Abstract
Escherichia coli chemoreceptors are type I membrane receptors that have a periplasmic sensing domain, a cytosolic signaling domain, and two transmembrane segments. The aerotaxis receptor, Aer, is different in that both its sensing and signaling regions are proposed to be cytosolic. This receptor has a 38-residue hydrophobic segment that is thought to form a membrane anchor. Most transmembrane prediction programs predict a single transmembrane-spanning segment, but such a topology is inconsistent with recent studies indicating that there is direct communication between the membrane flanking PAS and HAMP domains. We studied the overall topology and membrane boundaries of the Aer membrane anchor by a cysteine-scanning approach. The proximity of 48 cognate cysteine replacements in Aer dimers was determined in vivo by measuring the rate and extent of disulfide cross-linking after adding the oxidant copper phenanthroline, both at room temperature and to decrease lateral diffusion in the membrane, at 4°C. Membrane boundaries were identified in membrane vesicles using 5-iodoacetamidofluorescein and methoxy polyethylene glycol 5000 (mPEG). To map periplasmic residues, accessible cysteines were blocked in whole cells by pretreatment with 4-acetamido-4′-maleimidylstilbene-2, 2′ disulfonic acid before the cells were lysed in the presence of mPEG. The data were consistent with two membrane-spanning segments, separated by a short periplasmic loop. Although the membrane anchor contains a central proline residue that reaches the periplasm, its position was permissive to several amino acid and peptide replacements.
Escherichia coli has five chemoreceptors that guide cells to favorable environments (41, 47). Four of these are methyl-accepting chemoreceptors (MCPs) that bind periplasmic ligands and transmit this information across the membrane to the cytosolic two-component chemotaxis cascade. The fifth receptor, Aer, is an aerotaxis, energy, and redox sensor containing N-terminal PAS sensing and C-terminal signaling domains separated by a putative membrane anchor (1, 2, 52, 56).
Although not proven, several lines of evidence indicate that both PAS sensor and C-terminal signaling domains of Aer are cytosolic. (i) All known PAS domains are intracellular sensors (62, 68). (ii) Native folding of the N-terminal PAS domain requires HAMP domain residues that are C terminal to the membrane anchor (18). (iii) Mutations in the HAMP domain are suppressed by mutations in the PAS domain (65). (iv) GFP fusions to Aer N termini (D. Salcedo and M. S. Johnson, unpublished data) or C termini (11) fluoresce. Since GFP fluoresces in the cytosol but not in the periplasm (11), both N and C termini are likely cytosolic. Thus, a topology similar to that of MCPs, where the sensor is periplasmic and the signaling region is cytosolic, is not likely.
Aer has just one hydrophobic segment long enough to span the membrane. The region exhibits several hallmarks consistent with two membrane-spanning segments separated by a hairpin loop. These include ∼38 consecutive hydrophobic residues, a central proline (P186), and flanking N- and C-terminal arginines (Fig. 1). It is known that successive positively charged residues near the boundaries of a transmembrane (TM) segment tend to be cytoplasmic, whereas negatively charged residues near boundaries of a TM segment tend to be exported (10, 14, 29, 59). Furthermore, central proline residues in model systems can convert a 40-residue single membrane-spanning hydrophobic polymer into two membrane-spanning helical segments (44). However, the central Pro186 present in E. coli Aer is not conserved in other Aer proteins, so its influence may not be important for membrane topology. From a survey of known membrane protein structures (19), the length of the Aer membrane anchor meets the minimum requirements to span the membrane once, extrude into the periplasm, and return to the cytosol (Fig. 1). However, a number of other conceivable structures (e.g., parallel to the bilayer surface) could occur if this segment were unable to span the membrane twice (54, 55).
FIG. 1.
Hypothetical membrane anchor topology for the dimeric Aer receptor. (A) A helix-loop-helix membrane anchor would accommodate a cytosolic placement for both PAS and signaling domains. (B) An expanded view of the membrane region (highlighted in gray) for one Aer monomer, including the residues examined in this study.
To determine the overall topology of the Aer homodimeric protein, we used a cysteine-scanning approach with whole cells (33) and with membrane vesicles (3, 23, 35). We estimated the proximity of cognate cysteine replacements by measuring the rate and extent of dimer formation after the addition of the oxidant copper phenanthroline. Membrane boundaries were identified by using a series of sulfhydryl-reactive probes (3, 23, 35). We show that the Aer membrane anchor spans the membrane twice and contains a central, flexible loop that faces the periplasmic space. However, the orientation between cognate transmembrane helical faces could not be identified, due to the sparse cross-linking within the membrane core.
MATERIALS AND METHODS
Reagents.
Methoxy polyethylene glycol 5000 (mPEG) was purchased from Nektar Therapeutics (Huntsville, AL); 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid, disodium salt (AMS) was from Molecular Probes, Inc. (Eugene, OR); 5-iodoacetamidofluorescein (5-IAF) was from Pierce (Rockford, IL); PEFAbloc was from Centerchem (Norwalk, CT); and N-ethyl maleimide (NEM) was purchased from Sigma (St. Louis, MO).
Membrane prediction algorithms.
The membrane-spanning segments of Aer were predicted by analyzing the entire Aer sequence with the following programs: MEMSAT (25, 27), DAS (8), PHDhtm (57, 58), TopPred (7, 64), TMHMM (30), TMpred (21), HMMtop (version 2.0) (63), TMAP (50, 51), TMfinder (12), PRED-TMR (49), SPLIT 4.0 (28), and SOSUI (20).
Bacterial strains and plasmids.
BT3312 (aer tsr) (56) is a derivative of E. coli strain RP437, which is the wild type for aerotaxis (48). Plasmid pMB1 is a cysteine-negative (C-less; Aer-C193S/C203A/C253A) derivative of pGH1 (wild-type Aer) (52) and was derived by digesting pGH1 with SmaI and SalI and replacing Aer by a C-less Aer construct previously engineered in pSB20 (2, 38). Both pGH1 and pMB1 are derivatives of pTrc99A, under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ptrc promoter (52).
Single-cysteine replacements in the membrane anchor region (residues 163 to 210) of Aer were made in pMB1 using the Quik Change site-directed mutagenesis kit from Stratagene (La Jolla, Calif.). Each plasmid was transformed by heat shock into BT3312. Expression of the Aer protein was confirmed by Western blot analysis using antisera against Aer2-166 (56), and the mutation was confirmed by DNA sequencing. All cysteine replacement constructs were inoculated on semisolid succinate swarm plates containing ampicillin (100 μg ml−1) to assess aerotactic behavior as described previously (2).
In vivo cross-linking using copper phenanthroline.
Cells expressing single-cysteine replacements were grown in H1 minimal salts medium supplemented with 30 mM succinate, 0.1% Casamino Acids, and ampicillin (100 μg ml−1) and induced with 50 μM IPTG. Cysteine cross-linking using copper phenanthroline was performed as described by Lee et al. (32) with the following modifications. Unless otherwise stated, the standard reaction was carried out at 23°C for various time intervals (0, 2, 5, 10, and 15 min) and quenched with a stop solution containing a 2.5 mM final concentration of NEM (32). To limit lateral diffusion of Aer in the membrane, a parallel reaction was carried out at 4°C with cells and oxidant that had been precooled separately at 4°C for 10 min prior to initiation of the reaction. After 20 min at 4°C, the reaction was quenched with stop solution containing NEM (as above) and incubated for an additional 10 min at 4°C before being boiled. Samples were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions and Western blotted. A control was used to test for artifactual cross-linking during the denaturation step by incubating cells with 2.5 mM NEM prior to oxidation with copper phenanthroline. The percentage of cross-linking was calculated by dividing the intensity of the cross-linked dimer band by the sum of the intensities of the monomer and dimer bands, multiplied by 100. BT3312/pGH1 (38) and BT3312/pMB1 were used as positive and negative cross-linking controls, respectively. The extent of cross-linking for 10 min at 23°C and 20 min at 4°C was compared for all cysteine replacements.
Preparation of membrane vesicles.
Bacterial membranes containing wild-type or mutated Aer receptors expressed at approximately 10% of the total membrane protein were prepared as previously described by Butler and Falke (4) with several modifications. Five milliliters of overnight cultures was inoculated into 250 ml of H1 minimal salts medium. Cultures were shaken at 30°C, induced with 0.6 mM IPTG at an optical density at 600 nm of 0.4, and grown for an additional 3 h. Cells were harvested by centrifugation at 10,000 × g for 10 min at 4°C and resuspended in 4 ml of low-salt buffer (100 mM sodium phosphate [pH 7.0], 10% glycerol, 10 mM EDTA, 1 mM 1,10-phenanthroline containing freshly added 46 mM dithiothreitol [DTT], and 2 mM PEFAbloc). Cells were disrupted by three freeze-thaw cycles, lysed by sonication (Branson Sonifier cell disrupter 200) at 60% power (three 15-s bursts with 45-s pauses) in an ice-salt bath, and centrifuged at 12,000 × g for 20 min at 4°C. Membranes were pelleted by centrifugation at 485,000 × g for 20 min and washed three times as follows. Membrane pellets were resuspended by sonication in high-salt buffer 1 (20 mM sodium phosphate [pH 7.0], 2 M KCl, 10% glycerol, 10 mM EDTA, 1 mM 1,10 phenanthroline containing freshly added 5 mM DTT, and 2 mM PEFAbloc), pelleted, resuspended in high-salt buffer 2 (without DTT or 1,10-phenanthroline), pelleted, resuspended in final buffer (with no DTT; 1,10-phenanthroline; or KCl), pelleted, resuspended in 200 μl of final buffer, aliquoted, frozen in a dry ice-ethanol bath, and stored at −80°C. The concentration of protein in the membrane was measured by a BCA (bicinchoninic acid) assay (Pierce Chemical) using bovine serum albumin as the standard. Frozen membrane preparations were diluted to the required concentration with final buffer prior to use.
Accessibility studies with 5-IAF.
These experiments were modifications of the protocol described by Boldog and Hazelbauer (3). Membrane vesicles containing between 5 and 15 μg of protein in 12 μl of final buffer were incubated with 5-IAF (500 μM in dimethyl formamide) in the presence and absence of 1% SDS to label the denatured and native forms of the protein, respectively. The SDS-treated reaction mixture was boiled for 5 min, whereas the native sample was incubated at 23°C for 5 min. Both reactions were stopped by the addition of 10 μl of SDS sample buffer containing 5% β-mercaptoethanol. Samples were boiled for 5 min and subjected to SDS-PAGE. Immediately after electrophoresis, wet gels were analyzed for fluorescein fluorescence with an Alpha Innotech gel documentation system with a UV light box. The total protein in each Aer-5-IAF band was estimated by staining the gel with Coomassie blue. The percent accessibility for each residue was calculated by taking the ratio of native to denatured Aer fluorescence, dividing by the ratio of native to denatured Aer protein, and multiplying by 100.
Accessibility studies with mPEG.
Between 4 and 6 μg of membrane vesicles in 13 μl of final buffer was incubated with mPEG (5 mM final concentration). Parallel reactions of native proteins were carried out at 23°C and at 4°C for 1 h. The reactivity of the denatured forms was determined in the presence of 1% SDS at ∼100°C for 5 min. Reactions were stopped by the addition of 6.75 μl of SDS sample buffer containing 5% β-mercaptoethanol. Samples were boiled for 5 min, run on SDS-PAGE, and Western blotted. Accessibility to mPEG was measured by comparing the accessibilities under native and denaturing conditions.
Preblocking accessible cysteines with AMS in intact cells.
Whole intact cells with single-cysteine replacements in the membrane anchor were incubated with or without AMS (8 mM final concentration) in final buffer for 45 min at 23°C. Unreacted AMS was removed by three washes in final buffer before the cells were disrupted for 4 min at 100°C, immediately after the addition of SDS and mPEG to a final concentration of 1% and 10 mM, respectively. The lysate was incubated for another 15 min at 23°C before the reaction was stopped with 10 μl of SDS sample buffer containing 5% β-mercaptoethanol. Samples were boiled for 5 min, run on SDS-PAGE, and Western blotted.
RESULTS
TM segments and secondary structure prediction of the membrane anchor region.
Twelve membrane protein prediction programs were used to predict Aer membrane topology, but there was no consensus among these algorithms. Nine programs forecasted just one transmembrane helix: MEMSAT, residues 183 to 204 (25, 27); DAS, residues 168 to 203 (8); PHDhtm, residues 173 to 197 (57, 58); TopPred, residues 167 to 187 (7, 64); TMHMM, residues 172 to 194 (30); TMpred, residues 169 to 188 (21); HMMtop, version 2.0, residues 171 to 195 (63); TMAP, residues 164 to 192 (50, 51); and TMfinder, residues 166 to 204 (12). Three of the programs predicted two TM helices: PRED-TMR, residues 167 to 185 (TM1) and residues 187 to 204 (TM2) (49); SPLIT 4.0, residues 165 to 183 (TM1) and 187 to 207 (28); and SOSUI, residues 166 to 188 (TM1) and 196 to 218 (TM2) (20). The secondary structure for this region was also analyzed. Jpred2 (9) and PSIpred (26), which predict secondary structures most reliably from multiple sequence alignments such as those from PSI-BLAST searches (39, 40), predicted a helix-loop-helix for this membrane anchor region.
In vivo cross-linking of single-cysteine replacements in the membrane anchor.
To analyze the structure of the membrane anchor of Aer, we employed a cysteine disulfide cross-linking approach (31, 43, 46). Wild-type Aer has three native cysteines at positions 193, 203, and 253. A construct missing the three native cysteines (C-less), encoding Aer-C193S/C203A/C253A (38), was cloned into pTrc99A to create plasmid pMB1 (Materials and Methods). C-less Aer mediated aerotaxis on succinate swarm plates and was therefore functional. Plasmid pMB1 was used to introduce a single cysteine at desired positions, to create a series of Aer mutants with cysteines that spanned residues 163 to 210. This segment included the predicted membrane anchor and the bordering residues. All 48 single-cysteine replacements, including residues 163 to 210, mediated aerotaxis in E. coli BT3312 (aer tsr) when inoculated on semisolid succinate agar, indicating that these cysteine replacements did not significantly alter receptor function (data not shown).
The rates and extent of cysteine cross-linking in response to the oxidant copper phenanthroline reflect the proximity of these residues in cognate subunits of Aer and/or the flexibility of the region (16, 22, 31). Once formed, the dimers can be separated from non-cross-linked monomers by SDS-PAGE under nonreducing conditions and visualized after Western blotting.
Initially, the rates of in vivo cross-linking after copper phenanthroline treatment at 23°C were determined. The rate plots for cross-linked cysteines fell into two major categories, distinguished by the presence (residues 184 to 188, 205, 208, and 209), or absence (residues 171, 173, 176, 182, 183, 191, 197, and 203) of visible cross-linking within 2 min. Once visible, however (with the exception of A184C and V187C) (Fig. 2A and B), the formation of the dimer product increased linearly until approximately 15 min (see, e.g., F182C and V209C) (Fig. 2A). Residues A184C and V187C were unique in both the rate and extent of cross-linking. A184C cross-linked most rapidly, and the reaction was nearly complete within 2 min (Fig. 2A and B). V187C exhibited noticeable cross-linking in the absence of oxidant (Fig. 2A and B, 0-min time point), suggesting that this residue might be located in an oxidative environment. With these exceptions (A184C and V187C), the 10-min time point was in the linear range for all constructs, and this incubation time was used for overall comparisons, as represented by the Western blots shown in Fig. 2C. The positive control, Aer-WT, contained three native cysteines, two of which can cross-link (C203 in the membrane anchor and C253 in the HAMP domain) (38); the negative control, Aer-pMB1, had no cysteines.
FIG. 2.
Rates and extent of Aer cysteine cross-linking in intact cells after the addition of the oxidant copper phenanthroline. (A) The rate of cross-linking for representative classes of cysteine replacements, as discussed in the text. Except for Aer-A184C (□) (left) and Aer-V187C (▪) (left), the 10-min time point was in the linear range of the reaction for all cross-linking cysteine replacements, e.g., F182C (▵) and V209C (▴) (right). (B) Western blots showing (i) that the dimerization reaction for Aer-A184C was nearly complete within 2 min (2′) and (ii) the presence of a dimer band in Aer-V187C before the addition of copper phenanthroline (0′). (C) Representative Western blots comparing the extent of cross-linking at 10 min, which was the time point chosen to compare the extent of cross-linking for each cysteine replacement in the membrane anchor region. Abbreviations: M, monomer; D, dimer.
A contour plot generated by the PSA server at Boston University (Fig. 3A) (60, 61, 66) is a convenient way to show the probability of the helix-loop-helix secondary structure in the membrane anchor region (37). The bar graphs below this contour plot (Fig. 3B and C) are summaries of the cross-linking data from three or more independent experiments. At 23°C (Fig. 3B), multiple residues cross-linked between cognate monomers (within a dimer) with no obvious periodicity. However, there was strong cross-linking in consecutive residues from F182C to V188C, consistent with close proximity and/or high flexibility in this region and supporting the loop structure predicted in the PSA contour plot (Fig. 3A).
FIG. 3.
Summary of cysteine cross-linking in Aer mutants at 23°C and 4°C. The extent of Aer dimers with cross-linked cysteines in the membrane anchor demonstrates a central region of high proximity and flexibility, consistent with the PSA server secondary structure prediction. (A) Contour plot of secondary-structure probabilities (PSA server) (60, 61, 66). Rows indicate the secondary structure state; columns indicate each residue position. The probability of each structural state is depicted with contour lines in probability increments of 0.1. The β-strand prediction value was <0.1 and therefore is not included. (B) Extent of in vivo cross-linking for all 48 cysteine replacements after incubation of cells with copper phenanthroline for 10 min at 23°C. (C) Extent of in vivo cross-linking for the same cells shown in panel B, incubated with copper phenanthroline for 20 min at 4°C.
In reality, cross-linking between monomers might not represent close proximity between cognate residues but could occur from random collisions of receptors diffusing laterally through the lipid bilayer. To limit lateral diffusion, we repeated the cross-linking analysis at 4°C (Fig. 3C), which is well below the lipid-phase transition temperature of approximately 18°C in E. coli (45). To accommodate the decrease in kinetics at this temperature, the reaction time was increased from 10 min to 20 min. At 4°C, there was a low level of cross-linking in the putative TM segments, but similar cross-linking (to that of 23°C) in consecutive residues of the central loop (184 to 188), consistent with flexibility in this region. Notably, A184C and V187C still cross-linked at high levels, indicating close proximity between neighboring residues at these positions.
Mapping membrane boundaries with 5-IAF and mPEG.
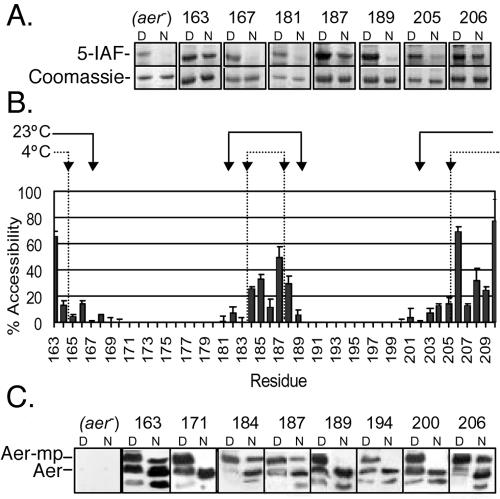
To map the boundaries of the Aer protein in the E. coli cytoplasmic membrane, we measured the surface accessibility of substituted cysteine residues by using sulfhydryl-reactive probes. The reagent 5-IAF is membrane impermeable and should not therefore react with integral membrane residues (3). Mixed membrane vesicles expressing Aer single-cysteine replacements at residues 163 to 210 were reacted with 5-IAF for 5 min at 23°C, and the percentage of fluorescence associated with native Aer relative to denatured Aer was determined (Fig. 4).
FIG. 4.
Surface accessibility of Aer cysteine replacements to the sulfhydryl-reactive reagents 5-IAF and mPEG in membrane vesicles. (A) SDS-PAGE gels showing the reactivity of strategic cysteine replacements towards 5-IAF. (Top) Negative fluorescent images of samples reacted with 5-IAF under native (N) and denaturing (D) conditions for 5 min. (Bottom) The same bands stained with Coomassie blue to estimate the total protein in each band. Note the unknown labeled protein in the aer control lane under denaturing conditions. (B) Bar graph summarizing the percent accessibility of cysteine replacements to 5-IAF during incubations for 5 min at 23°C. The results are the average of three or more independent experiments. The arrows at the top of the graph represent the accessibility boundaries for mPEG at 23°C (solid lines) and at 4°C (dotted lines). (C) Representative Western blots showing the presence (residues 163, 184, 187, and 206) or absence (residues 171, 189, 194, and 200) of a mobility shift in Aer (Aer-mp) after incubation of membrane vesicles with mPEG under native (N) conditions for 1 h. All cysteine replacements were PEGylated under denaturing (D) conditions.
Representative gels showing the extent of 5-IAF labeling of single-cysteine replacements under denaturing and native conditions are shown in Fig. 4A. Coomassie blue-stained gels, showing the relative levels of protein in each lane, are displayed below the respective negative image of the same 5-IAF-labeled gel. There was a notable artifact under denaturing conditions with 5-IAF-treated membranes. The negative control lacking Aer (aer negative) showed apparent 5-IAF labeling under denaturing conditions and considerably less (but observable) labeling under native conditions (Fig. 4A). This indicated that another unidentified membrane protein(s) comigrated with Aer. With this caveat in mind, we estimated the accessibility of each cysteine to 5-IAF and have summarized these data in Fig. 4B. As shown, there were large changes in accessibility between residues 163 to 164, 183 to 184, 188 to 189, and 205 to 206, indicating that these regions are near the boundaries of the membrane and aqueous phases. From these data, putative transmembrane segments would include residues 164 to 183 and 189 to 205.
To overcome the inherent background labeling with the 5-IAF probe, membrane boundaries were reanalyzed in membrane vesicles by using another hydrophilic sulfhydryl-reactive reagent, mPEG (35). This reagent has an actual molecular mass of 5 kDa, but Aer-mPEG complexes exhibited an apparent molecular mass increase of ∼10 kDa higher than that of unreacted Aer (Fig. 4C), so the two forms could be separated by SDS-PAGE and analyzed on Western blots. As shown in the representative Western blots of Fig. 4C, the Aer monomer and Aer-mPEG forms were easily distinguishable (e.g., residues 184, 187, and 206), and there was no background signal from membranes that lacked Aer. However, as previously reported (38), an ever-present Aer proteolytic fragment was visible (Fig. 4C).
The lower background noise of this method allowed us to increase the protein loaded into each lane to maximize the threshold at which we could visualize Aer-mPEG complexes. As shown in Fig. 4C, residues predicted by the 5-IAF studies to be membrane embedded, such as T189C, 193C, and A194C, showed no reactivity to mPEG in native membranes. However, residues predicted to be near the membrane aqueous interface, such as A184C, V187C, and W206C, had high levels of reactivity with mPEG. To eliminate the possibility of slow diffusion into the membrane, experiments were performed in parallel with mPEG at 23°C and at 4°C for 1 h. At 4°C, mPEG does not cross membranes when incubated for 24 h (35).
A summary of the membrane boundaries determined by PEGylation at 23°C and 4°C are marked above the bar graph in Fig. 4B. As expected, more residues were accessible to mPEG at 23°C than at 4°C, and boundary demarcations were similar but not identical to those identified by 5-IAF. However, the combined data were consistent with a membrane anchor that has two membrane-spanning segments and a short periplasmic loop. The N- and C-terminal boundaries would represent the cytosolic membrane interface and the central loop boundaries would delineate the periplasmic membrane interface.
Depending on the temperature and the particular sulfhydryl-reactive probe (Fig. 4B), the first membrane-embedded residue after the N-terminal cytoplasmic domain was 164 (5-IAF; 23°C), 165 (mPEG; 4°C), or 167 (mPEG; 23°C), and the last membrane-embedded residue before the C-terminal cytoplasmic domain was 202 (mPEG; 23°C) or 205 (5-IAF, 23°C; mPEG, 4°C). The higher accessibility of mPEG at 23°C suggested that the maleimide group of mPEG might penetrate further into the membrane than 5-IAF. Whether this was due to an inherent difference in hydrophobicity or caused by the difference in incubation time, which was 1 h for mPEG and 5 min for 5-IAF, is not known.
Similar temperature and reagent differences were evident for the central loop region, which was expected to be periplasmic. The last membrane-embedded residue before the loop was 181 (mPEG; 23°C) or 183 (5-IAF, 23°C; mPEG, 4°C) and the first membrane-embedded residue after the loop was 188 (mPEG; 4°C), 189 (5-IAF; 23°C), or 190 (mPEG; 23°C). Taken together, the maximum lengths of the transmembrane segments include residues 164 to 183 for TM1 and 188 to 205 for TM2.
The central flexible loop is periplasmic.
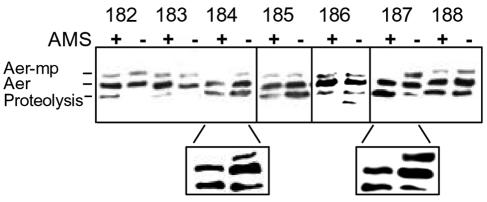
Although the accessibility data and the predicted secondary structure of the membrane anchor indicated that the central loop was periplasmic, we could not exclude unusual topologies wherein the flexible loop was cytoplasmic, since both sides of these vesicles are accessible to sulfhydryl reagents (3). To verify that the flexible loop was periplasmic, we used an in vivo approach with AMS (23, 35), a small hydrophilic sulfydryl-reactive reagent that can transverse the outer membrane porins but cannot cross the inner membrane. We pretreated intact cells with AMS to block accessible periplasmic cysteines from subsequent reactions with mPEG under denaturing conditions.
As shown in Fig. 5, residues A184C and V187C were completely blocked from PEGylation by AMS, indicating that these residues are fully exposed to the periplasmic environment. Thus, the central flexible loop was periplasmic and not cytosolic. However, residues A185C and P186C, which were accessible to both 5-IAF and mPEG in membrane vesicles, were not totally blocked by AMS (Fig. 5).
FIG. 5.
Pretreatment with AMS in intact cells blocks periplasmic cysteine replacements from subsequent PEGylation. Cells were reacted with (+) and without (−) AMS for 45 min before being washed and probed with mPEG under denaturing conditions as described in the text. Aer-mPEG adducts (Aer-mp) showed a mobility shift. The enlarged lanes in the lower panel are higher exposures taken from different Western blots; these highlight replacements 184 and 187, which were reproducibly blocked by AMS.
The sequence of the Aer periplasmic loop is not critical for function.
The Aer-P186C replacement was functional, indicating that Pro186 was not absolutely required at this position. To further investigate amino acid stringency at this position, Pro186 was replaced by amino acid residues Arg, Ser, Phe, Trp, Ala, and Asp. All replacements were functional in aerotaxis on semisoft succinate agar, indicating that this site was not critical for activity (data not shown). The site was also permissive to short insertions, as a Ser-Gly-Ser replacement of Pro186 and an Arg-Pro-Arg-Ile insertion between residues 185 and 186 (36) showed aerotaxis. The low level of amino acid stringency and permissiveness to insertions at this site are consistent with a region that is both near the membrane (for shielding hydrophobic residues) and accessible to the periplasm (when accommodating charged residues or insertions).
DISCUSSION
These data are consistent with a model in which the membrane anchor of Aer forms two membrane-spanning regions (TM1 and TM2) flanking a central periplasmic loop. Such a model supports previous studies indicating that both the N-terminal PAS domain and the C-terminal HAMP and signaling domains are cytosolic (11, 18, 62, 65, 68).
Nine of the 12 membrane protein structure prediction programs that were tested failed to predict two transmembrane segments for the 38 residues that were previously predicted to form the Aer membrane anchor (1, 2, 52, 56). The best prediction methods can correctly identify all membrane helices in only 50% to 70% of proteins with known structure (6). Moreover, developers may overestimate the accuracy of their methods by 15% to 50% (5). PRED-TMR (49), SPLIT 4.0 (28), and SOSUI (20) were the only programs tested that predicted two transmembrane segments, although SOSUI predicted spans that were inconsistent with this study. Of note, a recent study in which the C termini of 601 E. coli inner membrane proteins were tagged with alkaline phosphatase and green fluorescent protein correctly predicted a cytosolic location for both the N-terminal and C-terminal ends of Aer (11).
An in vivo cysteine cross-linking strategy was used to study the proximity of the transmembrane segments between cognate monomers within the Aer dimer (31, 38, 43, 46). Several consecutive residues (182 to 188) flanking Pro186 in the central region of the membrane anchor cross-linked strongly at 23°C (Fig. 3B), indicating that this segment was highly flexible. Moreover, residues 184 to 187 cross-linked strongly at 4°C (Fig. 3C), suggesting that the cross-linking was due to proximity between monomers and not to random collisions from laterally diffusing receptors in the membrane. In E. coli, the lateral diffusion of membrane proteins decreases markedly below 18°C, which is the lipid-phase transition temperature (45). An example of this temperature effect was shown for lactose permease, where strong cross-linking that occurred from random collisions between monomers at room temperature was markedly inhibited at 0°C during 1-h incubations (17).
Unlike the central loop region, there was sparse cross-linking in the TM1 and TM2 segments. Moreover, residues that cross-linked did not show the periodicity one would expect for close interactions between cognate helices (33). Several explanations for low cross-linking levels and lack of periodicity are possible. (i) The relative tilt of the cognate transmembrane helical segments, driven by the solvation of side chains at the membrane boundaries (54, 55), may limit cross-linking. (ii) The nearest helix-helix interactions might occur through TM1-TM2 or TM1-TM2′ segments rather than via TM1-TM1′ and TM2-TM2′ segments. That said, the sequence of amino acids in the transmembrane segments of Aer did not show periodic variations in polarity, as one might expect for TM-TM interactions. This is consistent with the low level of sequence conservation found in the membrane anchor region. In general, residues facing outward toward the lipids are considerably more hydrophobic than those facing inward, toward the other helix(es) in the membrane (53), opposite of that seen in aqueous environments. (iii) Helix-helix interactions may not be exclusively intradimeric; some residues may cross-link within a dimer while others cross-link between dimers in a trimer of dimers unit. Preliminary evidence indicates that this is the case (D. N. Amin, unpublished data). (iv) Membrane-spanning segments might form a β-sheet rather than a helix. This scenario is unlikely, as secondary structure programs predicted α-helices for this region, and β-sheets are generally found in pore proteins, as well as being amphipathic. Helices form spontaneously when a hydrophobic sequence inserts into a membrane bilayer, due to the free energy of hydrogen bonding between the polar backbone carbonyls and amide groups (15).
Membrane boundaries were mapped in vitro by determining the accessibility of the cysteine replacements to the membrane-impermeable sulfhydryl-reactive probes 5-IAF and mPEG. The mPEG probe had the advantage of specificity, as PEGylated Aer could be discriminated from unmodified Aer on Western blots. For in vivo studies, we preblocked accessible cysteines with the periplasm-accessible, membrane-impermeable, sulfhydryl-reactive probe AMS before reacting solubilized cells with mPEG. The accessibility of residues in the flexible loop to 5-IAF and mPEG and the blocking of residues 184 and 187 in vivo with AMS indicate that this loop is in the periplasm. The combination of strong cross-linking and accessibility indicates that the loop is dynamic. Otherwise, an inverse relationship between cross-linking and accessibility would be expected. Interestingly, residues 185 and 186 were accessible to 5-IAF and mPEG but not to AMS. One possible explanation for this difference is that AMS is more selectively excluded from the membrane surface than is 5-IAF or mPEG. In this case, residues 185 and 186 would be proximal to the periplasm but would not protrude into the periplasm. Alternatively, the presence of a membrane potential in whole cells or other changes occurring during the preparation of membrane vesicles could alter the local topology and shield these residues from the aqueous phase.
Accessibility studies indicated that the membrane cytosolic boundaries lay between residues 164 and 167 for TM1 and between residues 202 and 205 for TM2. Residues 163 and 206 were accessible to all probes under all conditions (23°C and 4°C). Notably, residues 163 and 206 are tryptophans in native Aer. Tryptophans are well known for stabilizing membrane-aqueous boundaries (13, 67). Given their hydrophobic nature, it is likely that these native tryptophan residues reside at the membrane boundary interface rather than protrude into the cytosol, as occurred for their cysteine replacements.
From the cysteine accessibility data, the maximum lengths of the transmembrane regions would include 20 residues for TM1 (from 164 to 183) and 18 residues for TM2 (from 188 to 205). The value for TM2 is close to the average length (17.7 residues or 27 Å) (19) for a membrane helix that spans the hydrophobic part of the bilayer. The absolute minimum lengths for these segments would include 15 residues for TM1 (167 to 181) and 13 residues for TM2 (190 to 202). These lengths are untenable if a helical structure is assumed, as they would not span the membrane (27 Å). As stated for the AMS probe, variations in accessibility between 5-IAF and mPEG sulfhydryl probes may also represent differences in the ability of these probes to penetrate the membrane surface. The estimates for the maximum TM1 and TM2 lengths appear reasonable and, as stated, are supported by the presence of tryptophans at residues 163 and 206.
The central loop between TM1 and TM2 contains a nonconserved proline residue; prolines are established helix breakers in globular proteins (34). In model membrane systems, a single proline residue can change a 40-mer hydrophobic peptide from a single membrane-spanning helix into two membrane-spanning helices (44). However, in membrane segments, prolines may often stabilize the helical structure (34), and bends within membrane regions generally require more than one proline unless a glycine residue is spaced four residues away (24). Previously, we found that Aer was functional after a tetrapeptide (RPRI) had been inserted between residues Ala185 and Pro186 (36). In the present study, we found that amino acid replacements at Pro186 in Aer did not abolish the function. In addition to the Cys replacement, Aer was functional when Pro186 was replaced by polar (S), charged (D and R), or aromatic (F and W) residues, as well as with a tripeptide (SGS). These data and the fact that Pro186 is not conserved in other Aer receptors indicate that proline is not necessary to form the two transmembrane segments. Moreover, since the loop was permissive to short peptide inserts, the region must not be directly involved with signaling.
In summary, the Aer membrane anchor in E. coli forms two transmembrane segments that flank a central short periplasmic loop. The loop itself does not appear to be involved in signal transduction. Whether the function of the membrane anchor is to localize Aer to the membrane, maintain registry between N-terminal PAS and C-terminal signaling regions, or be actively involved in signaling like the MCPs (13, 42) remains to be determined.
Acknowledgments
We thank Maxwell Brandon for creating pMB1, Gordon Harding for developing the PEGylation protocol, and Kylie Watts for critical analysis and helpful discussions. We are grateful to Sheena Fry and Nathan Abraham for technical assistance.
This work was supported by grants from Loma Linda University to M.S.J. and the National Institute of General Medical Sciences (GM29481) to B.L.T.
REFERENCES
- 1.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boldog, T., and G. L. Hazelbauer. 2004. Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues. Protein Sci. 13:1466-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. P., A. Kernytsky, and B. Rost. 2002. Transmembrane helix predictions revisited. Protein Sci. 11:2774-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. P., and B. Rost. 2002. State-of-the-art in membrane protein prediction. Appl. Bioinformatics 1:21-35. [PubMed] [Google Scholar]
- 7.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 8.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 9.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 10.Dalbey, R. E. 1990. Positively charged residues are important determinants of membrane protein topology. Trends Biochem. Sci. 15:253-257. [DOI] [PubMed] [Google Scholar]
- 11.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 12.Deber, C. M., C. Wang, L. P. Liu, A. S. Prior, S. Agrawal, B. L. Muskat, and A. J. Cuticchia. 2001. TM Finder: a prediction program for transmembrane protein segments using a combination of hydrophobicity and nonpolar phase helicity scales. Protein Sci. 10:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draheim, R. R., A. F. Bormans, R. Z. Lai, and M. D. Manson. 2005. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry 44:1268-1277. [DOI] [PubMed] [Google Scholar]
- 14.Ehrmann, M., D. Boyd, and J. Beckwith. 1990. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc. Natl. Acad. Sci. USA 87:7574-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eilers, M., S. C. Shekar, T. Shieh, S. O. Smith, and P. J. Fleming. 2000. Internal packing of helical membrane proteins. Proc. Natl. Acad. Sci. USA 97:5796-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falke, J. J., A. F. Dernburg, D. A. Sternberg, N. Zalkin, D. L. Milligan, and D. E. Koshland, Jr. 1988. Structure of a bacterial sensory receptor. A site-directed sulfhydryl study. J. Biol. Chem. 263:14850-14858. [PubMed] [Google Scholar]
- 17.Guan, L., F. D. Murphy, and H. R. Kaback. 2002. Surface-exposed positions in the transmembrane helices of the lactose permease of Escherichia coli determined by intermolecular thiol cross-linking. Proc. Natl. Acad. Sci. USA 99:3475-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hildebrand, P. W., R. Preissner, and C. Frommel. 2004. Structural features of transmembrane helices. FEBS Lett. 559:145-151. [DOI] [PubMed] [Google Scholar]
- 20.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 22.Hughson, A. G., and G. L. Hazelbauer. 1996. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc. Natl. Acad. Sci. USA 93:11546-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwaki, S., N. Tamura, T. Kimura-Someya, S. Nada, and A. Yamaguchi. 2000. Cysteine-scanning mutagenesis of transmembrane segments 4 and 5 of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a permeability barrier in the middle of a transmembrane water-filled channel. J. Biol. Chem. 275:22704-22712. [DOI] [PubMed] [Google Scholar]
- 24.Javadpour, M. M., M. Eilers, M. Groesbeek, and S. O. Smith. 1999. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys. J. 77:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. T. 1998. Do transmembrane protein superfolds exist? FEBS Lett. 423:281-285. [DOI] [PubMed] [Google Scholar]
- 26.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 27.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 28.Juretic, D., L. Zoranic, and D. Zucic. 2002. Basic charge clusters and predictions of membrane protein topology. J. Chem. Inf. Comput. Sci. 42:620-632. [DOI] [PubMed] [Google Scholar]
- 29.Kimbrough, T. G., and C. Manoil. 1994. Role of a small cytoplasmic domain in the establishment of serine chemoreceptor membrane topology. J. Bacteriol. 176:7118-7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 31.Lee, G. F., G. G. Burrows, M. R. Lebert, D. P. Dutton, and G. L. Hazelbauer. 1994. Deducing the organization of a transmembrane domain by disulfide cross-linking. The bacterial chemoreceptor Trg. J. Biol. Chem. 269:29920-29927. [PubMed] [Google Scholar]
- 32.Lee, G. F., D. P. Dutton, and G. L. Hazelbauer. 1995. Identification of functionally important helical faces in transmembrane segments by scanning mutagenesis. Proc. Natl. Acad. Sci. USA 92:5416-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, G. F., M. R. Lebert, A. A. Lilly, and G. L. Hazelbauer. 1995. Transmembrane signaling characterized in bacterial chemoreceptors by using sulfhydryl cross-linking in vivo. Proc. Natl. Acad. Sci. USA 92:3391-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, S. C., N. K. Goto, K. A. Williams, and C. M. Deber. 1996. Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. USA 93:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, J., and C. Deutsch. 2001. Pegylation: a method for assessing topological accessibilities in Kv1.3. Biochemistry 40:13288-13301. [DOI] [PubMed] [Google Scholar]
- 36.Ma, Q. 2001. HAMP domain and signaling mechanism of the Aer protein. Ph.D. dissertation. Loma Linda University, Loma Linda, CA.
- 37.Ma, Q., M. S. Johnson, and B. L. Taylor. 2005. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes flavin adenine dinucleotide-binding determinants to the AS-2 helix. J. Bacteriol. 187:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma, Q., F. Roy, S. Herrmann, B. L. Taylor, and M. S. Johnson. 2004. The Aer protein of Escherichia coli forms a homodimer independent of the signaling domain and flavin adenine dinucleotide binding. J. Bacteriol. 186:7456-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 40.McGuffin, L. J., and D. T. Jones. 2003. Benchmarking secondary structure prediction for fold recognition. Proteins 52:166-175. [DOI] [PubMed] [Google Scholar]
- 41.Miller, A. F., and J. J. Falke. 2004. Chemotaxis receptors and signaling. Adv. Protein Chem. 68:393-444. [DOI] [PubMed] [Google Scholar]
- 42.Miller, A. S., and J. J. Falke. 2004. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry 43:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milligan, D. L., and D. E. Koshland, Jr. 1988. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 263:6268-6275. [PubMed] [Google Scholar]
- 44.Nilsson, I., and G. von Heijne. 1998. Breaking the camel's back: proline-induced turns in a model transmembrane helix. J. Mol. Biol. 284:1185-1189. [DOI] [PubMed] [Google Scholar]
- 45.Overath, P., M. Brenner, T. Gulik-Krzywicki, E. Shechter, and L. Letellier. 1975. Lipid phase transitions in cytoplasmic and outer membranes of Escherichia coli. Biochim. Biophys. Acta 389:358-369. [DOI] [PubMed] [Google Scholar]
- 46.Pakula, A. A., and M. I. Simon. 1992. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc. Natl. Acad. Sci. USA 89:4144-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116-121. [DOI] [PubMed] [Google Scholar]
- 48.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasquier, C., V. J. Promponas, G. A. Palaios, J. S. Hamodrakas, and S. J. Hamodrakas. 1999. A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng. 12:381-385. [DOI] [PubMed] [Google Scholar]
- 50.Persson, B., and P. Argos. 1994. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol. 237:182-192. [DOI] [PubMed] [Google Scholar]
- 51.Persson, B., and P. Argos. 1996. Topology prediction of membrane proteins. Protein Sci. 5:363-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rees, D. C., L. DeAntonio, and D. Eisenberg. 1989. Hydrophobic organization of membrane proteins. Science 245:510-513. [DOI] [PubMed] [Google Scholar]
- 54.Ren, J., S. Lew, J. Wang, and E. London. 1999. Control of the transmembrane orientation and interhelical interactions within membranes by hydrophobic helix length. Biochemistry 38:5905-5912. [DOI] [PubMed] [Google Scholar]
- 55.Ren, J., S. Lew, Z. Wang, and E. London. 1997. Transmembrane orientation of hydrophobic alpha-helices is regulated both by the relationship of helix length to bilayer thickness and by the cholesterol concentration. Biochemistry 36:10213-10220. [DOI] [PubMed] [Google Scholar]
- 56.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 58.Rost, B., R. Casadio, P. Fariselli, and C. Sander. 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci. 4:521-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seligman, L., J. Bailey, and C. Manoil. 1995. Sequences determining the cytoplasmic localization of a chemoreceptor domain. J. Bacteriol. 177:2315-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stultz, C. M., R. Nambudripad, R. H. Lathrop, and J. V. White. 1997. Predicting protein structure with probabilistic models, p. 447-506. In N. Allewell and C. Woodward (ed.), Protein structural biology in bio-medical research, vol. 22B. JAI Press, Greenwich, Conn. [Google Scholar]
- 61.Stultz, C. M., J. V. White, and T. F. Smith. 1993. Structural analysis based on state-space modeling. Protein Sci. 2:305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 64.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 65.Watts, K. J., Q. Ma, M. S. Johnson, and B. L. Taylor. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J. Bacteriol. 186:7440-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White, J. V., C. M. Stultz, and T. F. Smith. 1994. Protein classification by stochastic modeling and optimal filtering of amino-acid sequences. Math. Biosci. 119:35-75. [DOI] [PubMed] [Google Scholar]
- 67.Yau, W. M., W. C. Wimley, K. Gawrisch, and S. H. White. 1998. The preference of tryptophan for membrane interfaces. Biochemistry 37:14713-14718. [DOI] [PubMed] [Google Scholar]
- 68.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]