Abstract
In Escherichia coli, the response to oxidative stress due to elevated levels of superoxide is mediated, in part, by the soxRS regulon. One member of the soxRS regulon, nfsA, encodes the major oxygen-insensitive nitroreductase in Escherichia coli which catalyzes the reduction of nitroaromatic and nitroheterocyclic compounds by NADPH. In this study we investigate the regulation of nfsA in response to the superoxide generating compound paraquat. The transcription start site (TSS) of nfsA was located upstream of the ybjC gene, a small open reading frame of unknown function located directly upstream of nfsA, suggesting that these two genes form an operon. The activity of the promoter associated with this TSS was confirmed with lacZ fusions and was shown to be inducible by paraquat. Footprinting and band shift analysis showed that purified His-tagged SoxS protein binds to a 20-base sequence 10 bases upstream of the −35 promoter sequence in the forward orientation, suggesting that the ybjC-nfsA promoter is a class I SoxS-dependent promoter.
Nitrosubstituted compounds have been used for many years in the health industry as antimicrobial agents (43) and more recently have been examined for use in enzyme-prodrug cancer chemotherapies (3). In addition, a large number of nitroaromatic and nitroheterocyclic derivatives are potent mutagens and suspected human carcinogens, and therefore their presence in environmental samples is of considerable health concern (33, 50, 51). The mutagenic and antimicrobial activity of these compounds is mediated by short-lived intermediates formed during their reduction by nitroreductases (31, 32). Two types of nitroreductases have been identified in Escherichia coli (6, 10, 32). Oxygen-insensitive nitroreductases are flavoproteins that mediate the transfer of two electrons from NAD(P)H to the nitro moiety of nitrosubstituted compounds (10); biologically active intermediates produced through this pathway include nitroso and hydroxylamine derivatives that are further reduced to yield biologically inactive products (32). Oxygen-sensitive nitroreductases mediate single-electron transfers, producing nitro-anion free radicals which, in the presence of oxygen, can be rapidly reoxidized in a futile redox cycle through which superoxide is generated (37).
Two oxygen-insensitive nitroreductases, NfsA and NfsB, have been characterized in E. coli (10, 30, 54, 56, 57). NfsA is the major oxygen-insensitive nitroreductase and uses NADPH as an electron source, while NfsB is a minor nitroreductase that can use either NADH or NADPH as a source of reducing equivalents (10, 56, 57). The nfsA gene (mdaA) is located at 19.2 min on the E. coli chromosome (56), upstream of the rimK gene (9) that encodes a protein responsible for posttrans lational modification of the S6 ribosomal protein (20), and downstream from ybjC, a small open reading frame (ORF) of unknown function (9). A role for NfsA in the bacterial oxidative stress response has been suggested by a recent study demonstrating that nfsA is upregulated by the redox cycling compound, paraquat, in a soxRS-dependent manner, suggesting that it is part of the soxRS regulon (27). A macroarray study demonstrating that nfsA, rimK, and ybjC are all members of the closely related marRAB regulon suggests that these genes may form an operon (7).
The soxRS regulon includes at least 15 genes that are upregulated in response to superoxide formed by redox-cycling compounds such as paraquat (8, 15, 24, 39, 52). The regulon includes several genes whose products play a direct role in responding to oxidative stress, including manganese super oxide dismutase (sodA), exonuclease IV (nfo), glucose 6-phosphate dehydrogenase (zwf) (15), fumarase C (fumC) (25), and NADPH ferredoxin reductase (fpr) (26). Upregulation of these genes occurs in a two-step process in which the SoxR protein acts both as a sensory protein to detect elevated levels of superoxide within the cell and as a positive regulator of soxS transcription (2, 15, 16, 53). The SoxR protein contains two binuclear iron-sulfur clusters [2Fe-2S] that remain in the reduced state under normal (nonstress) conditions. In the presence of elevated levels of superoxide, the SoxR [2Fe-2S] clusters become oxidized (16). While both the oxidized and reduced forms of SoxR bind equally well to the promoter sequence of soxS, only the oxidized form activates transcription of soxS, presumably by enhancing open-complex formation by RNA polymerase (12). The SoxS protein, in turn, is a positive regulator of the members of the soxRS regulon (2, 22).
The SoxS protein is a member of the XylS/AraC family of transcription activators that includes two other proteins, MarA and Rob, that share sequence identity (41 and 55%, respectively) with SoxS. The marRAB locus was first described as a multiple-antibiotic-resistance locus in E. coli responsible for increased bacterial resistance to a wide range of unrelated antibiotics and organic solvents after exposure to compounds such as sodium salicylate (1). Over 40 genes in E. coli, including 9 of the 15 genes upregulated by SoxS, are upregulated by the MarA protein (7). The Rob protein is constitutively expressed and binds to the right arm of the E.coli origin of chromosome replication (oriC), but its function is still unknown (46). Many studies have demonstrated a considerable overlap in the genes regulated by each of these three proteins, strongly suggesting that the genes involved should be considered part of the same Sox/Mar/Rob regulon which is regulated, to various degrees, by the three closely related regulatory proteins (4, 19, 23, 29, 38).
A comparison of 16 sequences associated with SoxS/MarA-regulated promoters and the oriC, shown by footprinting and gel shift analysis to be involved in SoxS/MarA/Rob binding, have identified a 20-bp “soxbox” with the consensus sequence: AYNGCACNNWNNRYYAAAYN (where Y = C or T, W = A or T, R = A or G, and N = any nucleotide) (28). SoxS is an ambidextrous activator in that it is able to interact with the promoter sequence and RNA polymerase in two different ways comparable to the well-studied catabolite activator protein (CAP; or cAMP receptor protein [CRP]) (11, 13, 18, 28, 34, 55). In class I SoxS-regulated promoters, the soxbox is located upstream of the −35 sequence and, with the exception of the zwf promoter, is in the reverse orientation relative to the promoter. Class I promoters include fldA, fpr, mar, poxB, acrAB, ribA, and zwf (28, 55). Activation at these promoters is believed to require interaction between SoxS and C-terminal domain of the α-subunit of RNA polymerase (αCTD) (18). Class II promoters include fumC, micF, nfo, sodA, ara1, inaA, oriC, pgi, and tolC (28, 55). In these promoters, the SoxS binding site is in the forward orientation and overlaps the −35 sequence. SoxS activation of the class II promoters fumC, micF, nfo and sodA has been shown to be independent of αCTD (18).
In this study, we examined regulation of nfsA gene expression by the redox-cycling agent paraquat. We have identified both the transcription start site (TSS) and the SoxS binding site of the nfsA gene and have demonstrated that SoxS regulates both nfsA and the small ybjC gene directly upstream of nfsA in a coordinated manner. From the location of the soxbox, it would appear that the ybjC-nfsA promoter is a class I SoxS promoter in which SoxS binds 10 bp upstream of the −35 site in the forward orientation comparable to the zwf promoter.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Cloning was conducted in E. coli DH5α [supE44 lacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1]. β-Galactosidase assays were conducted in E. coli S90C[ara Δ(lac-pro) strA thi] (36). His6-SoxS protein was prepared from E. coli HB310 [W3110 Δ(argF-lac)U169] carrying pBADHis6-SoxS, a plasmid that expresses a SoxS protein tagged with six histidine residues at the amino terminus (55). All cultures were grown at 37°C in Luria broth supplemented with 30 μg of ampicillin per ml where necessary. Plasmid pMP28.5 is a pUC18Not derivative carrying a lacZ gene lacking both transcriptional and translational signals (40).
Construction of lacZ fusions.
Regions upstream of the nfsA gene were amplified by PCR by using primers depicted in Fig. 1. PCRs contained 10 mM Tris-HCl (pH 9), 50 mM KCl, 1% Triton X-100, 2 mM MgCl2, 0.2 mM deoxynucleoside triphosphate (dNTP), 0.3 mM primers, and 2 U of Taq polymerase (BRL) and consisted of a 1-min, 94°C predwell, followed by 25 cycles of 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C, and finally a 5-min postdwell at 72°C. The PCR products were purified by Wizard column (Promega) prior to restriction digestion. PCR products are referred to by the primer set used in their amplification. Plasmids pH3X1, pH 2X1, and pS4X1 were constructed by cloning H3X1, H2X1, and S4X1 PCR products cut with XhoI and HindIII or SphI into plasmid pMP28.5 cut with the same enzymes. Plasmids pH3X3 and pS4X3 were constructed by replacing the HindIII-XhoI or SphI-XhoI fragment of pH3X1 or pS4X1 with the H3X3 or S4X3 PCR products. Plasmid pHXΔ was constructed by recircularizing pH2X1 after it had been digested with XhoI and HindIII and the ends were made blunt with Klenow and dNTPs. Plasmid pH3B1 was constructed by cloning the H3B1 PCR product cut with HindIII and BamHI into pMP28.5 cut with the same enzymes. Recombinant plasmids were confirmed by gel electrophoresis, and the junctions were verified by dideoxy sequencing with Sequenase as previously described (54).
FIG. 1.
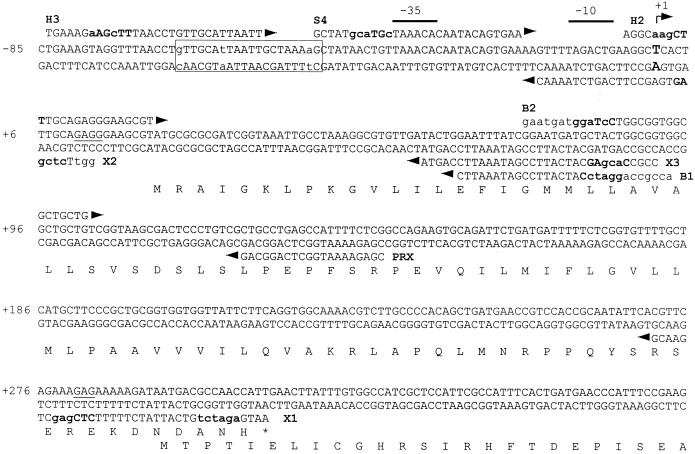
nfsA regulatory region. The nucleotide sequence of the region upstream of nfsA is presented with the numbering based on the TSS indicated by a bent arrow and +1 above the sequence and in a large, bold font within the sequence. The −35 and −10 promoter sequences associated with the TSS are indicated by lines above the sequence. Primer sequences used for PCR and primer extension are indicated above or below the DNA sequence, depending on their orientation. Restriction sites are in boldface, and mismatches to DNA sequence are in lowercase. The soxbox identified by footprinting is boxed with mismatches to the consensus sequence (28) in lowercase. The amino acid sequence of YbjC and the N terminus of NfsA are indicated below the nucleotide sequence and the Shine-Dalgarno sequences are underlined within the sequence.
Lac assays.
Assays for β-galactosidase activity were conducted according to the method of Miller (35). Cultures were grown at 37°C to an optical density at 600 nm (OD600) of 0.2, split into two tubes, and paraquat (0.2 mM) was added to one. After an additional hour of incubation, the OD600 was determined. Duplicate 1-ml samples of these cells were pelleted and washed with Z buffer (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4 · 7H2O, 50 mM β-mercaptoethanol). The cells were resuspended in 100 μl of Z buffer and permeabilized by vortexing for 1 min with 5 μl of acetone-toluene (1:9 [vol:vol]). Aliquots (5 to 50 μl) of permeabilized cells were diluted to a volume of 1 ml with Z buffer. After 5 min at 37°C, reactions were initiated by the addition of 200 μl of ONPG (o-nitrophenyl-β-d-galactopyranoside; 4 mg/ml) and allowed to proceed for 5 to 20 min until a yellow color was visible. Reactions were stopped with 500 μl of 1 M Na2CO3, and the OD550 and OD420 values were determined. The activity in Miller units was calculated by using the following formula: activity = 1,000 × [OD420 − (1.72 × OD550)]/[OD600 × time (min) × volume of cells (ml)].
Primer extension.
Total RNA for primer extension experiments was extracted as described by Rothmel et al. (42) from a 50-ml culture of DH5α/pH3X1 grown at 37°C for 5 h. The culture was induced for the last hour of incubation by the addition of paraquat (0.2 mM). PRX primer (see Fig. 1) was end labeled with [γ-32P]ATP by using T4 DNA polymerase, and 1.7 pmol was annealed to 12 μg of RNA in a 10-μl reaction containing 4 U of RNasin (Boehringer Mannheim) by heating to 80°C for 2 min, followed by incubation at 50°C for 99 min. After this, the primer was extended in a reaction mixture containing 50 mM Tris-HCl (pH 8.3), 10 mM dithiothreitol, 30 mM KCl, 8 mM MgCl2, 25 mM dNTP, and 25 U of Moloney murine leukemia virus reverse transcriptase by incubation at 42°C for 99 min. The reaction was stopped with the addition of 10 μl of sequencing loading buffer. Primer extension reaction mixtures were boiled for 3 min, and a 3-μl portion was run on an 8% denaturing polyacrylamide gel together with mixtures from sequencing reactions carried out with pH3X1 plasmid DNA template and PRX primer.
Purification of His6-SoxS protein.
Purification of His-tagged SoxS protein was conducted according to the method of Wood et al. (55) by Ni-nitrilotriacetic acid (NTA) affinity chromatography under denaturing conditions. Briefly, an 85-ml culture of HB301/pBADHis6-SoxS was grown at 37°C with shaking to an OD600 of 0.6 to 0.7 and induced with 0.02% arabinose for a further 4 h at 37°C. The cells were pelleted and resuspended in 1 ml of solubilization buffer (6 M guanidine-HCl; 20 mM Tris, pH 7; 300 mM NaCl; 5% glycerol [vol/vol]; 5 mM imidazole) and disrupted by sonication. The cell lysate was collected after a 30-min spin in a microcentrifuge at 4°C and incubated, with mixing, for 1 h at 4°C with 150 μl of Ni-NTA resin (BioLabs). The resin was collected in a drip column and washed five times with 2 ml of solubilization buffer. The His6-SoxS protein was eluted with four washes of 200 μl of elution buffer (4 M guanidine-HCl; 20 mM NaHPO4, pH 5.5; 300 mM NaCl; 40% glycerol; 300 mM imidazole). The eluted protein was diluted to 0.5 mg/ml in elution buffer and renatured by dialysis (20 mM NaHPO4, pH 5.5; 300 mM NaCl; 40% glycerol). The protein concentration was determined by Coomassie blue staining assay (Bio-Rad) against a bovine serum albumin standard. A total of 2.6 mg of protein was purified from the 85-ml culture. The purity was estimated to be >95%.
Gel shift mobility assays.
DNA for binding assays was generated by PCR as described above. In each PCR, one primer was end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Binding assays were conducted in 10-μl reaction mixtures containing 20 mM NaCl, 50 mM KCl, 5 mM Tris, 2.4 mM HEPES (pH 7.9), 0.4 mg of bovine serum albumin/ml, 18% glycerol (14), 40 fmol of labeled DNA, and 12.5 to 50 ng of His6-SoxS protein. Binding was allowed to proceed for 30 min at 37°C. Reaction mixtures were chilled 10 min before loading on a prerun, 5% nondenaturing polyacrylamide gel and then run against TGE buffer (25 mM Tris, 190 mM glycine, 0.1 M EDTA [pH 8.3]) (14) at 80 V for 2 to 4 h at 4°C. Gels were dried and autoradiographed.
DNase I footprinting.
DNA for footprinting experiments was generated by PCR with the primers PRX (see Fig. 1) and GRX (5′-ACGCCAGATCTTTTGCACGCACA-3′, nucleotides [nt] −300 to −278); the GRX primer was end labeled with [γ-32P]ATP by using T4 polynucleotide kinase. The PCR product was purified on a Sephadex column, and His6-SoxS binding assays were conducted as described for the gel shift assays. After binding, the reaction mixtures were equilibrated to 20°C before the addition of 1 mM MgCl2, 0.5 mM CaCl2 and 0.03 U of DNase I. The reactions were allowed to proceed for 3 min and stopped with the addition of 100 μl of STOP solution (100 mM EDTA, pH 8; 0.2 μg of sheared salmon sperm DNA/ml; 0.6 M ammonium acetate). The DNA was precipitated with 200 μl of ethanol, washed twice with 70% ethanol, dried, and resuspended in 3 μl of distilled H2O and 3 μl of sequencing loading buffer. The samples were boiled and loaded onto an 8% denaturing polyacyrlamide gel next to mixtures from sequencing reactions carried out with GRX-PRX template DNA and the GRX primer.
RESULTS
Identification of the transcription start site of nfsA by primer extension.
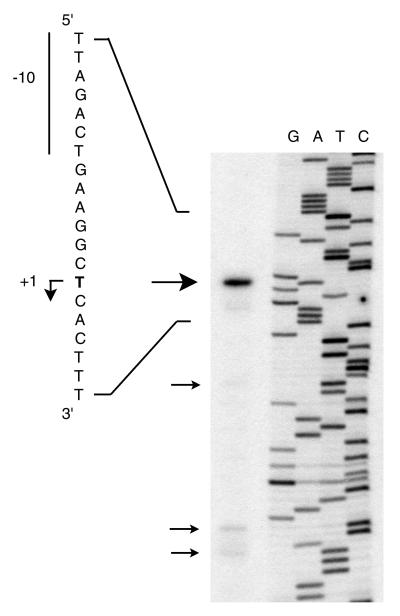
The sequence directly upstream of the nfsA coding region contains the ybjC coding region such that the 3′ end of ybjC overlaps 17 bp of the 5′ end of nfsA (Fig. 1). Based on the nucleotide sequence, a putative promoter had been identified 226 bases upstream of the nfsA start codon within the ybjC coding region (56). Primer extension was conducted to determine whether this promoter sequence is associated with the TSS. As seen in Fig. 2, the strongest primer extension product observed corresponds to a point 292 bp upstream of the nfsA start codon rather than to the putative promoter at 226 bases upstream. Sequences consistent with E. coli −10 and −35 promoter sequences are located upstream of the primary TSS (see Fig. 1). This putative promoter is upstream of both the ybjC and the nfsA coding regions, suggesting that ybjC and nfsA form an operon. In addition to the primary TSS, several fainter primer extension products, corresponding to start sites within the ybjC coding region, were visible on the autoradiograph, including one corresponding to the putative promoter sequence (nt +40 to +67 in Fig. 1) previously identified from the nucleotide sequence.
FIG. 2.
Primer extension analysis to determine the transcription start site of nfsA and ybjC. The primary extension product (large arrow) was generated from RNA isolated from paraquat-induced E. coli carrying pH3X1 by using the PRX primer. Secondary primer extension products are indicated by small arrows. The sequence shown to the left corresponds to the upper DNA strand depicted in Fig. 1 and is complementary to the sequence ladder (GATC) generated by dideoxy sequencing of pH3X1 by using the PRX primer. The transcription start site (+1) is in boldface and is indicated by a bent arrow. The −10 sequence is indicated.
Expression of lacZ fusions.
To confirm the activity of the promoter associated with the TSS identified by primer extension, sequences upstream of nfsA were cloned into plasmid pMP28.5 to create lacZ fusions in which the ninth codon of the lacZ gene is fused in frame with the second codon of nfsA (Fig. 3A). The expression of β-galactosidase from the lacZ fusions was measured in the Lac− E. coli strain S90C (Fig. 3B).
FIG. 3.
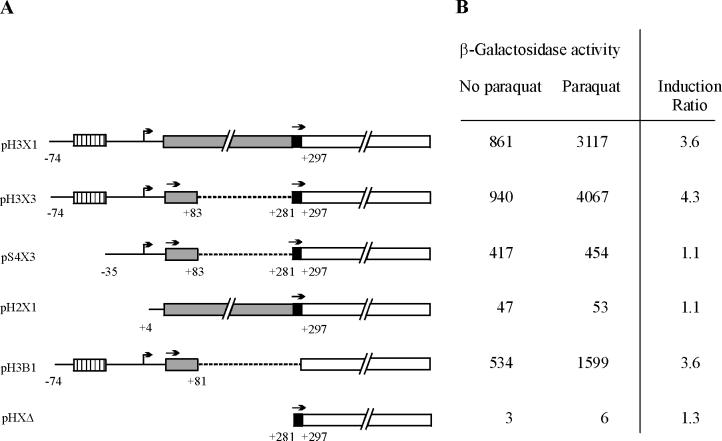
Expression of lacZ fusions. (A) Diagram of lacZ fusions. Numbering is based on the TSS site at +1 (bent arrow) and indicates the end of deletions (broken lines) and fusion junctions between noncoding regions (solid lines), ybjC (shaded bar), nfsA (solid bar), and lacZ (open bar). The drawing is not to scale as indicated by “//”. The soxbox is indicated by a hatched bar. Start codons in frame with the lacZ coding region in each fusion are indicated by straight arrows above the bars. Note that in X3 constructs, both the ybjC and nfsA start codons are in frame with the lacZ coding region, while in X1 constructs lacZ is in frame with nfsA and in H3B1 it is in frame with ybjC. (B) β-Galactosidase activity (in Miller units) of E. coli S90C strains carrying lacZ fusions. Results are the average of at least three independent trials run in duplicate. The induction ratio is the fold increase in activity of paraquat-induced strains relative to uninduced strains.
The nfsA-lacZ fusion, pH3X1, which carries the entire nfsA upstream region (nt −74 to +297), demonstrated a level of β-galactosidase activity 200 times the background level observed with pHXΔ, which carries only the nfsA ribosomal binding site and start codon (nt +281 to +297). Furthermore, the activity of the pH3X1 fusion increased an additional 3.6-fold in the presence of paraquat, indicating that this construct contains not only the promoter sequence of nfsA but also the sequences necessary for activation by SoxS. Deletion of the distal portion of the upstream region (nt −74 to +3) in pH2X1 resulted in a 20-fold decrease in β-galactosidase activity relative to pH3X1, confirming the activity of the promoter identified by primer extension. The 10-fold increase in activity of this construct over pHXΔ may be due to weak promoter activities within the ybjC coding region associated with the weak primer extension products observed in Fig. 2. It should be noted, however, that any weak promoter activity within the ybjC coding region is not inducible by paraquat.
As seen in a comparison of the results with pH3X1 and pH3X3, deletion of most of the ybjC coding region (nt +84 to +280) had little effect on the expression or induction of β-galactosidase. Construct pS4X3 is identical to pH3X3 except for a deletion of 38 bp directly upstream of the promoter identified by primer extension. While β-galactosidase activity increased over threefold in the presence of paraquat in strains carrying pH3X3, no such increase was observed in strains carrying pS4X3. Lack of induction in pS4X3 places the SoxS activation site either overlapping or just upstream of the ybjC-nfsA promoter identified by primer extension. Higher basal levels of β-galactosidase activity observed with pH3X3 over pS4X3 in the absence of paraquat may reflect a small stimulatory effect of basal levels of SoxS, MarA, or Rob at the intact binding site of pH3X3.
Placement of the nfsA promoter upstream of the ybjC coding region suggests that this ORF is transcribed in vivo. In order to test whether this ORF is translated in vivo, a ybjC-lacZ fusion, pH3B1, was constructed in which lacZ is fused to the 20th codon of ybjC such that the fusion protein is translated solely from the putative ybjC translation signals. The β-galactosidase activity obtained with this ybjC-lacZ fusion was ca. 100 times greater than the background level, demonstrating that YbjC can be expressed in vivo. Furthermore, as would be expected for a gene transcribed from the same promoter as nfsA, the activity of the ybjC-lacZ fusion increased more than threefold in the presence of paraquat, indicating that ybjC and nfsA constitute a functional operon that is inducible by paraquat.
Gel shift mobility assays.
Examination of the nucleotide sequence upstream of the ybjC-nfsA promoter identified two overlapping sequences (nt −66 to −47 and nt −50 to −31), each sharing sequence identity at 17 of 20 bases of the soxbox consensus sequence AYNGCACNNWNNRYYAAAYN (28). Both of these sequences are in the forward orientation. However, one is 10 bp upstream of the −35 sequence comparable to the Class I zwf promoter, while the other overlaps the −35 sequence in a manner consistent with class II promoters (14, 28).
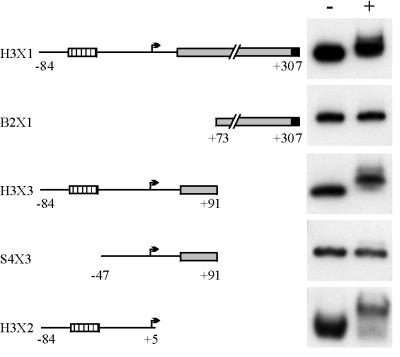
Gel shift mobility assays with His-tagged SoxS protein were conducted with radiolabeled PCR DNA fragments generated with the primers depicted in Fig. 1. It should be noted that, while the sequences carried by the PCR fragments are similar to those cloned into the lacZ fusion constructs, the latter are slightly smaller (by 6 to 10 bp) due to cleavage of the ends at restriction sites in the PCR primers prior to cloning. The upstream region of nfsA extending from nt −84 to +307 is contained on the H3X1 PCR fragment. Gel shift mobility assays with H3X1 confirmed that His6-SoxS does indeed bind to the region upstream of nfsA (Fig. 4). Gel shift assays with two smaller fragments representing either half of this region demonstrated that the right portion carried by fragment B2X1 (nt +73 to +307) was not able to bind the His6-SoxS protein and that binding was restricted to the left portion present in fragment H3X3 (nt −84 to +91). The binding site was further localized to the 89 left-most nucleotides based on the binding of the protein to PCR fragment H3X2 that carries nt −84 to +5. More telling are the results obtained with the S4X3 fragment which, lacks 38 nt from the left-most side (nt −84 to −48). Deletion of these 38 bases eliminated binding to the S4X3 fragment and clearly places all or part of the His6-SoxS binding site within the sequence immediately upstream or overlapping the −35 promoter sequence. These results are consistent with the induction results obtained with the lacZ fusions.
FIG. 4.
Gel shift mobility assays of the nfsA upstream region with His6-SoxS. Schematics to the left depict the region carried on the PCR fragments used in the gel shift assays shown to the right in the absence (−) or presence (+) of 50 ng of His6-SoxS protein. The symbols are as described in the legend to Fig. 3.
Based on the nucleotide sequence, a third putative soxbox containing 17 of 20 bases of the soxbox consensus sequence was identified within the ybjC coding region (nt +21 to +40) in the forward orientation, overlapping one of the weak putative promoters in a manner consistent with a class II promoter. Lack of binding of His6-Sox to S4X3 clearly demonstrates that this putative soxbox is not functional, a finding consistent with the lack of induction of the pS4X3 or pH2X1 nfsA-lacZ fusions by paraquat.
DNase I protection studies.
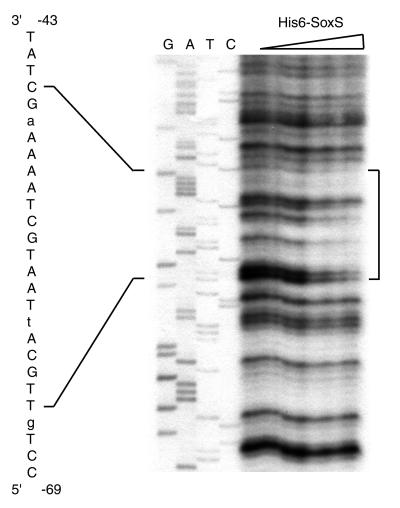
DNase I footprinting assays were conducted to determine which of the two putative soxboxes identified by gel shift experiments is involved in SoxS binding. As seen in Fig. 5, His6-SoxS protects the sequence extending from nt −65 to −46 which corresponds to the upstream soxbox sequence. From these results, we can conclude that the ybjC-nfsA promoter is a class I promoter in which the soxbox lies 10 bases upstream of the −35 site in the forward orientation in a manner comparable to the class I zwf promoter (14, 28).
FIG. 5.
DNase I footprinting analysis of His6-SoxS binding to the ybjC-nfsA promoter region. PCR-generated DNA incubated with increasing amounts of His6-SoxS protein (0, 10, 25, 50, 100, and 200 ng) was digested with DNase I and run on an 8% denaturing polyacrylamide gel. The region protected by His6-SoxS is indicated by a vertical bracket to the right. The sequence of the protected region is given to the left and corresponds to the upper strand shown in Fig. 1. Mismatches to the soxbox consensus sequence (28) are indicated in lower case. The sequencing ladder (GATC) was generated by dideoxy sequencing of template DNA by using the GRX primer.
DISCUSSION
The results of primer extension and lacZ fusion experiments presented in this study place the nfsA promoter upstream of both the nfsA and ybjC coding regions rather than within the ybjC coding region as predicted from the nucleotide sequence alone (56). Gel shift and footprinting analysis identified the regulatory sequence associated with this promoter that is responsible for SoxS-dependent induction of NfsA by paraquat (27). Coregulation of nfsA and ybjC from the same SoxS-dependent promoter confirms an earlier suggestion that these two genes are part of an operon, consisting of ybjC, nfsA, rimK, and b0853, that is upregulated by MarA (7). Inclusion of rimK and b0853 in this operon would indicate that the putative transcription termination sequence identified between nfsA and rimK is not functional (56). Further evidence for the inclusion of rimK in this operon can be taken from two earlier studies. Analysis of transcription products of rimK identified a transcript carrying both rimK and the nfsA upstream region (20). Similarly, two-dimensional gel analysis of proteins in E. coli identified an increase in the size of the small subunit ribosomal protein S6 concurrent with paraquat induction (15). The size increase was attributed to the addition of two to six glutamic acid residues to the carboxyl terminus of the S6 ribosomal protein by the rimK gene product (20).
Examination of sequence data available in the unfinished microbial genome NCBI database indicated that the order of the ybjC-nfsA-rimK genes found in E. coli is conserved in closely related enteric bacteria Klebsiella pneumoniae, Salmonella enterica serovar Typhi, Salmonella enterica serovar Enteritidis, and Salmonella enterica serovar Typhimurium. Another member of the Enterobacteriaciae with available sequence data is Yersinia pestis. Interestingly, although Y. pestis does not encode either an nfsA or a rimK homologue, it does retain a ybjC homologue upstream of a gene that encodes a protein with sequence similarity to the E. coli YbjN (b0853 gene product) (9). The YbjC homologue appears to be unique to the Entero bacteriaciae since no similar proteins have been identified in any of the other bacterial genomes available in the unfinished genome database despite wide distribution of NfsA and RimK homologues among both gram-positive and gram-negative bacteria.
Examination of the sequence associated with the paraquat-inducible ybjC-nfsA promoter revealed two potential soxbox sequences, each matching 17 of the 20 bases of the soxbox consensus sequence described by Martin et al. (28). However, footprinting analysis demonstrated that SoxS binds only to the sequence 10 bases upstream of the −35 sequence in a manner characteristic of class I promoters and not to the sequence overlapping the −35 sequence in a class II manner. Within the soxbox consensus sequence, AYNGCACNNWNNRYYAAAY, two functional domains (nt 4 to 7 and nt 13 to 19, underlined) have been identified from an analysis of aligned SoxS/Mar/Rob binding sites (28) by molecular information theory (44, 45, 55) and from the protein-DNA interaction sites identified from the crystal structure of the MarA (41) and Rob (21) proteins complexed with the mar and micF promoter sequences, respectively. The crystal structure for SoxS-DNA interaction has not yet been determined.
In the functional ybjC-nfsA soxbox, mismatches to the soxbox consensus sequence lie at positions 1 (G), 7 (T), and 19 (A). Adenosine is conserved at position 1 in 15 of the 16 known Sox/Mar/Rob binding sites and hence contributes high information content. However, this first nucleotide has not been shown to have any direct interaction with the MarA or Rob proteins, and systematic replacement of it with any other nucleotide has been reported to have no effect on activity in micF, fpr or zwf promoters (28). The pyrimidine at position 19 of the second domain of the soxbox consensus sequence has low sequence information content, and there is no evidence of interaction between it and either MarA or Rob. The third mismatch in the ybjC-nfsA soxbox is nucleotide T7, which lies within the highly conserved GCAC sequence. Both Gln45 of MarA and Gln39 of Rob make van der Waals contacts with the complement of C7 in the mar and micF soxboxes; however, a T is found at this position in three of the four other soxboxes with mismatches at this position (28), suggesting that such a change is tolerated. Thus, the three mismatches found in the functional ybjC-nfsA soxbox appear to be either unimportant, as in G1 and A19 or, in the case of T7, represents a change found in other functional soxboxes. The second, nonfunctional, soxbox contains mismatches at positions 2 (A), 6 (T), and 7 (A) of the soxbox consensus sequence. While the A2 is in a position that is not highly conserved and does not interact directly with either the MarA or Rob protein, the T6 and A7 mismatches lie within the GCAC domain at positions that have been shown to be protein contact sites. The complement of A6 of the mar soxbox forms van der Waals interactions with both Gln45 and Trp42 residues of MarA, while the complement of C7 interacts with Gln45 of MarA. Similar interactions are found in the Rob-micF complex. Only one known soxbox contains a T at position 6, and none contain an A at position 7. No functional soxbox contains two mismatches within this domain, and it is probably the combined effect of these two adjacent bases that prevents interaction with the protein.
The ybjC-nfsA promoter is the second class I SoxS-dependent promoter, after the zwf promoter, in which the soxbox is in the forward orientation (relative to the promoter). In the other six class I promoters characterized to date, the soxbox is in the reverse orientation 14 to 16 bases (fpr, fldA, mar, poxB) or 26 or 27 bases (acrAB, ribA) upstream of the −35 site (28, 55). A study by Wood et al. (55) has clearly demonstrated the correlation between the orientation and the distance of the soxbox from the −35 sequence in class I SoxS-dependent promoters. They observed a complete loss of induction with either the fpr (reverse orientation, 15 bp upstream) or the zwf (forward orientation, 7 bp upstream) soxbox if the spacing from the promoter was maintained but the orientation was switched to that found in the other promoter, or if the orientation was conserved but the position was changed. It has been proposed that, in class I promoters, a change in the orientation of the soxbox from reverse to forward at distances closer to the −35 sequence may facilitate interaction of SoxS with αCTD (28, 55). Thus, the forward orientation of the ybjC-nfsA soxbox is consistent with its relatively close proximity (10 bp) to the −35 sequence.
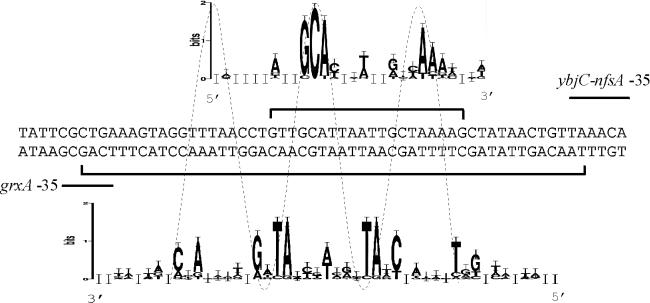
As seen in Fig. 6, the SoxS binding site identified in this study overlaps the OxyR binding site (47) of the divergently transcribed grxA (glutaredoxin-1) gene that lies directly upstream of the ybjC sequence. Grx1 has been shown to act as a hydrogen donor for ribonucleotide reductase (17). OxyR is the activator protein for a second oxidative-stress regulon in E. coli that is induced by hydrogen peroxide (49). Oxidized OxyR binds as a tetramer to a sequence of degenerate twofold symmetry, making contact at four adjacent major grooves on one face of the DNA (5, 48, 52). The SoxS protein is also predicted to bind to one face of the DNA, making contact with two adjacent major grooves (41). Overlap of the OxyR and SoxS binding sites suggests the possibility of interference between these two proteins under oxidative-stress conditions, particularly in the presence of superoxide which would be dismutated to hydrogen peroxide in the cell. Molecular information theory analysis of aligned OxyR (44) and SoxS (55) binding sites (Fig. 6) predict that the major grooves facing each of these two proteins are off-center by ca. 5 nt, suggesting that SoxS and OxyR bind to opposite faces of the grxA-ybjC intergenic region. This may allow simultaneous interaction of SoxS and OxyR with their respective regulatory sequences under conditions of oxidative stress.
FIG. 6.
SoxS and OxyR binding in the grx-ybjC intergenic region. SoxS and OxyR binding sites are indicated by square brackets above and below the sequence, respectively. The −35 sequences of the ybjC-nfsA and grxA promoters are indicated by lines above and below the sequence, respectively. The SoxS sequence logo is above the sequence in the 5′-3′ orientation and corresponds to the upper DNA strand. The OxyR sequence logo is below the sequence in the 3′-5′ orientation and corresponds to the lower DNA strand. Both logos were generated by using the web-based program available online (http://www.bio.cam.ac.uk/seqlogo/). The SoxS logo was generated from an alignment of the ybjC-nfsA sequence and 14 other sequences taken from Wood et al. (55). The OxyR logo was generated from an alignment of the grxA sequence and its complement and seven other OxyR binding sites and their complement taken from Schneider (44). The OxyR logo was generated from both strands of DNA sequence due to the symmetrical nature of the binding site (45). A dotted line represents the helical nature of the DNA strand. Peaks corresponding to nucleotides in major grooves (containing >1 bit of information) in each logo are spaced ca. 10 bases apart; peaks of the SoxS logos correspond to the troughs of the OxyR logo.
The observation that divergently transcribed genes from two different oxidative-stress regulons possess overlapping regulatory sequences is intriguing, since there is no obvious relationship between the physiological roles of the gene products. In addition to its role in the reduction of ribonucleotide reductase disulfide bonds, Grx1 may also play an important role in the bacterial cytoplasm by reducing non-native disulfide bonds that might arise during oxidative stress. In this way, Grx1 has been proposed to act in an autoregulatory circuit by reducing an internal disulfide bond in OxyR, thus inactivating it and downregulating grxA (5, 58). The physiological role of nitroreductase during oxidative stress remains unclear. Liochev et al. (27) have speculated that increased levels of oxygen-insensitive nitroreductase may increase the number of nitrosubstituted substrates that are processed through two-electron pathways, compared to the one-electron pathways that are characteristic of oxygen-sensitive nitroreductases. This would result in a diminution of active oxygen species arising concomitant with the reoxidation of nitro-anion radical species. Despite the lack of any obvious functional relationship between nfsA and grxA, overlapping of regulatory sequences is unlikely to be coincidental and warrants further investigation.
Acknowledgments
We thank R. E. Wolf, Jr., and K. L. Griffith for providing the pBADHis6-SoxS plasmid and for their helpful instruction and suggestions for the purification of the His6-SoxS protein.
This work was supported by a research grant to I.B.L. from the Natural Science and Engineering Research Council of Canada.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amabile-Cuevas, C. F., and B. Demple. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anlezark, G. M., R. G. Melton, R. F. Sherwood, B. Coles, F. Fiedlos, and R. J. Knox. 1992. The bio-activation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954). I. Purification and properties of a nitroreductase enzyme for Escherichia coli: a potential enzyme for antibody-directed enzyme pro-drug therapy (ADEPT). Biochem. Pharmacol. 44:2289–2295 [DOI] [PubMed] [Google Scholar]
- 4.Ariza, R. R., Z. Li, N. Ringstad, and B. Demple. 1995. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 177:1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslund, F., M. Zheng, J. Beckwith, and G. Storz. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thio-disulfide status. Proc. Natl. Acad. Sci. USA 96:6161–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asnis, R. E. 1957. The reduction of furacin by cell-free extracts of furacin-resistant and parent-susceptible strains of Escherichia coli. Arch. Biochem. Biophys. 66:208–216 [DOI] [PubMed] [Google Scholar]
- 7.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53:495–523 [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayher, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, G. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 227:1453–1462 [DOI] [PubMed] [Google Scholar]
- 10.Bryant, D. W., D. R. McCalla, M. Leeksma, and P. Laneuville. 1981. Type I nitroreductases of Escherichia coli. Can. J. Microbiol. 27:81–86 [DOI] [PubMed] [Google Scholar]
- 11.Busby, S., and R. H. Ebright. 1997. Transcription activation at class II CAP-dependent promoters. Mol. Microbiol. 23:853–859 [DOI] [PubMed] [Google Scholar]
- 12.Ding, H., E. Hidalgo, and B. Demple. 1996. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J. Biol. Chem. 271:33173–33175 [DOI] [PubMed] [Google Scholar]
- 13.Ebright, R. H. 1993. Transcription activation at class I CAP-dependent promoters. Mol. Microbiol. 8:797–802 [DOI] [PubMed] [Google Scholar]
- 14.Fawcett, W. P., and R. E. Wolf, Jr. 1994. Purification of a MalE-SoxS fusion protein and identification of the control sites of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 14:659–679 [DOI] [PubMed] [Google Scholar]
- 15.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:6181–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidalgao, E., J. M. Bollinger, Jr., T. M. Bradley, C. T. Walsh, and B. Demple. 1995. Binuclear [2Fe-2S] clusters in Escherichia coli SoxR protein and role of the metal centers in transcription. J. Biol. Chem. 270:20908–20914 [DOI] [PubMed] [Google Scholar]
- 17.Holmgren, A. 1976. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. USA 73:2275–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jair, K.-W. W. P. Fawcett, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 19:307–317 [DOI] [PubMed] [Google Scholar]
- 19.Jair, K.-W. X. Yu, K. Skarstad, B. Thony, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, W.-K. T. Icho, S. Isono, M. Kitakawa, and K. Isono. 1989. Characterization of the gene rimK responsible for the addition of glutamic acid to the C-terminus of ribosomal protein S6 in Escherichia coli K12. Mol. Gen. Genet. 217:281–288 [DOI] [PubMed] [Google Scholar]
- 21.Kwon, H. J., M. H. J. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424–430 [DOI] [PubMed] [Google Scholar]
- 22.Li, Z., and B. Demple. 1994. SoxS, an activator of superoxide stress genes in Escherichia coli. J. Biol. Chem. 269:18371–18377 [PubMed] [Google Scholar]
- 23.Li, Z., and B. Demple. 1996. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol. Microbiol. 20:937–945 [DOI] [PubMed] [Google Scholar]
- 24.Liochev, S. I., L. Benov, D. Touati, and I. Fridovich. 1999. Induction of the soxRS regulon of Escherichia coli by superoxide. J. Biol. Chem. 274:9479–9481 [DOI] [PubMed] [Google Scholar]
- 25.Liochev, S. I., and I. Fridovich. 1992. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc. Natl. Acad. Sci. USA 89:5892–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liochev, S. I., A. Hausladen, W. F. Beyer, Jr., and I. Fridovich. 1994. NADPH:ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc. Natl. Acad. Sci. USA 91:1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liochev, S. I., A. Hausladen, and I. Fridovich. 1999. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc. Natl.Acad. Sci. USA 96:3537–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, R. G., W. K. Gillette, S. Rhee, and L. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431–441 [DOI] [PubMed] [Google Scholar]
- 29.Martin, R. G., W. K. Gillette, and J. L. Rosner. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 35:623–634 [DOI] [PubMed] [Google Scholar]
- 30.McCalla, D. R., C. Kaiser, and M. H. L. Green. 1978. Genetics of nitrofurazone resistance in Escherichia coli. J. Bacteriol. 133:10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCalla, D. R., P. Olive, Y. Tu, and M. L. Fan. 1975. Nitrofurazone-reducing enzymes in E. coli and their role in drug activation in vivo. Can. J. Microbiol. 21:1484–1491 [DOI] [PubMed] [Google Scholar]
- 32.McCalla, D. R., A. Reuvers, and C. Kaiser. 1970. Mode of action of nitrofurazone. J. Bacteriol. 104:1126–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCalla, D. R., and D. Voutsinos. 1974. On the mutagenicity of nitrofurans. Mut. Res. 26:3–16 [DOI] [PubMed] [Google Scholar]
- 34.Meng, W., N. J. Savery, S. J. W. Busby, and M. S. Thomas. 2000. The Escherichia coli RNA polymerase α subunit linker: length requirements for transcription activation at CRP-dependent promoters. EMBO J. 19:1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Pineview, N.Y.
- 36.Miller, J. H., D. Ganem, P. Lu, and L. Schmitz. 1977. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J. Mol. Biol. 199:275–301 [DOI] [PubMed] [Google Scholar]
- 37.Peterson, F. J., R. P. Mason, J. Hovsepian, and J. L. Holtzman. 1979. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J. Biol. Chem. 254:4009–4014 [PubMed] [Google Scholar]
- 38.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109–114 [DOI] [PubMed] [Google Scholar]
- 40.Providenci, A. M. 2000. Identification and characterization of CbaR, a repressor of cbaABC, the 3-chlorobenzoic acid catabolic genes of Comamonas testosteroni BR60 (pBRC60). Ph.D dissertation. Carleton University, Ottawa, Canada.
- 41.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc.Natl. Acad. Sci. USA 95:10413–10418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothmel, R. K., A. M. Chakrabarty, A. Berry, and A. Darzins. 1991. Genetic systems in Pseudomonas. Methods Enzymol. 204:485–514 [DOI] [PubMed] [Google Scholar]
- 43.Samuelson, J. 1999. Why metronidazole is active against both bacteria and parasites. Antimicrob. Agents Chemother. 43:1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider, T. D. 1996. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 274:445–455 [DOI] [PubMed] [Google Scholar]
- 45.Schneider, T. D., G. D. Stormo, L. Gold, and A. Ehrenfeucht. 1986. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 188:415–431 [DOI] [PubMed] [Google Scholar]
- 46.Skarstad, K., B. Thony, D. S. Hwang, and A.Kornberg. 1993. A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268:5365–5370 [PubMed] [Google Scholar]
- 47.Tao, K. 1997. oxyR-dependent induction of Escherichia coli grx gene expression by peroxide stress. J. Bacteriol. 179:5967–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tartaglia, L. A., C. J. Gimeno, G. Storz, and B. N. Ames. 1992. Multidegenerate DNA recognition by the OxyR transcriptional regulator. J. Biol. Chem. 267:2038–2045 [PubMed] [Google Scholar]
- 49.Tartaglia, L. A., G. Storz, and B. N. Ames. 1989. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 210:709–719 [DOI] [PubMed] [Google Scholar]
- 50.Tazima, Y., T. Kada, and A. Murakami. 1975. Mutagenicity of nitrofuran derivatives, including furylfuramide, a food preservative. Mutat. Res. 32:55–80 [DOI] [PubMed] [Google Scholar]
- 51.Tokiwa, H., and Y. Ohnishi. 1986. Mutagenicity of nitroarenes and their sources in the environment. CRC Crit. Rev. Toxicol. 17:23–60 [DOI] [PubMed] [Google Scholar]
- 52.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897–909 [DOI] [PubMed] [Google Scholar]
- 53.Tsaneva, I. R., and B. Weiss. 1990. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 172:4197–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteway, J., P. Koziarz, J. Veall, N. Sandhu, P. Kumar, B. Hoecher, and I. B. Lambert. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood, T. I., K. L. Griffith, W. P. Fawcett, K.-W. Jair, T. D. Schneider, and R. E. Wolf, Jr. 1999. Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol. Microbiol. 34:414–430 [DOI] [PubMed] [Google Scholar]
- 56.Zenno, S., H. Koike, A. N. Kumar, R. Jayaraman, M. Tanokura, and K. Saigo. 1996. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 178:4508–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zenno, S., H. Koike, M. Tanokura, and K. Saigo. 1996. Gene cloning, purification and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FraseI, the major flavin reductase in Vibrio fischeri. J. Biochem. 120:736–744 [DOI] [PubMed] [Google Scholar]
- 58.Zheng, M., F. Aslund, and G. Storz. 1998.Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718–1721 [DOI] [PubMed] [Google Scholar]