Abstract
Ku is an abundant nuclear protein with an essential function in the repair of DNA double-strand breaks. Various observations suggest that Ku also interacts with the cellular transcription machinery, although the mechanism and significance of this interaction are not well understood. In the present study, we investigated the subnuclear distribution of Ku in normally growing human cells by using confocal microscopy, chromatin immunoprecipitation, and protein immunoprecipitation. All three approaches indicated association of Ku with RNA polymerase II (RNAP II) elongation sites. This association occurred independently of the DNA-dependent protein kinase catalytic subunit and was highly selective. There was no detectable association with the initiating isoform of RNAP II or with the general transcription initiation factors. In vitro protein-protein interaction assays demonstrated that the association of Ku with elongation proteins is mediated, in part, by a discrete C-terminal domain in the Ku80 subunit. Functional disruption of this interaction with a dominant-negative mutant inhibited transcription in vitro and in vivo and suppressed cell growth. These results suggest that association of Ku with transcription sites is important for maintenance of global transcription levels. Tethering of double-strand break repair proteins to defined subnuclear structures may also be advantageous in maintenance of genome stability.
The Ku protein is part of the DNA-dependent protein kinase (DNA-PK), which is required for the nonhomologous end-joining pathway of DNA double-strand break (DSB) repair (53, 59). When a break occurs, Ku binds avidly to the DNA ends, translocates inward, and recruits the 470-kDa DNA-PK catalytic subunit (DNA-PKcs) to form an active protein kinase complex (19, 26, 74). Proteins in this complex cooperate with XRCC4, DNA ligase IV, and other factors to carry out DSB repair (reviewed in reference 22). Ku-deficient cells are sensitive to ionizing radiation and are unable to complete the process of V(D)J recombination, which involves a DSB intermediate.
In addition to its well-documented role in DSB repair, Ku appears to have some interaction with the RNA polymerase II (RNAP II) transcription apparatus. Several reports describe the binding of Ku to promoter regions or its ability to regulate transcription of individual genes (reviewed in reference 21 and also see references 11, 25, 31, and 72). Other reports describe interaction of Ku with the general transcription machinery. DNA-PK efficiently phosphorylates RNAP II in vitro (50), and a fraction of Ku resides in RNAP II-containing complexes (41). The importance of these interactions is suggested by the finding that nuclear extracts of Ku-deficient cells exhibit a characteristic transcription defect (70). Transcription of several different promoters is decreased two- to fivefold relative to that of extracts from a matched Ku-containing cell line (70). The defect is entirely at the level of reinitiation and is not seen in assays in which transcription is limited to a single round. Mechanistic studies suggest that the effect is mediated by direct interaction of Ku with transcription proteins. Neither DNA-PK-dependent protein phosphorylation nor stable association of Ku with the template seems to be required (70, 71). One hypothesis is that Ku accelerates recycling of limiting transcription factors to the promoter by influencing the higher-order organization of the transcription apparatus (71). For example, recruitment of a preformed complex is inherently more rapid than recruitment of the same proteins in a stepwise manner.
Previous work was performed in vitro with cell extracts in which the native organization of the transcription apparatus had been extensively disrupted. It was important to extend studies in vivo to determine whether Ku is associated with the transcription apparatus under physiological conditions in normally growing cells and whether disruption of this interaction interferes with transcription.
Transcription occurs in vivo within discrete structures, or “transcription factories” (15, 51, 75). RNAP II itself exists in dynamic equilibrium between an elongating isoform, RNAP IIO, and an initiating isoform, RNAP IIA, which differ in the phosphorylation states of their large subunits (subunits known as IIo and IIa, respectively) (reviewed in references 16 and 46). Phosphorylation of the RNAP II large subunit occurs cooperatively within a C-terminal domain composed of tandem copies of a heptad repeat, YSPTSPS (10). Early studies of isoform-specific functions suggested a relatively simple model in which phosphorylation was coupled to initiation of transcription (1, 10, 13, 35). More recent work, using phosphospecific antibodies to characterize isoform function in vivo, provides a complex picture involving multiple kinases, coupling of transcription to RNA processing, and dynamic relocalization of RNAP II within the nucleus (8, 33, 45, 55, 65, 69, 75). In the present study, we characterize the association of Ku with two RNAP II populations defined by reactivity with isoform-specific monoclonal antibodies (MAbs). MAb H5 recognizes the IIo subunit when it is phosphorylated at serine 2 of the heptad repeat. This form is present in transcription complexes that have cleared the promoter and are engaged in efficient, processive elongation (14, 34). MAb 8WG16 recognizes the nonphosphorylated IIa subunit, which is present in preinitiation complexes. Studies with model peptides indicate that both MAbs are highly isoform specific (48).
Initiating and elongating isoforms of RNAP II interact with different constellations of transcription factors. One of the factors associated with elongating RNAP II is DRB sensitivity-inducing factor (DSIF), isolated originally on the basis of its ability to sensitize a cell-free transcription system to the effects of the elongation inhibitor DRB (66). DSIF is composed of the human homologues of the Spt4 and Spt5 proteins, which have been studied in several other organisms (4, 28, 30, 32, 39). In Drosophila, the Spt4/5 complex colocalizes with RNAP IIO on polytene chromosomes and associates with transcribed DNA in a chromatin immunoprecipitation (ChIP) assay (4).
We show here that Ku associates with RNAP IIO and DSIF, but not with RNAP IIA or other markers of the preinitiation complex. Association with the elongation complex is mediated, in part, by a discrete structural domain of the Ku80 subunit. The isolated domain acts as a dominant-negative mutant, inhibiting transcription in vitro and in vivo. This suggests that the association between Ku and RNAP II is important for maintenance of global transcription levels.
MATERIALS AND METHODS
Materials.
HCT116 cells were a gift of E. Hendrickson (University of Minnesota), and MO59K and MO59J cells were a gift of J. Allalunis-Turner (University of Alberta). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Polyclonal antibodies against TFIIB, TFIID, TFIIE, TFIIF, and TFIIH were purchased from Santa Cruz Biotechnology. MAbs S10B1 (anti-Ku80), 25-4 (anti-DNA-PKcs), and N3H10 (anti-Ku70) were purchased from Neomarkers. MAb H5 (anti-RNAP IIo) was purchased from Covance. The anti-RNAP IIa 8WG16-producing cell line was a gift from N. Thompson and R. Burgess (60). Mouse anti-bromodeoxyuridine (BrdU) and rat anti-hemagglutinin (HA) MAbs were obtained from Roche. Rabbit anti-mouse immunoglobulin G2a (IgG2a) and anti-IgG2b and biotin-conjugated rat anti-mouse IgG1 were purchased from Zymed. Fluor 594- and 488-conjugated goat anti-mouse IgG, Fluor 594-conjugated goat anti-mouse IgM, Fluor 488-conjugated goat anti-rabbit IgG, and Fluor 594-conjugated goat anti-rat IgG were purchased from Molecular Probes. Cy3-avidin was obtained from Sigma. Plasmid pGEX-2T was obtained from Amersham Pharmacia Biotechnology, and pcDNA3.1(Zeocin), pD1Red N1(Neo), and pEGFP N1 were obtained from Invitrogen.
Immunostaining and in situ run-on transcription assay.
Cells were grown on glass coverslips, washed briefly with phosphate-buffered saline (PBS), fixed in PBS containing 4% formaldehyde for 10 min at room temperature, and permeabilized with acetone for 3 min at −20°C or with 0.5% Triton X-100 in PBS for 5 min at room temperature. Samples were blocked with PBS containing 3% bovine serum albumin (BSA) for 1 h at room temperature. Antibodies were diluted in the same buffer. Sequential incubation was performed with each primary antibody, with secondary antibodies, and, when necessary, with tertiary antibody or avidin. Antibodies were incubated at room temperature for 1 h. Detection of transcription by in situ run-on assay (Fig. 1) was performed as described previously (75). Imaging was performed with a Nikon confocal microscope. Images from each channel were recorded separately and then merged.
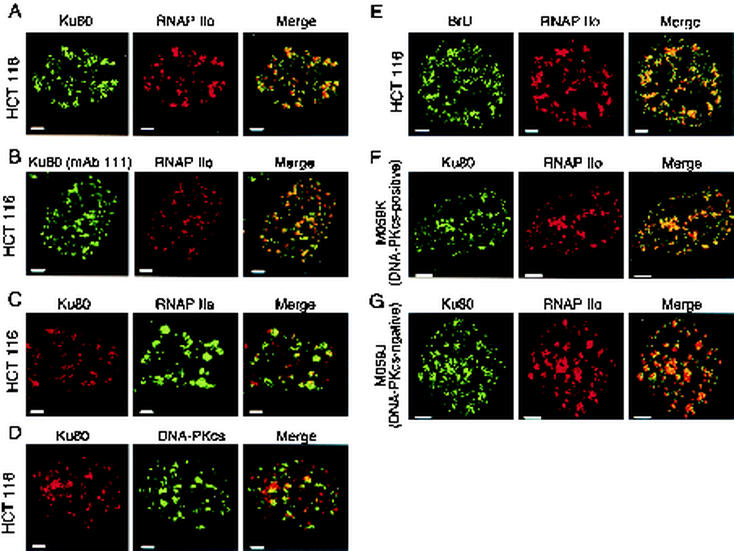
FIG. 1.
Colocalization of Ku with other nuclear components. (A) Colocalization of Ku with RNAP IIO. HCT 116 cells were stained with anti-Ku80 (MAb S10B1, IgG1) and anti-RNAP IIo subunit primary (MAb H5, IgM) antibodies and with class-specific secondary antibodies (Fluor 488-conjugated goat anti-mouse IgG and Fluor 594-conjugated goat anti-mouse IgM). Each panel depicts a confocal section of a single cell nucleus. Areas of overlap between Ku80 and RNAP IIo appear as yellow in the merged image. Panel B is the same as panel A, except with a different anti-Ku primary antibody (MAb 111). (C) Colocalization of Ku with RNAP IIA. Cells were treated as in panel A, except with primary antibody against the IIa subunit (MAb 8WG16, IgG2a). Cells were stained with isotype-specific secondary antibodies (biotin rat anti-mouse IgG1 and rabbit anti-mouse-IgG2a) and dye-conjugated tertiary reagents (Cy3-avidin and Fluor 488-conjugated goat anti-rabbit IgG). (D) Colocalization of Ku80 with DNA-PKcs. HCT116 cells were stained with anti-Ku80 (MAb S10B1, IgG1) and anti-DNA-PKcs (MAb 25-4, IgG2a), with isotype-specific secondary antibodies (biotin-conjugated rat anti-mouse IgG1 and rabbit anti-mouse-IgG2a), and with dye-conjugated tertiary reagents as in panel C. (E) Colocalization of sites of BrU incorporation with RNAP IIo. HCT116 cells were permeabilized and incubated with bromouridine triphosphate as described in Materials and Methods. After fixation, cells were stained with anti-BrdU (mouse MAb), which cross-reacts with BrU (75), and with anti-RNAP IIo (MAb H5, IgM). Cells were stained with secondary antibodies as in panel A. (F) Colocalization of Ku80 with RNAP IIo in human MO59K cells (DNA-PKcs positive). Cells were stained as in panel A. Panel G is the same as panel F, but with M059J cells (DNA-PKcs negative). Size bars are 2 μm in panels A to E and 5 μm in panels F and G. Dilutions were optimized for each primary antibody in order to best visualize the contrast between areas of high and low antigen concentration. Three slides were stained in each experiment, and 500 cells were scanned visually on each slide. Representative results are shown.
ChIP assay.
For each experiment, 10 plates (150 mm in diameter) of HCT116 cells were used. ChIP was performed as described (7). Briefly, medium was adjusted to 1% formaldehyde. After 10 min, the reaction was quenched, cells were harvested and lysed, and chromatin was sheared by sonication. Immunoprecipitation was performed, precipitates were washed extensively, and cross-linking was reversed by heating overnight at 65°C. Nucleic acids were precipitated with ethanol, and pellets were resuspended in 600 μl of water. For PCR analysis, 1 to 9 μl of DNA was incubated in the presence of 1 pmol (each) of primers, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 1 mM deoxynucleoside triphosphates (dNTPs), 2 μl of dimethyl sulfoxide, and 0.5 U of Taq DNA polymerase in a final volume of 20 μl. PCR was performed for 30 cycles of incubation at 94°C for 1 min, at the designated annealing temperature for each primer pair for 30 s, and at 68°C for 90 s, with a final extension at 68°C for 10 min. Products (20 μl) were analyzed on a 2% agarose gel and detected by staining with ethidium bromide.
Design of PCR primers for human heat shock protein 70 (HSP70) and HSP89α were adapted from reference 4. The primers and annealing temperatures were as follows: fragment 1, d(GTGAATCCCAGAAGACTCTGGA) and d(TCACTCTCGAAAAAGGTAGTGGA), annealed at 58°C; fragment 2, d(CTGTGCGGCTGCAGGCACCGGC) and d(TGGTGCGGTTGCCCTGGTCGTT), annealed at 65°C; fragment 3, d(GAGATCTCGTCCATGGTGCTGA) and d(GATCAGGACGTTGCGCTCCCCC), annealed at 62°C; fragment 4, d(ATCTCGTGGCTGGACGCCAACA) and d(AAGTCTTGAAGCTCCAAAACAA), annealed at 53°C; fragment 5, d(AAGGCGCGGGGGCGGGGTGC) and d(CCACAACCACCCGTCACCTTGG), annealed at 68°C; fragment 6, d(CCTCTGTAGACGTCCTGCAAGGT) and d(ATCCGATTCTGGGTTAATAAGTG), annealed at 60°C; fragment 7, d(TAGCTGGCTTTAAGAAATCTTTG) and d(TAAAGAAAAACATCCTTGAAAAT), annealed at 51°C, fragment 8 (a 300-bp sequence upstream of the c-myc gene on chromosome 8), d(GTACAGACTGGCAGAGAGCAGG) and d(AGCAACGCATTGCCACGTATACT), annealed at 68°C; fragment 9, d(TGAGTGGTCACCATGGTGGTGATGG) and d(TCCTGCTCCAGCCAGGTGTGGA), annealed at 61°C; fragment 10, d(AGGTAGAGCAGATCCTGGCAGAG) and d(TGAGCCTTCTGGGGTGGAGCGCA), annealed at 63°C; fragment 11, d(GGCTGGACAGCGTGGTGACG) and d(CTGGACCAGGCGCGCCTCGA); and fragment 12, d(TTTGCTCACTAAACCTGTTGGCC) and d(ACTAAGTAGGATGACAGGCTGCC), annealed at 58°C.
Immunoprecipitation and GST pull-down assays.
HeLa cell nuclear extracts were prepared as described previously (20). HCT116 nuclear extracts were prepared by sonication of cells in 10 mM Tris-HCl, 100 mM KCl, 2.5 mM MgCl2, 10 μg of aprotinin per ml, 2 μg of pepstatin per ml, 2 μg of leupeptin per ml, and 0.1% NP40. For immunoprecipitation, protein A-Sepharose beads were coated with anti-RNAP IIo, anti-RNAP IIa, anti-Ku, or anti-DSIF antibody (2 μg per 3 mg of dry beads) and washed to remove unbound antibody. Rabbit or goat anti-mouse IgG or anti-mouse IgM was also present in the incubations to facilitate collection of mouse antibodies on protein A-labeled beads. In some experiments, antibodies were cross-linked to beads prior to incubation with extract. Extracts were incubated with beads (30 to 50 μl of beads per 50 μl of extract) in a final volume of 1 ml of immunoprecipitation buffer (10 mM Tris-HCl [pH 7.9], 100 mM KCl, 5 mM MgCl2) for 3 h at 4°C. After incubation with extract, beads were washed three times in wash buffer (10 mM Tris-HCl [pH 7.9], 150 mM KCl, 5 mM MgCl2, 1% Triton X-100), and bound proteins were eluted with 30 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were resolved by SDS-PAGE and transferred to a nitrocellulose membrane for immunoblotting. The membrane was blocked with 3% BSA and then incubated with primary and secondary antibodies. Except as noted, immunoblots were developed with the ECL enhanced chemiluminescence system (Amersham Pharmacia Biotech). Glutathione S-transferase (GST) pull-down assays were performed similarly, except that cell extract was incubated for 16 h at 4°C in 1 ml of buffer containing 0.5 μg of GST or GST-Ku80C, 20 mM Tris-HCl, 150 mM KCl, 6.25 mM MgCl2, and 1 mM dithiothreitol. Complexes were collected by incubation with glutathione-agarose (20 μl) in the same buffer for 4 h. Beads were washed four times in the same buffer containing 1% Triton X-100 and eluted with this buffer containing 10 mM glutathione.
Cell-free transcription assays.
In vitro runoff transcription was performed as described previously (70), except with HeLa cell nuclear extract (5). Supercoiled AdMLP template consisted of the adenovirus major late core promoter upstream of a 380-bp G-less cassette. Samples were resolved on a prerun 5% urea-polyacrylamide gel and visualized by PhosphorImager analysis (Molecular Dynamics).
Transient transfection assays for effect of Ku80C on RNA synthesis and cell proliferation.
Constructs expressing Ku80-HA and Ku80C-HA were prepared by PCR amplification of Ku80 fragments with N-terminal primer d(GAGGATCCATGGTGCGGTCGGGGAATAAGGCAGCT) or d(GAAGAT GGATCCACAGCTATGAAATTAAAG) and a common C-terminal primer, d(CCTAAGCGTAATCTGGAACATCGTATGGGTAACGCGTTATCATGTCCAATAAATCGTCCAC), that encodes the HA epitope (YPYDVPDYA). Products were subcloned in the PCR2.1-TOPO vector (Invitrogen), excised with BamHI and EcoRI, and inserted into the corresponding sites of pcDNA3 (for constitutive expression) or pcDNA4TO (for tetracycline-inducible expression [described below]). HCT116 cells were grown on coverslips, and the pcDNA3-Ku80-HA and pcDNA3-Ku80C-HA constructs were introduced by transfection with Lipofectamine (Gibco BRL). Pulse-labeling of RNA transcripts with bromouracil (BrU) was performed as described previously (49). At 48 h after transfection, fresh medium containing 2 mM BrU (Sigma-Aldrich) was added, cells were incubated for 1 h at 37°C, washed briefly three times with PBS, and then fixed and stained with anti-HA and anti-BrdU antibodies.
For cell proliferation assays, a construct expressing a Ku80C-enhanced green fluorescent protein (EGFP) fusion protein was created by PCR amplification of the Ku80C fragment with primers d(AAAAGCTTATGAAATTAAAGACTGAGCAA) and d(TGGATCCCGTATCATGTCCAATAAATCGTC). The product was digested with HindIII and BamHI and inserted at the corresponding sites of pEGFP N1. The resulting fusion cDNA was excised with HindIII and StuI and inserted between the HindIII and EcoRV sites of pcDNA 3.1 (Zeocin). The cell proliferation assay was performed as described previously (61). HCT116 cells were grown on coverslips, and Ku80C-EGFP or EGFP vectors were introduced by transfection with Lipofectamine (Gibco BRL) After 3 days, green colonies were counted and categorized, based on whether they contained one, two, or three or more cells. The percentage of colonies in each category, relative to the total number of colonies counted, was determined.
Stable transfection assays.
For colony suppression assays, HCT116 cells were cultured in six-well plates. Cells were cotransfected with EGFP, Ku80C-EGFP, or empty vector, together with pD1Red N1 vector carrying the neomycin resistance gene at a ratio of 30:1. Cells were cultured in medium supplemented with 100 μg of G418 per ml for 14 days and then fixed and stained with hematoxylin. The number of colonies in EGFP and Ku80C-EGFP transfections was expressed as a ratio relative to the number of colonies obtained from transfection with empty vector. For expression of Ku80C under tetracycline-inducible control, HCT116 cells were transfected with pcDNA6/TR vector by using Lipofect Plus (GIBCO BRL). This plasmid expresses the natural tetracycline repressor under control of a cytomegalovirus (CMV) promoter. Blasticidin (5 μg/ml) was added 48 h after transfection. Individual blasticidin-resistant colonies were tested in transient transfection assays for the ability to direct tetracycline-dependent gene expression with a pcDNA4-EGFP reporter plasmid. Cells from a tetracycline-responsive clone were transfected with a pcDNA4-Ku80C-HA plasmid and subjected to selection in 50 μg of Zeocin per ml. Individual cell clones that showed tetracycline-dependent Ku80C-HA expression were identified by immunoblotting and immunofluorescence.
RESULTS
Colocalization with RNAP II elongation sites.
Staining of human HCT116 cells with anti-Ku MAb S10B1 revealed a punctate distribution, with Ku concentrated at 50 to 100 sites within a given optical section (Fig. 1A, left panel). Similar results were obtained with another anti-Ku antibody, MAb 111 (Fig. 1B, left panel). We compared this staining pattern with those of three other nuclear proteins: the elongating RNAP IIO isoform, detected with isoform-specific MAb H5; the initiation-competent RNAP IIA isoform, detected with isoform-specific MAb 8WG16; and DNA-PKcs. All three of these proteins had punctate distributions, but only RNAP IIO was coincident with Ku protein (Fig. 1A to D). The finding that Ku and DNA-PKcs showed little overlap is consistent with biochemical observations that Ku and DNA-PKcs do not interact in the absence of free DNA ends (20, 58). In control experiments, no staining was seen in the absence of primary antibody (data not shown). Similar colocalization of Ku with RNAP IIO was also seen in preliminary work with HeLa cells, MCF7 breast cancer cells, and human fibroblasts (data not shown).
To confirm that sites of Ku and RNAP IIO localization corresponded to actual sites of RNAP elongation in our system, we performed in situ run-on transcription in the presence of bromouridine triphosphate (75). As expected, staining with isoform-specific anti-RNAP IIo and anti-BrU antibodies was nearly coincident (Fig. 1E). In a separate experiment, there was little overlap between anti-RNAP IIa and anti-BrU staining (data not shown). Because our anti-Ku and anti-BrU antibodies were of the same isotype, we did not examine the distribution of endogenous Ku relative to BrU directly, although later experiments with epitope-tagged Ku showed a high degree of overlap (see Fig. 6). Together, the immunostaining data show a remarkable colocalization of Ku, RNAP IIO, and sites of RNA synthesis.
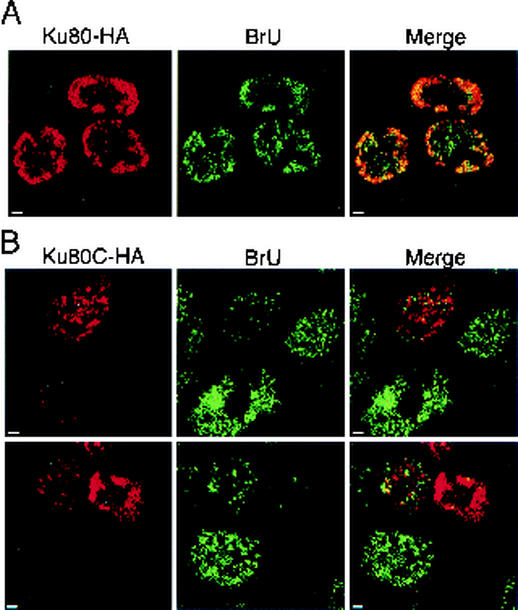
FIG. 6.
Ku80C inhibits transcription in vivo. Ku80-HA and Ku80C-HA were expressed in HCT116 cells under the control of a CMV promoter. At 48 h posttransfection, cells were incubated with BrU for 1 h to label nascent RNA, fixed, and stained with anti-HA (rat MAb; Roche) and anti-BrU (MAb; Roche), as well as with species-specific secondary antibodies. (A) Transfection with Ku80-HA vector. Each field shows a confocal section with nuclei of three or more cells. Areas of overlap between Ku80-HA and BrU signals appear yellow in the merged image (right panels). Panel B is the same as panel A, except that transfection was with Ku80C-HA vector. Size bar, 2 μm. The experiment was repeated three times with 200 to 400 cells examined each time. Results are representative.
It was of interest to determine whether DNA-PKcs was required for the association of Ku with transcription elongation sites. Although there was only modest overlap between Ku and DNA-PKcs signals, it was possible that DNA-PKcs influenced localization of Ku at elongation sites by some indirect mechanism. We therefore performed immunostaining experiments with the matched human glioblastoma cell lines M059J and M059K (2). M059J lacks DNA-PKcs and is radiation sensitive, whereas M059K is its DNA-PKcs-containing, radiation-resistant counterpart (3, 36). Control immunostaining experiments confirmed the presence of DNA-PKcs in our cultures of M059K cells and its absence from M059J cells, consistent with published reports (data not shown). Immunostaining for Ku and RNAP IIo showed extensive colocalization in both cell lines (Fig. 1F and G). Thus, the association of Ku80 with sites of transcriptional elongation occurs independently of DNA-PKcs.
Cross-linking to coding regions of active genes.
To corroborate the results of immunostaining, we examined whether Ku protein was associated with actively transcribed genes by using a ChIP assay (7). In this assay, HCT116 cells were treated with formaldehyde to cross-link chromatin proteins to DNA, chromatin was extracted and sheared, and the resulting preparations were subjected to immunoprecipitation with anti-Ku80 and anti-RNAP IIo. After reversal of the cross-linking, precipitates were analyzed for DNA content by PCR. Four genes were analyzed, as shown in Fig. 2A. Two of these are HSP genes, which have been widely used as model systems for study of transcriptional elongation (4, 24, 40, 45, 69); the others were genes coding for glucokinase and PCNA.
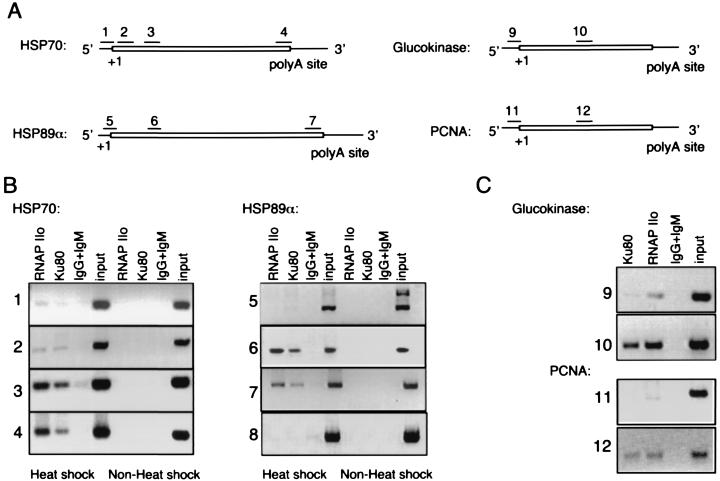
FIG. 2.
ChIP. (A) Schematic representations of HSP70 and HSP89α, glucokinase, and PCNA genes showing location of primer sets, transcription start site (+1), and polyadenylation sites. Boxed sequence denotes transcribed region. Introns are not shown. Primer set 8 amplifies a nontranscribed region of chromosome 8 (not shown). (B) ChIP. HCT116 cells were incubated at 43°C for 1 h and allowed to recover at 37°C for 1 h. Cell lysates were analyzed by ChIP, as described in Materials and Methods, with anti-RNAP IIo (MAb H5), anti-Ku80 (MAb S10B1), or a mixture of control mouse IgG and IgM as indicated. Precipitated DNA was analyzed by PCR with the indicated primer pairs. A negative image of the ethidium bromide-stained gel is shown. Input lanes represent 0.1% of material used for immunoprecipitation. Panel C is the same as panel B, but without heat treatment.
As shown in Fig. 2B and C, anti-Ku antibody precipitated chromatin fragments corresponding to the coding regions of all four genes (primer sets 2, 3, 4, 6, 7, 10, and 12). Precipitation of the HSP70 and HSP89α genes was dependent on heat shock induction (Fig. 2B). In contrast, anti-Ku antibody gave less, and in some cases undetectable, precipitation of chromatin fragments corresponding to the promoter regions of the same genes (primer sets 1, 5, 9, and 11). A control reaction was performed with a pair of primers that amplify a region of nontranscribed DNA (primer set 8). Anti-Ku80 antibody did not precipitate chromatin corresponding to this region of the genome.
Control reactions were included for all primer sets (Fig. 2B and C). Positive control reaction mixtures, containing a small fraction of input chromatin, yielded a single PCR product of the expected size. Negative control reaction mixtures, containing material immunoprecipitated with a mixture of nonspecific IgG and IgM, showed little or no product. In all cases, PCR products were detected by direct ethidium bromide staining, excluding the possibility that binding of a hybridization probe to contaminating endogenous mRNA influenced results.
Parallel immunoprecipitation reactions were performed with anti-RNAP IIo antibody. As expected, these reactions showed selective precipitation of the coding regions of transcriptionally active genes. The overall pattern was similar to that obtained with anti-Ku antibody.
Together, the ChIP results indicate that molecular contacts between Ku and transcribed regions of DNA are present at significant steady-state levels. It is important to note that cross-linking presents a snapshot of the contacts present at any given time. These contacts may be short-lived and dynamic and should not be equated with the high-affinity complexes formed at broken DNA ends. Nevertheless, ChIP data provide an important confirmation that Ku protein is localized at or near sites of transcription elongation.
Coimmunoprecipitation with protein markers of elongation.
We next investigated whether soluble complexes containing Ku and transcription elongation proteins could be detected in cell extracts. Immunoprecipitation was performed either with anti-Ku MAb or with antibodies against various transcription proteins. Precipitation and washing were performed at physiological salt concentrations (0.1 to 0.15 M KCl), immune complexes were collected, and analysis was performed by immunoblotting with the same or different antibodies. The experiment was performed with extracts from both HCT116 cells and HeLa cells.
In Fig. 3, panels A and C show immunoblots that were probed with antibodies against markers of the elongation complex. Anti-Ku antibodies coprecipitated both RNAP IIO and the elongation factor, DSIF. Reciprocal coimmunoprecipitation experiments with anti-DSF and anti-RNAP IIo were also performed. Each of these antibodies precipitated Ku protein with nearly the same efficiency as they precipitated their own target antigens.
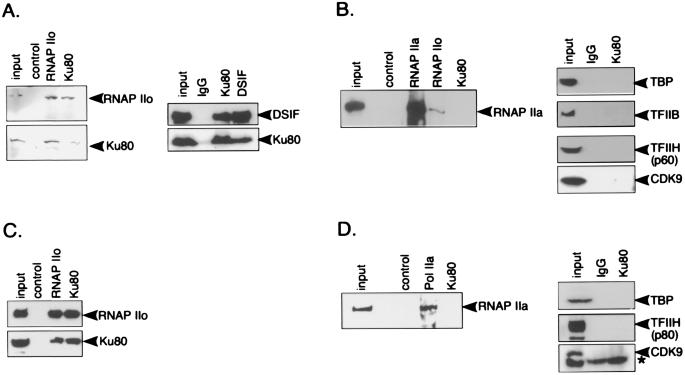
FIG. 3.
Coimmunoprecipitation of Ku with transcription proteins. Immunoprecipitation was performed with anti-RNAP IIo (MAb H5), anti-RNAP IIa (MAb 8WG16), anti-Ku80 (MAb S10B1), anti-DSIF (MAb; Pharmingen), or control (mixture of mouse IgG and IgM, unless otherwise indicated) antibody. Antibodies were cross-linked to agarose beads as described in Materials and Methods. Immunoblots were developed with the ECL enhanced chemiluminescence system (Amersham Pharmacia Biotech) unless otherwise indicated. In all panels, input represents 5 to 10% of material used for immunoprecipitation. (A) HCT116 cell immunoprecipitates probed with antibodies to markers of the elongation complex as indicated. Immunoblots were developed with bromochloroindolyl phosphate and nitroblue tetrazolium (left panel) or with the ECL system (right panel). (B) HCT116 cell immunoprecipitates probed with antibodies to markers of the initiation complex. (C) HeLa cell immunoprecipitates probed with markers to the elongation complex. (D) HeLa cell immunoprecipitates probed with markers to the initiation complex. An asterisk by the CDK9 panel indicates a nonspecific background band.
Very different results were obtained when immunoblots were probed with markers of the initiation complex (Fig. 3B and D). Anti-Ku antibody did not precipitate detectable amounts of RNAP IIA or the general transcription factors TBP, TFIIB, and TFIIH, which act at the initiation step of transcription. Anti-Ku80 also failed to precipitate CDK9, which is the catalytic subunit of P-TEFb and which phosphorylates RNAP II during the transition to active elongation (52).
Control lanes were included in each analysis. Immunoblotting of the input material showed that the antibodies were specific for their target antigens (with the exception of anti-CDK9 in HeLa cells, which detects a background band slightly below the authentic CDK9). Immunoprecipitates obtained with nonspecific control antibodies showed little or no background signal. In addition, anti-RNAP IIo precipitated little material detectable with anti-RNAP IIa (Fig. 3B), in agreement with the results of prior studies demonstrating the specificity of this antibody (48).
The observed specificity of the interaction of Ku with RNAP IIO and DSIF and the apparent absence of interaction with RNAP IIA and other general transcription factors corroborates immunostaining and ChIP results showing association of Ku with elongation sites. We note that immunoprecipitation results should not be interpreted as evidence of direct molecular contacts between Ku and individual transcription proteins. Indeed, precipitates are likely to represent soluble remnants of the structures visualized by confocal microscopy in Fig. 1. Consistent with this, analysis of immunoprecipitates by SDS-PAGE with silver staining revealed a complex mixture of components (data not shown).
Role of Ku80 C-terminal domain.
The Ku70 and Ku80 subunits share common folds in their N-terminal and central domains, which together form the high-affinity DNA end-binding site (67). In addition to these shared domains, both subunits have discrete, nonhomologous C-terminal domains, which do not directly contact the free DNA end in the Ku-DNA complex. We have previously described a mutant bearing a small deletion in the Ku80 C-terminal (Ku80C) domain, which exhibits wild-type function in a cell-free repair assay, but inhibits transcription in a cell extract (71). This result led to the hypothesis that the Ku80C domain was important for interactions with the transcription apparatus.
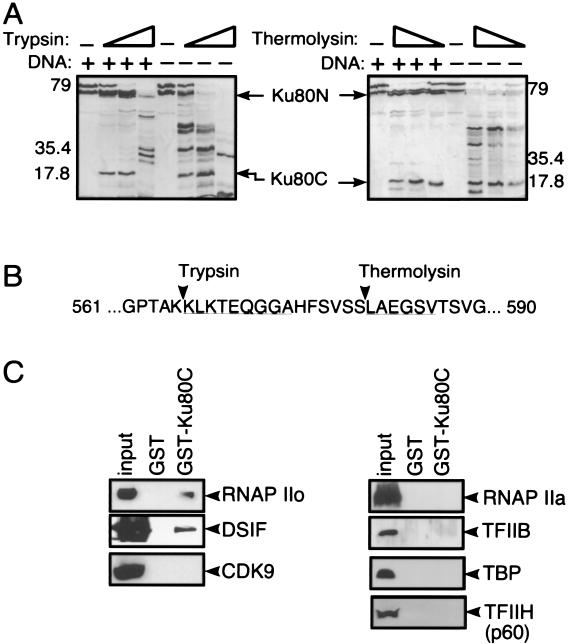
To further investigate the role of the Ku80C domain, it was necessary to establish its exact boundary. The linker segment joining the Ku80C domain to the remainder of the protein is highly susceptible to proteases (29, 43, 47). To determine the precise cleavage sites, we subjected Ku-DNA complexes to limited proteolysis and performed N-terminal sequencing of the products (Fig. 4A). The results showed that trypsin and thermolysin cleaved DNA-bound Ku after residues 565 and 580, respectively (Fig. 4B). The C-terminal trypsin product was subcloned for expression as a GST fusion protein (GST-Ku80C) in Escherichia coli.
FIG. 4.
Ku80 contains a discrete C-terminal domain that binds to transcription factors. (A) Limited proteolysis of Ku80. Trypsin (left) and thermolysin (right) were incubated with Ku in the presence and absence of DNA. Open triangles denote increasing ratio of protease to Ku. The trypsin/Ku ratio varied from 1:100 to 1:10, and the thermolysin/Ku ratio varied from 1:300 to 1:30. Arrowheads denote major Ku80 N-terminal and Ku80 C-terminal fragments released by cleavage in the presence of DNA. (B) Exact sites of cleavage by trypsin and thermolysin. Underlined sequence denotes amino acid residues identified by N-terminal Edman degradation of proteolytic fragments. (C) The Ku80 C-terminal domain binds to RNAP IIO and DSIF. A GST fusion protein containing sequences found in the Ku80 C-terminal trypsin fragment was incubated with proteins from a HeLa nuclear extract. Bound proteins were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Input represents 5% of the material used for pull-down assays.
To investigate the interaction between the Ku80C domain and transcription proteins, pull-down experiments were performed in which purified GST-Ku80C fusion protein was incubated with HeLa cell nuclear extract in transcription buffer, and the resulting complexes were collected for analysis by SDS-PAGE and immunoblotting (Fig. 4C). Results were similar to those obtained with full-length endogenous Ku in the coimmunoprecipitation experiments (Fig. 3). GST-Ku80C pulled down the immunoreactive RNAP IIo subunit, but not RNAP IIa. It also pulled down DSIF, but not the general transcription initiation factors TFIIB, TBP, and TFIIH. It also did not pull down CDK9. GST alone, used as a control, did not pull down any of the transcription proteins. These results suggest that the Ku80C domain accounts, at least in part, for the interactions that are observed between endogenous Ku protein and the transcription apparatus. As in the coimmunoprecipitation experiments, interactions detected in the pull-down assay may be indirect. In a separate experiment, GST-Ku80C did not pull down a biochemically purified preparation of RNAP II containing a mixture of RNAP IIO and RNAP IIA components (data not shown).
Isolated Ku80C inhibits transcription in vitro.
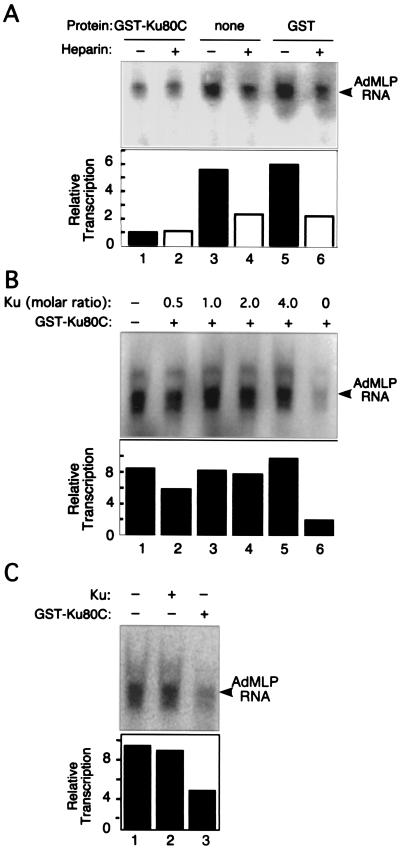
The ability of isolated Ku80C to interact with transcription elongation proteins suggested that it might be able to interfere with transcriptional activity by competing with endogenous Ku. To test this, we added GST-Ku80C to an in vitro transcription system containing HeLa nuclear extract and supercoiled AdMLP template. In a multiple round assay, addition of GST-Ku80C led to a fourfold decrease in accumulation of the promoter-specific 380-nucleotide RNA product (Fig. 5A). GST alone had no effect.
FIG. 5.
Effect of GST-Ku80C in a cell-free transcription system. AdMLP template was preincubated with HeLa nuclear extract in the presence and absence of approximately 100 ng of GST-Ku80C or GST control protein for 30 min at 30°C. Transcription was initiated by addition of a mixture of ATP, UTP, and [α-32P]CTP. An arrowhead denotes the 380-nucleotide full-length AdMLP transcript. RNA was analyzed by urea-PAGE and visualized by PhosphorImager analysis. (A) Transcription assay performed under single and multiple round conditions. Where indicated, 10 μg of heparin per ml was added after initiation to prevent the initiation of additional rounds. (B) Reversal of inhibition by full-length Ku. Reactions were performed as in panel A, except that Ku heterodimer (prepared as in reference 71) and GST-Ku80C were incubated with nuclear extract for 10 min at 0°C prior to addition of template. (C) Lack of effect of full-length Ku alone. Reactions were performed in panel A in the presence and absence of approximately 100 ng of each of the indicated proteins.
As discussed previously, Ku acts primarily to increase the efficiency of the reinitiation step of the transcription cycle (70, 71). In control reactions lacking GST-Ku80C, addition of heparin, which limits transcription to a single round, decreased transcription by about 2.5-fold (Fig. 5A, lanes 3 to 6). This indicates that the HeLa cell extract is capable of directing multiple rounds of transcription under the conditions used. In contrast, in reaction mixtures containing GST-Ku80C, heparin produced no further decrease in the already low level of transcription (Fig. 5A, lanes 1 and 2). These results indicate that GST-Ku80C inhibits reinitiation.
To further investigate the mechanism of GST-Ku80C inhibition, a transcription experiment was performed in which GST-Ku80C and the purified Ku heterodimer were added together to the in vitro transcription system. Full-length Ku restored transcription to levels seen in the absence of inhibitor (Fig. 5B, lanes 2 to 5). Full-length Ku alone, in the absence of GST-Ku80C, had no effect (Fig. 5C). Taken together, the effect of GST-Ku80C at the level of reinitiation and the reversal of inhibition by full-length protein are consistent with the idea that the isolated Ku80C domain inhibits AdMLP transcription by antagonizing the activity of endogenous Ku protein.
Expression of Ku80C inhibits transcription in vivo.
To extend these studies in vivo, experiments were performed in which HA-tagged Ku80 derivatives were expressed transiently in HCT116 cells, under the control of the CMV promoter. At 48 h posttransfection, nascent RNA was pulse-labeled by incubation in BrU-containing medium, and cells were immunostained with anti-HA and anti-BrdU antibodies.
Both the full-length Ku80-HA and the Ku80C-HA derivatives were expressed at comparable levels, as judged by the brightness of anti-HA staining, and each assumed a punctate distribution in the nucleus, similar to that seen with native Ku (Fig. 6A). With the exception of several regions that correspond to nucleoli, sites of full-length Ku80-HA expression coincided with sites of BrU incorporation, confirming earlier findings with endogenous Ku (Fig. 1). A strikingly different result was obtained with Ku80C-HA. Expression of this construct and BrU incorporation were mutually exclusive (Fig. 6B). In the fields shown, cells are present that expressed different amounts of transgene. In the cell expressing the largest amounts of the Ku80C-HA transgene, BrU incorporation was almost completely inhibited. In cells expressing intermediate amounts of Ku80C-HA, BrU incorporation was partially inhibited and was restricted to areas of the nucleus where Ku80C was absent. Several cells did not express Ku80C-HA, apparently because they did not take up the expression plasmid, and these showed strong BrU incorporation. As an additional control, an N-terminal fragment of Ku80, specifically lacking the Ku80C domain, was also tested. Like the full-length Ku80-HA, this had no apparent effect on BrU incorporation (data not shown), confirming that transcriptional inhibition is specifically attributable to the isolated Ku80C domain.
Expression of Ku80C inhibits cell proliferation.
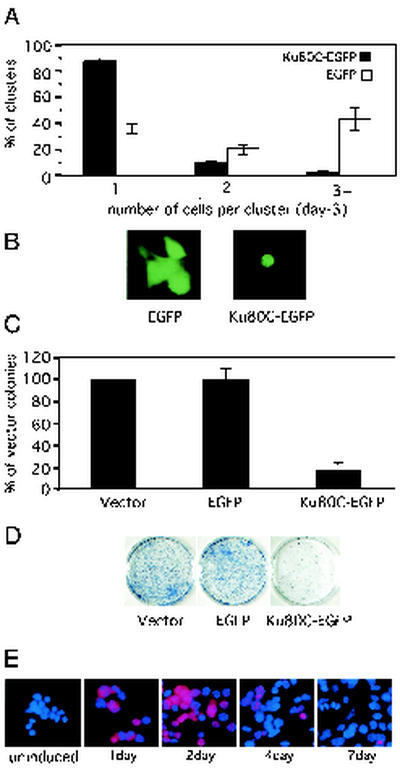
It was of interest to determine whether sustained expression of Ku80C affected cell growth and whether it resulted in cell death. In the first of several complementary approaches, we constructed a vector expressing Ku80C joined to EGFP. This vector and a control vector expressing EGFP alone were introduced into HCT116 cells by transient transfection. Fluorescent cell clusters were scored after 3 days (61). With Ku80C-EGFP, almost all of the fluorescent clusters consisted of only a single cell (Fig. 7A and B). In contrast, with EGFP alone, the majority of clusters contained two, three, or more cells (Fig. 7A and B). At times beyond 3 days, very few cells expressing Ku80C-EGFP remained, whereas cells expressing EGFP alone showed continued growth (data not shown).
FIG. 7.
Expression of the Ku80 C-terminal domain inhibits cell proliferation. (A) EGFP and Ku80C-EGFP were expressed in HCT116 cells by transient transfection. Green fluorescent cell clusters were scored after 3 days. The graph shows the percentage of the fluorescent cluster containing one, two, or three or more cells. Values are mean of three independent experiments. A minimum of 200 fluorescent clusters were scored in each experiment. Error bars denote standard error. (B) Typical appearance of fluorescent cell clusters in cultures transfected with EGFP or Ku80C-EGFP. (C) Colony suppression assay. HCT116 cells were transfected with empty vector, EGFP expression vector, or Ku80C-EGFP expression vector. A neomycin resistance vector was also present at a 1:30 ratio to expression vector. After 14 days of selection in G418 (200 μg/ml), colonies were counted: the results are expressed as a percentage of colonies seen with expression vector alone. Values are means of three independent experiments. A minimum of 5,000 colonies were scored in each experiment. Bars denote standard error. (D) Typical appearance of stained plates from panel C. (E) Extinction of Ku80C expression in an inducible cell line. A stably transfected clone of HCT116 cells was prepared that expressed HA-Ku80C under the control of a tetracycline-inducible pro-moter. Tetracycline (1 μg/ml) was added, and the cells were fixed and stained at the indicated times with anti-HA (red) and DAPI (4′,6′-diamidino phenylindole) (blue).
These results were confirmed in a colony suppression assay (62, 68). HCT116 cells were cotransfected with the Ku80C-EGFP expression vector and a neomycin resistance vector. After 2 weeks of selection in G418, plates were fixed, stained, and scored for colony formation. Expression of Ku80C-EGFP resulted in an 80% reduction in colony formation, relative to control populations transfected with EGFP or empty vector (Fig. 7C and D).
A third approach involved expression of HA-Ku80C under tetracycline-inducible control. The Ku80C-HA protein was subcloned for expression under the control of the tetracycline promoter. This was introduced into an HCT116 cell line expressing a tetracycline repressor protein, and stable clones were isolated. After addition of tetracycline to the medium, most of the cells began to express Ku80C within 1 day (Fig. 7E). Expression reached a peak at 2 days and then declined, and expression was almost entirely absent at 7 days, suggesting that Ku80C-expressing cells are overgrown by others in the population or that Ku80C expression is extinguished (Fig. 7E). Together, the results of these three complementary assays indicate that expression of Ku80C causes arrest of cell growth.
DISCUSSION
We have shown that endogenous Ku protein is closely associated with sites of transcriptional elongation, as evidenced by colocalization in the light microscope, cross-linking to transcribed regions of DNA, and the ability to coprecipitate Ku with protein markers of the elongation complex. Physical association of Ku with elongation sites does not, in itself, imply a specific function. However, the dominant-negative effect of Ku80C overexpression argues that the association of Ku with transcription sites fulfills a specific purpose. In principle, the dominant-negative effect could arise either at the level of transcription or RNA processing, since these events are closely linked in space and time (reviewed in reference 42). However, the finding that the dominant-negative effect also occurs in a cell-free system, where gene expression is independent of RNA processing, suggests that Ku influences transcription itself.
Pull-down assays suggest that the Ku80C domain is, in part, responsible for interaction of Ku with the transcription apparatus. However, the dominant-negative effect of Ku80C overexpression implies that the isolated domain lacks an additional function present elsewhere in the native Ku protein. Indeed, in previous work, we have also seen a dominant-negative effect on transcription by using a different Ku mutant containing an intact DNA binding domain and a deletion in the Ku80C domain (71). Together, these results suggest that stimulation of transcription by native Ku requires concerted action of sequences in the Ku80C domain and elsewhere in the protein.
In considering potential mechanisms by which Ku may influence transcription, it is worth noting the DNA binding domain has an appreciable nonspecific affinity for RNA (73). Although this affinity is 2 orders of magnitude weaker than the affinity for DNA ends, one would nevertheless expect significant binding to nascent RNA at the high concentrations that prevail in vivo in “transcription factories.” It could be that simultaneous binding of Ku to elongation proteins and to nascent RNA, mediated by separate domains in the protein, allows elongation proteins to remain associated with the transcript after template release. We speculate that this may promote transport of the elongation complex to a location where RNAP II can be dephosphorylated and reassembled into an initiation-competent holoenzyme form. Although unproven, such a mechanism could account for the distinctive, and otherwise unexplained, effect of Ku on the reinitiation step of the transcription cycle.
In preliminary work (unpublished results), we found that anti-Ku and anti-RNAP IIo staining remained coincident in cells treated with the transcription inhibitor α-amanitin and that both proteins relocalized into “speckles” similar to those observed by previous workers (75). This is consistent with a model in which Ku is stably associated with elongation factors through protein-protein interactions, but the entire complex is capable of dynamic relocalization in response to changes in cell state.
The positive contribution of Ku to global transcription must be reconciled with several reports that Ku negatively regulates transcription when bound to specific sites in promoter regions (11, 25, 31, 72). The mechanism by which Ku recognizes specific sequences in DNA is unclear, because the closed topology of the DNA binding site is incompatible with binding to duplex DNA lacking a free end (67). It is possible that sequence-specific binding involves an independent DNA binding site in the Ku70 C terminus (76) or an alternative DNA secondary structure. In either case, it is easy to envision that static association of Ku with a specific sequence in the promoter region might be incompatible with a dynamic role of Ku in the normal transcription cycle.
It is uncertain whether the function of Ku in transcription is independent of its function in DSB repair, or whether the two are related. There is conflicting information about the requirement for the Ku80C domain in repair. A Ku80 C-terminal peptide binds to the DNA-PKcs in vitro (23). In addition, genetic rescue studies have defined a role for C-terminal sequences in repair (57). However, a more exact comparison of genetic and structural data suggests that the residues that are essential for repair function, which lie between positions 554 and 560 (57), are in the linker sequence that joins the Ku80C domain to the remainder of the molecule rather than the Ku80C domain proper (67) (Fig. 4). Other mutations that affect only the Ku80C domain proper have no effect in genetic rescue experiments (57) or in cell-free end-joining assays (71).
Even if Ku80C has no direct enzymatic role in repair, its association with the transcription apparatus could help indirectly to promote genome stability. Mammalian cells tolerate several grays of radiation, which is sufficient to induce dozens of breaks simultaneously (12, 54). The ability of Ku to anchor newly formed ends to an underlying structure could serve an important function in suppressing illegitimate rejoining of mismatched DNA ends. It may be that subtle effects of Ku80C on genome stability, reflecting its association with an underlying structure, will become more evident when repair assays are extended to longer-term or whole-animal systems.
Ku-deficient mice show a pleiotropic phenotype, which includes dwarfism and accelerated aging (27, 38, 44, 64). A question arises of whether this phenotype is attributable to defects in transcription, repair, or both. An interesting precedent is provided by Cockayne syndrome, a human premature aging disease caused by molecular defects that affect both transcription and transcription-coupled repair (6, 9, 18, 56, 63). Genetic analysis of a mouse model suggests that the transcription defect, rather than loss of repair function, is the direct cause of the Cockayne syndrome phenotype (17). It is thus plausible that a transcription defect underlies the premature aging defect in the Ku-deficient mice as well.
Ku is far more abundant in humans than in rodents, and human cells exhibit a much more severe defect when the Ku80 gene is disrupted. In HCT116 cells, homozygous gene disruption results in a massive onset of apoptosis after only a limited number of cell divisions (37). It is notable that large numbers of apoptotic cells were not evident in our Ku80C-overexpressing populations (Fig. 7). Thus, the apoptotic phenotype associated with loss of Ku in human cells may reflect loss of yet additional functions, beyond those disrupted by Ku80C expression.
Acknowledgments
We thank Dimitrios Moskiphidis, Rhea-Beth Markowitz, and Shuyi Li for comments and other assistance with the manuscript; Juren Huang and Robin Woodard for providing nuclear extracts; Farlyn Hudson for protein purification; Eric Hendrickson (University of Minnesota) for providing HCT116 cells and sharing unpublished results; and the Medical College of Georgia Molecular Biology Core Facility for technical services.
This work was supported by Public Health Service grant GM 35866. W.S.D. received support as an Eminent Scholar of the Georgia Research Alliance.
REFERENCES
- 1.Akoulitchev, S., T. P. Makela, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557-560. [DOI] [PubMed] [Google Scholar]
- 2.Allalunis-Turner, M. J., G. M. Barron, R. S. Day III, K. D. Dobler, and R. Mirzayans. 1993. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat. Res. 134:349-354. [PubMed] [Google Scholar]
- 3.Anderson, C. W., J. J. Dunn, P. I. Freimuth, A. M. Galloway, and M. J. Allalunis-Turner. 2001. Frameshift mutation in PRKDC, the gene for DNA-PKcs, in the DNA repair-defective, human, glioma-derived cell line M059J. Radiat. Res. 156:2-9. [DOI] [PubMed] [Google Scholar]
- 4.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias, J. A., and W. S. Dynan. 1989. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J. Biol. Chem. 264:3223-3229. [PubMed] [Google Scholar]
- 6.Balajee, A. S., A. May, G. L. Dianov, E. C. Friedberg, and V. A. Bohr. 1997. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl. Acad. Sci. USA 94:4306-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, K. E., and P. J. Farnham. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19:8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bregman, D. B., R. Halaban, A. J. van Gool, K. A. Henning, E. C. Friedberg, and S. L. Warren. 1996. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. USA 93:11586-11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadena, D. L., and M. E. Dahmus. 1987. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J. Biol. Chem. 262:12468-12474. [PubMed] [Google Scholar]
- 11.Camara-Clayette, V., D. Thomas, C. Rahuel, R. Barbey, J. P. Cartron, and O. Bertrand. 1999. The repressor which binds the −75 GATA motif of the GPB promoter contains Ku70 as the DNA binding subunit. Nucleic Acids Res. 27:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cedervall, B., R. Wong, N. Albright, J. Dynlacht, P. Lambin, and W. C. Dewey. 1995. Methods for the quantification of DNA double-strand breaks determined from the distribution of DNA fragment sizes measured by pulsed-field gel electrophoresis. Radiat. Res. 143:8-16. [PubMed] [Google Scholar]
- 13.Chesnut, J. D., J. H. Stephens, and M. E. Dahmus. 1992. The interaction of RNA polymerase II with the adenovirus-2 major late promoter is precluded by phosphorylation of the C-terminal domain of subunit IIa. J. Biol. Chem. 267:10500-10506. [PubMed] [Google Scholar]
- 14.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook, P. R. 1999. The organization of replication and transcription. Science 284:1790-1795. [DOI] [PubMed] [Google Scholar]
- 16.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 17.de Boer, J., and J. H. Hoeijmakers. 1999. Cancer from the outside, aging from the inside: mouse models to study the consequences of defective nucleotide excision repair. Biochimie 81:127-137. [DOI] [PubMed] [Google Scholar]
- 18.Dianov, G. L., J. F. Houle, N. Iyer, V. A. Bohr, and E. C. Friedberg. 1997. Reduced RNA polymerase II transcription in extracts of Cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 25:3636-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dvir, A., S. R. Peterson, M. W. Knuth, H. Lu, and W. S. Dynan. 1992. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl. Acad. Sci. USA 89:11920-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dvir, A., L. Y. Stein, B. L. Calore, and W. S. Dynan. 1993. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem. 268:10440-10447. [PubMed] [Google Scholar]
- 21.Dynan, W. S., and S. Yoo. 1998. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26:1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Featherstone, C., and S. P. Jackson. 1999. DNA double-strand break repair. Curr. Biol. 9:R759-R761. [DOI] [PubMed] [Google Scholar]
- 23.Gell, D., and S. P. Jackson. 1999. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 27:3494-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerber, M., J. Ma, K. Dean, J. C. Eissenberg, and A. Shilatifard. 2001. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. EMBO J. 20:6104-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giffin, W., W. Gong, C. Schild-Poulter, and R. J. Hache. 1999. Ku antigen-DNA conformation determines the activation of DNA-dependent protein kinase and DNA sequence-directed repression of mouse mammary tumor virus transcription. Mol. Cell. Biol. 19:4065-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlieb, T. M., and S. P. Jackson. 1993. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72:131-142. [DOI] [PubMed] [Google Scholar]
- 27.Gu, Y., K. J. Seidl, G. A. Rathbun, C. Zhu, J. P. Manis, N. van der Stoep, L. Davidson, H. L. Cheng, J. M. Sekiguchi, K. Frank, P. Stanhope-Baker, M. S. Schlissel, D. B. Roth, and F. W. Alt. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7:653-665. [DOI] [PubMed] [Google Scholar]
- 28.Guo, S., Y. Yamaguchi, S. Schilbach, T. Wada, J. Lee, A. Goddard, D. French, H. Handa, and A. Rosenthal. 2000. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408:366-369. [DOI] [PubMed] [Google Scholar]
- 29.Han, Z., C. Johnston, W. H. Reeves, T. Carter, J. H. Wyche, and E. A. Hendrickson. 1996. Characterization of a Ku86 variant protein that results in altered DNA binding and diminished DNA-dependent protein kinase activity. J. Biol. Chem. 271:14098-14104. [DOI] [PubMed] [Google Scholar]
- 30.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeanson, L., and J. F. Mouscadet. 2002. Ku represses the HIV-1 transcription: identification of a putative Ku binding site homologous to the MMTV NRE1 sequence in the HIV-1 LTR. J. Biol. Chem. 277:4918-4924. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, E., L. Du, D. B. Bregman, and S. L. Warren. 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laybourn, P. J., and M. E. Dahmus. 1990. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J. Biol. Chem. 265:13165-13173. [PubMed] [Google Scholar]
- 36.Lees-Miller, S. P., R. Godbout, D. W. Chan, M. Weinfeld, R. S. Day III, G. M. Barron, and J. Allalunis-Turner. 1995. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science 267:1183-1185. [DOI] [PubMed] [Google Scholar]
- 37.Li, G., C. Nelsen, and E. A. Hendrickson. 2002. Ku86 is essential in human somatic cells. Proc. Natl. Acad. Sci. USA 99:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, G. C., H. Ouyang, X. Li, H. Nagasawa, J. B. Little, D. J. Chen, C. C. Ling, Z. Fuks, and C. Cordon-Cardo. 1998. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell 2:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792-803. [PMC free article] [PubMed] [Google Scholar]
- 41.Maldonado, E., R. Shiekhattar, M. Sheldon, H. Cho, R. Drapkin, P. Rickert, E. Lees, C. W. Anderson, S. Linn, and D. Reinberg. 1996. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature 381:86-89. [DOI] [PubMed] [Google Scholar]
- 42.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 43.Muller, C., C. Dusseau, P. Calsou, and B. Salles. 1998. Human normal peripheral blood B-lymphocytes are deficient in DNA-dependent protein kinase activity due to the expression of a variant form of the Ku86 protein. Oncogene 16:1553-1560. [DOI] [PubMed] [Google Scholar]
- 44.Nussenzweig, A., C. Chen, V. da Costa Soares, M. Sanchez, K. Sokol, M. C. Nussenzweig, and G. C. Li. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382:551-555. [DOI] [PubMed] [Google Scholar]
- 45.O'Brien, T., S. Hardin, A. Greenleaf, and J. T. Lis. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370:75-77. [DOI] [PubMed] [Google Scholar]
- 46.Oelgeschlager, T. 2002. Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control. J. Cell Physiol. 190:160-169. [DOI] [PubMed] [Google Scholar]
- 47.Paillard, S., and F. Strauss. 1993. Site-specific proteolytic cleavage of Ku protein bound to DNA. Proteins 15:330-337. [DOI] [PubMed] [Google Scholar]
- 48.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273:4689-4694. [DOI] [PubMed] [Google Scholar]
- 49.Pellizzoni, L., B. Charroux, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 152:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson, S. R., A. Dvir, C. W. Anderson, and W. S. Dynan. 1992. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 6:426-438. [DOI] [PubMed] [Google Scholar]
- 51.Pombo, A., M. Hollinshead, and P. R. Cook. 1999. Bridging the resolution gap: imaging the same transcription factories in cryosections by light and electron microscopy. J. Histochem. Cytochem. 47:471-480. [DOI] [PubMed] [Google Scholar]
- 52.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rathmell, W. K., and G. Chu. 1994. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 91:7623-7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruiz de Almodovar, J. M., G. G. Steel, S. J. Whitaker, and T. J. McMillan. 1994. A comparison of methods for calculating DNA double-strand break induction frequency in mammalian cells by pulsed-field gel electrophoresis. Int. J. Radiat. Biol. 65:641-649. [DOI] [PubMed] [Google Scholar]
- 55.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selby, C. P., and A. Sancar. 1997. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:11205-11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singleton, B. K., M. I. Torres-Arzayus, S. T. Rottinghaus, G. E. Taccioli, and P. A. Jeggo. 1999. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 19:3267-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suwa, A., M. Hirakata, Y. Takeda, S. A. Jesch, T. Mimori, and J. A. Hardin. 1994. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc. Natl. Acad. Sci. USA 91:6904-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taccioli, G. E., T. M. Gottlieb, T. Blunt, A. Priestley, J. Demengeot, R. Mizuta, A. R. Lehmann, F. W. Alt, S. P. Jackson, and P. A. Jeggo. 1994. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265:1442-1445. [DOI] [PubMed] [Google Scholar]
- 60.Thompson, N. E., D. B. Aronson, and R. R. Burgess. 1990. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J. Biol. Chem. 265:7069-7077. [PubMed] [Google Scholar]
- 61.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 62.Umek, R. M., A. D. Friedman, and S. L. McKnight. 1991. CCAAT-enhancer binding protein: a component of a differentiation switch. Science 251:288-292. [DOI] [PubMed] [Google Scholar]
- 63.van Gool, A. J., E. Citterio, S. Rademakers, R. van Os, W. Vermeulen, A. Constantinou, J. M. Egly, D. Bootsma, and J. H. Hoeijmakers. 1997. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 16:5955-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogel, H., D. S. Lim, G. Karsenty, M. Finegold, and P. Hasty. 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. USA 96:10770-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Mikecz, A., S. Zhang, M. Montminy, E. M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker, J. R., R. A. Corpina, and J. Goldberg. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607-614. [DOI] [PubMed] [Google Scholar]
- 68.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roesler, and N. A. Timchenko. 2001. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 69.Weeks, J. R., S. E. Hardin, J. Shen, J. M. Lee, and A. L. Greenleaf. 1993. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 7:2329-2344. [DOI] [PubMed] [Google Scholar]
- 70.Woodard, R. L., M. G. Anderson, and W. S. Dynan. 1999. Nuclear extracts lacking DNA-dependent protein kinase are deficient in multiple round transcription. J. Biol. Chem. 274:478-485. [DOI] [PubMed] [Google Scholar]
- 71.Woodard, R. L., K. J. Lee, J. Huang, and W. S. Dynan. 2001. Distinct roles for Ku protein in transcriptional reinitiation and DNA repair. J. Biol. Chem. 276:15423-15433. [DOI] [PubMed] [Google Scholar]
- 72.Yang, S.-H., A. Nussenzweig, L. Li, D. Kim, H. Ouyang, P. Burgman, and G. C. Li. 1996. Modulation of thermal induction of hsp70 expression by Ku autoantigen or its individual subunits. Mol. Cell. Biol. 16:3799-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoo, S., and W. S. Dynan. 1998. Characterization of the RNA binding properties of Ku protein. Biochemistry 37:1336-1343. [DOI] [PubMed] [Google Scholar]
- 74.Yoo, S., and W. S. Dynan. 1999. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 27:4679-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng, C., E. Kim, S. L. Warren, and S. M. Berget. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16:1401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, Z., L. Zhu, D. Lin, F. Chen, D. J. Chen, and Y. Chen. 2001. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J. Biol. Chem. 276:38231-38236. [DOI] [PubMed] [Google Scholar]