Abstract
mSin3A and Transducin-Like Enhancer of Split (TLE) are two histone deacetylase (HDAC)-containing corepressors that function to repress transcription at targeted genes. Pf1 is a plant homeodomain zinc finger protein that interacts with both mSin3A and TLE, suggesting that it coordinates their function. Here we show that mSin3A and TLE interact with members of the mortality factor (MORF) family of putative transcriptional regulators. This family comprises MORF on chromosome 4 (MORF4) and MORF-related genes on chromosomes X and 15 (MRGX and MRG15, respectively) and is proposed to contribute to cellular senescence. Consistent with a role in transcription, we demonstrate that Gal4 fusions to each MORF family member repress transcription from a Gal4-dependent luciferase reporter. By using both mapping experiments and a dominant negative form of TLE, we show that repression by MORFs requires associations with mSin3A and TLE. Therefore, common functions of the MORFs are likely elicited through the action of a MORF/mSin3A/TLE complex. While the MORFs may have common functions, MRG15, but not MRGX or MORF4, interacted with Pf1. Therefore, MRG15 may have functions that are distinct from those of MRGX and MORF4. Consistent with this hypothesis, Pf1 reduced transcriptional repression by Gal4-MRG15 but it had no effect on repression by MRGX and MORF4. Pf1 has independent binding sites for MRG15 and mSin3A. In addition, Pf1 and MRG15 bind different domains on mSin3A. Together, these data suggest that the unique functions of MRG15 are elicited through the action of an MRG15/Pf1/mSin3A complex.
The acetylation state of core histones in nucleosomes influences the transcription of eukaryotic genes. In general, genes bearing hyperacetylated histones are more transcriptionally active than genes whose histones are hypoacetylated. The enzymes that acetylate and deacetylate lysines in core histone tails are histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively (23, 25, 40). Typically, HATs and HDACs are components of multiprotein complexes, and the associated proteins appear to regulate the activities and targeting of these classes of enzymes (4, 13, 23, 29). Targeting of HAT complexes to genes results in transcriptional activation, while targeting of HDAC complexes results in transcriptional repression (30). The mammalian mSin3A complex was among the first corepressors demonstrated to require HDACs to repress transcription (24, 33, 43, 49). mSin3A was initially shown to function as a corepressor for the Mad family of transcriptional repressors (6, 42); however, it is now known that mSin3A is utilized by a plethora of transcriptional repressors (1, 4, 29). As such, the mSin3A corepressor provides an excellent model for studying the function of HDAC-dependent corepressors.
The identification and characterization of the proteins associated with mSin3A has revealed how it functions to repress transcription (4, 29). Purification of a highly stable core mSin3A complex from a number of cell types identified 8 to 12 polypeptides tightly associated with mSin3A (24, 31, 47, 49; T. Fleischer and D. E. Ayer, submitted for publication). The components of the mSin3A core complex include HDAC1 and HDAC2, the retinoblastoma-associated proteins 46 and 48 (RbAp46 and RbAp48), mSin3A-associated protein 30 (SAP30), and retinoblastoma binding protein 1 (Rbp1). Experiments using deacetylase inhibitors and mutagenesis demonstrated that most mSin3A-dependent transcriptional repression is due to the activities of HDAC1 and HDAC2 (24, 26, 32, 33). RbAp46 and RbAp48 are likely involved in targeting HDACs to nucleosomes (46). SAP30 is an adaptor protein that specifies mSin3A involvement in repression by a subset of nuclear hormone receptors and is required for transcriptional repression by Rb through Rbp1 (32, 34, 35). The mSin3A protein is the scaffold upon which the complex assembles and has four paired amphipathic alpha helix domains (PAH1 to -4) and the HDAC interaction domain (HID), which function as highly conserved protein-protein interaction surfaces (4, 29, 45).
The core mSin3A complex can associate with other complexes that regulate chromatin structure, including the Transducin-Like Enhancer of Split (TLE), which is orthologous to the Drosophila melanogaster Groucho corepressor (17, 19, 48). TLE is not a component of the core mSin3A complex but can be tethered to mSin3A through association with proteins that are not part of the core complex. One protein demonstrated to link TLE to mSin3A is the plant homeodomain (PHD) zinc finger protein Pf1 (48). Pf1 has two separate mSin3A interaction domains that directly bind mSin3A; Pf1SID1 binds PAH2 and Pf1SID2 binds PAH1 (48). Although Pf1 does not contain a known Groucho/TLE binding motif, binding of TLE to Pf1 does not require mSin3A, suggesting that TLE and Pf1 may interact directly through a novel domain (48). TLE proteins also appear to interact with mSin3A through Pf1-independent mechanisms (17, 19). Thus, these data suggest that there may be functional cross talk between the mSin3A and TLE corepressors.
Here, we demonstrate that one member of the mortality factor (MORF) family, MORF-related gene on chromosome 15 (MRG15), is a Pf1-interacting protein. The MORF family includes MORF on chromosome 4 (MORF4), MORF-related gene on chromosome X (MRGX), and MRG15 and is implicated in the transcriptional regulation of cellular senescence (7, 8). Pf1 interaction is restricted to MRG15 despite the high sequence identity among the MORFs. We show that when fused to the Gal4 DNA binding domain, MORF4, MRGX, and MRG15 repress transcription and associate with both mSin3A and TLE. Together, these findings suggest that MORFs are components of multiple and distinct corepressor complexes and that this family of transcription factors has both shared and unique functions.
MATERIALS AND METHODS
Plasmids.
The full-length Pf1 cDNA was amplified by PCR with Pfu Turbo polymerase (Stratagene) and inserted into pBTM-116 ura3 (9) to generate LexA-Pf1. pcDNA3.1-HA-MRG15, pcDNA3.1-FLAG-MORF4, and pcDNA3.1-HA-MRGX were provided by O. M. Pereira-Smith. FLAG-AES1 (where AES1 stands for Amino Enhancer of Split 1) was provided by S. Stifani. PCR was used to amplify full-length cDNAs for mrg15, morf4, and mrgx, and the products were inserted into the pFA vector (Stratagene) to generate Gal4 DNA binding domain fusion proteins. PCR was used to generate the mrg15 deletion series, and the products were inserted into pFA. Similarly, PCR was used to generate the pf1 deletion series, and the products were inserted into pBTM-116 ura3. The construction of FLAG-Pf1 is described elsewhere (48). The FLAG epitope sequence, DYKDDDDK, was engineered onto the amino terminus of MRG15 by PCR, and the product was inserted into pcDNA3.1 to generate FLAG-MRG15. The Myc epitope-tagged mSin3A deletion plasmids were provided by R. N. Eisenman (33). Constructs and mutations were verified by sequencing.
Yeast two-hybrid screen.
LexA-Pf1 was used to screen a VP16 fusion cDNA library made from RNA derived from day 9.5 and 10.5 mouse embryos as described previously (10, 44) with the following modifications. The Saccharomyces cerevisiae strain DY5736 (9) was transformed with LexA-Pf1 to make the bait strain. Into this strain, the VP16 library was transformed, and the yeast cells were plated on selective media containing 5 or 10 mM 3-amino-1,2,4-triazol (Sigma). Yeast cells were cured of LexA-Pf1 with 5-flouroorotic acid (Sigma). β-Galactosidase (β-Gal) assays were performed after the original LexA-Pf1 fusion or the control plasmids were reintroduced to test for binding specificity. The cDNAs fused to VP16 from 217 positive clones were isolated and sequenced. Of these, 17 contained mrg15 sequences that were aligned with Sequencher software. For the yeast two-hybrid assays, different LexA-Pf1 deletion constructs were cotransformed with a VP16-MRG15 construct that contained the minimal overlapping region from the mrg15 clones isolated in the two-hybrid screen, which corresponds to amino acids 149 through 303 [VP16-MRG15 (149-303)] (see below), and β-Gal assays (3) were performed on colonies transferred to nitrocellulose filters (Osmotics).
Transfections, immunoprecipitations, and Western blotting.
In vivo interaction assays were performed by transfection of HEK293 cells (American Type Culture Collection) and HEK293:FLAG-Pf1 cells (48) with the plasmids indicated in the figure legends. Coimmunoprecipitation of proteins from cellular extracts, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blot analysis were performed as described previously (48). Primary antibodies used to detect immunoprecipitated proteins include anti-mSin3A (24), anti-Pf1 (48), anti-MRG15 (36), anti-FLAG (Sigma), antihemagglutinin (anti-HA) (Boehringer Mannheim), anti-Myc (Santa Cruz), anti-Gal4 (Santa Cruz), and anti-pan TLE (kind gift from S. Stifani). Protein A-Sepharose (Sigma) was used in all immunoprecipitations with the exception of the FLAG immunoprecipitations, which were performed with M2 anti-FLAG agarose (Sigma). Secondary antibodies conjugated to horseradish peroxidase (Amersham) and ECL reagent (Amersham) were used to detect blotted proteins.
Transcription assays.
Plasmids encoding Gal4 fusion proteins, cytomegalovirus (CMV) β-Gal, and the 14DG4-Luc reporter (5) were cotransfected into 5 × 105 HEK293 cells. Twenty-four hours later, luciferase and β-Gal activities were detected with the luciferase assay system (Promega) and an MLX microtiter plate luminometer (Dynex). Typically, 100 ng of the reporter, 25 ng of CMV β-Gal, and 200 ng of the expression construct were cotransfected. Values are reported as relative light units divided by the value for β-Gal to control for transfection efficiency. Each sample was tested in triplicate to calculate the mean standard deviation of error. Assays with the dominant negative TLE construct, AES1, were performed as described above except that 1 μg of the FLAG-AES1 plasmid was included in the cotransfection mixtures.
RESULTS
Pf1 interacts with MRG15.
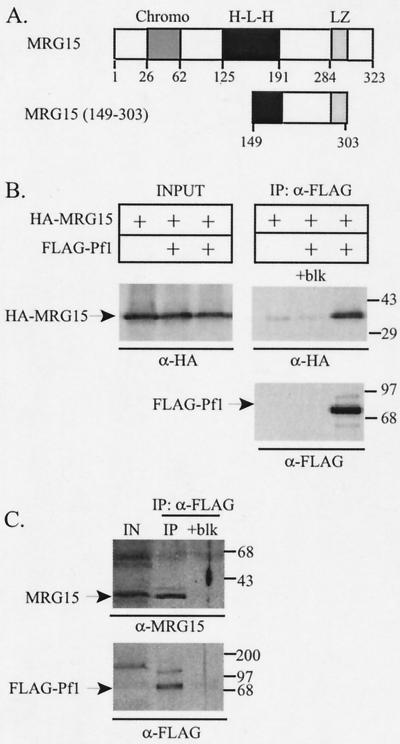
To identify Pf1-interacting proteins, Pf1 was fused to the LexA DNA binding domain and used as bait in a yeast two-hybrid screen. We obtained 217 candidate cDNAs, 15 of which encoded regions of mSin3A known to interact with Pf1, namely, the PAH1 or PAH2 domain (48). Of the remaining 202 interacting clones, 17 cDNAs encoded regions of the MRG15 protein. The minimal overlapping region of all mrg15 cDNAs isolated corresponded to amino acids 149 through 303 (Fig. 1A). In vitro-transcribed and -translated FLAG-Pf1 and VP16-MRG15 (149-303) interacted in a coimmunoprecipitation assay (data not shown). This result confirms the two-hybrid interaction and suggests that Pf1 and MRG15 interact directly.
FIG. 1.
Pf1 interacts with MRG15. (A) Schematic of MRG15 (top) and the region of MRG15 isolated in the Pf1 two-hybrid screen as a VP16 fusion (bottom). The chromatin modifier (Chromo), HLH (H-L-H), and leucine zipper (LZ) domains of MRG15 are indicated. (B) Western blots of FLAG immunoprecipitates (IP) from HEK293 cells cotransfected with HA-MRG15 and FLAG-Pf1. Proteins were detected with anti-HA and anti-FLAG antibodies. Molecular weight markers, with masses in kilodaltons, are indicated at the right. (C) Anti-MRG15 and anti-FLAG Western blots of FLAG immunoprecipitates from HEK293:FLAG-Pf1 cells, a cell line that stably expresses FLAG-Pf1. IN or INPUT, 1/15 of the input lysate used in each immunoprecipitation; +blk, block (FLAG agarose incubated with FLAG peptide prior to immunoprecipitation).
MRG15 is a member of the MORF family of proteins implicated in regulating cellular senescence (7, 8). The founding member of this family, MORF on chromosome 4, or MORF4, was identified as a factor that induced immortalized cell lines to senesce in chromosome transfer assays (7). This characteristic was restricted to MORF4, as other members, including MRG15 and MRGX, did not cause senescence in similar assays (7). MORFs are hypothesized to function as regulators of transcription based upon their nuclear localization and because they contain domains typically found in transcription factors, including leucine zippers, helix-loop-helix (HLH) motifs, and chromodomains (7, 8, 38).
To determine whether Pf1 interacts with full-length MRG15 in mammalian cells, we performed coimmunoprecipitation assays. Expression plasmids encoding FLAG epitope-tagged Pf1 and HA-tagged MRG15 were cotransfected into HEK293 cells. Pf1 and associated proteins were purified from cell extracts with anti-FLAG M2 agarose, and HA-MRG15 association was determined by Western blotting with anti-HA antibodies. Pf1 coimmunoprecipitated MRG15, and this interaction was blocked in a parallel sample containing FLAG agarose preincubated with FLAG peptide (Fig. 1B). Interaction between Pf1 and MRG15 may have resulted from high levels of expression of the two proteins, so we modified the coimmunoprecipitation assay to reflect conditions that may occur in vivo. We utilized the HEK293:FLAG-Pf1 cell line (48), which stably expresses much lower levels of FLAG-Pf1 than do transfected cells, and looked for an association of endogenous MRG15 protein with FLAG-Pf1. Endogenous MRG15 specifically associated with FLAG-Pf1 in cell lysates prepared from HEK293:FLAG-Pf1 cells (Fig. 1C), demonstrating that MRG15 and Pf1 associate in vivo.
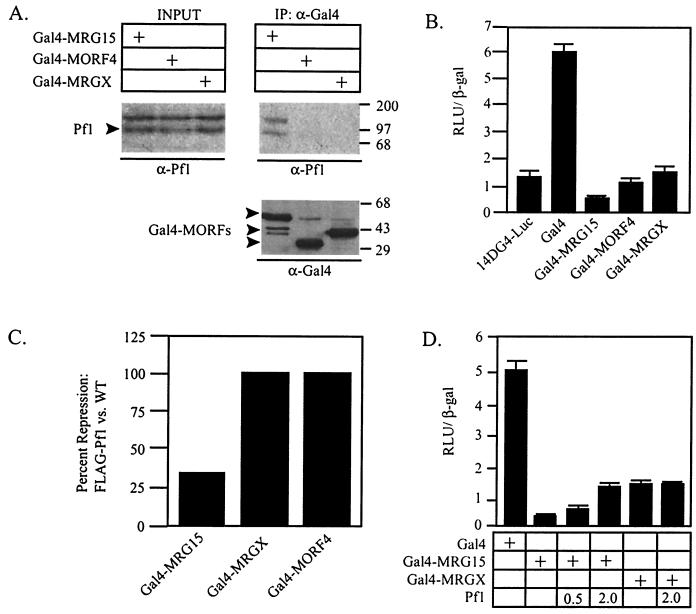
MRG15 is highly similar to MRGX and MORF4. In fact, the three proteins are nearly identical over a 200-amino-acid region (7). Because the fragment of MRG15 that interacted with Pf1 in the two-hybrid screen contains this region of high similarity, it is possible that Pf1 also interacts with MRGX and MORF4. However, we did not recover MRGX or MORF4 clones in the two-hybrid screen, suggesting either that the clones were not represented in the library or that Pf1 interacts only with MRG15. We therefore determined whether MRGX and/or MORF4 interacted with FLAG-Pf1 in the HEK293:FLAG-Pf1 cells. To perform these and subsequent experiments, the MORF family members were fused to the Gal4 DNA binding domain (Gal4). The Gal4-MORF fusions were transfected individually into the HEK293:FLAG-Pf1 cells, and Pf1 binding was determined by Western blotting of Gal4 immunoprecipitates with a Pf1 antibody. All the MORFs were expressed to similar levels; however, only Pf1 interacted with Gal4-MRG15, demonstrating that, in spite of high sequence similarity among MORFs, Pf1 association is restricted to MRG15 (Fig. 2A). Endogenous MRGX did not associate with FLAG-Pf1 in the HEK293:FLAG-Pf1 cell line, further supporting a specific interaction between Pf1 and MRG15 alone (data not shown).
FIG. 2.
The Pf1 interaction is restricted to MRG15, and targeted MORFs repress transcription. (A) Western blot of anti-Gal4 immunoprecipitates (IP) from HEK293:FLAG-Pf1cells transfected with the indicated Gal4-MORF plasmid. Pf1 association was determined by Western blotting with anti-Pf1 antibodies. (Bottom panel) Anti-Gal4 Western blot of immunoprecipitates, demonstrating that equal amounts of Gal4-MORFs were precipitated. Molecular weight markers, with masses in kilodaltons, are indicated at the right. INPUT, 1/15 of the input lysate used in each immunoprecipitation. (B) HEK293 cells were cotransfected with a minimal Gal4-responsive luciferase reporter (14DG4-Luc), CMV β-Gal, and the indicated Gal4-MORF fusion proteins, and transcriptional activities were measured. (C) HEK293:FLAG-Pf1 cells and HEK293 cells were cotransfected with 14DG4-Luc, CMV β-Gal, and the indicated Gal4-MORF fusion proteins. Data are presented as percentages of repression relative to the transcriptional activity of the Gal4 DNA binding domain in HEK293:FLAG-Pf1 cells versus that in HEK293 cells. WT, wild type. (D) HEK293 cells were transfected as described for panel B except that Pf1 was added to the transfection mix in the indicated microgram amounts. For all transcription assays, luciferase and β-Gal activities were measured 24 h after transfection. RLU, relative light units.
MORF and mSin3A interactions repress transcription.
MORF proteins are hypothesized to function in transcriptional regulation; however, they lack a recognizable DNA binding domain (7, 8). We determined the direct transcriptional potential of MORFs by testing the activities of the Gal4 fusions on a Gal4-dependent luciferase reporter (14DG4-Luc) (5). The activity of 14DG4-Luc was measured in HEK293 cells cotransfected with the different Gal4-MORF plasmids. All three Gal4-MORF proteins repressed transcription relative to the transcription the activity of the Gal4 DNA binding domain alone (Fig. 2B). Therefore, MORF4, MRGX, and MRG15 have the capacity to interact with transcriptional repression machinery when they are directly targeted to a reporter plasmid. However, because HEK293 cells do not express endogenous Pf1 protein and neither MRGX nor MORF4 interacted with Pf1 (Fig. 2A), this repression must be independent of Pf1.
We next determined the effect of Pf1 on the transcriptional repression activity of each Gal4-MORF. To do so, we performed transcription assays using the 14DG4-Luc reporter and Gal4-MORF fusions in wild-type HEK293 cells and HEK293:FLAG-Pf1 cells. In the HEK293 cells, each MORF protein repressed transcription approximately fourfold as before (Fig. 2B, data not shown). However, transcriptional repression by Gal4-MRG15 was reduced nearly fourfold in HEK293:FLAG-Pf1 cells relative to its repression activity in HEK293 cells (Fig. 2C). Levels of repression by Gal4-MORF4 and Gal4-MRGX were the same in both cell types, suggesting that the effect of Pf1 on Gal4-MRG15 repression was specific and not due to a defect in the transcriptional repression machinery in the HEK293:FLAG-Pf1 cells (Fig. 2C). Pf1 reduced transcriptional repression by Gal4-MRG15 in HEK293 cells in a dose-dependent manner, whereas it had no effect on repression by Gal4-MRGX (Fig. 2D). Together, these results are consistent with the restricted interaction between Pf1 and MRG15 and suggest that Pf1 may specifically alter the corepressor complexes associated with MRG15.
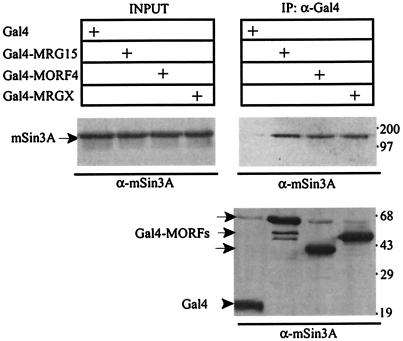
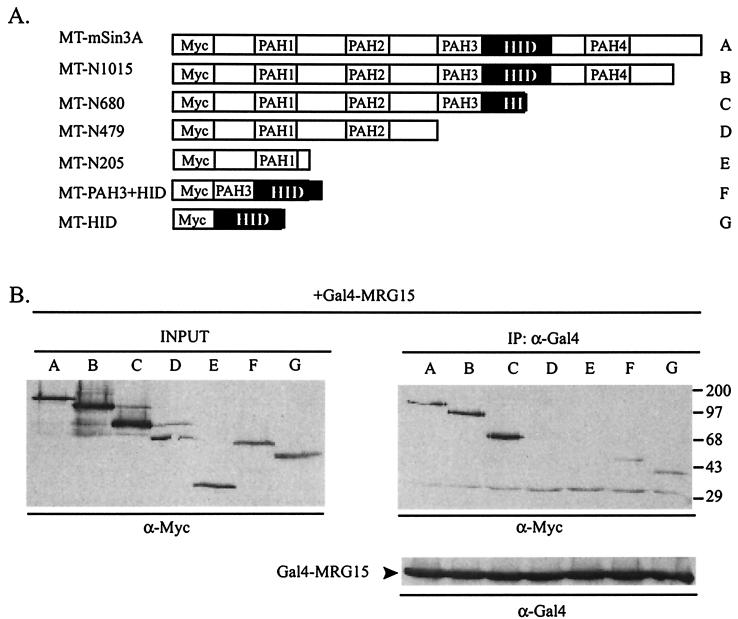
We were interested in determining the molecular mechanism(s) underlying the transcriptional repression activities of the Gal4-MORFs. Because the mSin3A corepressor complex is ubiquitously expressed, interacts with Pf1, and is used by a wide variety of transcriptional repressor proteins, we determined whether MORFs could interact with endogenous mSin3A in HEK293 cells. Plasmids encoding Gal4-MRG15, Gal4-MRGX, and Gal4-MORF4 were transfected into HEK293 cells, and binding to endogenous mSin3A was determined by Western blotting of anti-Gal4 immunoprecipitates. All three mortality factors bound to mSin3A (Fig. 3A). The binding site on mSin3A for MRG15 was determined with a series of Myc-tagged mSin3A deletion constructs (33) (diagrammed in Fig. 4A). Gal4-MRG15 coimmunoprecipitated a fusion protein containing only the HID (construct G), thereby defining this region as the minimal MRG15 interaction domain on mSin3A (Fig. 4B). Gal4-MORF4 also interacted with the HID, suggesting that the binding site for mSin3A is conserved among MORFs (data not shown).
FIG. 3.
MORFs interact with mSin3A. Western blots of Gal4 immunoprecipitates (IP) from HEK293 cells transfected with the indicated Gal4-MORF are shown. mSin3A interaction was detected by Western blotting with anti-mSin3A antibodies. (Bottom panel) Anti-Gal4 Western blot of immunoprecipitates, demonstrating that equal amounts of Gal4-MORFs were precipitated. Molecular weight markers, with masses in kilodaltons, are indicated at the right.
FIG. 4.
MRG15 binds the HID of mSin3A. (A) Schematic of the Myc epitope-tagged mSin3A deletion constructs used in panel B. The black box indicates the HID. MT, Myc tag. (B) Anti-Myc Western blots of anti-Gal4 immunoprecipitates (IP) from HEK293 cells cotransfected with Gal4-MRG15 and the indicated Myc-tagged mSin3A deletion construct. Molecular weight markers, with masses in kilodaltons, are indicated at the right. (Bottom panel) Anti-Gal4 Western blot of immunoprecipitates, demonstrating that equal amounts of Gal4-MRG15 were precipitated. INPUT, 1/15 of the input lysate used in each immunoprecipitation.
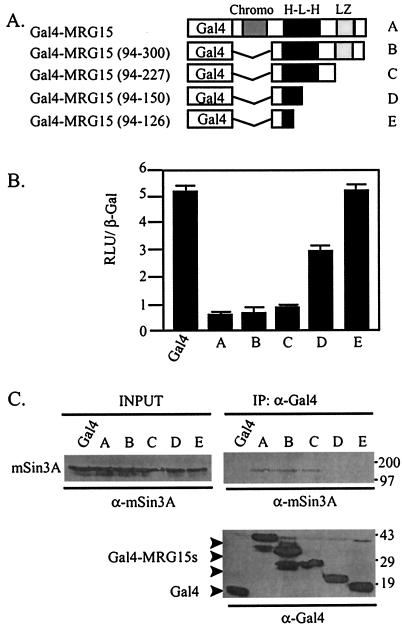
A series of Gal4-MRG15 deletion constructs was generated to determine which region of MRG15 interacts with mSin3A and whether the MRG15-mSin3A interaction was necessary for transcriptional repression (Fig. 5A). Gal4-MRG15 (94-227) repressed transcription and bound mSin3A similarly to Gal4-MRG15, suggesting that this fragment contained the mSin3A binding site (Fig. 5B and C). In contrast, neither Gal4-MRG15 (94-150) nor Gal4-MRG15 (94-126) repressed transcription nor bound mSin3A (Fig. 5B and C). Together, these results suggest that the HLH domain of MRG15 binds mSin3A and that Gal4-MRG15 assembles into functional mSin3A complexes to repress transcription.
FIG. 5.
Gal4-MRG15 interacts with mSin3A to repress transcription in HEK293 cells. (A) Schematic of Gal4-MRG15 deletions used in panels B and C. Chromo, chromatin modifier domain; H-L-H, HLH domain; LZ, leucine zipper domain. (B) HEK293 cells were transfected with the 14DG4-Luc reporter, CMV β-Gal, and the indicated Gal4-MRG15 fusion proteins. Luciferase activity was measured 24 h later. RLU, relative light units. (C) Anti-mSin3A and anti-Gal4 Western blots of Gal4 immunoprecipitates (IP) from HEK293 cells transfected with the indicated Gal4-MRG15 deletion constructs. (Bottom panel) Anti-Gal4 Western blot of immunoprecipitates, demonstrating that equal amounts of the Gal4-MRG15 proteins were precipitated. Molecular weight markers, with masses in kilodaltons, are indicated at the right. INPUT, 1/15 of the input lysate used in each immunoprecipitation.
Pf1 has separate binding surfaces for MRG15 and mSin3A.
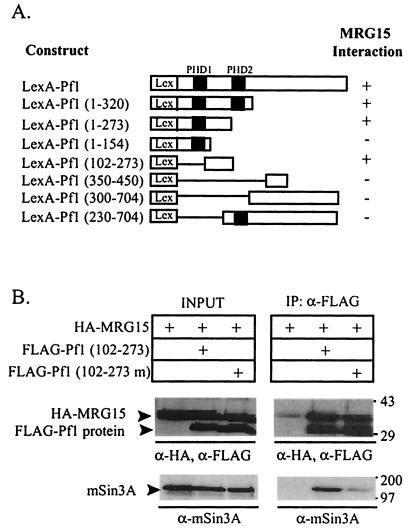
Pf1 binds both mSin3A and MRG15. Previously, we localized two independent mSin3A interaction domains on Pf1 (Pf1SID1 and Pf1SID2) (48). To determine whether mSin3A and MRG15 have independent or overlapping interaction domains on Pf1, we mapped the MRG15 binding site on Pf1 using a yeast two-hybrid assay (Fig. 6A). A series of Pf1 deletion mutants were fused to the LexA DNA binding domain and cotransformed into yeast with the VP16-MRG15 (149-303) clone isolated in the original two-hybrid screen (Fig. 1A). LexA-Pf1 (102-273) was the smallest protein that interacted with MRG15 in this assay, demonstrating that the binding site on Pf1 for MRG15 lies between amino acids 102 and 273 (Fig. 6A). Pf1SID1 contains amino acids 212 and 216 (48). To determine if MRG15 interacts with Pf1 independently of mSin3A or whether the binding sites overlap, we tested whether HA-MRG15 interacts with a Pf1 mutant construct that reduces mSin3A binding, Pf1 (102-273) with the L212P and A216P mutations (48), in a coimmunoprecipitation assay. For simplicity, the construct with these two point mutations will be referred to as Pf1 (102-273 m). Pf1 (102-273 m) coimmunoprecipitated the same amount of HA-MRG15 as Pf1 (102-273), demonstrating that MRG15 does not require mSin3A to bind Pf1 (Fig. 6B). Reprobing of this blot with anti-mSin3A antibodies showed that Pf1 (102-273) bound mSin3A but that the binding of mSin3A to Pf1 (102-273 m) was greatly reduced, consistent with previously published results (48) (Fig. 6B). Together, these results demonstrate that mSin3A and MRG15 have independent binding domains on Pf1 and suggest that a ternary complex may form between these three proteins.
FIG. 6.
MRG15 and mSin3A have independent binding sites on Pf1. (A) Yeast cells were cotransformed with the indicated LexA-Pf1 fusion proteins and VP16-MRG15 (149-303). +, β-Gal activity indicating a positive interaction; −, no β-Gal activity and, therefore, no interaction. Black boxes denote the PHD zinc fingers of Pf1. Lex denotes the LexA DNA binding domain. (B) Anti-FLAG and anti-HA Western blots of FLAG immunoprecipitates (IP) from HEK293 cells cotransfected with HA-MRG15 and the indicated FLAG-Pf1 proteins. FLAG-Pf1 (102-273) contains Pf1SID1, while FLAG-Pf1 (102-273 m) has the L212P and A216P mutations that disrupt mSin3A binding. Molecular weight markers, with masses in kilodaltons, are indicated at the right. INPUT, 1/15 of the lysate used in each immunoprecipitation.
MORFs associate with TLE.
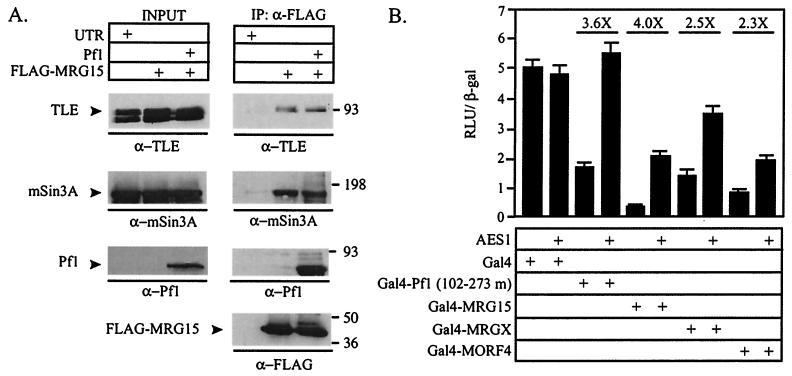
The mSin3A complex has been shown to interact with the TLE corepressor by Pf1-dependent and Pf1-independent mechanisms (17, 19, 48; D. E. Ayer, unpublished observations). Therefore, it is possible that MRG15 also associates with TLE. To test this, FLAG-MRG15 was transfected into HEK293 cells, MRG15 and associated proteins were immunoprecipitated, and endogenous TLE association was detected by Western blotting. FLAG-MRG15 did coprecipitate TLE, demonstrating that MRG15 and TLE associate in vivo (Fig. 7A). To determine whether Pf1 could affect the interaction between MRG15 and TLE, it was cotransfected with FLAG-MRG15. Pf1 had no effect on the association of TLE with MRG15, suggesting that MRG15 and TLE bind independently of Pf1 (Fig. 7A).
FIG. 7.
MORFs associate with TLE. (A) Anti-mSin3A, anti-TLE, and anti-Pf1 Western blots of anti-FLAG immunoprecipitates (IP) from HEK293 cells transfected with FLAG-MRG15. Cotransfection with Pf1 is indicated. INPUT, 1/15 of the lysate used in each immunoprecipitation; UTR, untransfected. Molecular weight markers, with masses in kilodaltons, are indicated at the right. (B) HEK293 cells were cotransfected with the 14DG4-Luc reporter, CMV β-Gal, and the Gal4 fusion constructs indicated. Addition of 1 μg of a dominant negative TLE construct, AES1, to the transfection is indicated. Fold derepression in the presence of AES1 relative to derepression in the absence of AES1 is indicated above the bars. RLU, relative light units.
To determine the contribution of TLE to the transcriptional repression of the Gal4-MORFs, we used a member of the TLE family, AES1, in transcription assays. AES1 is highly related to the tetramerization domain of TLE and likely functions as a dominant negative inhibitor of TLE by disrupting functional TLE tetramers (14, 16, 48). Therefore, if MORFs interact with endogenous TLE to repress transcription, this repression should be sensitive to AES1. As previously reported, AES1 relieved repression by Pf1 (102-273 m) but did not affect the activity of Gal4 (48) (Fig. 7B). Transcriptional repression by each Gal4-MORF was relieved by AES1, demonstrating that TLE contributes to transcriptional repression by each Gal4-MORF fusion. This trend was also seen in transcription assays conducted with Gal4-MORFs in the HEK293:FLAG-Pf1 cells, further suggesting that Pf1 does not affect TLE association (data not shown). Together, these data suggest that MORFs interact with the functional TLE corepressor.
DISCUSSION
Members of the MORF family are implicated as regulators of cellular senescence (7). MORFs have motifs common to transcription factors and localize to the nucleus, suggesting that they likely function in transcriptional regulation (7). In support of this hypothesis, we demonstrate that MRG15, MRGX, and MORF4 associate with multiple corepressors. MORF4, MRGX, and MRG15 all associate with mSin3A and TLE, whereas only MRG15 interacts with Pf1. Therefore, the MORFs are likely to have both shared and unique functions. We have uncovered a restricted interaction between MRG15 and Pf1, but we cannot rule out the possibility of the existence of specific partners for MORF4 and MRGX.
MORF4, MRGX, and MRG15 all bind mSin3A and TLE, suggesting that one shared characteristic of the MORFs is to contribute to the functions of these abundant corepressor complexes. TLE has been shown to interact with mSin3A (17, 19, 48), and we report that MRG15 and MORF4 interact with the HID of mSin3A. As such, we propose that the interaction between the MORFs and TLE is bridged by mSin3A as opposed to being direct and independent. Repression by mSin3A is thought to occur close to the site of targeting (28, 41), whereas TLE can likely repress transcription at a distance (18). Whereas both mSin3A and TLE depend, in part, on HDAC activity to drive transcriptional repression (11, 15, 24, 26, 32, 33), TLE also binds to the N-terminal tails of the histones to repress transcription (39). Therefore, a ternary complex between mSin3A, TLE, and the different MORFs may be capable of repressing transcription by different mechanisms at short and long ranges. As TLE and the MORFs do not appear to be stoichiometric components of an mSin3A complex (Fleischer and Ayer, submitted) we propose that this ternary complex will be involved only in regulating a subset of mSin3A-dependent genes. Pf1 can also tether TLE to mSin3A, but surprisingly, Pf1 overexpression did not alter the interaction between MRG15, TLE, and mSin3A (Fig. 7A), suggesting that a quaternary complex does not form. The binding sites for both MRG15 and TLE are located between amino acids 102 and 273 of Pf1, raising the possibility that their interaction with Pf1 is mutually exclusive.
Pf1 binding is restricted to MRG15, expanding the number of regulatory complexes formed by MRG15 relative to those formed by MRGX and MORF4. MRG15 and Pf1 have independent binding sites on mSin3A, suggesting the formation of a ternary complex. In the presence of Pf1, Gal4-MRG15 repression is reduced whereas repression by the other MORFs is unaffected (Fig. 2). These data are consistent with differential roles for MRG15/Pf1/mSin3A complexes and MRG15 (or MORF4 or MRGX)/mSin3A/TLE complexes in transcriptional regulation. Because MORFs are implicated in senescence, we examined pf1 mRNA levels in young and old fibroblasts and found that pf1 expression increased in senescent cells compared to that in presenescent cells (data not shown). Therefore, one function of Pf1 may be to specialize MRG15 transcriptional regulatory complexes during cellular senescence.
Transcriptional repression by Gal4-MRG15 correlated with its binding to mSin3A (Fig. 5); however, this activity was insensitive to the HDAC inhibitor trichostatin A (TSA) (data not shown). Therefore, it is likely that MRG15 can interact with HDAC-independent corepressors in addition to the HDAC-dependent mSin3A. Transcriptional repression by mSin3A is not solely dependent on HDAC activity (24, 33), suggesting the possibility that mSin3A itself may provide MRG15 with an HDAC-independent repression capability. Whether the HDAC-independent functions of MRG15 can be attributed to interactions with mSin3A, TLE, or another at-present-unidentified corepressor remains to be determined.
The MORFs are found within multiple transcriptional complexes; however, their functions within these complexes are currently unknown. All MORFs have putative protein-protein interaction motifs, including HLH and leucine zipper domains (7, 8). MORFs may contribute to the stability or assembly of the corepressor complex or mediate interactions between their different corepressor complexes and other transcriptional regulators. Furthermore, only MRG15 interacts with Pf1, suggesting that it has unique functions compared to MORF4 and MRGX. In addition to binding Pf1, MRG15 is the only MORF with a chromodomain (7, 8). The chromodomain of MRG15 is most closely related to the chromodomain of the male specific lethal (MSL) proteins (8). Within the MSL-3 protein, the chromodomain has been shown to bind roX2 RNA and contribute to the regulation of dosage compensation (2). These findings raise the intriguing possibility that MRG15-containing complexes also regulate transcription via interactions with RNA.
In addition to binding the mSin3A and TLE complexes, MRG15 binds the Rb transcriptional corepressor (36). MRG15 relieved E2F-mediated repression of the myb promoter, presumably by affecting Rb function (36). Whether this effect was unique to MRG15 or shared with the other MORFs was not tested. Transcriptional repression by Rb depends in part on association with HDACs (12, 20, 37), and recent reports demonstrate that mSin3A is tethered to Rb through interactions with SAP30 and RBP1 (34, 35). As such, it is possible that MRG15 is recruited to Rb indirectly by interactions with the mSin3A complex.
While most of the existing data suggest a role for the MORFs in transcriptional repression, recent data from studies of S. cerevisiae suggest a potential role in activation as well. For example, the yeast homolog of the MORF proteins, Eaf3p, is a component of the NuA4 HAT complex (8, 21). The catalytic acetyltransferase subunit of the NuA4 complex is Esa3p, and Esa3p is the yeast homolog of mammalian TIP60 acetyltransferase. Therefore, the TIP60 HAT complex and the NuA4 complex might function analogously. Not all of the components of the TIP60 complex have been characterized, but these findings suggest that the MORFs are likely to be components of the human TIP60 complex and contribute to its transcriptional activation function (8, 21, 27).
mSin3A, TLE, and Rb are abundant transcriptional corepressors that control diverse cellular programs. Interaction between the MORFs and these corepressors implies that they have widespread functions as well. Unraveling the contribution of each MORF will require a careful analysis of the distinct MORF-containing corepressor complexes and the transcriptional targets of these complexes. Recently, SIN3 complexes that contain the yeast orthologs of TLE, HDAC1, MRG15, and Pf1 were purified from S. cerevisiae (22). As such, S. cerevisiae provides an attractive model system to address the functional roles of MRG15-containing complexes in transcriptional regulation.
Acknowledgments
We thank O. Pereira-Smith for MORF expression plasmids and MORF antibodies, R. Eisenman for Myc-tagged mSin3A expression plasmids, and M. Rubio and J. Campisi for cDNA from young and old fibroblasts. We thank Stefano Stifani for the pan-TLE antibody. We also thank Bradley Cairns, Elizabeth Leibold, Christopher Pickett, and Tracey Fleischer for critical reviews of the manuscript.
DNA sequencing and oligonucleotide synthesis were supported by Cancer Center support grant 2P30 CA42014. This work was supported by NIH grants GM55668 and funds from the Huntsman Cancer Foundation. D.E.A. is a scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar, A., D. Zink, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules . Nature 407: 405-409. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 4.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 5.Ayer, D. E., C. D. Laherty, Q. A. Lawrence, A. Armstrong, and R. N. Eisenman. 1996. Mad proteins contain a dominant transcription repression domain. Mol. Cell. Biol. 16:5772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767-776. [DOI] [PubMed] [Google Scholar]
- 7.Bertram, M. J., N. G. Berube, X. Hang-Swanson, Q. Ran, J. K. Leung, S. Bryce, K. Spurgers, R. J. Bick, A. Baldini, Y. Ning, L. J. Clark, E. K. Parkinson, J. C. Barrett, J. R. Smith, and O. M. Pereira-Smith. 1999. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 19:1479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram, M. J., and O. M. Pereira-Smith. 2001. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene 266:111-121. [DOI] [PubMed] [Google Scholar]
- 9.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15:2457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billin, A. N., A. L. Eilers, C. Queva, and D. E. Ayer. 1999. Mlx, a novel max-like BHLHZip protein that interacts with the max network of transcription factors. J. Biol. Chem. 274:36344-36350. [DOI] [PubMed] [Google Scholar]
- 11.Brantjes, H., J. Roose, M. van De Wetering, and H. Clevers. 2001. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29:1410-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 13.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 14.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1-16. [DOI] [PubMed] [Google Scholar]
- 15.Chen, G., J. Fernandez, S. Mische, and A. J. Courey. 1999. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13:2218-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, G., P. H. Nguyen, and A. J. Courey. 1998. A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol. 18:7259-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi, C. Y., Y. H. Kim, H. J. Kwon, and Y. Kim. 1999. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 274:33194-33197. [DOI] [PubMed] [Google Scholar]
- 18.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 19.Dasen, J. S., J. P. Barbera, T. S. Herman, S. O. Connell, L. Olson, B. Ju, J. Tollkuhn, S. H. Baek, D. W. Rose, and M. G. Rosenfeld. 2001. Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev. 15:3193-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira, R., L. Magnaghi-Jaulin, P. Robin, A. Harel-Bellan, and D. Trouche. 1998. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. USA 95:10493-10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 22.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 23.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 24.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 25.Hassig, C. A., and S. L. Schreiber. 1997. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1:300-308. [DOI] [PubMed] [Google Scholar]
- 26.Hassig, C. A., J. K. Tong, T. C. Fleischer, T. Owa, P. G. Grable, D. E. Ayer, and S. L. Schreiber. 1998. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. USA 95:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 28.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoepfler, P. S., and R. N. Eisenman. 1999. Sin meets NuRD and other tails of repression. Cell 99:447-450. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 31.Kuzmichev, A., Y. Zhang, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell. Biol. 22:835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laherty, C. D., A. N. Billin, R. M. Lavinsky, G. S. Yochum, A. C. Bush, J. M. Sun, T. M. Mullen, J. R. Davie, D. W. Rose, C. K. Glass, M. G. Rosenfeld, D. E. Ayer, and R. N. Eisenman. 1998. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 2:33-42. [DOI] [PubMed] [Google Scholar]
- 33.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 34.Lai, A., B. K. Kennedy, D. A. Barbie, N. R. Bertos, X. J. Yang, M. C. Theberge, S. C. Tsai, E. Seto, Y. Zhang, A. Kuzmichev, W. S. Lane, D. Reinberg, E. Harlow, and P. E. Branton. 2001. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 21:2918-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, A., J. M. Lee, W. M. Yang, J. A. DeCaprio, W. G. Kaelin, Jr., E. Seto, and P. E. Branton. 1999. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol. 19:6632-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung, J. K., N. Berube, S. Venable, S. Ahmed, N. Timchenko, and O. M. Pereira-Smith. 2001. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 276:39171-39178. [DOI] [PubMed] [Google Scholar]
- 37.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 38.Marin, I., and B. S. Baker. 2000. Origin and evolution of the regulatory gene male-specific lethal-3. Mol. Biol. Evol. 17:1240-1250. [DOI] [PubMed] [Google Scholar]
- 39.Palaparti, A., A. Baratz, and S. Stifani. 1997. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 272:26604-26610. [DOI] [PubMed] [Google Scholar]
- 40.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 41.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber-Agus, N., L. Chin, K. Chen, R. Torres, G. Rao, P. Guida, A. I. Skoultchi, and R. A. DePinho. 1995. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80:777-786. [DOI] [PubMed] [Google Scholar]
- 43.Sommer, A., S. Hilfenhaus, A. Menkel, E. Kremmer, C. Seiser, P. Loidl, and B. Luscher. 1997. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr. Biol. 7:357-365. [DOI] [PubMed] [Google Scholar]
- 44.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 45.Wang, H., I. Clark, P. R. Nicholson, I. Herskowitz, and D. J. Stillman. 1990. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol. Cell. Biol. 10:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolffe, A. P., F. D. Urnov, and D. Guschin. 2000. Co-repressor complexes and remodelling chromatin for repression. Biochem. Soc. Trans. 28:379-386. [PubMed] [Google Scholar]
- 47.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 48.Yochum, G. S., and D. E. Ayer. 2001. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 21:4110-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]