Abstract
DRASTIC––Database Resource for the Analysis of Signal Transduction In Cells (http://www.drastic.org.uk/) has been created as a first step towards a data-based approach for constructing signal transduction pathways. DRASTIC is a relational database of plant expressed sequence tags and genes up- or down-regulated in response to various pathogens, chemical exposure or other treatments such as drought, salt and low temperature. More than 17700 records have been obtained from 306 treatments affecting 73 plant species from 512 peer-reviewed publications with most emphasis being placed on data from Arabidopsis thaliana. DRASTIC has been developed by the Scottish Crop Research Institute and the University of Abertay Dundee and allows rapid identification of plant genes that are up- or down-regulated by multiple treatments and those that are regulated by a very limited (or perhaps a single) treatment. The INSIGHTS (INference of cell SIGnaling HypoTheseS) suite of web-based tools allows intelligent data mining and extraction of information from the DRASTIC database. Potential response pathways can be visualized and comparisons made between gene expression patterns in response to various treatments. The knowledge gained informs plant signalling pathways and systems biology investigations.
INTRODUCTION
Recovering value from the burgeoning mass of genomics and gene expression data now being accumulated is a major task for biologists and computer scientists (1). Increasing amounts of gene sequence, expressed sequence tag (EST), northern blot and microarray data provide fertile ground for the mining of expression data, extracting information and adding value by evaluating how gene expression is regulated and biochemical pathways function (2). DRASTIC (Database Resource for the Analysis of Signal Transduction in Cells) and the INSIGHTS (INference of cell SIGnaling HypoTheseS) web-based suite of tools bring together data on plant responses to pathogens, environmental stresses and chemicals (treatments) from refereed journal publications. Presenting these data in a unified, searchable format allows the user to extract information beyond single genes, or clusters of similar expression patterns by browsing multiple treatments at once, identifying potential regulatory relationships between multiple treatments and genes. DRASTIC–INSIGHTS overcomes the limitations of other plant expression databases by allowing for updating of information from previous publications, by directly linking to publications and through the tracking of genes with unknown function that have the same accession or AGI (Arabidopsis genome initiative) number, which would otherwise be difficult to link between publications (3,4). Additionally, genomic, EST, northern data and information derived from microarrays from multiple plant species are included, after human curation, to ensure accuracy and to standardize the nomenclature of data (5). The INSIGHTS tools encourage comparison of gene expression patterns, intelligent mining of information, testing and formulation of novel hypotheses on the complex signal transduction and response pathways used by plants (6). Identifying common elements in pathways affected by different treatments permits the formation of hypotheses previously opaque to the user (7).
Database content
DRASTIC is a gene expression relational database developed by SCRI (Scottish Crop Research Institute) and UAD (University of Abertay Dundee) to record responses to treatments, which are defined as exposure to experimental conditions such as pathogens, chemicals and other environmental stresses. More than 17700 records are included with information on 73 species and 306 treatments obtained from 512 references. Each record contains expression data for a single gene, from a single host, subjected to a treatment obtained from a single refereed journal publication. Manually curated records include data from plant northern blots, ESTs, cDNA-AFLPs, quantitative RT–PCR, massively parallel signature sequencing and information derived from microarrays. These expression data are recorded as up- or down-regulated compared with control values, and, where applicable, the time and magnitude of expression are also recorded. DRASTIC makes it possible, for example, to rapidly identify plant genes that are up-regulated by multiple treatments and those that are up-regulated by a single treatment (see http://www.scri.sari.ac.uk/TiPP/PPS/DRASTIC/mpage/countoftreatment.asp for numbers of records per treatment). Such information represents important knowledge to assist in constructing putative signalling pathways for systems biology research (8). Database requirements were elicited using semi-structured interviews with computer scientists and bioscientists. The complex ERM (entity relationship model) consists of over 20 tables (see explanatory tables and ERM diagram in Supplementary Data). The ERM is implemented in Microsoft RDBMS (relational database management system) and is searched using the public web-based interface hosted by SCRI on Microsoft 2000 Advanced Server. The web toolkit was developed using SQL (structured query language) embedded in ASP (active server pages) to dynamically create HTML result pages based on user queries. All records in the DRASTIC–INSIGHTS database are accessible through the publically available website http://www.drastic.org.uk or can be freely downloaded in a comma delimited text file from the website download page (http://www.scri.sari.ac.uk/TiPP/PPS/DRASTIC/mpage/downloads.asp).
Data quality
Several methods have been implemented to ensure that the data stored in DRASTIC are of high quality. Inclusion in the database is solely following expert human curation of expression data from refereed publications. No expression data have been included by direct submission from laboratories. Accession numbers for ESTs are preferred, as the nomenclature of such sequences can be updated in the future. Information from some papers has not been included because accession numbers were not provided. The need to standardize nomenclature is important (5), thus the names used in the database correspond with current Unigene classification (9) rather than those cited in the original publication, unless a more recent primary publication indicates otherwise. Sometimes changes in gene identification are small but in other cases they can be dramatic and critical if signal transduction pathways are to be correctly understood. For example, one plant gene originally described as senescence-associated is now described as inositol-1,4,5-triphosphate 5-phosphatase, and another gene originally described as ‘no homology’ is now known to be a protein kinase. In addition, with genes from Arabidopsis, the Unigene system has been used to provide AGI (Arabidopsis genome initiative) numbers where known. This has proven particularly useful for genes classified as ‘unknown’, as ‘unknowns’ from different publications and with different accession numbers can be shown to be the same gene. For example, At2g36220 is classified as a gene of unknown function but by using information from many references we can see that it is up-regulated by abscisic acid, brassinosteroid, benzothiadiazol (BTH), cold, flagellin-22, hydrogen peroxide, low oxygen, Peronospora parasitica and sodium nitroprusside treatments. A backtracking facility has been included for historical gene names as all updates are stored in a data dictionary. In addition, a software routine called AGIDetect has been developed to check for mismatches between AGI, gene names and accession numbers to assist in maintaining data accuracy.
INSIGHTS data tools
At a simple level the web interface (http://www.drastic.org.uk) permits users to find published information on expression data for plant genes of interest. More importantly, INSIGHTS offers a number of tools to mine further information and create new knowledge. Some mining tools use AGI numbers where expression data correctly identify a specific member of a gene family. Through the INSIGHTS integrated toolkit users may investigate data in the following ways:
General database search provides a basic query function for the database. The user can select the following parameters: treatments, species, gene, regulation and date. The search returns the results in tabular format which can be sorted on all parameters and provides links to the primary references.
DRASTIC statistics provides an up-to-date list of statistics for the database including the total number of records, species and treatments. It also provides a breakdown of both records per species and records per treatment, which can be ordered alphabetically or numerically. To gain a more in-depth view, a table of data providing statistics on the number of records by species or treatment can be obtained. These can be further mined to view individual records with bibliographic references.
Accession number search provides a query function specifically for the accession numbers in the DRASTIC database. Selectable parameters include accession number, treatment, regulation type and date. The results are displayed in tabular format which can be sorted, providing links to references.
Arabidopsis genome initiative search provides a query function specifically for AGI numbers in the DRASTIC database. The user can select from AGI number, treatment, regulation and date.
Venn diagrams enables the creation of Venn diagrams using the Arabidopsis thaliana data from the DRASTIC database. The user can select two or three treatments and the tool will process the selections and output the results as a Venn diagram. The Venn diagram tool displays the number of genes regulated by each individual treatment or by multiple treatments based on the DRASTIC data. Records where genes have been up-regulated, down-regulated or both (up or down) can be included. The diagrams can be mined further by clicking on a segment of the diagram to view the individual records and relevant bibliographies.
TAIR AGI search enables the user to search records that include the AGI number and directly use them with the TAIR (the Arabidopsis information resource) chromosome mapping and functional categorization tools, which are specifically designed to analyse AGI data (10). The user can select a subset of records from DRASTIC using a search on a treatment, multiple treatment or gene group (such as kinases) and regulation type. The selected data are then formatted for use with the TAIR tools.
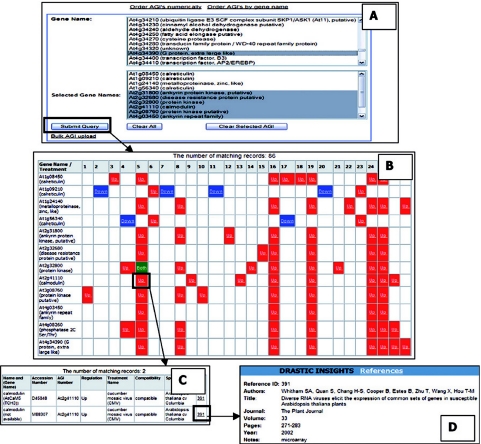
Pathway tool enables the user to extract and visualize knowledge from the database to hypothesize potential relationships between signalling elements. It includes a search facility to allow selection of a number of A.thaliana genes by AGI numbers. A ‘pathway’ is produced to display the regulation of selected genes in response to different treatments (Figure 1). Any groups of genes that are always co-regulated are identified, suggesting that they are likely to occur in the same signal transduction pathway. The pathway tool can be used to indicate the relatedness of induction patterns for selected genes. For instance, it can be shown that up-regulation of calreticulin 3 (At1g08450) in Arabidopsis has been shown to be associated with the up-regulation of a number of potential signalling genes (including kinases), which does not occur if calreticulin 1 (At1g56340) and calreticulin 2 (At1g09210) are down-regulated. The pathway tool can also be used in a hypothesis testing manner or as a quality control check tool for data in known signal transduction pathways (11).
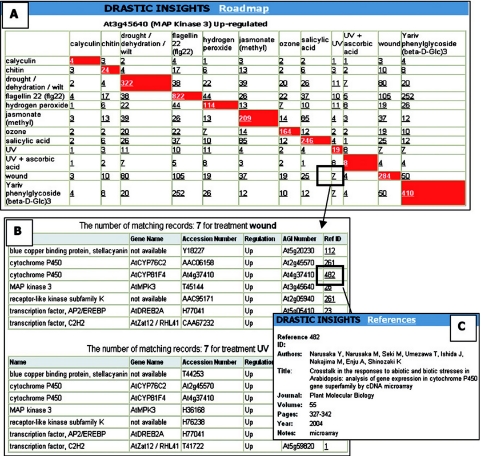
Roadmap tool creates lookup tables to find genes that are co-regulated by different treatments. The user can ‘drill down’ through the map to investigate individual genes and view all references that support each data point providing a level of confidence for each result. To operate the roadmap, the user selects an AGI number and a regulation (up-, down- or both) to include in the search. The tool establishes which treatments regulate expression of the selected gene and then displays in a map all the genes in DRASTIC that are regulated by these treatments (Figure 2). This tool demonstrates that it is possible to identify groups of treatments that appear to produce similar regulatory results in A.thaliana. Roadmap results can be used in conjunction with the Pathway tool.
Unique genes tool identifies all the A.thaliana genes that are regulated by a single treatment. Full details including references for each gene are linked to each record.
Figure 1.
Web interface for the Pathway tool. (A) The search page for a set of AGI numbers. The pathway result is shown in (B). Up-regulated genes are shown in red. Down-regulated genes are shown in blue. Green cells indicate that both up- and down-regulation record(s) are held in DRASTIC. The pathway can be further mined by choosing any coloured cell which will display all the records for the AGI/treatment combination as shown in (C). The references for each record can be selected as shown in (D).
Figure 2.
Web interface for the Roadmap tool. In this example, treatments up-regulating At3g45640 (MAP Kinase 3) were selected for investigation. (A) The resulting roadmap. From the DRASTIC data, 12 treatments up-regulate Atg45640. Using these treatments as the ‘lookup co-ordinates’, the map displays the total number of unique AGIs up-regulated by these treatments. The shaded squares hold the total number of genes up-regulated by a single treatment, and the numbers in the unshaded squares show the number of genes co-regulated by treatments. This map can be further mined by clicking on any of the squares to display the supporting records (see (B) where the co-ordinates wound and UV have been selected). Each record has a link to the reference it was curated from as shown in (C).
Genes for proteins involved in the same signal transduction pathway are likely to be co-regulated and show the same response to a range of treatments. Thus, to find e.g. kinases, transcription factors and calcium-binding proteins that are in the same signal transduction pathway expression patterns should be compared. Verification that identified genes are truly associated within signal transduction or metabolic pathways requires experimental confirmation, but the database and associated diagrams promote more targeted hypothesis formation. This type of analysis is useful in providing a framework for understanding signal transduction responses and to assist with identifying regulatory gene networks. It is also useful for finding genes associated with plant pathogen infection that are also affected by environmental stresses such as drought and cold in differing ways (12,13). A downloadable guide to using DRASTIC–INSIGHTS has been developed to assist users and is available at http://www.scri.sari.ac.uk/TiPP/PPS/DRASTIC/helpfiles/index.html.
Future work
DRASTIC–INSIGHTS will in the future be extended by linking both public and private domain data to enable scientists to hypothesize using personal and published data. Optional access to Nottingham Arabidopsis Stock Centre (NASC) microarray data will also be made available via individual user domains. Development of text mining and data capture tools to automatically identify suitable publications and datasets for inclusion in DRASTIC is currently being undertaken.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The contributions of Bruce Marshall, Adrian Newton, Peter Ghazal, Ishbel Duncan and Michael Idowu to the conceptual development of DRASTIC are acknowledged. DRASTIC–INSIGHTS is supported by funding from the Scottish Executive Environment Rural Affairs Department (SEERAD), Carnegie Trust, the Forestry Commission and the University of Abertay Dundee (UAD). The website was funded by Mylnefield Research Services Ltd. Infrastructure support was provided by Abertay Centre for the Environment (ACE). Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tian Q., Uhlir N.J., Reed J.W. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazic T. Biochemical Databases: Challenges and Opportunities, New Data Challenges in Our Information Age. In: Glaeser P.S., Millward M.T.L., editors. Proceedings of the 13th International CODATA Conference; Paris: CODATA Secretariat; 1994. pp. C133–C140. [Google Scholar]
- 3.Mueller L.A., Zhang P., Rhee S.Y. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thimm O., Blaesing O., Gibon Y., Nagel A., Meyer S., Krueger P., Selbig J., Mueller L.A., Rhee S.Y., Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 5.Lyon G.D., Newton A.C., Marshall B. The need for a standard nomenclature for gene classification (a Nucleotide Function code) and an automated data-based tool to assist in understanding the molecular associations in cell signalling in plant-pathogen interactions. Mol. Plant Pathol. 2002;3:103–109. doi: 10.1046/j.1464-6722.2002.00101.x. [DOI] [PubMed] [Google Scholar]
- 6.Cheong Y.H., Chang H.-S., Gupta R., Wang X., Zhu T., Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel B.N., Brooks D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis thaliana microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler D.L., Church D.M., Federhen S., Lash A.E., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Sequeira E., Tatusova T.A., et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzali S., Loreti E., Novi G., Poggi A., Alpi A., Perata P. The use of microarrays to study the anaerobic response in Arabidopsis. Ann. Bot. 2005;96:661–668. doi: 10.1093/aob/mci218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norman-Setterblad C., Vidal S., Palva E.T. Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant Microbe Interact. 2000;13:430–438. doi: 10.1094/MPMI.2000.13.4.430. [DOI] [PubMed] [Google Scholar]
- 13.McDowell J.M., Woffenden B.J. Plant disease resistance genes: recent insights and potential applications. TRENDS Biotechnol. 2003;21:178–183. doi: 10.1016/S0167-7799(03)00053-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.