Abstract
For a pathogen such as Streptococcus pyogenes, ecological success is determined by its ability to sense the environment and mount an appropriate adaptive transcriptional response. Thus, determining conditions for analyses of gene expression in vitro that are representative of the in vivo environment is critical for understanding the contributions of transcriptional response pathways to pathogenesis. In this study, we determined that the gene encoding the SpeB cysteine protease is up-regulated over the course of infection in a murine soft-tissue model. Conditions were identified, including growth phase, acidic pH, and an NaCl concentration of <0.1 M, that were required for expression of speB in vitro. Analysis of global expression profiles in response to these conditions in vitro identified a set of coregulated genes whose expression patterns showed a significant correlation with that of speB when examined during infection of murine soft tissues. This analysis revealed that a culture medium that promotes high levels of SpeB expression in vitro produced an expression profile that showed significant correlation to the profile observed in vivo. Taken together, these studies establish culture conditions that mimic in vivo expression patterns; that growth phase, pH, and NaCl may mimic relevant cues sensed by S. pyogenes during infection; and that identification of other environmental cues that alter expression of speB in vitro may provide insight into the signals that direct global patterns of gene expression in vivo.
With its remarkable ability to adapt to a variety of human tissues, Streptococcus pyogenes (group A streptococcus) provides a unique opportunity to investigate the complex regulatory systems responsible for sensing and responding to environmental changes in the dynamic host environment. Numerous virulence factors have been described that allow this single species of bacterium to produce a wide range of degrees of disease severity and a wide range of clinical manifestations, including pharyngitis and impetigo and invasive diseases such as necrotizing fasciitis, septicemia, and toxic-shock-like syndrome (15). How different streptococcal virulence factors interact with the host to produce these diverse diseases is unknown. However, it is likely that the development of any of these diseases requires that virulence factor expression be highly regulated in an ordered spatial and temporal fashion. Consistent with this, several regulatory factors have been identified which modulate transcription of various virulence genes in response to different environmental cues (reviewed in reference 31). However, the specific signals that are sensed in tissue to control the regulatory network remain largely unknown.
Insight into the types of signals that may be sensed in vivo has come mainly from analyses of virulence gene expression by use of in vitro models. Typically, cultures are grown in an artificial medium and the affect of alterations of a specific medium component or growth condition on transcription of genes controlled by a known regulatory pathway is monitored. For S. pyogenes, this strategy has been used to show that Mg2+ is sensed by the CovRS (CsrRS) two-component system (24) and that this pathway also responds to changes in pH, temperature, and osmolarity (16). Similarly, expression of genes regulated by the transcription activator Mga is altered by changes in iron limitation, temperature, O2, and CO2 conditions (5, 36, 52). Similar cues regulate genes controlled by members of the RofA/Nra family of transcription factors (21, 41, 52). The challenge in using these in vitro culture models has been to establish whether similar cues influence these regulatory pathways during the course of infection in tissue.
A complication for this analysis is that many virulence genes are also regulated by growth phase, including those regulated by CovR, Mga, and RofA/Nra (19, 37, 41). Growth-phase cues may be sensed as population densities (34, 60), as cell cycle status (48), or as a consequence of growth-induced alterations to the bacterium's immediate environment, including the production of metabolic byproducts or the consumption of nutrients (53, 54, 61). How growth-phase cues are integrated into the individual signaling pathways that control S. pyogenes virulence is not understood. However, it has been hypothesized that individual regulatory elements sense environmental cues that are altered by growth and that this behavior globally controls the temporal progression of gene expression in vivo (31). Testing this idea requires a more thorough understanding of the types of environmental cues that are sensed by the organism in tissue.
One approach to the search for relevant signals is to focus on a single virulence gene that is known to respond to multiple environmental signals in vitro. In addition, the ideal target gene should be expressed at levels that allow sensitive monitoring in vivo, and it should be transcribed in a growth-phase-dependent pattern. An attractive candidate for this analysis is speB, which encodes the SpeB cysteine protease. Although its role is controversial, SpeB may affect the severity and dissemination of streptococcal infections (1, 2, 25, 28, 30, 32, 55). Existing data derived from murine, primate, and teleost models of infection indicate that the speB message is produced during the course of infection of soft tissue (23, 58, 59) and muscle (12). It is also known that the biogenesis of SpeB proteolytic activity is tightly regulated, both at the transcriptional and posttranscriptional levels (reviewed in reference 14). A number of environmental factors modulate protease activity during in vitro culture growth, including bacterial cell density, atmospheric conditions, nutrient availability, carbon source depletion, temperature, and pH (7, 34, 39, 40, 52). Also, in the presence of the appropriate cues, speB is expressed in a strictly growth-phase-dependent pattern during the transition from the logarithmic to the stationary phase of in vitro growth, and it may be the most highly expressed gene at this time. However, how the temporal and environmental cues interact is not understood.
It is known that transcription of speB requires activation by the Rgg family member RopB (6, 33), which binds to sequences in the speB promoter region (38). The transcription of ropB itself is also subject to growth-phase control; however, disregulation of ropB transcription does not uncouple speB from its growth-phase pattern of expression (38). It is also not clear whether the temporal cues act independently of environmental cues or are a product of alterations to the environment produced by bacterial growth. For example, it has not been possible to uncouple expression from its temporal pattern through alteration of culture medium composition, including addition of spent culture medium (7), or by modification of regulatory protein expression (23, 38). Other regulators, including CovR and Mga, have also been implicated in speB regulation (19, 27, 42). However, how these regulatory pathways are integrated with RopB and temporal control is also not understood. Taken together, the observations that speB expression can be detected in vivo and that it responds to growth phase and multiple independent and overlapping regulatory pathways suggests that speB expression could serve as a useful probe for understanding streptococcal virulence gene regulation in vivo.
In this work, we examined the relationship between transcription of speB in vitro and transcription within infected tissue. Comparison of gene expression profiles of speB and of genes coregulated with speB under various in vitro growth conditions and in infected tissue defined a set of in vitro conditions that showed a strong correlation with profiles observed during infection of murine subcutaneous tissue. These studies establish an in vitro model for further investigation of virulence gene regulation that reflects expression patterns observed in vivo and suggests that environmental cues that promote expression of speB in vitro, including growth phase, pH, NaCl concentration, and a carbohydrate-poor and peptide-rich nutritional environment, may be conditions S. pyogenes encounters during infection of soft tissue. Further analyses of speB regulation in this in vitro model will likely provide additional insight into virulence gene regulatory programs in vivo.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Streptococcus pyogenes strain HSC5 was used for these experiments (26). Selected experiments utilized strain JWR100 derived from HSC5, where the wild-type speB gene was replaced with a version encoding a C192S amino acid substitution that ablates enzymatic activity by allelic replacement (J. Rosche and M. Caparon, unpublished). Bacteria were grown at 37°C without agitation in C medium as described previously (33); unless otherwise indicated, unmodified medium was adjusted to pH 7.5 with NaOH. Buffered medium was prepared by adding 1 M HEPES (pH 7.5) to achieve a final concentration of 0.1 M (Sigma) prior to autoclaving. For buffered acidic media, 1 M MES (morpholineethanesulfonic acid) (Sigma) adjusted to pH 6.5, 6.3, or 6.0 with HCl was added to achieve a final concentration of 0.1 M prior to autoclaving. In selected experiments NaCl was added to the final concentrations described in the text prior to autoclaving. For salt specificity experiments, NaCl, KCl, MgCl2, NaHPO4, or glycerol (all obtained from Sigma) was added to media to achieve a final concentration of 0.15 M prior to autoclaving.
Protease activity assays.
Analysis of SpeB proteolytic activity in cell-free supernatants with the substrate fluorescein isothiocyanate-casein was conducted as described previously (33). Sample volumes were adjusted for differences in culture density based on absorbance measured according to optical density at 600 nm (OD600), and uninoculated C medium was used to determine background values. Proteolytic activity is presented as relative to the activity of HSC5 in unmodified C medium. As described previously (33), the cysteine protease inhibitor E64 was added to selected samples to confirm that the protease activity measured was dependent on SpeB. Values reported represent the means and standard errors of the means for at least two independent experiments.
RNA isolation and real-time RT-PCR.
Overnight cultures of S. pyogenes were diluted 1:100 into 30 ml of unmodified or modified C medium and grown at 37°C until midexponential phase (∼4 h; OD600 ∼ 0.3) or the onset of stationary phase (∼6 h; OD600 ∼ 0.5). Cells were harvested by centrifugation, and total RNA was isolated as described elsewhere (3). For isolation of RNA from in vivo-grown bacteria, groups of outbred, immunocompetent, hairless female mice (Jackson Laboratories) were used for subcutaneous injection of midexponential phase inocula of strain HSC5 (approximately 107 CFU) as described previously (4). At 1, 2, or 3 days postinoculation the mice were sacrificed and the dermis and underlying soft tissues at the site of infection from at least five animals were harvested, pooled, and partially homogenized as described previously (4). Extracts were prepared by organic extraction and chaotropic disruption (RNeasy lipid tissue kit; QIAGEN). The extract was further homogenized using a reciprocal shaking device and a commercial extraction reagent, FastPrep lysing matrix B (Qbiogene), and total RNA was purified as described above. Real-time reverse transcription-PCR (RT-PCR) was conducted as described elsewhere (3) by use of the primers listed in Table S5 in the supplementary material. For the in vitro and in vivo conditions described in this study, the range of abundance of recA transcript in samples with similar amounts of total RNA was less than twofold; hence, transcript abundance was normalized to the abundance of recA and relative transcript abundance was calculated as described previously (3). Data represent the means from a minimum of two experiments performed on different days in which RNAs from at least two independent cultures were analyzed in triplicate. For comparison of in vitro and in vivo gene expression results, the values for individual genes in each data set were plotted in the x (in vitro) and y (in vivo) dimensions and the best-fit straight line was determined by the least-squares method as described previously (12). When correlation is high, R approaches 1.0.
Transcript analysis by DNA microarray.
Genomic arrays were designed and produced as described previously (12). Loci are numbered in accordance with the completed serotype M1 Streptococcus pyogenes SF370 genome (20). Bacterial growth and RNA isolation were as described above, and the synthesis, labeling, and hybridization of cDNA were performed as detailed elsewhere (12). Expression ratios are representative of duplicate experiments performed with RNA from two independent cultures where each sample was analyzed eight times. Fluorescence values from each array were measured and normalized as described previously (12). The output expression ratio (log2 ratio) was converted into severalfold change data for comparison with real-time RT-PCR data. This average value represents the amount of cDNA binding relative to a given probe in competitive hybridization and is presented as a severalfold difference in the results obtained with experimental (modified) C medium relative to control (unmodified) C medium results. For each probe, the null hypothesis of equal binding of control and experimentally derived cDNA was tested for significance computationally using Significance Analysis of Microarrays (SAM, version 1.21; http://www-stat.stanford.edu/∼tibs/SAM) (57) as described previously (12) with the following settings: log2-based transformation of severalfold change values and a one-class response format. Genes considered differentially expressed fulfilled the following criteria: the null hypothesis was rejected when the estimated false discovery rate at the 90th percentile was ≤0.1% and the severalfold change value was at least ±2.0. The complete list of positive and negative significant genes meeting these criteria for each experimental condition is detailed in the supplemental material.
RESULTS
Expression of speB is regulated during development of a cutaneous infection.
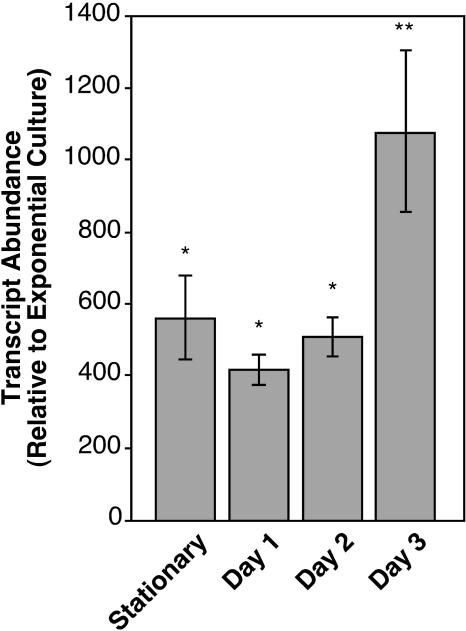
It has been shown that the speB transcript can be detected 48 h following inoculation in a murine model of cutaneous infection (23). However, temporal patterns of speB expression during the development of the ulcerous lesion in this model have not been examined. To investigate this issue, RNA was isolated from infected mouse tissue at various time points and transcript abundance of speB was determined by real-time RT-PCR. For S. pyogenes strain HSC5, the kinetics of ulcer development have been described in detail elsewhere (4). Briefly, at 8 to 12 h postinfection, this strain produces a well-defined area of inflammation that is characterized by the recruitment of large numbers of neutrophils. By 24 h postinfection, this region begins to ulcerate. At this time point, the speB transcript was readily detected in infected tissue and was expressed at levels about 600-fold higher than in the initial inoculum. This level was similar to levels observed in stationary in vitro culture (stationary and day 1; Fig. 1). Following 24 h, the ulcer expands in size to include a maximum area at day 3. At this time point the amount of speB transcript was more than twofold up-regulated relative to day 1 results to levels greater than 1,000-fold higher than the initial inoculum level (day 3; Fig. 1). These data establish that speB expression can be readily detected in infected tissue, that its expression is highly up-regulated versus the initial inoculum results, and that its expression increased over the course of ulcer formation. The observation that speB is highly up-regulated during infection of tissue suggests that further analysis of the environmental cues that promote speB regulation in vitro may be useful for understanding the regulatory cues that direct virulence gene expression in vivo.
FIG. 1.
Transcription of speB is regulated during infection. The figure presents relative transcript abundances of speB. Comparisons are shown of bacteria grown in broth culture to midexponential or early stationary phase or collected at 1, 2, and 3 days postinfection from mice infected subcutaneously. Relative transcript abundance was determined by real-time RT-PCR, and transcript levels are reported as the level of transcript relative to that detected in broth-grown bacteria at midexponential phase. Data represent the means and standard deviations of three independent experiments for samples analyzed in duplicate. Statistically significant (P < 0.001) differences from the exponential-phase samples are indicated by an asterisk. A significant (P < 0.001) difference between day 3 and day 1 expression is indicated by two asterisks.
Environmental pH influences the pattern of protease expression.
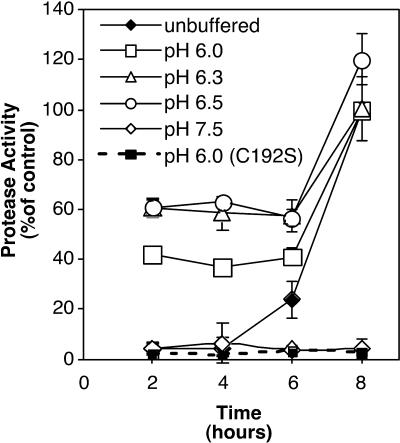
As discussed above, it has been proposed that alterations to the environment induced by streptococcal growth over time provide external cues that drive temporal patterns of gene expression. This predicts that these cues will regulate virulence gene expression similarly during in vitro and in vivo growth. A prominent growth-induced environmental alteration made by S. pyogenes is autoacidification resulting from the accumulation of organic acids generated as end products of its fermentative metabolism. For example, during culture of S. pyogenes HSC5, the pH of medium drops from an initial value of 7.5 to less than 6.0 at the onset of stationary phase. Consistent with this, it has long been known that the generation of SpeB protease activity is linked to low pH (13, 17, 22), although the relative importance of transcriptional versus posttranscriptional events in driving regulation has not been carefully examined. In medium buffered to maintain a consistent pH at a range of values, analysis of secreted proteolytic activities demonstrated that while supernatants harvested from unbuffered cultures had the expected growth-phase-dependent pattern of activity, bacteria grown in media buffered at pH 7.5 failed to produce activity at any later time points (Fig. 2). In contrast, buffering media to maintain a pH at acidic values less than 6.5 partially uncoupled the expression of protease activity from its temporal pattern, as these cultures expressed SpeB cysteine protease activity in early exponential phase at levels of up to 60% of that produced by unbuffered cultures at the onset of stationary phase (Fig. 2). There was no observable difference between the rate of growth under any of the conditions tested and that seen with unmodified medium, and when supernatants from all cultures were subjected to Western blot analysis, it was observed that the amount of proteolytic activity corresponded to the amount of detectable SpeB protein (data not shown). Lack of protease expression in a mutant strain with an inactive SpeB derivative (JWR100) under acidic conditions demonstrated that this proteolytic activity was SpeB specific (Fig. 2).
FIG. 2.
Protease activity is influenced by environmental pH. Protease activity present in culture supernatants was measured at various time points during the growth of the cultures by use of an assay based on the cleavage of fluorescein isothiocyanate-labeled casein. Media included unbuffered C medium, C medium buffered with 0.1 M HEPES (pH 7.5), and C medium buffered with 0.1 M MES (pH 6.0, 6.3, and 6.5). A derivative of the wild-type strain, JWR100 (C192S), which produces no SpeB proteolytic activity, was grown in acidic C medium (pH 6.0) and tested to control for the specificity of the reaction. Activity is presented as a percentage of the maximal protease activity produced by HSC5 at 8 h in unbuffered medium. Data represent the means and standard deviations for two independent experiments for samples analyzed in triplicate.
Protease expression is influenced by changes in NaCl concentrations.
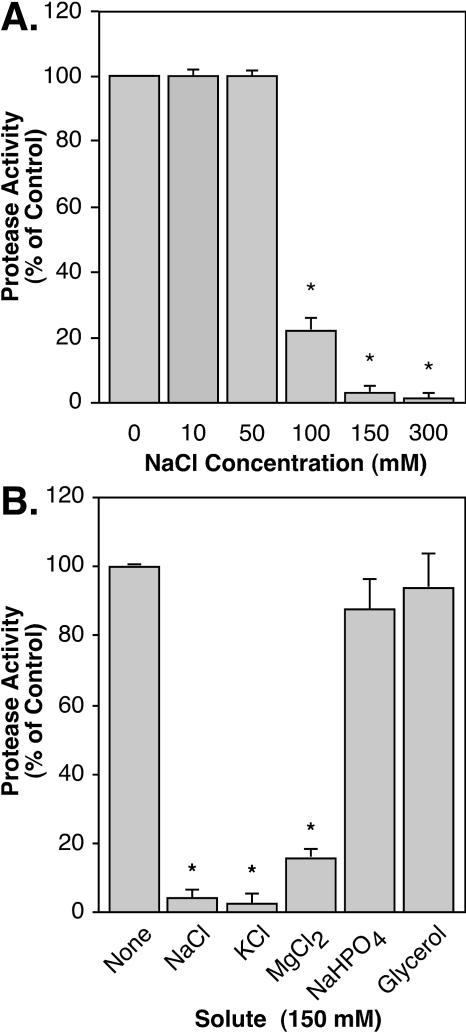
It has long been known that medium composition can alter expression of S. pyogenes virulence factors (7, 13). C medium is known to promote high-level speB expression (22). Among other characteristics, it is low in salt (0.017 M) relative to other streptococcal cultivation media. When examined, it was found that speB expression was sensitive to the concentration of NaCl in the medium. Analysis of secreted proteolytic activity revealed cultures grown in media containing greater than 100 mM added NaCl produced dramatically reduced amounts of proteolytic activity (Fig. 3A). At physiological concentrations of NaCl (150 mM) virtually no protease activity was detected (Fig. 3A). There was no observable difference between the rate of growth under any of the conditions tested and that seen with unmodified medium, and Western blot analysis revealed that failure to produce proteolytic activity was due to the absence of secreted SpeB polypeptide (data not shown).
FIG. 3.
Environmental chloride reduces protease activity. Protease activity present in culture supernatants was measured after 10 h using an assay based on the cleavage of fluorescein isothiocyanate-labeled casein. Media included unmodified C medium (“0” in panel A; “None” in panel B) and C medium supplemented as indicated. Activity is presented as a percentage of the protease activity produced by HSC5 in unmodified C medium. Data represent the means and standard deviations from three independent experiments for samples analyzed in triplicate. Statistically significant differences from unmodified medium are indicated by an asterisk (P < 0.001).
To gain insight into the mechanism of the NaCl-mediated repression of SpeB expression, a number of additional compounds were tested. The addition of glycerol, an osmolyte not fermented by S. pyogenes, did not inhibit protease expression (Fig. 3B), suggesting that alterations of medium osmolarity are not responsible for repression. Addition of Na+ in the form of NaHPO4 also did not reproduce the inhibitory effect (Fig. 3B). However, the addition of Cl− in the form of KCl or MgCl2 did mimic the inhibitory effect of NaCl (Fig. 3B), suggesting that the regulatory pathway is sensitive to the concentration of Cl− rather than Na+.
Neutral pH and increased NaCl influence transcription of speB but not ropB.
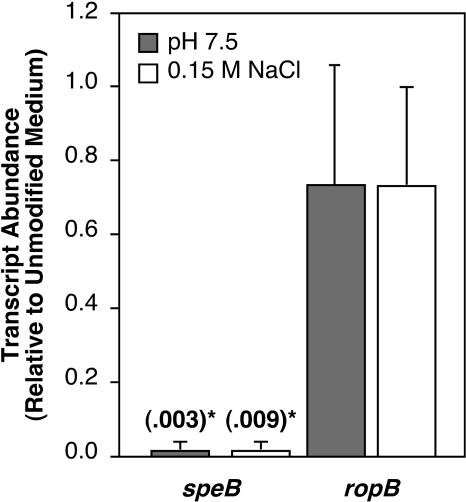
To determine whether the failure of S. pyogenes to secrete active SpeB in response to particular pH and salt conditions was at the level of speB transcription, we measured relative speB transcript levels in bacteria grown under conditions that repressed SpeB activity, including buffered C medium (pH 7.5) or C medium with added NaCl (0.15 M). Under each condition, the speB transcript was at least 100-fold less abundant relative to the unmodified medium results (Fig. 4). To determine whether the effect of pH and NaCl on speB expression was an indirect effect of repression of ropB, a transcriptional regulator essential for expression of speB, the relative transcript levels of ropB under each set of conditions were also measured. No significant differences in ropB expression were detected (Fig. 4). Furthermore, ectopic expression of ropB from a constitutive promoter did not affect speB repression under these conditions (data not shown). Combined with a previous observation (38), these data suggest that the major environmental speB regulatory cues are processed downstream of ropB transcription.
FIG. 4.
Transcription of speB is repressed at pH 7.5 and 0.15 M NaCl. The results shown represent relative levels of transcription of speB and ropB grown to early stationary phase. Expression levels are compared for unmodified C medium and C medium buffered to pH 7.5 with 0.1 M HEPES or C medium supplemented with an additional 0.15 M NaCl. Relative transcript abundances were determined by real-time RT-PCR. Transcript levels are reported relative to that detected in unmodified C medium. For selected bars, transcript abundance values are also shown in parentheses. Data represent the means and standard deviations of at least three independent experiments for samples analyzed in triplicate. Statistically significant (P < 0.001) differences from unmodified medium results are indicated by an asterisk.
Neutral pH and increased NaCl alter the global transcript profile of stationary phase cultures.
If pH and NaCl alter signaling pathways that are important for virulence in vivo, then it is likely that other genes are regulated in response to these signals. To examine this, differential expression profiles were determined for cultures grown to the onset of stationary phase in unmodified C medium for comparison with bacteria grown in both C medium buffered at pH 7.5 and C medium with 0.15 M NaCl added. These comparisons revealed that growth in buffered medium resulted in significant up-regulation of approximately 10% of the total number of genes in the genome (149 genes) and significant down-regulation of approximately 8% (115 genes) relative to growth in unmodified medium (Tables S1 and S2 in the supplemental material). To gain insight into the profile of the global response to pH, differentially regulated genes were assigned a code according to the COG (Clusters of Orthologous Groups) (56) system of functional classification (Table 1). Among the differentially regulated genes were many predicted to encode products associated with storage or processing of information (COGs J, K, and L; 76 genes in total) and with cellular metabolism (COGs C, G, F, H, I, and Q; 68 genes in total). These results are consistent with previous analysis of a related bacterium, Lactococcus lactis, for which it was determined that most genes differentially regulated by autoacidification were involved with glycolytic and fermentative pathways (18). Fewer differences were observed when the NaCl concentration was altered. In this case only 3% (43 genes) of the total number of genes in the genome were up-regulated and 3% (49 genes) were down-regulated (Tables S3 and S4 in the supplemental material). Again, gene products were mainly associated with information storage and processing (17 genes) or metabolism (31 genes), as determined as described above (Table 1).
TABLE 1.
Number of genes significantly up-regulated or down-regulated during growth in modified C medium versus unmodified C medium
| Gene code | Gene descriptiona (no. of genes in categoryb) | No. (%) of genes detected in C medium under the indicated conditionsc
|
|||
|---|---|---|---|---|---|
| pH 7.5
|
0.15 M NaCl
|
||||
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | ||
| J | Translation (140) | 39 (27.8) | 5 (3.6) | 1 (0.7) | 7 (5) |
| K | Transcription (93) | 9 (9.7) | 9 (9.7) | 1 (1.1) | 4 (4.3) |
| L | Replication, recombination, and repair (95) | 6 (6.3) | 5 (5.3) | 1 (1.1) | 3 (3.2) |
| C | Energy production and conversion (57) | 5 (8.8) | 6 (10.5) | 10 (17.5) | 0 (0) |
| E | Amino acid transport and metabolism (99) | 8 (8.1) | 0 (0) | 1 (1.0) | 0 (0) |
| F | Nucleotide transport and metabolism (59) | 10 (16.9) | 1 (1.7) | 2 (3.4) | 0 (0) |
| G | Carbohydrate transport and metabolism (124) | 12 (9.7) | 12 (9.7) | 6 (4.8) | 8 (6.5) |
| H | Coenzyme transport and metabolism (34) | 3 (8.8) | 1 (2.9) | 1 (2.9) | 0 (0) |
| I | Lipid transport and metabolism (45) | 5 (11.1) | 2 (4.4) | 1 (2.2) | 1 (2.2) |
| Q | Secondary metabolite biosynthesis, transport, and catabolism (11) | 1 (9.1) | 0 (0) | 0 (0) | 1 (9.1) |
| D | Cell cycle control, mitosis, and meiosis (16) | 2 (12.5) | 0 (0) | 0 (0) | 0 (0) |
| M | Cell wall/membrane biogenesis (64) | 2 (3.1) | 2 (3.1) | 0 (0) | 0 (0) |
| O | Posttranslational modification, protein turnover, chaperones (50) | 2 (4) | 8 (16) | 0 (0) | 7 (14) |
| P | Inorganic ion transport and metabolism (58) | 2 (3.4) | 5 (8.6) | 0 (0) | 1 (1.7) |
| T | Signal transduction mechanisms (37) | 3 (8.1) | 2 (5.4) | 0 (0) | 0 (0) |
| U | Intracellular trafficking and secretion (14) | 2 (14.3) | 1 (7.1) | 0 (0) | 0 (0) |
| V | Defence mechanisms (36) | 4 (11.1) | 4 (11.1) | 3 (8.3) | 1 (2.8) |
| R | General function prediction only (137) | 10 (7.3) | 5 (3.6) | 3 (2.2) | 3 (2.2) |
| S | Function unknown (126) | 7 (5.6) | 16 (12.7) | 0 (0) | 2 (1.6) |
| None | Not in COGs (222) | 17 (7.7) | 31 (14) | 13 (5.9) | 11 (5) |
| Totald | 149 (9.8) | 115 (7.6) | 43 (2.8) | 49 (3.2) | |
Genes were categorized according to a functional classification, COGs (clusters of orthologous groups) (56).
Structural RNA genes (tRNA and rRNA) and phage genes were omitted for clarity.
Number of genes displaying statistically significant differential expression in the modified medium as opposed to unmodified medium fulfilling the following criteria: average severalfold change of at least ±2.0 and a false discovery rate in SAM (57) of 90th percentile = 0.1%. Percentages represent numbers of statistically significant genes versus the total number of genes assigned to the category.
Total number of genes in all categories, 1,517.
Global transcript profile reveals a coordinated response to pH and NaCl.
The genes regulated by both pH and NaCl all followed the same trend of up-regulation under both conditions or down-regulation under both conditions (Fig. 5), highlighting the possibility that these signals are interpreted by overlapping pathways. All together, 22 genes were up-regulated under both the pH and the NaCl conditions examined (Table 2) and 28 genes were down-regulated by both conditions (Table 3). The differential regulation of a subset of these genes was confirmed by real-time RT-PCR, and results obtained with the two techniques were qualitatively similar (Table 2 and Table 3). Notably, transcription of a number of genes encoding stress adaptation and known or putative virulence factors responded to both the pH and the NaCl conditions (Table 2 and Table 3), suggesting that these conditions mimic cues encountered during infection.
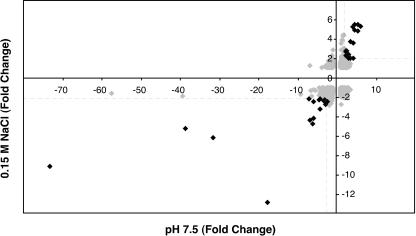
FIG. 5.
Expression of multiple genes is influenced by both pH 7.5 and 0.15 M NaCl. Total levels of RNA prepared from cultures grown to early stationary phase in C medium buffered with 0.1 M HEPES (pH 7.5) or in C medium supplemented with an additional 0.15 M NaCl were analyzed in competitive hybridization with cDNA from control cultures (unmodified C medium) by use of S. pyogenes DNA microarrays as described in Materials and Methods. Each gene included in the analysis is represented by a diamond; each value represents the average severalfold change in transcript abundance relative to results obtained with unmodified medium. Dashed lines denote twofold changes, and black diamonds represent genes that had statistically significant differential expression results in both experimental conditions from those seen under control conditions as described in Materials and Methods. See Table 1 and Table 2 for gene names and values.
TABLE 2.
Genes displaying increased transcript abundance under both pH 7.5 and 0.15 M NaCl conditionsa
| Locus | Gene | Putative function | Fold change in expression under the indicated conditionsb
|
|
|---|---|---|---|---|
| pH 7.5 | 0.15 M NaCl | |||
| SPy0145 | spy0145 | Putative endoribonuclease | 2.17 | 2.39 |
| SPy0165 | spn | NAD glycohydrolase precursorc | 5.73 (4.7) | 5.23 (7.4) |
| SPy0166 | spy0166 | Hypothetical proteinc | 6.56 | 5.02 |
| SPy0167 | slo | Streptolysin O precursorc | 4.37 (2.8) | 5.19 (3.7) |
| SPy0739 | sagB | Streptolysin S biogenesisd | 3.59 (3.4) | 3.61 (3.2) |
| SPy0740 | sagC | Streptolysin S biogenesisd | 3.42 | 3.39 |
| SPy0741 | sagD | Streptolysin S biogenesisd | 2.36 | 2.66 |
| SPy0742 | sagE | Streptolysin S biogenesisd | 2.57 | 2.80 |
| SPy0743 | sagF | Streptolysin S biogenesisd | 2.09 | 2.71 |
| SPy0744 | sagG | Streptolysin S biogenesisd | 2.45 | 2.79 |
| SPy0745 | sagH | Streptolysin S biogenesisd | 2.46 | 2.76 |
| SPy0746 | sagI | Streptolysin S biogenesisd | 2.29 | 2.26 |
| SPy0747 | spy0747 | Predicted extracellular nuclease | 2.15 | 2.40 |
| SPy0843 | spy0843 | Putative surface antigen | 2.81 | 2.03 |
| SPy1151 | ldh | Putative l-lactate dehydrogenase | 3.13 | 2.35 |
| SPy1378 | nrdF.2 | Ribonucleotide diphosphate reductase subunit | 4.19 | 2.01 |
| SPy1718 | spy1718 | Putative esterase | 2.49 | 2.36 |
| Spy1738 | manL | Putative mannose PTS IIABe | 4.96 (5.9) | 4.66 (2.3) |
| SPy1739 | manM | Putative mannose PTS IICe | 4.45 | 4.93 |
| SPy1740 | manN | Putative mannose PTS IIDe | 4.81 | 5.02 |
| SPy1939 | spy1939 | Hypothetical protein | 2.57 | 2.06 |
| SPy2019 | mga | M-protein trans-acting positive regulator | 2.88 | 2.36 |
Genomic loci, gene names, and predicted functions are based on current annotation of the genome of M1 SF370 of S. pyogenes (20) in the National Center for Biotechnology Information database. PTS, phosphotransferase system.
Severalfold change in expression in cultures grown to early stationary phase in pH 7.5 buffered medium or 0.15 M added NaCl medium versus cultures grown in unmodified control medium as determined by DNA microarray and real-time RT-PCR. Real-time RT-PCR data (shown in parentheses) were not collected for all genes that appeared to be organized in a monocistronic operon or for genes that were differentially regulated less than threefold as shown by microarray analysis.
Putative operon.
Putative operon.
Putative operon.
TABLE 3.
Genes displaying decreased transcript abundance under both pH 7.5 and 0.15 M NaCl conditionsa
| Locus | Gene | Putative function | Fold change in expression under the indicated conditionsb
|
|
|---|---|---|---|---|
| pH 7.5 | 0.15 M NaCl | |||
| SPy0912 | spy0912 | Hypothetical protein | −4.27 (−6.1) | −3.27 (−3.9) |
| SPy1060 | spy1060 | Putative PTS IID (mannose specific) | −2.51 | −2.57 |
| SPy1158 | spy1158 | Putative sphingosine kinase | −2.81 | −2.72 |
| SPy1405 | spy1405 | Hypothetical protein | −2.12 | −2.04 |
| SPy1557 | msrA | Putative peptide methionine sulfoxide reductase | −3.23 | −2.38 |
| SPy1559 | ccdA | Putative cytochrome c biogenesis protein | −2.28 | −2.51 |
| SPy1628 | fmt | Putative methionyl tRNA formyltransferase | −2.95 | −2.08 |
| SPy1724 | susA | Transcription termination-antitermination factor | −2.40 | −2.63 |
| SPy1725 | spy1725 | Conserved hypothetical protein | −2.77 | −2.79 |
| SPy1746 | fabZ | Putative beta-hydroxyacyl-ACP dehydratase | −3.10 | −2.46 |
| SPy1759 | dnaJ | Heat shock (chaperone) protein | −2.63 | −2.08 |
| SPy1760 | dnaK | Heat shock protein 70 | −2.12 | −2.45 |
| SPy1761 | grpE | Putative Hsp-70 cofactor | −2.93 | −2.57 |
| SPy1763 | hrcA | Putative heat shock transcription repressor | −2.70 | −2.65 |
| SPy1815 | scrA | Putative sucrose PTS IIc | −7.03 | −2.15 |
| SPy1816 | scrB | Putative sucrose-6-phosphate hydrolasec | −5.81 | −2.44 |
| SPy1915 | salA | Lantibiotic precursor | −5.94 (−9.4) | −4.49 (−5.1) |
| SPy2033 | spy2033 | Hypothetical protein | −2.74 | −2.45 |
| SPy2037 | spy2037 | Conserved hypothetical proteind | −73.07 | −9.10 |
| SPy2039 | speB | Pyrogenic exotoxin Bd | −31.63 (−363.3) | −6.19 (−273.2) |
| SPy2040 | spy2040 | Hypothetical proteind | −38.70 | −5.24 |
| SPy2041 | spy2041 | Hypothetical proteind | −17.75 | −12.86 |
| SPy2050 | spy2050 | Putative PTS IICe | −4.19 | −2.22 |
| SPy2051 | spy2051 | Putative PTS IIBe | −4.56 | −2.29 |
| SPy2147 | spy2174 | Hypothetical protein | −2.35 | −2.49 |
| SPy2169 | spy2169 | Putative membrane proteinf | −5.94 (−4.3) | −4.17 (−5.9) |
| SPy2170 | spy2170 | Hypothetical proteinf | −6.08 | −4.15 |
| SPy2193 | spy2193 | Conserved hypothetical protein | −2.32 | −2.36 |
Genomic loci, gene names, and predicted functions are based on current annotation of the genome of M1 SF370 of S. pyogenes (20) in the National Center for Biotechnology Information database. PTS, phosphotransferase system.
Severalfold change in expression in cultures grown to early stationary phase in pH 7.5 buffered medium or 0.15 M added NaCl medium versus cultures grown in unmodified control medium as determined by DNA microarray or real-time RT-PCR. Real-time RT-PCR data (shown in parentheses) were not collected for all genes that appeared to be organized in a monocistronic operon or for genes that were differentially regulated less than threefold as determined by microarray analysis.
Putative operon.
Putative operon.
Putative operon.
Putative operon.
In vitro growth conditions promote gene expression similar to expression in a model of soft-tissue infection.
We reasoned that if the environmental signals affecting the growth-phase-dependent regulation of speB were similar to signals encountered in the host, then genes regulated similarly to speB in vitro should also be similarly regulated in infected host tissue. To test this, real-time RT-PCR was used to measure expression of a panel of genes from RNA recovered from infected murine tissue relative to their expression in exponential culture. These data were then compared with expression in modified and unmodified media. Criteria for target gene selection included genes that were highly up- or down-regulated in stationary phase according to pH and/or NaCl conditions. This analysis revealed that for the panel of 10 genes analyzed, 9 demonstrated a greater than twofold level of regulation in unmodified medium (Table 4), indicative of growth-phase-dependent regulation of gene expression. Furthermore, there was a strong correlation between the relative expression levels obtained for tissue and those observed in unmodified medium (Runmodified = 0.996). Alteration of either pH or NaCl values produced expression profiles that correlated poorly with the tissue profile (RpH = 0.114, RNaCl = 0.007), suggesting that signals such as those detected in response to late stages of in vitro growth that alter speB expression are likely to participate in coordinate regulation in infected soft tissue.
TABLE 4.
Expression in stationary bacteria in C media and tissue relative to exponential culture resultsa
| Locus | Gene (putative function) | Fold change in expression under the indicated conditionsb
|
|||
|---|---|---|---|---|---|
| Tissue | Unmodified mediumc | pH 7.5d | 0.15 M NaCle | ||
| SPy2039 | speB (cysteine protease) | 1,083.10 | 588.56* | 1.10 | 1.99 |
| SPy1915 | salA (lantibiotic precursor) | 15.83 | 5.13* | −1.86 | 1.56 |
| SPy0739 | sagB (SLS biogenesis) | 2.22 | 2.00* | 5.53* | 5.80* |
| SPy1065 | spn (NAD glycohydrolase) | −1.11 | −1.41* | 3.30 | 3.91 |
| SPy1738 | manL (mannose PTS II) | −11.97 | −8.11* | −1.06 | −2.54* |
| SPy1378 | nrdF.2 (ribonucleotide diphosphate reductase) | −4.16 | −21.06* | −1.79 | −20.58* |
| Spy1228 | spy1228 (lipoprotein) | −6.58 | 2.26 | 7.14 | 1.85 |
| SPy1109 | malP (l-malate permease) | 2.10 | 42.47* | −3.73 | 25.22* |
| SPy1815 | scrA (sucrose PTS II) | 7.30 | 7.23* | 1.29 | 6.22* |
| SPy0149 | ntpK (sodium ATPase) | −58.21 | −10.14* | −9.94* | −1.58 |
Genomic loci, gene names, and predicted functions are based on current annotation of the genome of M1 SF370 of S. pyogenes (20) in the National Center for Biotechnology Information database.
Severalfold change in transcript abundance relative to unmodified medium exponential culture as determined by real-time RT-PCR. Broth cultures were harvested at early stationary phase, and tissue samples were harvested from the ulcer of infected hairless mice 3 days after inoculation of wild-type bacteria. An asterisk denotes genes for which the transcript profiles show the same trend as in the tissue sample (either both up- or both down-regulated).
R, 0.996. (The calculation used to determine correlation coefficient values presented in the table is described in Materials and Methods.)
R, 0.114.
R, 0.007.
DISCUSSION
Our examination of speB regulation revealed a possible role for pH and salt in coordinating a transcriptional response of other temporally regulated genes in S. pyogenes. Furthermore, in vivo expression of a panel of these genes fits a transcriptional profile characteristic of stationary-phase cultures grown in unmodified C medium. Taken together, these data suggest (i) that growth-phase-dependent regulation of virulence genes likely occurs during infection; (ii) that environmental cues that modulate expression of temporally regulated genes in vitro may be encountered at the site of infection; and (iii) that further analysis of the environmental cues and regulatory pathways that control speB expression under the in vitro conditions used in this study will likely reveal regulatory phenomena important for virulence gene regulation during infection.
The utility of using speB as a model virulence gene is enhanced by the fact that it is regulated by a large number of different signals in vitro. The regulatory pathways must integrate these signals and somehow interact with the essential speB activator RopB in order to control the output of the speB promoter. Signal integration also plays a major role in virulence gene regulation in the gram-positive pathogen Staphylococcus aureus. Like S. pyogenes, S. aureus produces numerous adhesins and toxins whose expression is coordinately regulated by both environmental and temporal cues, often in a growth-phase and tissue-specific pattern (reviewed in references 11 and 43). It is rare for any specific staphylococcal virulence gene to be under the control of a single linear pathway or of several pathways whose effects are additive. Rather, gene subsets are coordinately controlled by multiple overlapping and interacting feedback networks that appear to function in an ordered hierarchy involving two-component regulators, a regulatory RNA (RNAIII), an alternate sigma factor (sigma B), a peptide-based quorum-sensing system (Agr), and a large family of related transcription factors known as the SarA family. It is not at all clear that a similar hierarchal regulatory network functions in S. pyogenes. For example, the S. pyogenes genome is considerably smaller (∼1.8 versus ∼2.8 Mb), and it does not appear to encode either sigma B or a large family of related transcription factors that could act analogously to the SarA family. The S. pyogenes genome does encode numerous two-component regulators, of which one, CovRS (CsrRS), has been associated with global temporal regulation (19). However, CovRS acts as a repressor of a large number of genes, including speB (27), and mutation of the gene encoding the CovR response regulator only partially uncouples speB from its temporal signal (19) and does not appear to uncouple it from any of its necessary environmental cues (J. Loughman and M. Caparon, unpublished). Thus, in contrast to S. aureus results, coordinate virulence regulation in S. pyogenes may function through the cooperative effects of multiple linear pathways. Our finding that pH can partially, but not completely, uncouple speB expression from its temporal pattern also supports this mechanism for global control.
Additional support for a linear network model comes from observations that speB expression in biofilm, in muscle tissue in the zebrafish myositis model (12), and in the murine subcutaneous model (this study) is much higher than can be observed during in vitro planktonic culture. This may suggest that an additional cue(s) is present in these environments whose effects are additive with those generated by the other cues. On the other hand, it is possible that a different set of signals is sensed during growth in in vitro culture as opposed to growth in tissue. However, the data presented in this study do not support this contention in that it is unlikely that a large number of genes would demonstrate similar coordinate regulation results in vitro and in vivo if different cues and regulatory pathways were involved. The issue of in vitro versus in vivo modeling of gene regulation has become important for understanding how the hierarchical regulatory networks function in S. aureus, because some regulatory elements have been shown to be less important in controlling gene expression in several animal models of infection than has been predicted on the basis in vitro culture (for a review, see references 11 and 43). These observations highlight the importance of developing in vitro models for gene regulation that mimic as closely as possible the conditions encountered during infection.
Our finding that the tissue environment produced an expression profile most similar to that directed by unmodified medium in stationary-phase cultures and that the correlation was dependent upon acidification suggests that infected tissue is low in pH. In similarity to in vitro culture results, growth-induced autoacidification may be a mechanism by which a low pH environment is produced in tissue. The formation of an abscess or a necrotic lesion may also expose the organisms to low pH (51, 62). In addition, during infection S. pyogenes may encounter distinct microenvironments characterized by differences in local pH. For example, measurement of human skin pH has revealed an “acid mantle,” or lower pH at the surface, thought to be important for epidermal lipid organization and metabolism (45). Changes in surface pH are associated with conditions such as atopic dermatitis and may disturb skin barrier function and host defense mechanisms, and it has been shown that a number of conditions can lead to acidification of the lower levels of the stratum corneum (44). In this regard it is interesting that SpeB is an important virulence factor for infecting the stratum corneum in a humanized murine model of impetigo (55).
While pH is well studied, rather little is known about the biological function of chloride in prokaryotes. It has been speculated that some bacteria sense the salt concentration of their environment via chloride concentrations because of the dependence on this anion for growth and regulation of essential metabolic pathways (46, 47). Interestingly, chloride has also been implicated in the response to acid stress. For example, an Escherichia coli mutant lacking a CLC-type chloride channel was found to be acid sensitive (29, 35). The chloride channels are activated at low pH, and it has been proposed that the channels function as an electrical shunt for a proton pump linked to amino acid decarboxylation when bacteria encounter an acidic environment (29). A role for chloride as a cue for transcriptional regulation has been reported for Lactococcus lactis. In this circuit, genes encoding a glutamate-dependent acid resistance mechanism are transcribed by a chloride-inducible promoter (49, 50). Intriguingly, this regulation is under the control of GadR, a member of the Rgg-like family of transcriptional regulators that includes the speB regulator RopB (49). In similarity to the results seen with RopB regulation of speB, expression of gadR is not regulated by chloride (49). Thus, chloride or a chloride-induced factor may alter a function of both regulators, such as altering their affinity for binding to DNA. Chloride may be an important cue in vivo, where salt concentrations may differ depending on the location (skin surface versus subcutaneous), and alterations may result from inflammation at the site of infection (51, 62).
The mechanism by which the pH and chloride signals are integrated downstream of RopB is not understood. However, in addition to its role as an activator of speB transcription, RopB has been implicated in growth-phase-dependent regulation of genes associated with metabolism and stress responses via repression of amino acid catabolism (8-10). Autoacidification in lactic acid bacteria leading to cytoplasmic acidification has also been linked to perturbations in catabolic flux that result in alterations in transcription of metabolic enzymes (18). Thus, alterations to metabolism may link RopB with the environmental cues that are sensed to regulate speB expression. Whether environmental or nutritional cues are directly sensed by RopB or via the interaction between RopB and other regulator remains to be determined. However, this model predicts that metabolic flux is responsible for environmental and temporal patterns of speB regulation.
Differences in metabolic flux are induced by growth in specific microenvironments, and the environmental conditions that are present in those microenvironments may influence the subsets of genes that are expressed temporally during infection. In turn, these alterations in virulence gene expression may influence the manifestation of disease that develops in any one host compartment. This idea is supported by a recent longitudinal analysis of virulence-related gene expression in an experimental model of streptococcal pharyngitis in cynomolgus macaques, which revealed a complex pattern of changes in transcript abundance that could be correlated with phases of disease development (59). Consistent with this, there appears to be little overlap between the sets of genes temporally regulated in the primate pharynx and those observed in murine subcutaneous tissue in the present study. Further testing of this idea will require additional studies that combine the analysis of model genes in relevant in vitro models with in vivo transcriptional profiling to define signature transcriptional programs that may provide insight into the pathogenesis of individual S. pyogenes diseases.
Supplementary Material
Acknowledgments
We thank J. Rosch for providing strain JWR100. We also thank Wess Warren, Seth Crosby, and Michael Heinz in the microarray core facility at Washington University for technical assistance with the microarray analysis.
This work was supported by Public Health Service grant AI046433 from the National Institutes of Health to M.C.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ashbaugh, C. D., H. B. Warren, V. J. Carey, and M. R. Wessels. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Investig. 102:550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjorck, L., P. Akesson, M. Bohus, J. Trojnar, M. Abrahamson, I. Olafsson, and A. Grubb. 1989. Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature 337:385-386. [DOI] [PubMed] [Google Scholar]
- 3.Brenot, A., K. Y. King, and M. G. Caparon. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55:221-234. [DOI] [PubMed] [Google Scholar]
- 4.Brenot, A., K. Y. King, B. Janowiak, O. Griffith, and M. G. Caparon. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaussee, M. S., E. R. Phillips, and J. J. Ferretti. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Cho, K. H., and M. G. Caparon. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57:1545-1556. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, J. O. 1969. Effect of culture medium composition and pH on the production of M protein and proteinase by group A streptococci. J. Bacteriol. 99:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collin, M., and A. Olsen. 2003. Extracellular enzymes with immunomodulating activities: variations on a theme in Streptococcus pyogenes. Infect. Immun. 71:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalton, T. L., and J. R. Scott. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, S. D. 1950. The crystallization and serological differentiation of a streptococcal proteinase and its precursor. J. Exp. Med. 92:201-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Even, S., N. D. Lindley, P. Loubiere, and M. Cocaign-Bousquet. 2002. Dynamic response of catabolic pathways to autoacidification in Lactococcus lactis: transcript profiling and stability in relation to metabolic and energetic constraints. Mol. Microbiol. 45:1143-1152. [DOI] [PubMed] [Google Scholar]
- 19.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogg, G. C., and M. G. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlach, D., H. Knoll, W. Kohler, J. H. Ozegowski, and V. Hribalova. 1983. Isolation and characterization of erythrogenic toxins. V. Communication: identity of erythrogenic toxin type B and streptococcal proteinase precursor. Zentbl. Bakteriol. Mikrobiol. Hyg. A 255:221-233. [PubMed] [Google Scholar]
- 23.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gryllos, I., J. C. Levin, and M. R. Wessels. 2003. The CsrR/CsrS two-component system of group A streptococcus responds to environmental Mg2+. Proc. Natl. Acad. Sci. USA 100:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gubba, S., D. E. Low, and J. M. Musser. 1998. Expression and characterization of group A streptococcus extracellular cysteine protease recombinant mutant proteins and documentation of seroconversion during human invasive disease episodes. Infect. Immun. 66:765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanski, E., P. A. Horwitz, and M. G. Caparon. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holm, S. E., A. Norrby, A. M. Bergholm, and M. Norgren. 1992. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988-1989. J. Infect. Dis. 166:31-37. [DOI] [PubMed] [Google Scholar]
- 29.Iyer, R., T. M. Iverson, A. Accardi, and C. Miller. 2002. A biological role for prokaryotic ClC chloride channels. Nature 419:715-718. [DOI] [PubMed] [Google Scholar]
- 30.Kansal, R. G., A. McGeer, D. E. Low, A. Norrby-Teglund, and M. Kotb. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun. 68:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 32.Lukomski, S., C. A. Montgomery, J. Rurangirwa, R. S. Geske, J. P. Barrish, G. J. Adams, and J. M. Musser. 1999. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect. Immun. 67:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 35.Maduke, M., D. J. Pheasant, and C. Miller. 1999. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J. Gen. Physiol. 114:713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podbielski, A., and B. A. Leonard. 1998. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol. Microbiol. 28:1323-1334. [DOI] [PubMed] [Google Scholar]
- 40.Podbielski, A., B. Pohl, M. Woischnik, C. Korner, K. H. Schmidt, E. Rozdzinski, and B. A. Leonard. 1996. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol. Microbiol. 21:1087-1099. [DOI] [PubMed] [Google Scholar]
- 41.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 42.Podbielski, A., M. Woischnik, B. Pohl, and K. H. Schmidt. 1996. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med. Microbiol. Immunol. (Berlin) 185:171-181. (In German.) [DOI] [PubMed] [Google Scholar]
- 43.Pragman, A. A., and P. M. Schlievert. 2004. Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. FEMS Immunol. Med. Microbiol. 42:147-154. [DOI] [PubMed] [Google Scholar]
- 44.Rippke, F., V. Schreiner, T. Doering, and H. I. Maibach. 2004. Stratum corneum pH in atopic dermatitis: impact on skin barrier function and colonization with Staphylococcus aureus. Am. J. Clin. Dermatol. 5:217-223. [DOI] [PubMed] [Google Scholar]
- 45.Rippke, F., V. Schreiner, and H. J. Schwanitz. 2002. The acidic milieu of the horny layer: new findings on the physiology and pathophysiology of skin pH. Am. J. Clin. Dermatol. 3:261-272. [DOI] [PubMed] [Google Scholar]
- 46.Roessler, M., and V. Muller. 2002. Chloride, a new environmental signal molecule involved in gene regulation in a moderately halophilic bacterium, Halobacillus halophilus. J. Bacteriol. 184:6207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roessler, M., and V. Muller. 2001. Osmoadaptation in bacteria and archaea: common principles and differences. Environ. Microbiol. 3:743-754. [DOI] [PubMed] [Google Scholar]
- 48.Ryan, K. R., and L. Shapiro. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72:367-394. [DOI] [PubMed] [Google Scholar]
- 49.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 50.Sanders, J. W., G. Venema, J. Kok, and K. Leenhouts. 1998. Identification of a sodium chloride-regulated promoter in Lactococcus lactis by single-copy chromosomal fusion with a reporter gene. Mol. Gen. Genet. 257:681-685. [DOI] [PubMed] [Google Scholar]
- 51.Simmen, H. P., H. Battaglia, P. Giovanoli, E. Hanseler, and J. Blaser. 1995. Biochemical analysis of peritoneal fluid in patients with and without bacterial infection. Eur. J. Surg. 161:23-27. [PubMed] [Google Scholar]
- 52.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 54.Steiner, K., and H. Malke. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svensson, M. D., D. A. Scaramuzzino, U. Sjobring, A. Olsen, C. Frank, and D. E. Bessen. 2000. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol. Microbiol. 38:242-253. [DOI] [PubMed] [Google Scholar]
- 56.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Virtaneva, K., M. R. Graham, S. F. Porcella, N. P. Hoe, H. Su, E. A. Graviss, T. J. Gardner, J. E. Allison, W. J. Lemon, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2003. Group A streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect. Immun. 71:2199-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 61.Whitehead, K. E., G. M. Webber, and R. R. England. 1998. Accumulation of ppGpp in Streptococcus pyogenes and Streptococcus rattus following amino acid starvation. FEMS Microbiol. Lett. 159:21-26. [DOI] [PubMed] [Google Scholar]
- 62.Wiese, K. G. 1994. Electrolyte concentration, real and osmotic pressure in abscesses. Zentlbl. Chir. 119:54-59. (In German.) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.