of Las17p. pASΔΔ vectors containing different segments of LAS17 fused with GAL4-DB were transformed into Y190 cells expressing GAL4-AD in fusion with full-length BZZ1. Activity was assayed as in panel A.
Database and bibliographic searches showed that Bzz1p shares strong similarity with human CIP4 (Cdc42-interacting protein 4). CIP4 has been previously demonstrated to interact with the Rho-type GTPase Cdc42 and WASP and to be involved in cytoskeleton organization (3, 62). Bzz1p and CIP4 are both members of the PCH family, which is composed of proteins implicated in actin-based function (37). Bzz1p, like other PCH proteins, contains, from the amino terminus to the carboxyl terminus, an FCH domain (aa 5 to 60) (3), two potential coiled-coil regions (aa 130 to 210 and 305 to 355), a PEST sequence (aa 507 to 519), and two SH3 domains (aa 500 to 554 and 583 to 633). Most of the other PCH family proteins contain a unique SH3 domain. Like CIP4 and WASP (62), the fragment of Bzz1p found to bind Las17p in the two-hybrid screen (aa 494 to 633) contained only the two predicted SH3 domains.
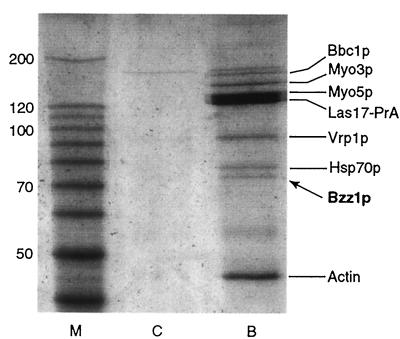
In parallel to the two-hybrid screen, Bzz1p was also identified as an Las17p-associated protein in the affinity purification first described by Lechler et al. (32). Briefly, protein extract prepared from yeast expressing a functional Bee1 protein tagged with protein A was enriched for Las17p by ammonium sulfate precipitation and then subjected to affinity chromatography on IgG-Sepharose beads. Bound proteins were identified by mass spectrometry. Besides the expected Myo3p and Myo5p, a number of other proteins were found to coimmunoprecipitate with Las17pBee1-PrA, including actin, Vrp1p, Bzz1p, Bbc1p/Mti1p, and Hsp70p (Fig. 2). Vrp1p and actin were previously described as interacting with Las17p (41). The amount of Bzz1p was roughly stoichiometric with the amounts of Vrp1p and Myo3p.
FIG. 2.
Bzz1p associates with Las17p in cell extracts. High-speed extracts from RLY1 wild-type cells or RLY650 cells carrying Las17-protein A were prepared as previously described (30). Extracts were bound to IgG-Sepharose, washed, eluted with 0.5 M acetic acid (pH 3.6), and then electrophoresed on an 8% polyacrylamide gel. The result shown is a Coomassie blue-stained gel that contains molecular weight markers (M), control untagged wild-type extract (C), or Las17-protein A extract (B). The identities of the proteins determined by mass spectroscopy analysis of gel band eluates are indicated.
Confirmation of direct physical interaction was obtained by overproducing the proteins Bzz1p and Las17p and testing their interaction in vitro. Plasmids that coded for GST-Bzz1p, GST-Bzz1p-SH3-SH3, and 6His-Las17p recombinant proteins were constructed. These different fusion proteins were then overproduced and purified from E. coli (see Materials and Methods). GST-Bzz1p associates with 6His-Las17p in GST pull-down experiments, whereas GST alone does not (Fig. 1B). Furthermore, similar experiments with GST-Bzz1p-SH3-SH3 confirm that the interaction is mediated through the two SH3 domains of Bzz1p (Fig. 1B). These data suggest that Bzz1p interacts directly with Las17p and that the two SH3 domains of Bzz1p are sufficient for this binding.
Bzz1p binds to the Las17p polyproline domain.
Like other WASP family proteins, Las17p is organized in a modular way: an amino-terminal WH1 domain, two carboxyl-terminal WH2/V (G-actin binding motif) and acidic regions, and a central polyproline region. SH3 domains are known to be protein-protein interaction modules that bind specifically to proline-rich ligands (42). Having found that Bzz1p SH3 domains were necessary and sufficient to bind with Las17p, we can hypothesize that Bzz1p binds Las17p through the polyproline motif of the latter. To test this hypothesis, we determined the region of Las17p required for the interaction with Bzz1p by a two-hybrid mapping approach. Two-hybrid plasmids expressing Gal4BD fused to different fragments of Las17p (see Materials and Methods) were constructed and transformed into Y190 yeast strains expressing Gal4AD fused to full-length Bzz1p. All Las17p fusion proteins were expressed in the strain, as revealed by Western blot analysis (unpublished data). Since Bzz1p fusion expression levels varied from one fusion to the other, β-galactosidase activity measured in the transformants must be considered as qualitative. Whereas WH1 or WH2/V and acidic regions alone were unable to interact with Bzz1p, Las17p constructs that contained polyproline motifs bound to Bzz1p (Fig. 1C). In a two-hybrid screen with Bzz1p as bait (described below), we also found Las17p as a partner, reinforcing the results presented above. Unexpectedly, the fusion protein (obtained three times) corresponded to aa 91 to 209 of Las17p (633-aa total length), a peptide sequence outside of the central polyproline (approximately aa 300 to 500) region. This peptide is located just after the WH1 domain and contains a proline-rich peptide at positions 178 to 190. This result, in addition to the interaction of Bzz1p with the central polyproline region of Las17p (Fig. 1C), suggests that Bzz1p interacts with at least two proline-rich motifs of Las17p (see Discussion). Similar results showing Bzz1p-Las17p binding mediated by several SH3-polyproline interactions have recently been reported in the genomic study by Tong et al. (63).
BZZ1 overexpression provokes a severe cell growth defect.
In order to understand its functions in S. cerevisiae, BZZ1 coding sequence was deleted in three different strains, YPH501, FY1679 (S288C derivatives), and W303, by gene replacement with the KanMX4 marker as described in Materials and Methods. Irrespective of the genetic background, BZZ1 is not an essential gene. Its deletion provoked no obvious phenotype under several growth conditions, such as elevated temperature, hyperosmotic medium, benomyl (microtubule-depolymerizing drug), or latrunculin A (actin-depolymerizing drug)-containing medium (data not shown). Considering that characterized PCH family proteins are known to be involved in actin cytoskeleton functions (37), we were interested in the effect of BZZ1 deletion on microfilament organization in yeast. Therefore, bzz1Δ strain FSW701K was grown to the exponential phase, fixed, and stained for polymerized actin (see Materials and Methods). Microscopic observations showed that BZZ1 deletion provoked no obvious effect on actin patch or cable organization or abundance throughout the cell cycle. Furthermore, no noticeable difference from the wild type was observed when fluid-phase endocytosis was examined by Lucifer yellow uptake (unpublished data).
To gain further information on Bzz1p interaction with Las17p, we looked for synthetic enhancement between bzz1Δ and las17Δ mutations. Deletion of LAS17 has been reported to cause temperature-sensitive growth or lethality depending on the strain's genetic background (34, 41). A thermosensitive las17Δ strain was crossed with a bzz1Δ strain, FSW701K (Table 1). Viable haploid spores after meiosis were tested for G418 resistance (KanMX4 marker for bzz1Δ) and for histidine prototrophy (HISMX6 marker for las17Δ). The presence of more than 95% viable double mutants showed that bzz1Δ is not synthetically lethal with las17Δ. Further analysis indicated that the deletion of BZZ1 did not appear to enhance either the thermosensitivity or osmosensitivity provoked by the deletion of LAS17.
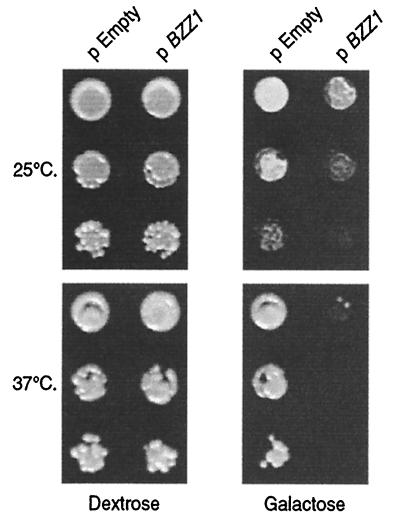
Another way to elucidate the function of a gene is to observe the effect of its overexpression on cell growth. To overexpress BZZ1, we cloned the corresponding ORF into p424-GAL1 vector under the control of the inducible GAL1 promoter. The resulting p424GAL1-BZZ1 plasmid and the empty vector were transformed into the FY1679 derivative cells of strain SLW001 (Table 1). The strains were first grown under repressive conditions and then spotted on galactose medium to initiate overexpression or onto dextrose medium to maintain repression. As shown in Fig. 3, Bzz1p overproduction provokes slow growth at 25°C compared to the strain with empty vector (compare pEmpty and pBZZ1 at 25°C on galactose). This effect was temperature dependent, because cells overexpressing BZZ1 were not able to grow at 37°C. Under repressive conditions, the presence of p424-GAL1-BZZ1 did not affect growth at either temperature (Fig. 3, dextrose column). To further characterize this phenotype, we also stained actin and the nuclei of these cells before and after induction of overexpression. No apparent actin or nuclear morphology defects were observed at different times after induction (unpublished data).
FIG. 3.
Bzz1p overexpression inhibits cell growth in a temperature-dependent manner. CLW001 wild-type strain containing either empty p424-GAL1 (pEmpty) or p424-GAL1-BZZ1 plasmid (pBZZ1) was grown overnight at 30°C in the selective liquid medium −Trp dextrose synthetic medium (SD). Ten-fold dilutions of about 106 cells containing empty proGAL vector plasmid or proGAL-BZZ1plasmid were spotted on SD-Trp or SG-Trp plates (dilutions from top to bottom for each temperature). Plates were incubated at 25 and 37°C for 2 days and then photographed.
GFP-Bzz1p localizes in cortical actin patches in a Las17p-dependent but F-actin-independent manner.
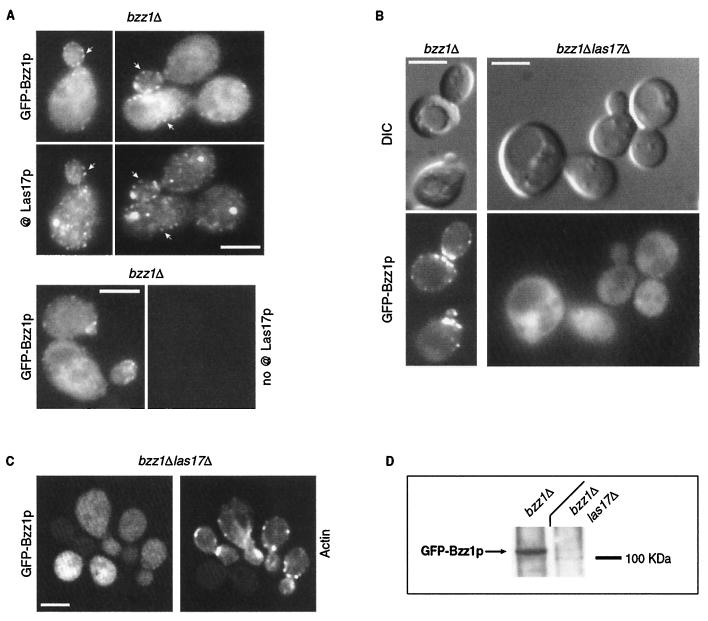
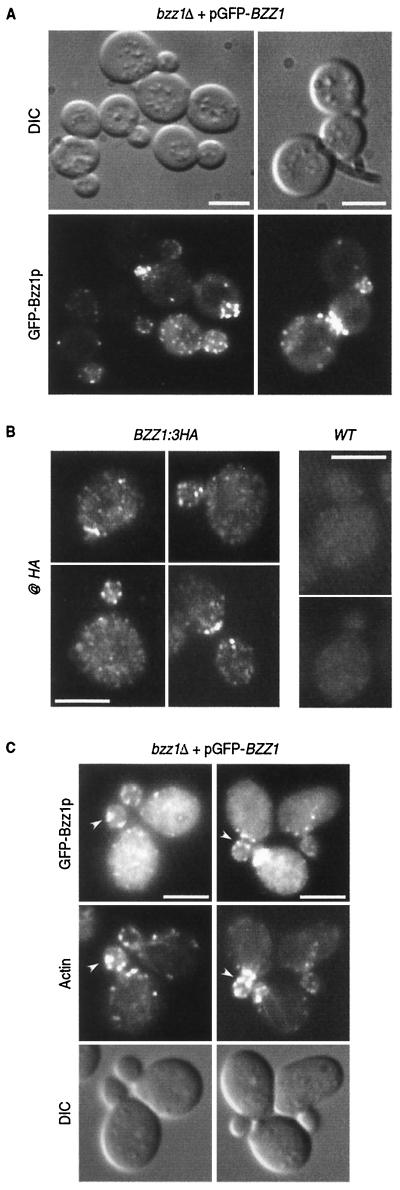
Las17p has been shown to be an important element of actin cytoskeleton that localizes in cortical actin patches independently of polymerized actin (34, 41). Since Las17p interacted directly with Bzz1p (Fig. 1), we examined Bzz1p localization and its dependence on Las17p and actin. To visualize Bzz1p in living cells, a plasmid expressing Bzz1p was fused to the carboxyl terminus of the GFP gene under the control of MET25 promoter. This vector was transformed into haploid FY1679-derived cells with BZZ1 deleted (strain FSW711K [Table 1]) in order to obtain a strain in which GFP-Bzz1p was the sole source of Bzz1p. The strain generated was grown under partial repression conditions to mimic endogenous Bzz1p expression as closely as possible. The presence of the recombinant protein was checked by Western blot analysis of a corresponding whole-cell extract with an anti-GFP antibody. GFP-Bzz1p was detected as an ∼110-kDa protein, which is consistent with the predicted molecular mass (Fig. 5D, left slot). The GFP-Bzz1p fusion protein is functional, since pGFP-Nfus-BZZ1 rescues the viability of a myo3Δ myo5Δ bzz1Δ triple mutant (see phenotype below). As shown in Fig. 4A, GFP-Bzz1p fluorescence in strain FSW711K localized in cortical patch structures concentrated at sites of bud emergence, in small buds, and at the bud neck before cytokinesis. Confocal time-lapse microscopy analysis showed that these GFP-Bzz1p-containing patches are localized in the cell cortex and are highly dynamic (unpublished data). However, this expression of GFP-Bzz1p fusion protein was not under the control of the endogenous BZZ1 promoter, and the possibility remained that localization of GFP-Bzz1p did not reflect the endogenous Bzz1p localization. To assess this possibility, we constructed a strain in which the stop codon of BZZ1 was replaced by the coding sequence of a 3HA tag (to give FSW71HAK [see Table 1 and Materials and Methods]). Western blot analysis of the protein extract prepared from this strain with anti-HA antibody showed that Bzz1-HAp was correctly expressed (unpublished data). The BZZ1:3HA gene was functional, since it did not provoke synthetic lethality when present in the myo3Δ myo5Δ double mutant strain. Bzz1-HAp was then localized by indirect immunofluorescence as described in Materials and Methods. Anti-HA immunostaining revealed that Bzz1-HAp localized in cortical patch structures like the GFP-Bzz1p fusion protein (Fig. 4B, BZZ1:3HA panel). The staining was specific, because a wild-type strain (without BZZ1:3HA) did not shown comparable signal when treated like the BZZ1:3HA strain (Fig. 4B, wild-type panel). Therefore, GFP-Bzz1p closely represents the endogenous protein. When FSW711K cells were fixed and stained with Alexa-phalloidin to visualize polymerized actin, GFP-Bzz1p and cortical actin patches showed clear but partial colocalization (Fig. 4C, arrows).
FIG. 5.
GFP-Bzz1p colocalization with Las17p and delocalization in a las17Δ mutant. (A) GFP-Bzz1p and Las17p colocalize in S. cerevisiae. A strain expressing GFP-Bzz1p (FSW711K) was grown to the exponential phase in dextrose synthetic medium (SD) −Ura liquid medium at 30°C. Cells were then fixed and incubated with polyclonal rabbit anti-Las17p primary antibody (@ Las17p), no antibody, or with Alexa-Fluor 568-labeled goat anti-rabbit secondary antibody. Cells were then visualized in a fluorescence microscope for GFP (GFP-Bzz1p) and Alexa 568 (@ Las17p, no @Las17p). The arrows show examples of overlapping fluorescence between GFP and Alexa 568. (B) FSW711K (bzz1Δ [left panel]) and FSW727KH (bzz1Δ las17Δ [right panel]) strains that contain pGFP-Nfus-BZZ1 were grown to the exponential phase in SD −Ura liquid medium at 25°C. An aliquot of harvested cells was washed in PBS buffer and observed with a fluorescence microscope. (C) Persistence of actin structures. An aliquot of the bzz1Δ las17Δ cells from panel A was washed in PBS, fixed, and stained with Alexa 568-phalloidin, and actin structures were visualized under a fluorescence microscope. (D) Anti-GFP Western blot analysis of whole-cell protein extracts from FSW711K (bzz1Δ) and FSW727KH (bzz1Δ las17Δ) strains. Strains were grown as in panel A, harvested, and washed in RIPA extraction buffer with protease inhibitors. Total protein extracts were prepared as described in Materials and Methods. Equal amounts of extract (20 μg) were subjected to SDS-PAGE. After transfer, the membrane was probed with rabbit anti-GFP antibody and revealed with HRP-conjugated goat anti-rabbit antibody. Bar, 5 μm.
FIG. 4.
GFP-Bzz1p localizes in cortical patches partially overlapping with actin. (A) Localization of Bzz1p in living FY cells. pGFP-Nfus-BZZ1 vector encoding an NH2-terminal GFP-Bzz1p fusion protein was introduced into strain FSW701K to obtain strain FSW711K.Cells were grown to the exponential phase in dextrose synthetic medium (SD) −Ura liquid medium at 25°C and then harvested. After washing, they were visualized under a fluorescence microscope. DIC, differential interference contrast. (B) Immunolocalization of Bzz1-HAp. FSW71HAK (Bzz1-HAp) and CLW001 (wild type [WT]) strains were grown to the exponential phase in liquid YPD medium at 30°C. Cells were then fixed and subjected to indirect immunostaining with mouse monoclonal anti-HA primary antibody (@ HA) and Alexa-Fluor 568-labeled goat anti-mouse antibody (see Materials and Methods). Cells were visualized under a fluorescence microscope with a rhodamine filter. (C) Colocalization of GFP-Bzz1p and cortical actin patches. FSW711K cells were grown as in panel A, fixed, and stained with Alexa-phalloidin (see Materials and Methods). The arrows show examples of overlapping fluorescence between GFP and Alexa-phalloidin. Bar, 5 μm.
Considering that Bzz1p and Las17p interact in vivo and in vitro (Fig. 1 and 2) and that GFP-Bzz1p localized in cortical actin patches (Fig. 4C) (63) like Las17p (34, 41), Bzz1p and Las17p colocalization was examined. To respond to this question, the strain expressing GFP-Bzz1p used in Fig. 4 was subjected to immunostaining with a rabbit polyclonal anti-Las17p primary antibody (68) and a goat-anti-rabbit Alexa-Fluor 568-labeled secondary antibody. Cells were observed for GFP fluorescence (Bzz1p) and Alexa 568 fluorescence (Las17). As shown in Fig. 5A, GFP-Bzz1p and Las17p colocalized (see arrows). The anti-Las17p immunostaining was specific, because the same cells treated with only secondary antibody did not show any Alexa 568 fluorescence (Fig. 5A, bottom panel).
We then tested whether the Bzz1p cortical patch localization depended on Las17p. Deletion of LAS17 provoked accumulation of aberrant actin aggregates, but not complete disappearance of actin patches and cables at permissive temperature (34, 41). If Bzz1p is recruited to cortical actin patches through its interaction with Las17p, then Bzz1p should no longer localize to patch structures in las17Δ cells. This was indeed the case, since in a strain with LAS17 deleted, GFP-Bzz1p was no longer localized in cortical patches, but showed diffuse staining throughout the cytoplasm (Fig. 5B), whereas patches and aberrant actin structures are present (Fig. 5C). However, in a strain with BZZ1 deleted, Las17p-GFP still localized in polarized patches (data not shown). This indicated that Las17p localized with Bzz1p in cortical patches. Moreover, deletion of LAS17 affected not only the GFP-Bzz1p localization, but also the stability of the fusion protein. When extracts made from the las17Δ strain were analyzed by anti-GFP Western-blot analysis, GFP-Bzz1p staining was much weaker and diffuse than extract from a LAS17 strain (Fig. 5D). This cannot be transcriptional regulation, because the promoter used to express GFP-Bzz1p was the inducible MET25 promoter and not the BZZ1 promoter. However, we could not distinguish whether GFP-Bzz1p degradation was due to its loss of localization or vice versa.
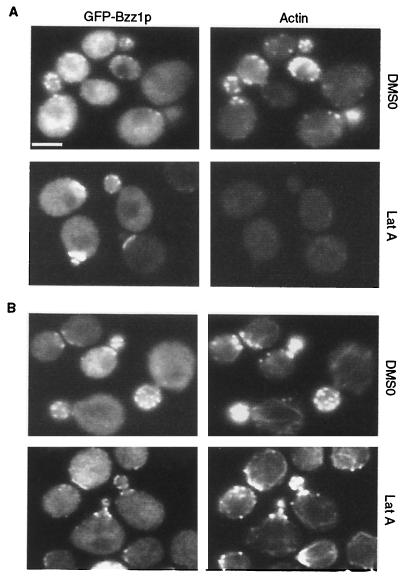
To determine whether Bzz1p was dependent on polymerized actin for its localization, we investigated the effects of latrunculin A, a drug that prevents actin polymerization (4), on Bzz1p localization. Since cortical patch localization of Las17p is independent of polymerized actin (41), it might also be the case for Bzz1p. To test this, FSW711K cells expressing GFP-Bzz1p were treated for 5, 15, or 30 min with 200 μM latrunculin A or DMSO solvent alone and then fixed and processed for Alexa-phalloidin staining (Fig. 6). GFP and actin visualization revealed that from the 5-min time point, polymerized actin was not detectable in latrunculin A-treated cells, whereas GFP-Lsb7p remained localized in polarized cortical structures, even after 30 min. (Fig. 6A shows the 15-min time point.) Moreover, when cells treated with latrunculin A for 45 min were washed and allowed to regenerate for 120 min in fresh medium, polarized cortical actin patches reassembled and colocalized with GFP-Bzz1p patchlike structures (Fig. 6B). Thus, maintenance of the intracellular polarized localization of Bzz1p is dependent upon Las17p, but not on the polarization or integrity of actin structures.
FIG. 6.
GFP-Bzz1p localized independently of actin. (A) Localization of Bzz1p after treatment of cells with latrunculin A (Lat A). Strain FSW711K was grown to the exponential phase at 30°C in liquid −Ura synthetic medium. An aliquot of these cells was then incubated at 30°C. in selective synthetic medium containing 200 μM (final concentration) latrunculin A in DMSO or in DMSO alone. Images in panel A show cells that were briefly fixed and stained with Alexa-phalloidin after 15 min of exposure to latrunculin A (or DMSO). (B) Localization of Bzz1p after latrunculin A removal. Cells from panel A were incubated for 45 min with latrunculin A (or DMSO), extensively washed, and allowed to regenerate for 120 min in fresh medium. Cells were then briefly fixed and stained for polymerized actin. All cells were visualized with the appropriate filters under a fluorescence microscope (see Materials and Methods). Bar, 5 μm.
Bzz1p interacts with type I myosin Myo5p.
To learn more about the interactions of Bzz1p and to determine its possible functions, the two-hybrid system was used to screen a yeast genomic DNA library for Bzz1p-interacting proteins (see Materials and Methods). Among the different potential Bzz1p partners, two were of particular interest: Las17p, as described above, and the type I myosin Myo5p (63; unpublished observation). Two different truncations of Myo5p were found to interact with Bzz1p: one comprising aa 939 to 1169 (found once) and the other comprising aa 853 to 1022 (detected 11 times). The fusion protein Myo5p(aa 853 to 1022) corresponded to approximately the TH1 domain and the first 20 aa of the TH2 domain, whereas the Myo5p(aa 939 to 1169) fusion protein included 61 aa before the TH2 domain, the TH2 and SH3 domains, and 47 aa following the SH3 domain. However, both fragments contain at least one proline-rich stretch. This finding was interesting, because Myo5p and its highly related isoform, Myo3p, have been shown to localize in actin patches and to play a critical role in cortical actin assembly in budding yeast (1, 14, 18, 21, 30, 32). Furthermore, yeast type I myosins were demonstrated to interact with Las17p and Vrp1p, with which they form a complex activating the Arp2/3 complex (1, 14, 30, 32). Given that Bzz1p interacts with Las17p (Fig. 1) and Myo5p and that Bzz1p and Myo5p coimmunoprecipitate together with Las17p (Fig. 2), Bzz1p could be part of the Las17p/Vrp1p/Myo3/5p complex (see Discussion).
Bzz1p and Myo5p act together in repolarization of actin cytoskeleton after salt stress.
In order to better understand the interaction between Bzz1p and the type I myosin Myo5p, we looked for synthetic enhancement between bzz1Δ and myo5Δ effects in the myo3Δ myo5Δ double mutant. Deletion of either MYO3 or MYO5 has little effect on cell growth, but the myo3Δ myo5Δ double mutant shows severe defects in growth and actin cytoskeleton organization (21). The thermosensitive myo3Δ myo5Δ strain RLY822 was crossed with the bzz1Δ strain FSW702K, giving rise to the recombinant strain SLW735HWK (Table 1). Viable haploid spores obtained after meiosis and tetrad dissection (37 tetrads) were tested for G418 resistance (KANMX4 marker for bzz1Δ) and for histidine and tryptophan prototrophy (HIS3 and TRP1 markers for myo3Δ and myo5Δ, respectively). Whereas almost all wild-type, single-mutant, and double-mutant spores grew after tetrad dissection at 25°C, no viable myo3Δ myo5Δ bzz1Δ triple mutants were obtained (Table 2). This genetic interaction between Bzz1p and the type I myosins was specific for Bzz1p. Indeed, a centromeric plasmid containing the BZZ1 ORF under its own promoter was able to rescue the viability of myo3Δ myo5Δ bzz1Δ triple mutant, partially restoring growth (comparable to that of the myo3Δ myo5Δ double mutant). Further analysis showed that deletion of BZZ1 and MYO5 together or BZZ1 and MYO3 together did not provoke any noticeable cell growth defects or thermosensitivity under normal conditions compared to wild-type cells or single mutants.
TABLE 2.
Synthetic effect of myo3Δ myo5Δ bzz1Δ triple deletion
| Spores | No. of spores with genotype:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild type | myo3Δ | myo5Δ | bzz1Δ | myo3Δ bzz1Δ | myo5Δ bzz1Δ | myo3Δ myo5Δ | myo3Δ myo3Δ bzz1Δ | |
| Total | 17 | 16 | 13 | 23 | 18 | 21 | 28 | 12 |
| Viable | 16 | 16 | 13 | 23 | 17 | 21 | 25 | 0 |
| Inviable | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 12 |
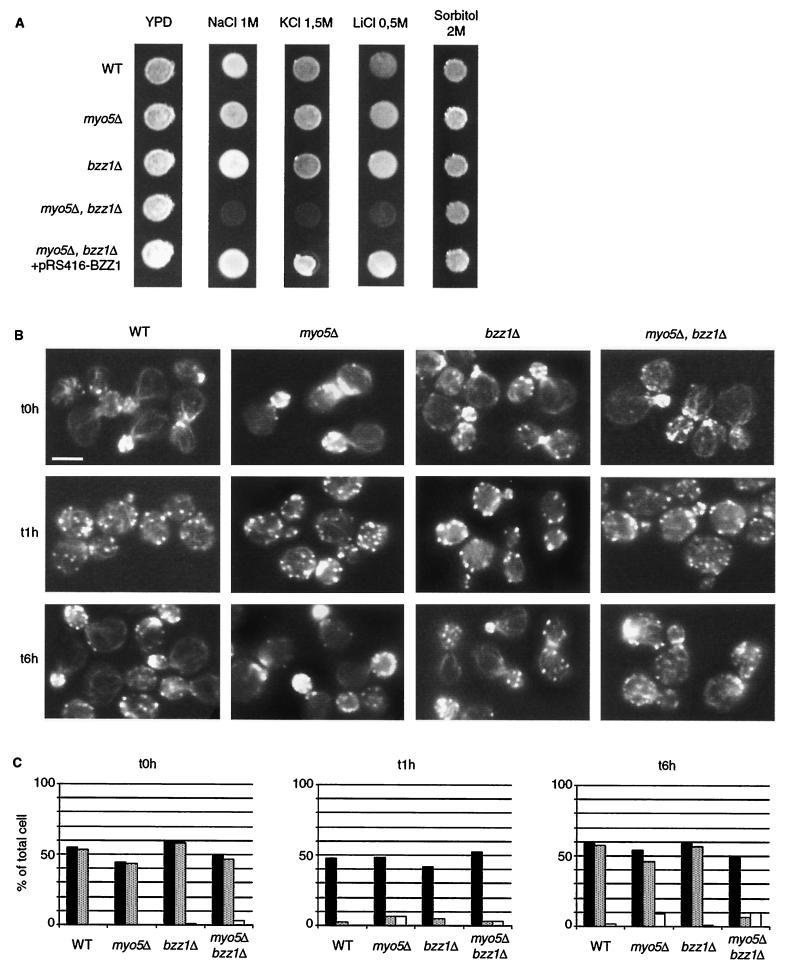
Phenotypic analysis of the different strains obtained after meiosis of SLW3570HKW cells showed that a myo5Δ bzz1Δ double-deletion strain was sensitive to salt stress. The double mutant myo5Δ bzz1Δ grew very poorly at 25°C on YPD medium containing 1 M NaCl (Fig. 7A) and not at all on 1.5 M NaCl, whereas it grew like the wild type on YPD alone. The single mutant bzz1Δ and myo5Δ strains were not affected by salt stress and grew at wild-type rates under these conditions. Furthermore, transformation of the myo5Δ bzz1Δ double mutant with a centromeric plasmid bearing BZZ1 under its own promoter restored normal growth at 25°C on YPD-1 M NaCl (Fig. 7A). Thus, the salt sensitivity observed was due to the loss of MYO5 and LSB7. The same effect was observed after addition of other salts, such as KCl (1.5 M) or LiCl (0.5 M). However on hyperosmotic medium containing 2 M sorbitol, the myo5Δ bzz1Δ double-mutant cells were able to grow like the wild type (Fig. 7A). Therefore, the growth inhibition was not due to osmosensitivity. Moreover, a strain with deletion of both BZZ1 and MYO3 was not sensitive to NaCl stress (unpublished data), indicating that the salt sensitivity was specifically due to the deletion of MYO5 and that MYO3 and MYO5 were not redundant in this phenomenon.
FIG. 7.
Salt stress sensitivity of a bzz1Δ myo5Δ double-deletion mutant. (A) SLW001 (wild type [WT]), SLW501T (myo5Δ), SLW701K (lsb7Δ), SLW571TK (myo5Δ lsb7Δ), and SLW572TK (myo5Δ bzz1Δ + pRS416-BZZ1) were grown in YPD to the exponential phase at 25°C. The same number of cells for each culture was then spotted on YPD, YPD-1 M NaCl, YPD-1.5 M KCl, YPD-0.5 M LiCl, and YPD-2 M sorbitol plates, which were incubated for 3 days at 25°C and then photographed. (B) Effect of NaCl on the actin cytoskeleton organization of a myo5Δ bzz1Δ double-disrupted strain. Strains used in panel A were grown in liquid YPD at 25°C. At 0 h, an equal volume of liquid YPD containing 2 M NaCl was added to each culture (final salt concentration, 1 M), and incubation continued at 25°C. The photos shown in panel B represent the four cell types fixed with formaldehyde and stained with Alexa-phalloidin at time points (t) 0, 1, and 6 h after addition of NaCl. Bar, 5 μm. (C) Quantification of actin cytoskeleton defects after salt stress. For each strain in panel B and at each time point (0, 1, and 6 h), more than 100 cells were counted. Black bars represent total budding cells, shaded bars represent budding cells with polarized actin patches, and white bars represent cells with aberrant actin structures. The results obtained were presented as a bar graph in which the horizontal axis represents different strains and the vertical axis represents the percentage of the total cell number.
Salt stress is known to provoke actin cytoskeleton depolarization and, among other effects, to delay growth of yeast cells. After 1 to 2 h, S. cerevisiae adapts to the high salt concentration, and its actin cytoskeleton repolarizes progressively and completely (10). To observe the effects of MYO5 and BZZ1 single or double deletions on the organization of the actin cytoskeleton before and after salt stress, wild-type (SLW001), myo5Δ (SLW501W), bzz1Δ (SLW701K), and myo5Δ bzz1Δ (SLW571WK) strains were grown to the exponential phase in rich medium at 25°C and then diluted into YPD medium containing a final concentration of 1 M NaCl. At 0, 1 (actin depolarization), and 3 and 6 (recovery) h after NaCl stress induction, aliquots of each culture were fixed and stained for actin. Before salt stress, all strains had a normal distribution of actin filaments with polarized actin patches and actin cables extending between the mother cell and bud (Fig. 7B, 0 h). One hour after salt stress induction, all cell types had fewer visible actin cables and a high density of actin patches that were completely depolarized (Fig. 7B, 1 h). Three or 6 h after stress induction, the myo5Δ bzz1Δ double mutant was unable to adapt to the high salt concentration and had not repolarized its actin cytoskeleton (Fig. 7B, 6 h [compared to wild-type and single-mutant cells]). Actin patches were still depolarized and formed aberrant aggregates (Fig. 7B, 6 h). This phenomenon was quantified by counting the total number of budding cells, budding cells with polarized actin patches, and cells with aberrant actin structures for each strain at each time point (0, 1, and 6 h). Before NaCl treatment, nearly all of the budding cells in each of the four strains presented polarized actin patches (Fig. 7C, 0 h). One hour after NaCl treatment, for each strain (wild type, myo5Δ, bzz1Δ, myo5Δ bzz1Δ), <10% of budding cells had polarized actin patches, whereas budding cells represented between 42 and 53% of total cell number (Fig. 7C; 1 h). After 6 h of salt stress, wild-type, bzz1Δ, and myo5Δ cells recovered actin patch polarization. Among 45 to 60% of the budding cells, almost all had polarized actin (Fig. 7C, 6 h). In the myo5Δ bzz1Δ double mutant, <10% of budding cells had polarized actin patches (Fig. 7C, 6 h); that is, there was little difference between the levels at 1 and 6 h. It is interesting that at 6 h, not all myo5Δ budding cells recovered polarized actin patches. Approximately 15% of budding cells remained depolarized. Quantification of cells with aberrant actin structures showed no difference between the myo5Δ single mutant and the myo5Δ bzz1Δ double mutant. These salt stress recovery experiments were also carried out in a W303 genetic background. Similar results, with more pronounced actin disorganization, were observed (unpublished data).
Bzz1p is able to recruit functional actin polymerization machinery through its two SH3 domains in an in vitro assay.
Recent studies with yeast highlight the role of the type I myosins, Vrp1p and Las17p as components of a high-molecular-mass complex implicated in regulation of polarized actin polymerization (14, 30, 32). This complex was proposed to contain two Arp2/3 complex activators in vitro: Las17p alone, and a second activity requiring Myo3p-Vrp1p interaction (30, 68). In an independent study, the Myo5p COOH-terminal tail region, containing part of the TH2, SH3, and acidic domains, was shown to recruit actin polymerization machinery and induce actin filament formation in vitro in a process that requires Myo5p-Vrp1p interaction (18).
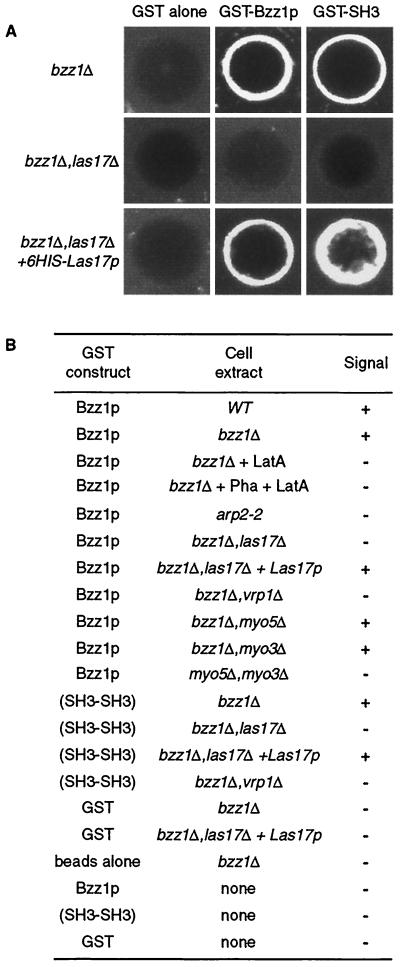
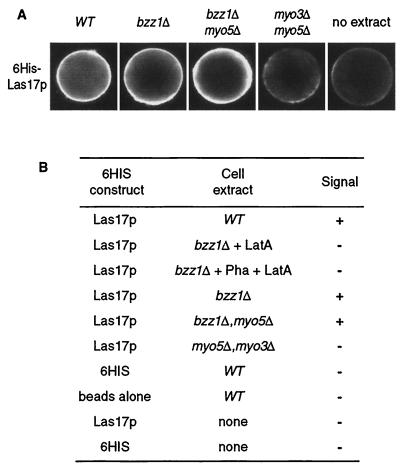
This information and our data showing that Bzz1p interacts with Las17p and Myo5p and participates with it in actin polarization (described above) suggest that Bzz1p might have a role in actin polymerization. In particular, we questioned whether the role of Bzz1p might be to recruit proteins of the polymerization machinery and/or induce localized actin polymerization in vitro. To test this hypothesis, we performed an in vitro visual actin polymerization assay as previously described by Ma et al. (38) and Geli et al. (18). In this assay, actin polymerization is triggered by the GST fusion protein GST-Bzz1p on fusion protein-coated glutathione-Sepharose beads. Beads are incubated with yeast extract in the presence of trace amounts of fluorochrome-labeled actin. If the GST fusion protein induces actin polymerization, a fluorescence signal accumulates around the beads and can be observed directly by fluorescence microscopy. We used extracts depleted of BZZ1 in order to obtain GST-Bzz1p as the sole source of Bzz1p in the reaction, as well as wild-type extracts. Strikingly, beads coated with GST-Bzz1p fusion protein, and not those coated with naked GST protein alone, accumulated a bright fluorescent signal when incubated in cell extracts from a bzz1Δ strain (FSW701K) containing small amounts of added Alexa-labeled actin (Fig. 8A, compare GST alone versus GST-Bzz1p). This was also the case for the extract made from the wild-type strain (SLW001), indicating that endogenous Bzz1p had no detectable effects on the reaction (Fig. 8B). This signal was abolished when the assay was performed in the presence of latrunculin A (Fig. 8B), indicating that it corresponds to an accumulation of actin filaments—that is, the process required actin polymerization. The accumulation of the observed fluorescence could be due to the binding of preformed actin filaments around GST-Bzz1p-coated beads rather than to induced actin polymerization. To dismiss this possibility, cell extracts were incubated with Alexa-actin in the absence of beads. F-actin was stabilized by the addition of phalloidin, and finally, beads were added in the presence of latrunculin A to prevent further polymerization. Under these conditions, no signal was detected on the GST-Bzz1p-coated beads, whereas polymerized actin aggregates were visible in the extract. This result clearly indicates that Bzz1p plays an active role in actin polymerization. Kinetic analysis of the reaction by time-lapse microscopy showed that actin polymerization was initiated in spot structures on the beads and that actin filaments elongated from these points, forming a fast-growing F-actin aggregate (unpublished data). This observation is in agreement with recent observations in an independent study on myosin I-induced actin polymerization (24). Bzz1p-induced actin polymerization was dependent on the presence of other cellular components, since minimal background signal was detected around GST-Bzz1p-coated beads when buffer was used in the assay instead of cellular extract (Fig. 8B). Furthermore, extract from an arp2-2 mutant was unable to accumulate polymerized actin on the GST-Bzz1p-coated beads (Fig. 8B). This showed that Bzz1p-triggered actin polymerization required a functional Arp2/3 complex. Thus, Bzz1p seems to be linked to the regulation of actin polymerization. To explore the need for the proteins of the Las17p/Vrp1p/Myo3/5p complex in this process, we performed the GST-Bzz1p-coated bead assay with different yeast extracts depleted of Las17p, Vrp1p, Myo3p, or Myo5p. Extracts from las17Δ or vrp1Δ singly deleted strain or the doubly deleted myo5Δ myo3Δ strains were unable to polymerize actin on GST-Bzz1p-coated beads, whereas extracts from myo3Δ or myo5Δ single-mutant strains did polymerize actin (Fig. 8A and B). To investigate whether a direct Bzz1p-Las17p interaction was the basis for lack of recruitment in a las17Δ extract, GST-Bzz1p-coated beads were incubated with purified 6His-Las17p prior to assay with the extract from a las17Δ strain (see Materials and Methods for protein preparation). Adding back the purified recombinant protein bountifully restored polymerization on beads (Fig. 8A, lower panel). This reconstitution clearly shows that, in the assay, the actin polymerization machinery requires Bzz1p-Las17p interaction.
FIG. 8.
Bzz1p recruits the actin polymerization machinery in vitro. (A) Glutathione-Sepharose beads coated with either GST or GST fused to full-length Bzz1p or the two C-terminal SH3 domains of Bzz1p were incubated with extracts from either the bzz1Δ cells (FSW701K, upper panel) or the bzz1Δ las17Δ cells (FSW717KH middle panel) in the presence of Alexa-actin. Samples were incubated at room temperature, and the fluorescent signal was visualized. The photos shown below indicate representative positive (+) and negative (−) signals. In the lower panel (bzz1Δ las17Δ + 6His-Las17p), GST fusion protein-coated beads were first incubated with an excess of purified 6His-Las17p (see Materials and Methods) for 20 min. After extensive washing, the beads were tested for the actin polymerization assay by using the lsb7Δ las17Δ-derived extract. Panel B summarizes the results obtained and shows the results from experiments with extracts from wild-type (WT [SLW001]), arp2-2 (FKW201), bzz1Δ vrp1Δ (FSW751KK), bzz1Δ myo5Δ (SLW571KT), bzz1Δ myo3Δ (SLW371HK), and myo5Δ myo3Δ (RLY822) strains. Latrunculin A (LatA) was added to the bzz1Δ + LatA sample at the 0-h incubation. For the bzz1Δ + Pha + LatA sample, actin filament polymerized in the absence of beads was stabilized with phalloidin (Pha) prior to the addition of GST-Bzz1p-coated beads and latrunculin A.
As demonstrated in Fig. 1, Bzz1p interacts directly with Las17p through its two SH3 domains. Therefore, it was attractive to determine whether the two SH3 domains of Bzz1p could promote actin polymerization in the visual assay. For this, a GST fusion protein containing solely the two SH3 domains of Bzz1p was constructed (see Materials and Methods) and assayed with the actin polymerization assay in same way as full-length GST-Bzz1p fusion protein. Strikingly, GST-SH3-SH3-coated beads served as well as GST-Bzz1p-coated beads in the visual actin polymerization assay. The SH3 domains induced actin polymerization around beads in the presence of yeast extract from bzz1Δ strains, and this polymerization depended on the Arp2/3 complex, Las17p, and Vrp1p (Fig. 8A and B). Clearly, the interaction between the SH3 domains of Bzz1p and Las17p appeared to be sufficient for this process, because Las17p add-back reconstituted the actin polymerization from the las17Δ extract.
Las17p is able to induce actin polymerization in vitro in a Bzz1p-independent way.
Given these results, we wondered whether the actin polymerization assay would be affected by the depletion of Bzz1p. For this, we replaced GST-Bzz1p on the beads with another component of the complex. Following the same principle described for Fig. 8, we used Ni-NTA agarose beads coated with 6His-Las17p fusion protein as a template for actin polymerization. Beads coated with 6His-Las17p that had been incubated in cell extract from a wild-type strain (SLW001) in the presence of Alexa-labeled actin accumulated a bright fluorescence signal (Fig. 9A). When the beads coated with 6His-Las17p were incubated without cell extract, only a very faint signal could be observed (Fig. 9A, no extract). With beads coated with six His alone and incubated with the wild-type extract, no signal was observed (Fig. 9B). The same controls used in Fig. 8 with latrunculin A and phalloidin were used (Fig. 9B). Faint background signal, similar to the one observed without extract, was detected around 6His-Las17p-coated beads under these conditions. This clearly indicated that the signal observed with wild-type extract was not provoked by accumulation of G-actin or preformed F-actin around the beads and that 6His-Las17p was also able to trigger actin polymerization in this assay. In the next step, we explored the need for Bzz1p and type I myosins in Las17p-triggered actin polymerization by using cell extracts from strains depleted of Bzz1p, both Bzz1p and Myo5p, or both Myo5p and Myo3p. Extracts from bzz1Δ and bzz1Δ myo5Δ strains polymerized actin around 6His-Las17p beads, whereas extract from the myo3Δ myo5Δ strain did not (Fig. 9A). Thus, actin polymerization in this system did not require Bzz1p or the shared function of Bzz1p and Myo5p, but did require the shared function of the type I myosins.
FIG. 9.
Las17p can also recruit the actin polymerization machinery in vitro. (A) Ni-NTA-agarose beads coated with six His fused to full-length Las17p were incubated in the presence of small amounts of Alexa-labeled actin with extracts from either the wild-type (WT) strain (CLW001), the bzz1Δ strain (FSW701K), the bzz1Δ myo5Δ strain (CLW571KT), or the myo3Δ myo5Δ strain (RLY822) or without yeast extract. After 10 to 20 min of incubation at room temperature, the fluorescence signal was visualized. The photos shown below indicate representative positive (+) and negative (−) signals. Panel B summarizes the results obtained.
DISCUSSION
Las17p, like other WASP/SCAR family proteins, is a key regulator of the cortical actin cytoskeleton that stimulates the actin nucleation activity of the Arp2/3 complex (for reviews, see references 20 and 39). Thus, investigation of cellular signals able to modulate the activity of Las17p is of particular interest to better understand the dynamics and function of the actin cytoskeleton. In a previous two-hybrid screen to identify potential Las17p partners, we found that Las17p could interact with several SH3 domain-containing proteins, including Rvs167p and four previously uncharacterized proteins, Lsb1p to -4p (41). Here, we report the characterization of Bzz1p, another SH3 domain-containing protein found in the extension of the two-hybrid screen and by affinity purification of Las17p-associated proteins.
Bzz1p, a new PCH family protein, interacts directly with Las17p in yeast extracts and in vivo.
BZZ1 codes for a 633-aa protein that is a member of the PCH family, containing proteins such as Cdc15p in S. pombe, Hof1p in S. cerevisiae, PSTPIP in M. musculus, and CIP4p in H. sapiens (37). All of the proteins of this family present a highly conserved organization of their predicted structural domains. They contain an N-terminal FCH domain, the nonkinase domain of the FER and Fes/Fps family of tyrosine kinases (3), and a putative coiled-coil region near their amino terminus, one or two SH3 domain(s) at the carboxyl terminus, and a proline-glutamic acid-serine-threonine-rich (PEST) sequence between the coiled-coil region and the SH3 domain (37). Most of the characterized proteins of the PCH family are known to be involved in actin cytoskeleton functions, such as cytokinesis, endocytosis, and actin organization, and to interact with WASP family proteins (for review, see reference 37). Syndapin, for example, has been linked to endocytosis, because it interacts and colocalizes with dynamin I, a protein implicated in endocytosis. Furthermore, syndapin I interacts with N-WASP through the proline-rich domain, and its overexpression has a marked effect on cortical actin organization, inducing filopodia in mammalian neural cells (52, 53). These findings are consistent with a direct role of syndapin I in actin dynamics. Another PCH family protein, the human protein CIP4p, was shown to interact with the Rho GTPase CDC42p and with WASP through an SH3 domain-proline-rich domain binding and thus participates in GTPase-dependent regulation of the actin cytoskeleton (3, 62). Finally, Hof1p/Cyk2p, an S. cerevisiae PCH protein, appears to be involved in cytokinesis. Indeed, Hof1p colocalizes with the acto-myosin ring and septin at the cleavage furrow during cytokinesis, and its deletion provokes a disassembly of this ring just when the contraction starts (26, 36). Moreover, Hof1p is able to interact with Vrp1p, the yeast homologue of WIP and Las17p, two interacting proteins implicated in cortical actin patch regulation (41, 48, 64).
Overall, the information cited above supports the idea that Bzz1p might participate in actin dynamics in yeast, despite the lack of an obvious actin phenotype in the deletion mutant or in wild-type cells overexpressing Bzz1p. This stimulated us to determine whether the Bzz1p-Las17p interaction found in the two-hybrid screen reflected its in vivo function. We found that Bzz1p was able to interact directly with purified Las17p in vitro and that Bzz1p coimmunoprecpitated with Las17p from yeast extracts, confirming the initial two-hybrid interaction. The demonstration that Bzz1p bound directly to Las17p through its SH3 domains in the two-hybrid and GST pull-down systems led us to map the regions of Las17p participating in the interaction with Bzz1p. The WASP/SCAR protein family presents highly conserved organization of their structural domains. They share a central proline-rich region with a variable number of polyproline stretches followed by a WH2 (WASP homology 2) domain and an acidic region in the C terminus. Additionally, WASP and N-WASP contain a GTPase binding domain (GBD) located between the WH1 and proline-rich regions. The amino-terminal region of the molecule defines functional subgroups based on the presence of either a WH1 domain found in WASP, N-WASP, Wsp1p (S. pombe), and Las17p (S. cerevisiae) or a SCAR homology (SH) domain specific to the known SCAR isoforms (reviewed in reference 9). When we analyzed (with a two-hybrid system) the interaction of Bzz1p with truncations of Las17p containing one or more of the different domains described, we found that Bzz1p interacted with the central polyproline region of Las17p. The central proline-rich domain of Las17p does not seem to be the unique site of interaction with Bzz1p SH3 domains. Indeed, in a two-hybrid screen with Bzz1p as bait, we identified another region of interaction in Las17p that is not part of this central region. This Las17p fragment corresponds to aa 91 to 209 and contains a polyproline stretch (182-HKAPPPPPPT-191) that could mediate the interaction with Bzz1p. During the preparation of the manuscript, the results of a general yeast SH3 domain interaction network study (63) also demonstrated that this Las17p polyproline stretch was capable of interacting with SH3 domains of Bzz1p. Their study shows that Bzz1p interacts with a polyproline stretch (aa 339 to 366) located in the central proline-rich region of Las17p and that Bzz1p and Las17p coimmunoprecipitate. These independent findings are in agreement with our directed two-hybrid and coprecipitation results. Thus, Bzz1p appears to interact through its SH3 domains with at least two and probably more proline-rich sequences in Las17p.
We also found that the proteins Bbc1p/Mti1p (myosin tail region-interacting protein) and Hsp70p could coimmunoprecipitate with Las17p. Thus, it is possible that these two proteins are part of the Las17p/Vrp1p/Myo3/5p complex. This possibility is corroborated for Bbc1p by very recent results showing that Bbc1p was a component of cortical actin patches in yeast and that it could interact physically with the type I myosin Myo5p and genetically with Vrp1p. Deletion of BBC1 suppressed phenotypes provoked by a VRP1 deletion (44, 63). The presence of Hsp70p in this biochemical complex raises the questions both of a chaperone role for this protein in the formation of the cortical complex and whether it influences the actin polymerization stimulatory activity of the complex.
Bzz1p is a cortical actin cytoskeleton component dependent on Las17p.
At the same time, we were interested in the localization of Bzz1p in yeast cells and its relationship to Las17p and actin organization. Employment of GFP-Bzz1p and the Bzz1-HAp fusion protein allowed us to demonstrate that Bzz1p localized in highly polarized cortical patch structures. These patches showed movements typical of actin patches (12, 66) in live cells and in fixed cells colocalized at least partially with actin patches and Las17p throughout the cell cycle. Furthermore, in the presence of the actin depolymerization drug latrunculin A, GFP-Bzz1p localized in polarized cortical patches in the absence of any detectable concentration of filamentous actin, as do Vrp1p (64), Las17p (41), Cdc42p, and other proteins important for polarity development in yeast (4). Compared to the polarized localization of other proteins important for actin cytoskeleton and secretion, such as Arp2p, Aip1p, or Sec4p (4), which are dependent on actin for cortical recruitment, Bzz1p must be upstream of actin with respect to the flow of polarity information. Furthermore, Bzz1p appears to be directly dependent on Las17p, since GFP-Bzz1p is not able to localize in polarized patch structures when cells show deletion of LAS17, even though some actin patches and aggregates are still present in the cell. The opposite is not true, because Las17p-GFP still localizes in polarized cortical patches in a strain with BZZ1 deleted. In a las17Δ strain, Bzz1p is unstable and must be rapidly degraded, since it is barely detectable in steady-state cell extracts. It is unlikely that this instability is due to the GFP moiety, since we found that GFP-Bzz1p was stable and functional in a wild-type background. Furthermore, the GFP moiety is fused to the N terminus and the SH3 domains interacting with Las17p are at the C terminus. Thus, these results place Bzz1p downstream of Las17p and upstream of actin. It has been recently reported that Las17p patches are not able to polarize at emerging bud sites in the absence of Vrp1p, but are distributed randomly in the cells (30). This is consistent with the earlier finding that in cells with VRP1 mutated, cortical actin patches are disorganized and depolarized (11). In light of this, it is interesting to point out that GFP-Bzz1p is still able to localize in cortical patches in cells lacking Vrp1p or lacking the type I myosins Myo3p and Myo5p (our unpublished data). However, these patches have lost polarity and are distributed randomly between mother and bud.
Is Bzz1p a component of the Las17p/Vrp1p/Myo3/5p complex?
Recent investigations have demonstrated that Las17p, the WIP homologue Vrp1p, and the type I myosins could be part of the same complex implicated in regulation of actin polymerization (14, 30, 32). These proteins interact with each other (1, 14, 32, 41). Particularly, type I myosins coimmunoprecipitate stoichiometrically with Las17p (32). Vrp1p and Las17p, which were previously found to interact in suppression and two-hybrid analyses (41, 49), cofractionate completely in a high-molecular-mass stable complex (30). According to our results presented above, we postulate that Bzz1p is part of this complex. Indeed, Bzz1p is able to interact directly with Las17p in vitro and in vivo. We found that Bzz1p interacted with Myo5p in the two-hybrid system and that it coimmunoprecipitated with the type I myosins and Las17p from cell extracts. Importantly, Vrp1p and actin also coimmunoprecipitated with Las17p in this experiment. In light of the stoichiometric precipitation of Myo5p with Las17p first reported by Lechler et al. (32) and confirmed here, our hypothesis that Lsb7 is part of this complex is reinforced. During the preparation of the manuscript, proteomic analyses in accord with our results have been reported. First, analysis of an interaction network of yeast SH3 proteins has shown that Bzz1p is able to interact directly with Las17p and with Myo5p in the two-hybrid system and by coprecipitation (63). In another large-scale study, systematic analyses of protein immunoprecipitates from yeast cell extracts and compilation of the resulting proteome complexes placed Bzz1p in a large protein complex that included Las17p, Vrp1p, and Sla1p (17).
Bzz1p is implicated in the regulation of the cortical actin cytoskeleton by the Las17p/Vrp1p/Myo3/5p complex.
We have demonstrated that the Bzz1p-Myo5p interaction is functionally significant. Consistent with this hypothesis, Bzz1p shares at least one redundant function with the two type I myosins. Indeed, deletion of BZZ1 is synthetically lethal with a myo3Δ myo5Δ double mutant. Furthermore, Bzz1p seems to be implicated specifically with Myo5p rather than Myo3p in another specific function. Codeletion of the BZZ1 and MYO5 genes strongly enhances the sensitivity to a high concentration of NaCl and salts of other monovalent cations. Such sensitivity was not observed in a strain with BZZ1 and MYO3 codeleted. Other functional differences between Myo3p and Myo5p also have been described. Indeed, myo5Δ mutant cells, but not myo3Δ cells, exhibit a partial receptor-mediated endocytosis defect at elevated temperature and show synthetic lethality with mutations in VRP1 or ARP3 genes (18, 19). NaCl stress and generally salt stress are known to provoke delayed growth in S. cerevisiae, during which the actin cytoskeleton completely depolarizes. After 1 to 2 h, adaptation to the high salt concentration occurs, and the actin cytoskeleton completely repolarizes (10). NaCl stress of a myo5Δ bzz1Δ double mutant results in a loss of the capacity of the cortical actin cytoskeleton to repolarize. This suggests that Myo5p and Bzz1p could be required together for the recognition of sites of repolarization after salt stress. Thus, they would be an important factor or target of the pathway needed for adaptation of the actin cytoskeleton to salt stress.
Recently, Lechler et al. (30) reported data supporting a model in which the Las17p/Vrp1p/MyoI complex could contain two sets of independent Arp2/3 complex activators. Las17p activates directly, whereas the other activation function is shared between Vrp1p and type I myosins (30). The Arp2/3 complex alone promotes inefficient polymerization, and kinetic studies have shown a concentration-independent initial lag in filament formation (45). The C terminus of the WASP/SCAR family proteins decreases the initial lag and increases the slope of polymerization curves (40). Our results indicate that Bzz1p is able to recruit Arp2/3 complex-dependent actin polymerization in an in vitro assay. Here we have also shown that the recruitment of the actin polymerization machinery is made through the SH3 domains of Bzz1p, since they function equally as well as the full-length Lsb7 protein. Accumulation of fluorescence on the GST-Bzz1p-coated beads depends not only on the Arp2/3 complex, but also on the presence of Las17p, Vrp1p, and type I myosins. The question arises of whether Bzz1p itself is necessary for the triggering of actin polymerization. This does not appear to be the case, at least in this assay. Effectively, with 6His-Las17p-coated beads, induction of actin polymerization depends on the presence of type I myosins, but not on the presence of Bzz1p, since a bzz1Δ extract supported actin polymerization as well as wild-type extracts. This may well represent functional redundancy of Bzz1p with other SH3 domain-containing proteins. The fact that actin polymerization depended on the presence of Myo5p and Myo3p in both cases is in agreement with the redundancy of type I myosins in stimulating actin polymerization in other S. cerevisiae cell-free systems (30).
Our in vitro results also raise the question of whether Bzz1p is involved solely in recruitment of the actin-polymerizing machinery or whether the accumulation of actin filaments around Bzz1p-coated beads also represents a stimulation of polymerization. A possible interpretation of our overall polymerization assay results is that both Bzz1p and Las17p are capable of recruiting a functional complex, which then triggers or induces actin polymerization. While this explanation seems most likely, our data do not exclude the possibility that Bzz1p, as part of a functional Las17p/Vrp1p/myosin I complex (containing other proteins as well) might be implicated not only in recruitment, but also in regulation or activation of actin polymerization. One may also note that a myo5Δ bzz1Δ extract was capable of supporting actin polymerization, whereas the myo5Δ bzz1Δ strain from which the extract was derived was incapable of recovery from salt-induced stress. Bzz1p thus appears to participate in at least two different functions of the actin cytoskeleton: one through Las17p and the other with Myo5p.
Acknowledgments
We thank M. Crouzet, M. Geli, L. Pon, and M. Fromont-Racine for strains and plasmids and M. White and P. Legrain for two-hybrid banks. We give particular thanks to Chantal Burgard for LAS17 plasmid constructions and members of the Winsor and Li laboratories for support and encouragement.
This work was financed by French National Research (Centre National de la Recherche Scientifique) funding to the FRE 2375 laboratory and by French Cancer Association (Association pour la Recherche sur le Cancer) grants to B. Winsor. A. Soulard was supported during this work by a scholarship from the French government (Ministère de la Recherche).
REFERENCES
- 1.Anderson, B. L., I. Boldogh, M. Evangelista, C. Boone, L. A. Greene, and L. A. Pon. 1998. The Src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polarization. J. Cell Biol. 141:1357-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anton, I. M., W. Lu, B. J. Mayer, N. Ramesh, and R. S. Geha. 1998. The Wiskott-Aldrich syndrome protein-interacting protein (WIP) binds to the adaptor protein Nck. J. Biol. Chem. 273:20992-20995. [DOI] [PubMed] [Google Scholar]
- 3.Aspenstrom, P. 1997. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7:479-487. [DOI] [PubMed] [Google Scholar]
- 4.Ayscough, K. R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D. G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399-416. (Erratum, 146:1201, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barylko, B., D. D. Binns, and J. P. Albanesi. 2000. Regulation of the enzymatic and motor activities of myosin I. Biochim. Biophys. Acta 1496:23-35. [DOI] [PubMed] [Google Scholar]
- 6.Bon, E., P. Recordon-Navarro, P. Durrens, M. Iwase, E. A. Toh, and M. Aigle. 2000. A network of proteins around rvs167p and rvs161p, two proteins related to the yeast actin cytoskeleton. Yeast 16:1229-1241. [DOI] [PubMed] [Google Scholar]
- 7.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50:643-650. [DOI] [PubMed] [Google Scholar]
- 8.Carlier, M. F., P. Nioche, L. H. I. Broutin, R. Boujemaa, C. Le Clainche, C. Egile, C. Garbay, A. Ducruix, P. Sansonetti, and D. Pantaloni. 2000. GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich syndrome protein (N-WASp) with actin-related protein (ARP2/3) complex. J. Biol. Chem. 275:21946-21952. [DOI] [PubMed] [Google Scholar]
- 9.Caron, E. 2002. Regulation of Wiskott-Aldrich syndrome protein and related molecules. Curr. Opin. Cell Biol. 14:82-87. [DOI] [PubMed] [Google Scholar]
- 10.Chowdhury, S., K. W. Smith, and M. C. Gustin. 1992. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J. Cell Biol. 118:561-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly, S. F., M. J. Pocklington, D. Pallotta, and E. Orr. 1993. A proline-rich protein, verprolin, involved in cytoskeletal organization and cellular growth in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 10:585-596. [DOI] [PubMed] [Google Scholar]
- 12.Doyle, T., and D. Botstein. 1996. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. USA 93:3886-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 14.Evangelista, M., B. M. Klebl, A. H. Tong, B. A. Webb, T. Leeuw, E. Leberer, M. Whiteway, D. Y. Thomas, and C. Boone. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148:353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401:926-929. [DOI] [PubMed] [Google Scholar]
- 16.Fromont-Racine, M., J. C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277-282. [DOI] [PubMed] [Google Scholar]
- 17.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 18.Geli, M. I., R. Lombardi, B. Schmelzl, and H. Riezman. 2000. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 19:4281-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geli, M. I., and H. Riezman. 1996. Role of type I myosins in receptor-mediated endocytosis in yeast. Science 272:533-535. [DOI] [PubMed] [Google Scholar]
- 20.Goode, B. L., and A. A. Rodal. 2001. Modular complexes that regulate actin assembly in budding yeast. Curr. Opin. Microbiol. 4:703-712. [DOI] [PubMed] [Google Scholar]
- 21.Goodson, H. V., B. L. Anderson, H. M. Warrick, L. A. Pon, and J. A. Spudich. 1996. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133:1277-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, New York, N.Y.
- 23.Hill, J., K. A. Donald, D. E. Griffiths, and G. Donald. 1991. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 19:5791.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idrissi, F. Z., B. L. Wolf, and M. I. Geli. Cofilin, but not profilin, is required for myosin-I-induced actin polymerization and the endocytic uptake in yeast. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 25.Johnson, D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamei, T., K. Tanaka, T. Hihara, M. Umikawa, H. Imamura, M. Kikyo, K. Ozaki, and Y. Takai. 1998. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273:28341-28345. [DOI] [PubMed] [Google Scholar]
- 27.Kandels-Lewis, S., and B. Seraphin. 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262:2035-2039. [DOI] [PubMed] [Google Scholar]
- 28.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lechler, T., G. A. Jonsdottir, S. K. Klee, D. Pellman, and R. Li. 2001. A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast. J. Cell Biol. 155:261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lechler, T., and R. Li. 1997. In vitro reconstitution of cortical actin assembly sites in budding yeast. J. Cell Biol. 138:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lechler, T., A. Shevchenko, and R. Li. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol 148:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, W. L., M. Bezanilla, and T. D. Pollard. 2000. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, R. 1997. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 136:649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, W., and H. She. 2000. The SH2 and SH3 adapter Nck: a two-gene family and a linker between tyrosine kinases and multiple signaling networks. Histol. Histopathol. 15:947-955. [DOI] [PubMed] [Google Scholar]
- 36.Lippincott, J., and R. Li. 1998. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143:1947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lippincott, J., and R. Li. 2000. Involvement of PCH family proteins in cytokinesis and actin distribution. Microsc. Res. Tech. 49:168-172. [DOI] [PubMed] [Google Scholar]
- 38.Ma, L., R. Rohatgi, and M. W. Kirschner. 1998. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc. Natl. Acad. Sci. USA 95:15362-15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machesky, L. M., and R. H. Insall. 1999. Signaling to actin dynamics. J. Cell Biol. 146:267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machesky, L. M., R. D. Mullins, H. N. Higgs, D. A. Kaiser, L. Blanchoin, R. C. May, M. E. Hall, and T. D. Pollard. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 96:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madania, A., P. Dumoulin, S. Grava, H. Kitamoto, C. Scharer-Brodbeck, A. Soulard, V. Moreau, and B. Winsor. 1999. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 10:3521-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer, B. J. 2001. SH3 domains: complexity in moderation. J. Cell Sci. 114:1253-1263. [DOI] [PubMed] [Google Scholar]
- 43.Miki, H., K. Miura, and T. Takenawa. 1996. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 15:5326-5335. [PMC free article] [PubMed] [Google Scholar]
- 44.Mochida, J., T. Yamamoto, K. Fujimura-Kamada, and K. Tanaka. 2002. The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott- Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics 160:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullins, R. D., J. A. Heuser, and T. D. Pollard. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95:6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munn, A. L., B. J. Stevenson, M. I. Geli, and H. Riezman. 1995. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 6:1721-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naqvi, S. N., Q. Feng, V. J. Boulton, R. Zahn, and A. L. Munn. 2001. Vrp1p functions in both actomyosin ring-dependent and Hof1p-dependent pathways of cytokinesis. Traffic 2:189-201. [DOI] [PubMed] [Google Scholar]
- 49.Naqvi, S. N., R. Zahn, D. A. Mitchell, B. J. Stevenson, and A. L. Munn. 1998. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr. Biol. 8:959-962. [DOI] [PubMed] [Google Scholar]
- 50.Niedenthal, R. K., L. Riles, M. Johnston, and J. H. Hegemann. 1996. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12:773-786. [DOI] [PubMed] [Google Scholar]
- 51.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 52.Qualmann, B., and R. B. Kelly. 2000. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J. Cell Biol. 148:1047-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qualmann, B., J. Roos, P. J. DiGregorio, and R. B. Kelly. 1999. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol. Biol. Cell 10:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramesh, N., I. M. Anton, N. Martinez-Quiles, and R. S. Geha. 1999. Waltzing with WASP. Trends Cell Biol. 9:15-19. [DOI] [PubMed] [Google Scholar]
- 55.Rivero-Lezcano, O. M., A. Marcilla, J. H. Sameshima, and K. C. Robbins. 1995. Wiskott-Aldrich syndrome protein physically associates with Nck through Src homology 3 domains. Mol. Cell. Biol. 15:5725-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rohatgi, R., P. Nollau, H. Y. Ho, M. W. Kirschner, and B. J. Mayer. 2001. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 276:26448-26452. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Shevchenko, A., O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, H. Boucherie, and M. Mann. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 93:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 60.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takenawa, T., and H. Miki. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801-1809. [DOI] [PubMed] [Google Scholar]
- 62.Tian, L., D. L. Nelson, and D. M. Stewart. 2000. Cdc42-interacting protein 4 mediates binding of the Wiskott-Aldrich syndrome protein to microtubules. J. Biol. Chem. 275:7854-7861. [DOI] [PubMed] [Google Scholar]
- 63.Tong, A. H., B. Drees, G. Nardelli, G. D. Bader, B. Brannetti, L. Castagnoli, M. Evangelista, S. Ferracuti, B. Nelson, S. Paoluzi, M. Quondam, A. Zucconi, C. W. Hogue, S. Fields, C. Boone, and G. Cesareni. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321-324. [DOI] [PubMed] [Google Scholar]
- 64.Vaduva, G., N. C. Martin, and A. K. Hopper. 1997. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J. Cell Biol. 139:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 66.Waddle, J. A., T. S. Karpova, R. H. Waterston, and J. A. Cooper. 1996. Movement of cortical actin patches in yeast. J. Cell Biol. 132:861-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wear, M. A., D. A. Schafer, and J. A. Cooper. 2000. Actin dynamics: assembly and disassembly of actin networks. Curr. Biol. 10:R891-R895. [DOI] [PubMed] [Google Scholar]
- 68.Winter, D., T. Lechler, and R. Li. 1999. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 9:501-504. [DOI] [PubMed] [Google Scholar]
- 69.Wu, C., V. Lytvyn, D. Y. Thomas, and E. Leberer. 1997. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 272:30623-30626. [DOI] [PubMed] [Google Scholar]