In the merciless world of natural selection, microorganisms have turned their most obvious characteristic, their small size, into their greatest advantage. Small size results in a large surface-to-volume ratio, which facilitates exchange of chemicals between the interior of bacterial cells and the external environment. The hallmark of prokaryotes (Archaea and Bacteria) is their metabolic diversity. Virtually every conceivable energy or nutrient source can be utilized by one microbial species or another, and viable microorganisms can be found practically anywhere on our planet that one bothers to look, whether at the bottom of the ocean, on the frozen landscape of Antarctica, or a mile underground. One measure of the remarkable success of prokaryotes is that bacteria are conservatively estimated to outnumber humans by an astronomical factor of at least 1021 (93). It is therefore not surprising that prokaryotes comprise the vast majority of phylogenetic diversity on Earth (68).
However, the same forces that have given bacteria their impressive metabolic properties also dictate that these capabilities be expressed in the form of enzyme synthesis or activity only when appropriate substrates are available. To do otherwise would be energetically wasteful, a significant competitive disadvantage. In conjunction with their metabolic flexibility, bacteria have necessarily evolved extensive and sophisticated sensory systems to monitor parameters of interest in their environment and to regulate gene expression and behavior accordingly. One strategy that has been particularly successful on an evolutionary scale is the use of two-component regulatory systems. For example, the complete genome sequence of Pseudomonas aeruginosa has revealed the presence of ∼65 such systems (72). In their simplest permutation, two-component regulatory systems consist of two proteins (reviewed in reference 84). Sensor histidine kinases (HKs) detect environmental stimuli and use this information to regulate their activities. The kinases catalyze autophosphorylation of a conserved histidine residue, with ATP acting as the source of phosphoryl groups. Response regulators (RRs) then transfer the phosphoryl group from the HK to a conserved aspartate residue within their own regulatory domain, with the degree of phosphorylation typically determining the extent to which the RR turns expression of appropriate genes on or off.
Approximately 200 scientists gathered in Cuernavaca, Mexico, in January 2001 to discuss the latest developments in bacterial locomotion and signal transduction (BLAST) at the sixth biennial BLAST meeting. This review summarizes highlights from the meeting and attempts to provide a sense of where our field is going. Three themes featured prominently in the reported experimental results and ensuing discussions. First, the availability of complete bacterial genome sequences has provided investigators with a much more complete cast of characters potentially responsible for any given function and also has greatly facilitated progress in the study of bacteria not easily amenable to genetic analysis. Second, fluorescence technology has been used to great advantage in tracking individual cells among a larger population, monitoring the subcellular localization of specific signaling proteins, and measuring protein-protein interactions in living cells. Third, in many cases, the level of analysis has moved beyond the traditional reductionist approach focusing on a single molecule or phenomenon and is headed towards an integrative approach that seeks comprehensive understanding of an entire pathway. In addition to figures presented in this article, readers are referred to videos that are available at a web site (http://www.uic.edu/orgs/blast/videos) that has been created to document the remarkable motile behaviors of cells and cell populations.
FUNCTIONAL GENOMICS OF BACTERIAL CHEMOTAXIS.
In order to fully exploit their metabolic capabilities, many bacteria have evolved the ability to direct their movement to an optimal chemical environment, a behavior termed chemotaxis. It has been apparent for several years that some bacterial species (e.g., Rhodobacter sphaeroides) encode multiple copies of proteins which have a high degree of amino acid sequence similarity to proteins previously demonstrated to control chemotaxis in other bacteria (35). The challenge has been to decipher the functions of the putative chemotaxis proteins and assemble them into signaling circuits. While some knockout mutations in tentatively identified chemotaxis genes eliminate chemotaxis as expected, other gene knockouts have failed to yield any observable phenotype, forcing investigators to postulate a role for these proteins in hypothetical chemotaxis processes which have not been reproduced under laboratory conditions. The recent proliferation of complete bacterial genome sequences has significantly exacerbated our unsatisfactory state of understanding, with many bacterial species possessing an even larger repertoire of chemotaxis proteins than was previously appreciated.
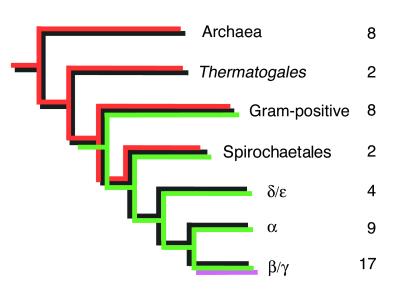
Igor Zhulin (Georgia Institute of Technology) gave a progress report on his phylogenetic analysis of chemotaxis genes from more than 50 sequenced prokaryotic genomes (Fig. 1). No such genes have been found in Eucarya. All bacteria with chemotaxis genes appear to possess an information processing pathway consisting minimally of methylated chemotaxis proteins (MCPs) (which are receptors for environmental signals), CheW (which physically couples MCPs to CheA), CheA (an HK whose phosphorylation state is controlled by the MCPs), and CheY (an RR whose phosphorylation state controls locomotive behavior rather than gene transcription) (Fig. 2) (104). Except for Helicobacter pylori, all sequenced prokaryotic species with chemotaxis genes also have CheR and CheB, which add and remove methyl groups, respectively, from MCPs in a delayed negative feedback loop that mediates behavioral adaptation. Zhulin’s phylogenetic analysis suggests that a chemotaxis system of ancient origin exists in Archaea and the older branches of Bacteria (e.g., gram-positive bacteria and spirochetes), with a more modern version of chemotaxis arising in proteobacteria. Two additional proteins, CheC and CheD, have widespread phyletic distribution, although CheC has been lost from the “modern” systems. CheD appears to mediate an as-yet-unidentified covalent modification (not methylation) of some MCPs as reported by Chris Kristich (G. Ordal’s lab, University of Illinois, Urbana-Champaign), whereas CheC appears to be involved in adaptation (45). CheC and CheD have been reported to interact with one another (73). CheV, a hybrid of CheW-like and RR-like domains that may mediate methylation-independent adaptation, is absent from the Archaea but is present in a subset of both “ancient” and “modern” lineages of Bacteria. It is important to recognize that the extensively characterized chemotaxis systems of Escherichia coli and Salmonella enterica serovar Typhimurium, referred to herein as S. enterica (reviewed in reference 18), do not possess CheC and CheD, and E. coli lacks CheV. In contrast, CheZ (which stimulates dephosphorylation of CheY) is restricted to β/γ proteobacteria, i.e., “modern” systems (59; I. Zhulin, unpublished observations). Zhulin therefore noted that Bacillus subtilis might be a good organism on which to focus future investigations, as this species possesses all known bacterial chemotaxis proteins except CheZ yet does not suffer from the complication of multiple copies of individual che genes.
FIG. 1.
Occurrence of chemotaxis proteins in prokaryotes. Black indicates the presence of MCPs, CheW, CheA, CheY, CheR, CheB, and CheD; red indicates CheC; green indicates hybrid Che proteins such as CheV; magenta indicates CheZ. Numbers to the right of each branch represent the number of completely or nearly completely sequenced genomes in which chemotaxis genes have been found. Alpha, beta, gamma, delta, and epsilon indicate subdivisions of Proteobacteria. Evolutionary distances are not drawn to scale. The cladogram represents general evolutionary trends. The particular set of chemotaxis genes in a given genome may be different due to gene loss and lateral gene transfer. For example, CheD and CheV are missing from E. coli but present in many other bacteria from the β/γ subdivision of Proteobacteria; laterally transferred CheC is found in 3 of 30 sequenced proteobacterial species. The figure was provided by I. Zhulin.
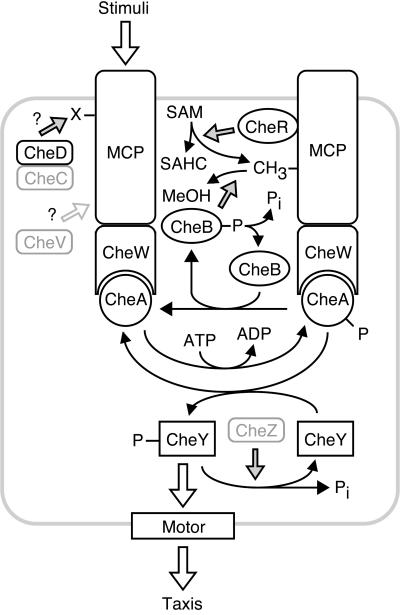
FIG. 2.
Signaling circuitry controlling bacterial taxis. This schematic diagram illustrates general functions of Che proteins and does not correspond to any species in particular. Proteins indicated in gray are not present in many bacterial species (Fig. 1), and multiple versions of individual Che proteins are present in some species. Solid arrows indicate chemical reactions, shaded arrows indicate catalysis or influence on a chemical reaction, and open arrows indicate binding or influence on a process. Note that stimuli can either enhance or diminish autophosphorylation of CheA, and the polarity of signal output (i.e., whether CheY or phospho-CheY interacts with the motor to control taxis) is arbitrary. Abbreviations: MeOH, methanol; SAHC, S-adenosylhomocysteine; SAM, S-adenosylmethionine; X, unidentified covalent modification of MCPs.
In one form or another, many presentations addressed the question of assigning roles to apparent chemotaxis proteins. The spirochete Borrelia burgdorferi has multiple periplasmic flagella attached at each end of the cell cylinder (Fig. 3A). Stuart Goldstein (University of Minnesota) reviewed the dynamics of B. burgdorferi motility (52) and presented an animated model (video 1) which illustrated that in order to work together to accomplish cell translation, the flagellar motors at either end of the cell (33) must rotate in opposite directions. Conversely, when both sets of flagella rotate in the same direction, the cell stops and may flex in the center. The primary established role for CheY is to control the direction of motor rotation. An obviously important question then is how the directions of rotation of motors at opposite ends of a 10- to 20-μm-long B. burgdorferi cell are appropriately coordinated on a time scale faster than CheY can diffuse the length of the bacterium. Previous studies have demonstrated that in some organisms lacking CheZ, some of the “extra” CheY proteins can take on the role of phosphate sinks, utilizing reverse phosphotransfer via CheA to drain phosphoryl groups off a primary CheY which interacts with the flagellar switch (81). Genome sequencing indicated that B. burgdorferi has three chemotaxis gene clusters, with three cheY genes and no cheZ (29). In an attempt to assign functional roles to each of the B. burgdorferi CheY proteins, Nyles Charon and colleagues (West Virginia University) constructed insertions in each cheY gene. Only one of the three cheY mutants had a pronounced phenotype—it swam exclusively in one direction and did not flex. Redundant functions might account for the lack of phenotypes for two mutants, but the mutant phenotype is difficult to explain. Regardless of whether the role of the CheY in question is to interact with motors at one or both ends of the cell or as a phosphate sink, removal of this CheY should result in all motors turning in the same direction at least some of the time, which would be manifested as a flexing motion. It therefore appears that additional roles for CheY remain to be discovered.
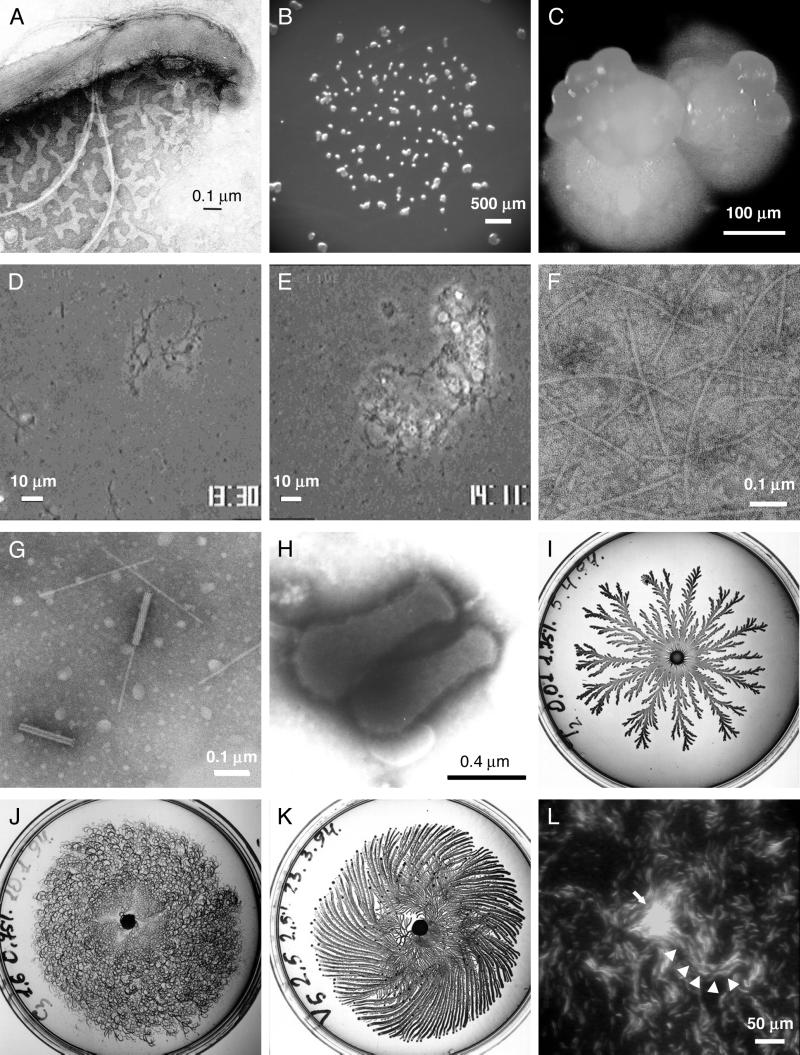
FIG. 3.
Motile bacteria and motility apparatus. (A) B. burgdorferi strain 297 with the outer membrane sheath disrupted with detergent to visualize the subpolarly attached periplasmic flagella. (B and C) M. xanthus fruiting bodies. Following starvation, approximately 100,000 cells aggregate to form a fruiting body within which cells convert to spores. The process usually takes 2 to 3 days. The images are of M. xanthus after 3 weeks of development and show the aggregation of cells into fruiting bodies and a higher-magnification image of a collection of several fruiting bodies. The fruiting bodies are orange due to photoprotective carotenoids. (D) S. grandis mixed with S. enterica wild-type cells at zero time. Long filamentous cells are S. grandis, and smaller rod-shaped cells are S. enterica. (E) After 40 min of incubation, S. grandis cells form large aggregates with S. enterica cells trapped within. (F) External filaments isolated from S. grandis cell surfaces. (G) Phage-like particles, so-called rhapidosomes, isolated from S. grandis. (H) M. mobile gliding in the direction of the tapered end. (I to K) Tip-splitting (I), chiral (J), and vortex (K) morphotypes exhibited by Paenibacillus dendritiformis on agar plates. (L) M. xanthus aggregation center visualized by fluorescence microscopy of GFP-expressing cells. Wild-type cells constitutively expressing GFP were mixed 1 to 10 with nonfluorescent wild-type cells and subjected to starvation. The fluorescent cells are seen on a background of nonfluorescent cells. The arrow points to an asymmetric aggregation center. Cells enter the aggregation center organized in streams (arrowheads). Images were provided by J. Kreiling and N. Charon (A), Z. Yang (B), J. Kirby (C), S. Aizwa (D to G), M. Miyata (H), D. Gutnick and E. Ben-Jacob (I to K), and L. Søgaard-Andersen (L). Images in panels I, J, and K are reproduced with permission from the Annual Review of Microbiology (7).
DNA sequence analysis indicates that multiple sets of chemotaxis genes within a single species can arise either by duplication within a genome or by horizontal transfer from other sources. The complete genome sequence of P. aeruginosa reveals the presence of four sets of apparent chemotaxis genes (85). One set has been shown to regulate flagellum-mediated swimming behavior (43), and a second set controls pilus-mediated twitching motility (25). Thus, one reason for multiple chemotaxis systems is to control multiple motor types in the same cell. The roles of the remaining two sets of putative chemotaxis genes were the subject of investigation by Abel Ferrandez (C. Harwood’s lab, University of Iowa). Knockout mutations of multiple putative chemotaxis genes mostly yielded no apparent phenotype. However, elimination of tlpB2, encoding a putative MCP, caused a significant reduction of chemotaxis. This is surprising, because chemotactic signaling pathways typically receive input from multiple receptors. A cell lacking one chemoreceptor is usually blind to the class of stimuli sensed by the missing receptor but competent to respond to stimuli recognized by other receptors, and the operon under study did include another MCP gene. It should be noted that TlpB2 is a somewhat atypical MCP, in that it does not contain any putative transmembrane segments and is therefore predicted to be located in the cytoplasm. Interestingly, Angela Martin (J. Armitage’s lab, University of Oxford) reported that deletion of tlpC, a gene encoding a putative cytoplasmic MCP in R. sphaeroides, also results in a general chemotaxis defect. The cytoplasmic location of TlpC was confirmed by using a green fluorescent protein (GFP) fusion protein. Perhaps cytoplasmic MCPs, which have also been discovered in other species, play a different role in chemotactic signaling than their better-characterized transmembrane counterparts.
Control of motility is critically important to the developmental cycle of Myxococcus xanthus, which upon starvation aggregates to form multicellular fruiting bodies (Fig. 3B and C). Two distinguishable types of motility are evident—individual cells engage in adventurous motility (A motility), whereas multiple cells engage in social motility (S motility). Although the complete genome sequence of M. xanthus is not publicly available, investigation of mutants with various developmental abnormalities previously revealed two sets of che-like genes. The frz genes (13) control cell reversal frequency during both A and S motility, whereas the dif genes are specifically needed to coordinate cell movement during S motility (98). John Kirby (D. Zusman’s lab, University of California, Berkeley) used oligonucleotide probes to search a library of M. xanthus DNA for additional MCP-like genes and found four such loci. Sequencing of adjacent DNA then revealed two previously unknown sets of che-like genes, designated che3 and che4, bringing the known M. xanthus total to four sets. Kirby constructed insertion mutations in four different che3 genes, which resulted in inappropriate sporulation on rich media and overexpression of several developmentally regulated proteins but did not affect either A or S motility. In order to account for these phenotypes, Kirby proposed that the Che3 proteins regulate transcription rather than motor function as previously assumed. In his model, the mutant cells would be unable to sense a factor that inhibits development. Such a hypothesis is plausible for several reasons. First, the vast majority of known two-component regulatory systems regulate transcription rather than behavior. Second, MCPs which sense amino acids or dipeptides for chemotaxis exist. Finally, some intercellular signals used in M. xanthus development are known to be amino acids. The generalization of Kirby’s hypothesis, that “extra” che genes in various bacterial species may regulate transcription rather than chemotaxis, could also explain the results reported by Zhaomin Yang (Auburn University). Yang noted that extracellular matrix fibrils are needed to link M. xanthus cells together during S motility. He found that mutations in difA (MCP-like), difC (cheW-like), and difE (cheA-like) blocked fibril biogenesis (99). By analogy to known chemotactic signaling systems, perhaps DifA senses a signal to initiate S motility and transmits this information through DifC to DifE, which changes the phosphorylation state of an as-yet-unidentified RR and hence initiates transcription of the genes required for fibril production. The role of the other dif genes remains to be explained.
SIGNAL TRANSDUCTION IN CHEMOTAXIS.
A small number of organisms with single sets of chemotaxis genes have provided model systems for dissecting the molecular basis of signaling in chemotaxis. In these systems, functional activities have been ascribed to each protein and structures are available for most of the components. Recent studies have focused primarily on probing molecular mechanisms of intermolecular and intramolecular signaling. One of the biggest challenges in the field is determination of the mechanism of transmembrane signaling utilized by chemoreceptor complexes.
The chemoreceptors (MCPs) bind small molecule or protein ligands and modulate the activity of the HK CheA. It has been previously established that kinase activity of CheA is influenced not only by receptor ligand occupancy but also by the methylation state of the cytoplasmic domains of the receptors, a reversible modification that contributes to adaptation (14, 67). Joshua Bornhorst (J. Falke’s lab, University of Colorado) has examined receptor-controlled CheA kinase activity as a function of attractant ligand binding and the modification state of the methylation region of the aspartate receptor Tar. Varying the modification state of Tar affects both attractant binding and kinase activity, suggesting that the sites for ligand binding, methylation, and kinase interaction, though spatially separated, are tightly coupled (15). The data led to postulation of a two-state model in which both ligand binding and methylation status affect the equilibrium between two receptor signaling states. Consistent with the model, certain disulfide cross-links in Tar partially disrupt both ligand and methylation signaling. A two-state model further predicts that modifications which trap a pure signaling state will fully block both the ligand and methylation signals, and a search for such modifications is in progress.
In E. coli, attractant addition promotes receptor methylation and removal causes demethylation. Michael Zimmer (G. Ordal’s lab) accounts for the different behavior of B. subtilis, in which both addition and removal of attractants result in receptor demethylation, by suggesting that hydrolysis occurs at a different subset of sites for the two stimuli. Conversely, methylation of Glu630 and Glu637 promotes adaptation to the removal and addition of attractant, respectively. A model for signaling based on selective modification of these two sites was proposed (105).
A detailed understanding of receptor signaling ultimately requires a description of the interactions between the receptor and downstream signaling components. Although three-dimensional structures are available for domains of MCPs (44, 61), the HK CheA (12, 58, 102), and the coupling protein CheW (I. Griswold and F. Dahlquist, unpublished data), structures of complexes have not yet been determined. Previous data (54) indicate that the receptor signaling complex contains vastly more subunits than the simple 2:2:2 stoichiometry originally suggested. Intriguingly, in crystals, the cytoplasmic domain of Tsr exists as a trimer of dimers with trimer contacts occurring at the distal cytoplasmic domain tips that are involved in CheA-CheW interactions (44). The physiological relevance of this trimer is still uncertain, but the trimeric interaction could potentially provide a structural scaffold for a network of receptor signaling complexes. Diverse approaches are being taken to model and directly probe interactions between protein components of the signaling complex.
Using recently developed three-dimensional printer technology, Thomas Shimizu and colleagues (D. Bray’s lab, University of Cambridge) generated plastic hand-held models of Tsr, CheA, and CheW from Protein Data Bank coordinates (78). These were manipulated to identify possible dockings between the components, which were then reconstructed in molecular graphics programs. Published data on mutant phenotypes helped to select an arrangement in which trimers of receptor dimers, complexed with CheA and CheW, can pack into a two-dimensional hexagonal lattice. The model also suggests a framework in which substoichiometric interaction of CheA/CheW units with the receptor network could provide for variable receptor-CheA-CheW stoichiometries.
A direct experimental approach was used by Ian Griswold (F. Dahlquist’s lab, University of Oregon) to define the surface of CheW that interacts with CheA. The solution structure of Thermotoga maritima CheW was determined by nuclear magnetic resonance (NMR), revealing a fold consisting of two five-stranded β barrels, similar to the P5 domain of CheA (Fig. 4A). Chemical shift perturbations of CheW upon addition of a P4-P5 domain fragment of CheA were used to identify the interacting residues in CheW. The residues map to a mostly contiguous surface in domain 2 of CheW. Residues previously implicated by genetic suppression studies to be involved in interaction with MCPs (53) form a putative MCP binding surface on the CheW structure which partially overlaps the CheA binding surface defined by NMR. Notably, the experimentally defined CheW-CheA interaction surface is distinctly different than that proposed by Shimizu (see above) and illustrates the need for caution in interpreting theoretical models.
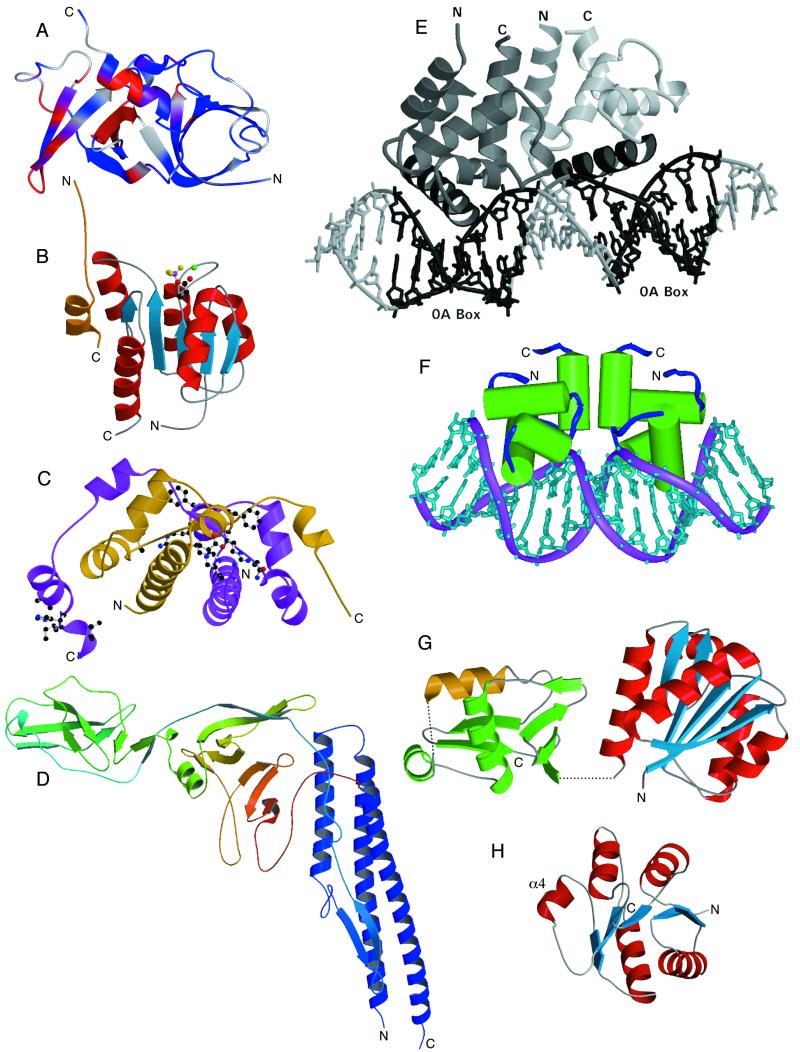
FIG. 4.
Proteins involved in chemotaxis, flagellar synthesis and assembly, and transcriptional regulation. (A) NMR structure of CheW from T. maritima. The ribbon diagram created with MOLMOL (47) calculated from the minimized average structure illustrates chemical shift changes in CheW induced by CheA (residues 354 to 671) binding, shown as a continuous color ramp from blue to red (0 to 100 Hz change). Gray indicates no available data. (B) Crystal structure of BeF3−-activated CheY from E. coli complexed with a peptide from its downstream target, FliM (50). An N-terminal 16-residue peptide derived from FliM (gold) binds to the α4-β5-α5 surface of CheY (blue, central β sheet; red, helices). BeF3− (magenta/gold atoms) bound to the active-site aspartate of CheY (Asp57; carbon and oxygen atoms for the side chain are black and red, respectively) mimics the active conformation of this RR protein. An essential Mg2+ ion is shown in green. (C) Crystal structure of the FlhD dimer from E. coli (20). Two molecules of FlhD (gold and purple) form a tightly associated dimer which, when complexed with another protein, FlhC, activates transcription of flagellar genes. Residues in FlhD that define a putative binding surface for FlhC, as identified by mutagenesis experiments (19), are shown in ball-and-stick representation. (D) Crystal structure of a flagellin monomer from S. enterica (75). Flagellar filaments are composed of a helical arrangement of flagellin subunits. The structure of a large portion of the flagellin monomer (the F41 fragment, 41,300 Da) can be described as a boomerang-shaped molecule, which provides structural insight into the assembly of the helical filament and the mechanism of supercoiling. (E) Crystal structure of the Spo0A-C dimer from B. subtilis bound to DNA. The C-terminal domain of the Spo0A RR forms a head-to-tail dimer (monomers shown in light and dark gray, respectively, with the DNA recognition helix in black) and is shown bound to DNA at two 0A boxes (black) separated by three base pairs. (F) Crystal structure of the NarL DNA-binding domain from E. coli. The C-terminal DNA-binding domain of the NarL RR dimerizes in a tail-to-tail fashion and binds with high affinity to an antiparallel “7-2-7” DNA binding site arrangement. (G) Crystal structure of the RR DrrD from T. maritima. The regulatory domain (blue/red) is positioned adjacent to the DNA-binding domain (green, with the DNA recognition helix in gold). Dashed lines represent two conformationally mobile regions that are disordered in the crystal lattice: a four-residue segment defining the interdomain linker and a five-residue segment adjacent to the recognition helix that is expected to interact with RNA polymerase during transcription initiation. (H) Crystal structure of the N-terminal receiver domain of PhoP from B. subtilis. The regulatory domain of PhoP is colored (blue/red) similarly to RR domains in panels B and G but is rotated approximately 90°. The regions in PhoP-N that differ from other RR structures are the α4 helix and β4-α4 loop. Images in panels A, E, F, G, and H were provided by I. Griswold, K. I. Varughese, A. Maris, D. Buckler, and C. Birck, respectively. Images in panels B, C, and D were generated with MOLSCRIPT (48) and Raster3D (60) using atomic coordinates obtained from the Protein Data Bank (1F4V, 1G8E, and 1IO1, respectively).
Experimental approaches are also being used to probe the physiological relevance of Tsr trimers. Sandy Parkinson and colleagues (University of Utah) have constructed proline, alanine, and tryptophan substitutions at 11 residues involved in trimer contacts. Cells expressing these mutant receptors, in either the presence or absence of other receptors, were characterized with respect to Tsr function, flagellar motor bias, and polar localization of receptor complexes. While most mutations altered motor bias, indicating improper control of CheA, most did not affect polar localization of receptor complexes. The bulky tryptophan substitutions were most epistatic, affecting the function of other receptors. One possible explanation of the data is that the mutated Tsr dimers disrupt the trimer interactions of heterologous receptor dimers.
Significant advances are being made in understanding intermolecular signaling events downstream from the receptor-kinase complex. One of the most exciting developments is the ability to monitor transient protein-protein interactions in vivo. Previous immunoelectron microscopic studies had established polar localization of chemotaxis proteins (55), and fluorescence technologies now allow examination of these interactions in living cells. Viktor Sourjik (H. Berg’s lab, Harvard University) has used fusions of yellow fluorescent protein to localize CheA, CheY, CheZ, and FliM in wild-type and mutant cells. CheA, CheY, and CheZ localize at the cell poles, and clustering is dependent on the presence of MCPs (80). CheY and CheZ localization is dependent on CheA, and CheA localization is dependent on CheW. Similar localization studies using a CheZ-GFP fusion were reported by Brian Cantwell (M. Manson’s lab, Texas A&M University). Sourjik also demonstrated colocalization of the motor protein with flagellar structures at the sides of the cell. These studies provided the foundation for fluorescence resonance energy transfer (FRET) experiments in which protein-protein interactions are detected by energy transfer between closely positioned fluorescent protein domains of fusion proteins. Cyan and yellow fluorescent protein fusions were used to monitor CheY-CheZ and CheY-FliM interactions in cells. Stimulation with aspartate causes a decrease in phospho-CheY, resulting in decreased CheY-CheZ and CheY-FliM interactions and a decreased FRET signal. A 50-fold-lower sensitivity to attractant was found in cheR cheB-deficient cells relative to wild-type cells. These data, like the in vitro experiments of Bornhorst (see above), demonstrate the influence of the methylation state of the chemoreceptors on their sensitivity to ligands. Flash-release of caged aspartate was used to address kinetic parameters of the interactions. The CheY-CheZ and CheY-FliM responses to aspartate addition have different lag times and decay times. Comparison of the CheY-FliM interaction to motor bias indicated that motor switching has a higher cooperativity than CheY-FliM binding. FRET experiments allow analysis of protein-protein interactions in living cells and hold the promise of correlating physiological and biochemical parameters to a greater extent than ever before.
CheY-CheZ and CheY-FliM interactions have been examined at a biochemical level by Martin Schuster (R. Bourret’s lab, University of North Carolina, Chapel Hill). The binding of CheZ and FliM peptides to CheY dramatically stimulates phosphorylation of CheY but has no effect on dephosphorylation (76). These results are analogous to the stimulation of OmpR phosphorylation by DNA binding (3), suggesting that conformational coupling between the phosphorylation site and the signaling surface may be a general feature of RRs.
The molecular details of the CheY-FliM interaction have been defined by the crystal structure of a BeF3−-activated CheY-FliM peptide complex (50), presented by David Wemmer (University of California, Berkeley). The structure of the 16-residue N-terminal peptide of FliM consists of a short β strand and a two-turn helix that pack against α4 and β5 of CheY (Fig. 4B). The β4-α4-β5-α5 surface was found to be the region of greatest conformational differences between structures of unphosphorylated CheY and a constitutively activated CheY-BeF3− complex, with magnitudes of structural changes similar to those previously observed for FixJ but less than those observed for NtrC. Residues of the peptide that are highly conserved in all FliM proteins are involved in structural roles or in direct contacts to CheY. Ihor Dzhaman (P. Matsumura’s lab, University of Illinois, Chicago) reported the selection of active mutants of CheY. The crystal structure of one mutant, CheY L24Q/E27G/A103V, was determined but showed no differences from the structure of wild-type unphosphorylated CheY. While phosphorylation or “activating” mutations may favor the active state, proteins are not locked in a single conformation, and any accessible conformation may potentially be observed in a crystal lattice.
Wemmer also reported the NMR solution structure of another activated RR domain, the NtrC receiver domain-BeF3− complex. NMR chemical shift perturbation analyses have localized the binding site of a peptide of the NtrC effector domain to the α4-β5 surface of the receiver domain, analogous to the surface observed in the CheY-FliM interaction. Wemmer also described the striking similarities between the active sites of RR receiver domains and the active site of Methanococcus jannaschii phosphoserine phosphatase (24, 92). Structures of phosphoserine phosphatase bound to BeF3−, AlF4−, and PO4−3 provide insight into the mechanism of phosphate hydrolysis that is likely to be conserved in RRs.
Bacteria sense and exhibit taxis towards numerous and diverse stimuli including oxygen and light. Aerotaxis in E. coli and phototaxis in the cyanobacterium Synechocystis sp. are mediated by receptors that contain cytoplasmic sensing domains rather than periplasmic ligand-binding domains as found in the extensively characterized E. coli chemoreceptors. The aerotaxis receptor, Aer, contains a PAS sensing domain (87), a HAMP domain (sometimes referred to as a linker domain) (4), and a signaling domain homologous to those of chemoreceptors. Mutagenesis studies presented by Qinhong Ma (B. Taylor’s lab, Loma Linda University) indicate that the HAMP domain is not merely a passive linker but is important for proper folding and FAD binding to the PAS domain (11, 71), for release from the chaperone GroEL, and for signaling.
Progress is also being made in characterizing phototaxis receptors in Synechocystis sp. Devaki Bhaya (A. Grossman’s lab, Carnegie Institution, Stanford University) described identification of genes involved in phototaxis, a process mediated by type IV pili (9). Phototaxis requires not only pil genes but also numerous other components, including two out of the three loci containing chemotaxis-like genes (10). Mutants altered in the Che1 locus show negative phototaxis, while mutants with changes in the Che3 locus are nonmotile and do not make type IV pili. CheD1, a transmembrane receptor encoded by this operon, lacks a periplasmic domain but contains an additional cytoplasmic domain (phy1) homologous to the chromophore-binding domains of plant phytochromes. Based on mutant analysis, a signaling pathway for Che components has been proposed.
TRANSCRIPTIONAL REGULATION, ASSEMBLY, AND STRUCTURE OF FLAGELLAR MOTOR AND FILAMENTS.
Construction and operation of locomotive organelles represent a substantial investment of precious resources for a bacterium, a circumstance which has several predictable consequences. First, although not all bacterial species are capable of movement, motile species are tactic, i.e., capable of guiding their movement in a beneficial direction. Second, when a motile bacterium is in an optimal environment, the need for locomotion (and hence for costly investment in propulsive machinery) ceases. Thus, transcription of genes encoding motor components is often environmentally regulated. Finally, at least in the case of flagella and their associated motors, the sequential assembly of complex structures composed of tens of different types of proteins is facilitated by a transcriptional hierarchy in which new elements are synthesized only after proper assembly of earlier substructures. This strategy also minimizes waste if something (e.g., a defective element) interferes with the assembly process.
In E. coli, there are three transcriptional classes of flagellar genes. The first group consists exclusively of flhC and flhD, whose products form a heterotetramer that is required for the synthesis of subsequent classes of flagellar components. Andres Campos and colleagues (P. Matsumura’s lab) described the crystal structure of the FlhD homodimer at 1.8-Å resolution (20) (Fig. 4C). Based on the FlhD structure, they made more than 60 different alanine substitutions, 12 of which interfered with function as assayed by motility (19). The loss-of-function mutant FlhD proteins were purified and assayed for the ability to form complexes with FlhC by using column chromatography. The substitutions that did not support complex formation defined a putative FlhC binding surface on FlhD. The substitutions that supported complex formation partially affected the ability of FlhD to bind DNA. This arrangement leads to a model of transcriptional activation in which FlhC binding rotates flexible arms of the FlhD dimer to bring them into contact with DNA.
The organization of flagellar transcription is somewhat different in Vibrio cholerae. Nidia Correa (K. Klose’s lab, University of Texas, San Antonio) reported that the availability of the complete genome sequence accelerated progress by revealing both flagellar structural and regulatory genes. Fusions of lacZ reporter genes to flagellar promoters were constructed in strain backgrounds from which various established and putative flagellar regulatory genes had been deleted. The resulting pattern of transcription was consistent with a four-tier hierarchy (70). The top level consists of FlrA alone, which with the help of ς54 is proposed to cause transcription of the class II genes, including FlrC (an RR) and FliA (ς28). FlrC, again with ς54, is believed to activate transcription of the class III genes, whereas ς28 appears to activate transcription of the class IV genes. The mechanism by which transcription of class IV genes is delayed with respect to transcription of class III genes, even though both activators are transcribed as class II genes, remains to be clarified.
As a result of decades of effort from many laboratories, extensive information is available concerning the architecture of the assembled flagellar motor and filament (Fig. 5). Nevertheless, gaps in our knowledge remain. For example, Chankyu Park (Korea Advanced Institute of Science & Technology) described experiments suggesting that some proteins involved in motor assembly and/or function have still not been identified. Deletion of the gene encoding H-NS, an abundant DNA binding protein in E. coli, results in bacteria that are flagellated but nonmotile. Park searched for genetic alterations that could restore motility, either by removing a negative function via a transposon insertion or restoring a positive function via a plasmid library. Each search yielded a single previously uncharacterized gene, ycgR and yhjH, respectively (46). Both genes proved to belong to the third and final transcriptional class of E. coli flagellar genes, but the roles played by their protein products are as yet unknown.
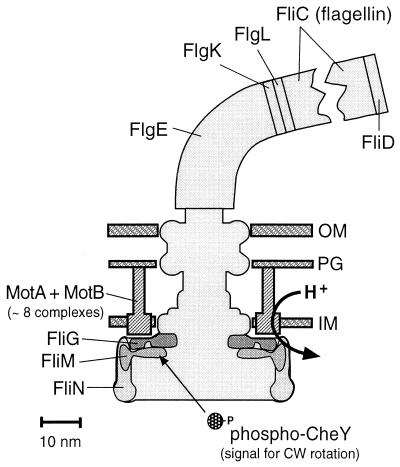
FIG. 5.
Key proteins of the flagellum. All of the proteins indicated are present in multiple copies in a flagellum. The MotA/MotB complexes are believed to be the stator, or nonrotating part, of the motor, whereas the other proteins indicated are part of the rotor. The actual length of a flagellar filament would be several hundred times greater than shown. The overall shape of the basal body (the membrane-embedded and cytoplasmic rings) is based on electron micrographic reconstructions by Francis et al. (28). OM, outer membrane; PG, peptidoglycan; IM, inner membrane. The figure was provided by D. Blair.
Debate also persists concerning the relative stoichiometries of some motor components. To address one such ambiguity, Perry Brown and colleagues (D. Blair’s lab, University of Utah) analyzed the FliM and FliN flagellar switch proteins of E. coli and T. maritima by analytical ultracentrifugation, with similar results for both species. Equilibrium centrifugation and velocity sedimentation both indicated that FliN forms a tetramer in solution. Analysis of a FliM/FliN mixture by the same techniques indicated a complex with a molecular mass consistent with either a FliM1FliN4 or FliM2FliN2 composition. The relative amounts of the two proteins determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the complex were most consistent with the former possibility.
Another limitation of our current understanding of the flagellar motor is that most of the structure is known at the resolution achieved with electron microscopy rather than the resolution achieved with X-ray crystallography. Determination of the unknown mechanism by which proton flow through the motor powers flagellar rotation would be facilitated by additional structural data. The proton channel is believed to be formed from a MotB dimer (one transmembrane segment per subunit) complexed with four MotA proteins (four transmembrane segments apiece). Tim Braun (D. Blair’s lab) has begun to use cysteine substitution mutants to work out the packing arrangement of the 18 transmembrane helices presumably present in this complex. Disulfide cross-linking patterns between many combinations of Cys substitutions, together with accessibility of individual sulfhydryl groups to modification by various chemical reagents, have previously been exploited to great advantage in analysis of MCP signaling (reviewed in reference 27). Braun’s cross-linking results suggest that the transmembrane helices of the MotB dimer are oriented with the two Asp32 residues, previously implicated in proton conduction (103), located on opposite rather than adjacent helical faces. The third transmembrane segment of MotA likely forms the rest of the proton channel.
Assembly of the flagellar filament, which can be an order of magnitude longer than the cell to which it is attached, is a significant engineering achievement because of the spatial separation between the site of protein synthesis inside the cell and the site of assembly at the distal tip of the growing filament outside the cell. Multiple flagellar components must enter the hollow core of the structure in the correct order, travel its entire length, and self-polymerize upon exiting. Frederic Auvray (C. Hughes’ lab, University of Cambridge) described the function of a set of cytoplasmic chaperones in S. enterica that are required for export of axial flagellar proteins through the flagellum-specific type III secretion apparatus. FlgN and FliT are chaperones specific for the hook-filament junction proteins FlgK and FlgL and the filament cap FliD, respectively (8, 30), whereas FliS is a chaperone for flagellin (5). FliS binds specifically to the C-terminal helical region of flagellin and prevents polymerization. The chaperones presumably facilitate secretion of flagellar axial subunits by inhibiting premature oligomerization. At the other end of the tunnel, a pentamer of the FliD protein forms a cap to prevent flagellin subunits from futile diffusion into the environment. Keiichi Namba and colleagues (Exploratory Research for Advanced Technology [ERATO]) discovered by using electron microscopy that the cap is not a passive structure but rather an active participant in flagellar assembly (101). The cap rotates and rises while a hole in its side guides each flagellin monomer into the proper position in the growing helical filament structure (video 2). Once assembled, the S. enterica flagellar filament can assume a left- or right-handed supercoil. The transition between these two forms, which involves changes in the packing of the flagellin subunits, is important because it determines the swimming behavior of the cell. Namba also reported the 2.0-Å structure of a fragment encompassing 80% of S. enterica flagellin from which the N- and C-terminal portions had been removed to prevent polymerization and allow crystallization (Fig. 4D). The structure allowed postulation of a possible basis for the left- and right-handed supercoils (75).
In contrast to the well-characterized E. coli and S. enterica flagellar filaments, which are primarily composed of a single flagellin protein, the complex flagella of Sinorhizobium meliloti and Rhizobium lupini are composed of four and three different flagellins, respectively. Birgit Scharf (R. Schmitt’s lab, Universitat Regensburg) constructed strains of both species containing various permutations of flagellin gene deletions and assayed the resulting mutants for motility and flagella. In both species, the major flagellin FlaA was necessary but not sufficient for flagellar function. The additional presence of at least one of the minor coflagellins restored function.
MOTILITY BEHAVIOR.
Over several decades, an increasingly detailed description of the motility of single cells of E. coli, S. enterica, and, to a lesser extent, B. subtilis has developed. This understanding now extends not only to the identification of participating proteins but also, on a molecular level, to sorting out specific amino acid residues involved in protein-protein interactions that promote cell movement. Considerably less is known regarding mechanisms of gliding motility found in other bacteria, such as Mycoplasma, myxobacteria, and Saprospira. Exciting results presented at the meeting indicate that we are now beginning to identify structures, genes, and proteins involved in the motility of some of these species.
Saprospira grandis is a marine flexibacterium that grows as a multicellular filament and moves on solid surfaces by gliding or by tangling with other objects (Fig. 3D). Shin-Ichi Aizawa (Teikyo University) described how S. grandis traps swimming bacteria of other species (Fig. 3E and videos 3 and 4). This entrapment, referred to as ixotropy (using stickiness, like flypaper), leads to eventual digestion of the trapped bacterium for nutrition. Aizawa and colleagues found that S. grandis was not able to trap nonflagellate mutants of S. enterica. Motile cells of S. enterica with either left-handed or right-handed flagella were readily trapped, whereas paralyzed mutants or mutants with straight flagella were trapped less efficiently. Taken together, the results indicate that both the motility and the flagella of the prey bacteria promote entrapment. Structural studies of S. grandis revealed the presence of thin, membranous sheath-covered filaments with a constant curvature (Fig. 3F). These filaments are postulated to play a role in trapping prey by entanglement.
The gliding motility of S. grandis ceases when the curved filaments are removed. Analysis of these structures indicated that they were not type IV pili, which are commonly involved in twitching motility in other bacteria. Thus, S. grandis may have a mechanism for moving over surfaces that is quite different from that of other bacteria. Aizawa suggests that the curved filaments are derived from internally located tobacco mosaic virus-like structures referred to as rhapidosomes (Fig. 3G).
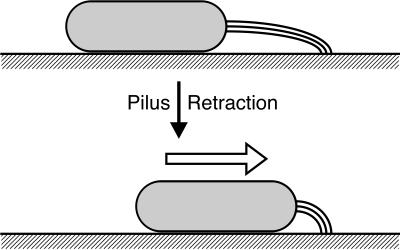
A model of gliding during S motility for the bacterium M. xanthus was proposed by Wenyuan Shi (University of California, Los Angeles) and colleagues (86). Upon starvation, these bacteria aggregate and form fruiting bodies. Cells first stream in chains by gliding motility to a central location and, with time, differentiate into fruiting bodies containing spores. A model for gliding is postulated in which cells extend type IV pili at cell poles, and the pili attach to a solid substrate (Fig. 6). The organism then retracts the pili, and the cell moves forward. Genetic evidence indicates that cellular reversal involves switching the site of pilus extrusion from one cell pole to the other, a process that is under the control of the frz chemotaxis gene cluster. Shi also proposes that chains of streaming cells occur when they attach to one another via other fibril materials located on the surface of the organism. The mechanism of gliding during A motility remains unknown.
FIG. 6.
Model of type IV pilus-dependent gliding motility. According to the work of Sun et al. (86), M. xanthus extends type IV pilus filaments at one cell pole and attaches the distal tips to the substrate. By pilus retraction, the cell moves toward the site of attachment. The figure was designed by H. Sun.
Several species of the wall-less, flagellumless, pilusless bacteria of the genus Mycoplasma glide on solid surfaces by a mechanism that is poorly understood. Mycoplasma mobile travels over surfaces as fast as 3 μm per s. A protrusion of the cell membrane, referred to as the attachment organelle (Fig. 3H), is involved in both adherence to the substrate and gliding (64). Makoto Miyata and coworkers (Osaka City University, in collaboration with H. Berg’s lab, Harvard University) found that M. mobile glides in the direction opposite fluid flow if a latex bead is attached to its tail (video 5). In one series of experiments, cells with attached latex beads were trapped with a laser trap and forces were measured. Their results suggest that some adhesion molecules remain bound to the substrate at all times and that the cells move under a wide range of loads.
The attachment organelle is also important in the adherence to host cell tissues. The Mycoplasma pneumoniae protein P1 is known to localize at the attachment organelle. Shintaro Seto (M. Miyata’s lab, Osaka City University) determined the distribution of the P1 protein during DNA replication and cell division using immunofluorescence (77). In dividing cells, there is duplication of the attachment organelle, with both copies adjacent to one another. One of the organelles migrates to the opposite pole of the cell before division. At least six other proteins were found to localize at the attachment organelle, and analysis of mutants indicated sequential assembly of these proteins.
A theme that emerged from several presentations is that bacterial interactions within a population lead to cooperativity in cell movement. Remarkable evidence was presented for cell-cell interactions in the complicated macroscopic growth and swarm pattern formation in Paenibacillus, cell aggregation in myxobacteria, and swarm cell differentiation in Salmonella. In some species, environmental factors, genes, and cell structures essential for cooperative cell movement have been identified. New techniques whereby tagged cells in a migrating population can be tracked have given this research a new dimension.
Many bacterial populations behave as communities. Work presented by David Gutnick (Tel Aviv University) emphasizes the sophistication of this interaction. Gram-positive bacteria belonging to the genus Paenibacillus swarm over agar plates, forming elaborate patterns (7, 88). Each type of pattern is produced by a specific morphotype. One morphotype forms branches with split ends (Fig. 3I), another produces chiral patterns (Fig. 3J), and a third produces vortexes (Fig. 3K and video 6). Tip splitting and chiral morphotypes actually are members of the same species which undergo adaptive reversible morphotype transitions in response to environmental conditions. Formation of the pattern is dependent on both cell density and motility and is also a function of agar concentration and richness of the media. During pattern formation, cells move through the agar as packs of cells, with cells often reversing direction. Gutnick recently isolated motile mutants deficient in pattern formation. Most remarkably, he isolated a low-molecular-weight protein from the supernatant fluid of wild-type cells that restored the ability of the mutants to form patterns. The results suggest that this protein plays an important role in cell-cell communication leading to pattern formation.
A complete understanding of development and differentiation in communities requires a description of the movement and positioning of individual cells. Roy Welch and colleagues (D. Kaiser’s lab, Stanford University) are applying such analyses to the early stages of fruiting body formation in M. xanthus. During the first few hours of development, opposing noninterfering traveling waves form on the surface of the population (video 7) (79). Each wave crest is a dense ridge of cells that travels at constant velocity. Welch mixed a small number of cells that express GFP with a large population that does not and tracked fluorescent cells for several hours under a microscope. He found that during rippling, tagged cells aligned themselves along the axis of rippling and moved back and forth in the direction of wave propagation. Fourier analysis indicated that there was a periodicity in the cell movements that correlated with wave propagation. The cell signaling csgA mutant generates waves of a different periodicity, and tagged csgA cells traveled back and forth with a different periodicity than that of wild-type cells. A computer model that can relate (and simulate) the movement of individual wild-type and mutant cells to wave propagation is being developed.
The cell surface-associated C signal promotes directional movement of M. xanthus cells resulting in cell aggregation and ultimately fruiting body formation. Lotte Søgaard-Andersen (University of Southern Denmark) addressed the question of which motility parameters are regulated by the C signal and how these changes in cell behavior may lead to fruiting body formation. The study involved quantitative analysis of single cells (40) and of individual cells in a population during starvation. The results indicate that C signal induces two chemokinetic responses: (i) cell density-independent changes, such as increased gliding speed and decreased stop frequency, and (ii) cell density-dependent changes involving decreased reversal frequency. Local cell-cell interactions with C-signal transmission were proposed to lead to global organization of cells in aggregation centers (Fig. 3L) (40, 41).
A model of swarming cell motility of S. enterica was presented by Rasika Harshey (University of Texas, Austin). S. enterica differentiates into hyperflagellated long swarmer cells that promote colonization on growth surfaces. This swarming has been shown to be cell density dependent, and genes involved in swarming have been identified by transposon mutagenesis (89). These genes encode components of the chemotaxis system, proteins of uncharacterized two-component systems, and enzymes involved in lipopolysaccharide synthesis, the latter being required for swarming but not for differentiation. Based on these observations, Harshey and colleagues proposed a model whereby the external slime, composed of O antigen among other polysaccharides, is both the signal and the milieu for swarming motility. When the concentration of O antigen is sufficiently high, cells initiate swarming. The O antigen also makes the agar surface more “wettable.” Additional signaling contributions from other two-component systems are suggested by the mutagenesis data.
The ability to swarm was also reported for two Bacillus species, Bacillus cereus and Bacillus thuringiensis. Emilia Ghelardi and coworkers (S. Senesi’s lab, University of Pisa) characterized nonswarming mutants of each of these species. Strains with mutations in a putative flagellar switch gene in B. cereus (fliY) (23) and flagellum assembly in B. thuringiensis (flhA) were unable to differentiate into swarmer cells. Each of the mutants also failed to produce a protein component of hemolysin. These and other results suggest that swarm cell differentiation and hemolysin production are coordinately controlled in these bacteria.
SIGNALING IN SOIL MICROBES.
The nitrogen-fixing soil bacterium Azotobacter vinelandii differentiates into cysts under adverse conditions. During encystment, A. vinelandii produces an extracellular polysaccharide (alginate) that becomes part of the cyst and concomitantly accumulates intracellular poly-β-hydroxybutyrate. Guadalupe Espín (Instituto de Biotechnologia) reported that in gacS or gacA mutants, which lack the HK or RR of the GacS-GacA two-component system, the expression of an essential enzyme for alginate synthesis that is encoded by algD was abolished in stationary phase but not in exponential phase (22). One of three algD promoters was found to be necessary for transcription in stationary phase, and mutations in gacS and gacA inhibited this transcription. Similarly, transcription of phbB, an enzyme essential for poly-β-hydroxybutyrate synthesis, was also inhibited by mutations in gacS and gacA. The results establish a major role for GacS-GacA in signal transduction controlling polymer synthesis and encystment in A. vinelandii.
Bacteria of the Rhizobiaceae infect the roots of many legume plants and, in doing so, produce nodules that enable the plant to fix nitrogen. The bacteria respond to many plant metabolites, such as flavonoids, by inducing several genes necessary for nodule formation. Otto Geiger and colleagues (Universidad Nacional Autonoma de Mexico) studied the role of phosphatidylcholine (PC) in nodule formation. S. meliloti was found to synthesize PC by either of two pathways. One involves methylation of phosphatidylethanolamine, and the other acquires choline provided by the plant for PC synthesis. Whereas single mutants in either pathway had no nodulation defects, double mutants in which both pathways were inactivated were unable to form nodules, implying that PC synthesis plays an essential role in nodule formation (26).
In another series of experiments, Esperanza Martínez-Romero (Universidad Nacional Autonoma de Mexico) observed that Rhizobium tropici and Rhizobium etli respond to environmental stimuli produced by plants by induction of periplasmic binding transport proteins. Using gene fusion analysis, they found that a crude glycoprotein fraction from the plant root (74) induced transcription of the transport genes and aggregation of the bacteria. Because motility and swarming of Rhizobium are necessary for aggregation and nodulation, they suggest that chemotaxis involving MCPs functions in plant recognition and eventual nodule formation. In addition, they reported some remarkable experiments related to antibiotic resistance (34). Plants are known to excrete phytoalexins, including flavonoids, which are toxic to bacteria. They find that R. etli survives these toxins by using multiple resistance genes that encode proteins that pump out the toxic flavonoids. The authors presented evidence that the flavonoids induce the antibiotic resistance gene by binding to and inactivating a specific repressor and noted that this system is homologous to the Tetr and emrAB systems found in the Enterobacteriaceae.
A different strategy for antibiotic resistance involving extracytoplasmic function (ECF) ς factors is utilized by B. subtilis. The ECF subfamily of ς factors, each with a corresponding stimulus-responsive anti-ς which is localized to the cytoplasmic membrane, provides a transcriptional regulatory mechanism with many parallels to two-component pathways (62). The work described by John Helmann (Cornell University) focused on two of the seven ECF ς factors in B. subtilis, specifically, the identification of regulons controlled by the ςX and ςW factors (37, 38). Using cDNA array and sequence analysis approaches, the Helmann group identified numerous genes regulated by ςX and ςW and proposed a role for these factors in B. subtilis antibiotic defense mechanisms. The ςW protein was shown to regulate fosfomycin resistance (21), while ςX controls genes that affect cell surface properties including d-alanylation of teichoic acids and membrane phospholipid composition. The modulation of cell surface charge by this regulon serves to protect the bacterium from antibiotics produced by other soil microbes.
TWO-COMPONENT REGULATORY SYSTEMS.
Two-component systems dominate bacterial signaling and have been extensively studied over the past decade. While many of the features of these systems are understood, a few fundamental questions are still unanswered, and many details relating to the adaptation of individual proteins to particular systems remain to be determined. Although the catalytic activities of transmembrane HK proteins have been well documented, less is known about the actual stimuli to which sensor HKs respond. At this meeting, several presentations focused on the sensing mechanism itself, that is, identification of the signal that specific HKs recognize and identification of specific amino acid residues within HKs that are important for signal reception and transmission.
Dimitris Georgellis (E. Lin’s lab, Harvard Medical School) reported on the E. coli ArcB/ArcA two-component system which regulates transcription of nearly 40 operons depending on the redox environment of the cell. Under reducing conditions, ArcB undergoes autophosphorylation, a process known to be stimulated by a number of allosteric effectors (31). ArcB transfers phosphoryl groups to the RR ArcA, which in turn binds to target promoters and modulates gene expression. A series of Tar-ArcB chimeras were constructed, and signaling activity was assessed using a lacZ reporter. Results from these and other experiments (49) indicate that the transmembrane domain of ArcB serves as an anchor that keeps the sensor close to the source of the signal. The actual signal was identified as the oxidized form of the quinone electron carriers ubiquinone and menaquinone (32). These were found to inhibit ArcB autophosphorylation.
The E. coli nitrate- or nitrite-dependent sensory transduction system regulates expression of genes involved in numerous respiratory and fermentation pathways. The sensor HKs of this system, NarX and NarQ, have a unique periplasmic sequence termed the P box that apparently forms part of the sensing domain responsive to nitrate and nitrite signals. Robert Gunsalus (University of California, Los Angeles) described in vitro phosphorylation assays of NarX-enriched membrane fractions in the presence or absence of nitrate or nitrite. NarX was 2 orders of magnitude more responsive to nitrate than nitrite, whereas NarQ was constitutively “on” (i.e., phosphorylated) and unresponsive to nitrate or nitrite. Interestingly, mutations in the P box resulted in a NarQ mutant that was more responsive to nitrate signals (i.e., more NarX-like). As for NarX, phosphotransfer from NarQ to the RR NarL apparently is not regulated by nitrate; however, it is dependent on the amount of phospho-NarX or phospho-NarQ present. Thus, in the NarXLQP system, stimuli directly control HK autophosphorylation activity and do not appear to regulate either the phosphotransfer or RR phosphatase activities of NarX and NarQ.
B. subtilis undergoes a complex differentiation process to produce dormant spores in response to starvation conditions. Initiation of sporulation is controlled by a multicomponent phosphorelay system involving several HKs that integrate multiple environmental signals and, via the phosphorelay, shuttle phosphoryl groups to the RR Spo0A, which in turn activates or represses expression of sporulation genes. KinA, a soluble HK that is the primary contributor of phosphoryl groups to the pathway, is composed of three PAS domains located near the N terminus and a typical C-terminal HK domain. Keith Stephenson (J. Hoch’s lab, Scripps Research Institute) reported a study in which the role of the PAS domains was probed by alanine-scanning mutagenesis (91). A total of 21 amino acid residues in the first PAS domain were identified as being important for KinA function. Mutations in most of these residues resulted in a lack of or a decrease in spore formation in vivo. One mutant, interestingly, exhibited a hypersporulation phenotype compared to the wild type.
For both HKs and RRs, a mechanistic understanding of the functioning of variable domains lags behind that of conserved domains. Phosphorylation of the conserved regulatory domain of an RR protein promotes an altered conformational state that in some way activates the variable effector domain. In the case of RRs that function as transcription factors, phosphorylation may promote higher affinity binding of the RR protein to DNA, or enhance the interaction of the RR with a specific ς factor and/or RNA polymerase. One example that illustrates this point is the ResD RR from B. subtilis, which activates transcription of genes required for aerobic respiration. ResD positively regulates the expression of the ctaA gene, which is required for synthesis of heme A. Salbi Paul (F. M. Hulett’s lab, University of Illinois, Chicago) presented data obtained from in vitro transcription experiments in which combinations of ς factors and core RNA polymerase (E) or RNA polymerase purified from different growth and developmental periods were employed (69). The data suggest different requirements for transcription of ctaA from two alternative promoters (Pv and Ps). Transcription from the upstream promoter (Pv) requires a single ResD binding site, phospho-ResD, and EςA. Transcription from the second promoter (Ps) requires two ResD binding sites, phospho-ResD, and RNA polymerase with ςE or a ς factor dependent on ςE. Thus, a single phosphorylated RR can participate in distinctly different transcription complexes.
Our understanding of how RRs bind to their target DNA sites was significantly advanced at this meeting by reports of structures of RR DNA-binding domains, two of which were solved in the presence of DNA. Sporulation in B. subtilis requires that the RR Spo0A activate expression of sporulation genes and repress genes for vegetative growth. The N-terminal domain of Spo0A inhibits the C-terminal domain (Spo0A-C) from binding to the recognition sequence (0A box), and phosphorylation relieves this inhibition. The X-ray structure of the Spo0A DNA-binding domain bound to 0A-box DNA was presented by K. I. Varughese (Scripps Research Institute). The fold of Spo0A-C consists of six α helices (51) and is not homologous to DNA-binding domains of other RRs. The DNA duplex contained two tandem repeats of the 0A boxes separated by three base pairs. Two Spo0A-C molecules bind to these sites and interact in a head-to-tail manner to form a dimer. Most of the interactions are mediated by the helix-turn-helix motif of Spo0A-C. The recognition helix is positioned in the major groove (Fig. 4E), and all the sequence-specific interactions are through this helix. Protein-protein interactions appear to be relevant for transcriptional activation.
The 2.1-Å crystal structure of the NarL C-terminal domain complexed with a 20-mer oligonucleotide was presented by Ann E. Maris (R. Dickerson’s lab, University of California, Los Angeles). The structure revealed a tail-to-tail dimer of NarL recognizing the antiparallel promoter half-sites (Fig. 4F). Residues involved in the dimer interface contact the receiver domain in the unactivated full-length protein structure (Protein Data Bank ID 1a04). Extensive DNA backbone contacts induce gradual DNA curvature and an overall bend of approximately 42°. In the DNA major groove, V189 of the helix-turn-helix recognition helix causes large distortions of a GC base pair.
The X-ray structure of DrrD from T. maritima, presented by David Buckler (A. Stock’s lab, University of Medicine and Dentistry of New Jersey), provides the first full-length structure for a member of the abundant OmpR/PhoB subfamily of RRs. The regulatory domain packs against the C-terminal DNA-binding domain, burying about 250 Å2 of surface area, far less than what has been observed in other multidomain RR structures solved to date (Fig. 4G). It seems likely that the two domains do not have a fixed orientation in the unphosphorylated protein. Modeling suggests that the N-terminal domain does not sterically block interaction of DNA with the recognition helix. However, with domains oriented as observed in the crystal structure, the intact protein cannot be placed as a tandem dimer on DNA without protein-protein collisions.
The B. subtilis PhoP protein, a member of the OmpR subfamily of RRs, binds to promoters in the Pho regulon to control the activation and repression of gene expression in response to phosphate starvation. The N-terminal phosphoreceiver domain of PhoP was crystallized in its unphosphorylated form in the presence of Mn2+, and its structure was determined at a resolution of 1.6 Å as reported by Catherine Birck (J.-P. Samama’s lab, IPBS-CNRS). Although the overall structure and the organization of the active site correspond well to structures of other regulatory domains, the α4 helix in PhoP is significantly shorter and the loop region between β4 and α4 differs from those of other RRs (Fig. 4H). In the crystal, two molecules of PhoP-N pack together, forming an extensive interface that buries about 1,000 Å2 of essentially hydrophilic surface area.
CELL DIFFERENTIATION AND CELL CYCLE REGULATION.
Many bacteria undergo various forms of differentiation in response to environmental stimuli or stresses. A few, such as Caulobacter crescentus, differentiate during normal progression through the cell cycle. In all cases, differentiation requires complex signaling cascades that coordinate differential gene expression associated with various stages of the developmental program. C. crescentus, a gram-negative aquatic bacterium, undergoes asymmetric cell division, which produces two distinct cell types: a stalked cell and a swarmer cell (57). The motile swarmer cell is capable of differentiating into a stalked cell. While multiple HKs and RRs are involved in regulation of cell division and differentiation in C. crescentus, CtrA is considered to be the global regulator for transcription of cell cycle and developmental genes.
Several years of investigation involving the isolation and characterization of bypass suppressors of HK and RR mutants defective in cell cycle progression and cell differentiation has led to a model in which CtrA phosphorylation is controlled by multiple parallel HK-dependent phosphorelay pathways (94). Noriko Ohta (A. Newton’s lab, Princeton University) undertook a yeast two-hybrid screen using DivK, an essential single-domain RR, as “bait” to investigate interactions between components of one of these phosphorelay pathways. Analysis of a large number of interacting clones identified 5 of the 61 Caulobacter HKs. Identification of DivJ and PleC supports their previous assignment as cognate HKs of DivK. The roles of the other interacting HKs in transmitting cell cycle signals to CtrA are being investigated.
During the transition from a swarmer to a stalked cell, the motility machinery is lost, including the single flagellum, which is replaced by a stalk at the same pole. It had been previously shown that the levels of the chemoreceptor McpA were modulated throughout the cell cycle (2). The Alley laboratory (Imperial College of Science Technology and Medicine) had identified a soluble chemoreceptor, McpB, which like McpA (90) is subject to cell cycle-regulated degradation by the ClpX protease pathway. Isabel Potocka described identification of the specific amino acid “signature sequence” recognized by the ClpX degradation pathway. Unlike the terminal signature sequences of other ClpX-degraded proteins, internal motifs near the C termini of McpA and McpB were found by mutagenesis studies to be necessary for degradation. The recently completed genome sequence of C. crescentus yields a large number of chemotaxis genes, including 16 other MCP genes (66). The presence of degradation motifs within these genes suggests that many of the MCPs and Che proteins may share the same fate as McpA and McpB.
Another complex developmental process occurs in the social bacterium M. xanthus. Under nutrient starvation conditions, this soil bacterium enters a developmental program that culminates with formation of a fruiting body and sporulation. During this process, rod-shaped vegetative cells differentiate into spherical spores. Cell-cell signaling and coordinated cell movement play an important role in the developmental program.
Early in development, an extracellular cell density signal known as A signal (composed of amino acids and small peptides) is required for developmental gene expression. Three regulatory proteins (AsgA, AsgB, and AsgC) are important for production of A signal (42). AsgA is a cytoplasmic hybrid HK composed of a receiver domain followed by a kinase domain. Using full-length AsgA as bait in a yeast two-hybrid system, Lynda Plamann and coworkers (University of Missouri, Kansas City) sought to identify other interacting partners that may constitute a phosphorelay system. Approximately 40 genomic fragments were identified as encoding domains that interact with AsgA. Of these 40, one was a homolog of a Ser/Thr kinase, several contained the highly conserved region of an MCP homolog, and 28 contained portions of one gene of unknown function. The rest had no similarities to each other or to known proteins. Interaction of AsgA with the S/T kinase homolog occurs through the kinase domain and not the receiver domain. A model was proposed in which the MCP homolog transmits extracellular signals to the cytoplasmic AsgA protein, which in turn interacts with the putative S/T kinase to control the AsgB transcription factor, which ultimately regulates the production of A signal and transcription of other developmental genes.
Another aspect of signaling via A signal was addressed in studies of the classic transmembrane HK SasS (97). SasS is responsible for sensing signals that reflect two environmental conditions, starvation and high cell density. SasS transfers phosphoryl groups to SasR, a positive regulator of transcription of early developmental genes. The N-terminal sensory domain of SasS bears a limited but significant sequence relationship to E. coli chemoreceptors. In particular, three arginine residues that are part of the binding site for attractant ligands in Tar and Tsr are conserved in SasS. The role of these arginines (R86, R89, and R93) in signal reception was addressed by Heidi Kaplan (University of Texas Medical School, Houston) using a site-directed mutagenesis approach. R86 and R89 appear to be important for negative and positive regulation of early developmental gene expression, respectively. Overall, the data support a model in which high cell density signals (A-signal amino acids) are sensed and transduced by SasS, with integration of starvation signals occurring via interaction with the membrane-localized negative regulator SasN (95).
A short time later in the developmental program, there is an increase in expression of CsgA, a protein required for production of C signal which promotes directional cell movement, resulting in cell aggregation and ultimately fruiting body formation. Cell aggregation and spore formation are induced by different concentrations of CsgA, and levels of CsgA are known to be two to three times higher in the fruiting body than in peripheral rod-shaped nondifferentiated cells. Anthony Garza (Washington State University) described a study in which a fusion protein was generated with GFP fused to a CsgA-dependent developmentally regulated gene. Fluorescence microscopy studies showed that this developmental protein is spatially localized. Furthermore, CsgA-dependent developmental genes are differentially expressed. Gene disruption experiments of the dev and exo genes abolish sporulation (phenotype similar to that of csgA-deficient cells), and expression of csgA was reduced (∼40%) in an actB mutant. Collectively, the data support a model in which CsgA is expressed on the cell surface and binds to a putative sensor on a nearby cell, triggering ActB expression. A positive feedback loop is formed via increased expression of the csgA gene, and finally, the dev and exo genes are expressed, resulting in the sporulation pathway.
FUTURE PROSPECTS.
The BLAST meetings convene in alternate years to consider progress in deciphering the mysteries of motility and signal transduction in prokaryotes. Given the wealth and breadth of subject matter, these topics seem likely to be vigorous and productive areas of basic research for the foreseeable future. Some currently prominent questions and technologies are outlined below.
Marine bacteria have been observed to “sprint” at more than 400 μm/s (63). When normalized for body length, this microscopic performance is faster than the macroscopic speed of a peregrine falcon in a gravity-assisted 200-mph dive. The remarkable propulsion system underlying bacterial swimming represents naturally engineered nanotechnology at its finest—bacterial flagellar motors have been observed to rotate at more than 100,000 rpm (56). How does ion flow power motor rotation? How does the motor switch direction of rotation? Determining the molecular architecture of the motor at high resolution is probably critical to answering both questions.
Flagella are not the only means of bacterial locomotion. Although many species of bacteria are capable of gliding over solid surfaces (reviewed in reference 83), the basis of this form of movement has eluded investigators for decades. The recent revelation that type IV pili are responsible for S gliding in M. xanthus (86) is a major breakthrough. What is the mechanism of A gliding motility in M. xanthus? For an even more challenging puzzle, one has only to look at cyanobacterial species that swim in the absence of flagella (16).
Although the His/Asp phosphorylation predominantly employed in prokaryotic signal transduction may at first glance seem quite different than the Ser/Thr/Tyr phosphorylation preferred by eukaryotes, biological signaling mechanisms actually have more fundamental similarities than differences across phylogenetic domains. Unicellular and multicellular organisms both exhibit intercellular and intracellular signaling. In all organisms, receptors first detect stimuli. Internal processing, which involves signal amplification and adaptation, then leads to appropriate responses. Until now, photolysis of caged stimuli has been the workhorse biophysical technique for real-time analysis of in vivo signal processing in bacteria (39). The pioneering use of fluorescence techniques to examine individual steps rather than the entire pathway, described at BLAST VI by Sourjik and Berg, is certain to be exploited for years to come.
All biological signaling mechanisms depend on an unstable signal to facilitate a timely response to stimuli. For pathways based on protein phosphorylation, the signal concentration is determined by the balance of kinase and phosphatase activities. The very instability of phosphorylated RRs hampered structural investigation of two-component regulatory systems for a decade. The 1999 discovery that BeF3− is a stable PO3−2 analog for RRs (96) had an immediate impact, but the experimental utility of BeF3−, for example, in stabilizing transient protein-protein interactions, has not yet been exhausted.
Biological signal transduction pathways are modular in design and employ different permutations of common elements to create unique circuits. The presence of multiple circuits with related elements and shared chemistry operating in parallel in a single cell generates the potential for inappropriate cross talk between pathways. A frequent solution to this design challenge appears to be multiprotein complexes, which presumably enhance both signal specificity and transmission speed (reviewed in reference 17). Given that high-resolution structures are now available for many bacterial signaling proteins, structures of multiprotein signaling complexes are a logical next target. How does the molecular architecture of a multiprotein complex determine its signaling properties?
Signal transduction mechanisms must cope with the cellular architecture within which they operate. In eukaryotic cells, for example, lipid modification is used to bring cytoplasmic signaling proteins to receptor complexes in the cytoplasmic membrane, and signals must penetrate the nucleus to alter gene transcription. Although we are relatively ignorant of subcellular organization in prokaryotes, it must exist. One example is the polar localization of chemoreceptors. What is the mechanism of polar localization? Is polar localization functionally important for chemotactic signaling? What other subcellular architectures constrain signaling in prokaryotes?
Once reductionist experimental attacks on individual elements of a biological information processing system yield sufficient detailed knowledge, it can profitably be integrated into comprehensive computer simulations, which in turn can be used to inform and inspire experimental design. The bacterial chemotaxis research community appears to have embraced this iterative strategy, with five different groups independently developing computer simulations in recent years (1, 6, 36, 65, 82, 100). Incorporation of computer simulations into signal transduction research is likely to increase in the future.
Finally, it is an accident of human history that studies of motility and signaling are relatively advanced for E. coli and S. enterica compared to other bacterial species. Genomics has begun to noticeably improve our phylogenetic perspective. What are the roles of Che proteins (CheC, CheD, and CheV) that happen to be absent from the most intensively studied model systems? What novel signaling strategies might be employed by bacteria grown in real-world environments rather than convenient but artificial laboratory conditions? What other motility and signaling mysteries await our attention in the incredibly diverse microbial world?
Acknowledgments
The participants of the BLAST meeting thank the following individuals for their efforts in coordinating scientific and administrative aspects of this and other BLAST meetings: E. Calva, F. Dahlquist, J. Falke, C. Harwood, M. Johnson, M. Manson, P. Matsumura, K. Oosawa, P. O’Neill, and J. Parkinson. We thank participants of the BLAST meeting for their assistance in editing text and providing figures and videos for this review and J. Dowd for creating the web site for video presentations.
Financial contributions for support of the meeting from the National Institutes of Health (1 R13 GM62997-01) and the National Science Foundation (MCB-0089139) are gratefully acknowledged.
REFERENCES
- 1.Abouhamad, W. N., D. Bray, M. Schuster, K. C. Boesch, R. E. Silversmith, and R. B. Bourret. 1998. Computer-aided resolution of an experimental paradox in bacterial chemotaxis. J. Bacteriol. 180:3757–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alley, M. R., J. R. Maddock, and L. Shapiro. 1993. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science 259:1754–1757. [DOI] [PubMed] [Google Scholar]
- 3.Ames, S. K., N. Frankema, and L. J. Kenney. 1999. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc. Natl. Acad. Sci. USA 96:11792–11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111–116. [DOI] [PubMed] [Google Scholar]
- 5.Auvray, F., J. Thomas, G. M. Fraser, and C. Hughes. 2001. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkai, N., and S. Leibler. 1997. Robustness in simple biochemical networks. Nature 387:913–917. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Jacob, E., I. Cohen, and D. L. Gutnick. 1998. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu. Rev. Microbiol. 52:779–806. [Online.] http://www.AnnualReviews.org. [DOI] [PubMed] [Google Scholar]
- 8.Bennett, J., J. Thomas, G. Fraser, and C. Hughes. 2001. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol. Microbiol. 39:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:941–951. [DOI] [PubMed] [Google Scholar]
- 10.Bhaya, D., A. Takahashi, and A. R. Grossman. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 98:7540–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131–141. [DOI] [PubMed] [Google Scholar]
- 13.Blackhart, B. D., and D. R. Zusman. 1985. “Frizzy” genes of Myxococcus xanthus are involved in control of the frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. USA 82:8767–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA 86:1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bornhorst, J. A., and J. J. Falke. 2000. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry 39:9486–9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brahamsha, B. 1999. Non-flagellar swimming in marine Synechococcus. J. Mol. Microbiol. Biotechnol. 1:59–62. [PubMed] [Google Scholar]
- 17.Bray, D. 1998. Signaling complexes: biophysical constraints on intracellular communication. Annu. Rev. Biophys. Biomol. Struct. 27:59–75. [DOI] [PubMed] [Google Scholar]
- 18.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campos, A., and P. Matsumura. 2001. Extensive alanine scanning reveals protein-protein and protein-DNA interaction surfaces in the global regulator FlhD from Escherichia coli. Mol. Microbiol. 39:581–594. [DOI] [PubMed] [Google Scholar]
- 20.Campos, A., R. G. Zhang, R. W. Alkire, P. Matsumura, and E. M. Westbrook. 2001. Crystal structure of the global regulator FlhD from Escherichia coli at 1.8 Å resolution. Mol. Microbiol. 39:567–580. [DOI] [PubMed] [Google Scholar]
- 21.Cao, M., B. A. Bernat, Z. Wang, R. N. Armstrong, and J. D. Helmann. 2001. FosB, a cysteine-dependent fosfomycin resistance protein under the control of ςW, an extracytoplasmic-function sigma factor in Bacillus subtilis. J. Bacteriol. 183:2380–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaneda, M., J. Guzman, S. Moreno, and G. Espín. 2000. The GacS sensor kinase regulates alginate and poly-beta-hydroxybutyrate production in Azotobacter vinelandii. J. Bacteriol. 182:2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celandroni, F., E. Ghelardi, M. Pastore, A. Lupetti, A. B. Kolsto, and S. Senesi. 2000. Characterization of the chemotaxis fliY and cheA genes in Bacillus cereus. FEMS Microbiol. Lett. 190:247–253. [DOI] [PubMed] [Google Scholar]
- 24.Cho, H., W. Wang, R. Kim, H. Yokota, S. Damo, S.-H. Kim, D. E. Wemmer, S. Kustu, and D. Yan. 2001. BeF3− acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF3− complex with phosphoserine phosphatase. Proc. Natl. Acad. Sci. USA 98:8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darzins, A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 11:137–153. [DOI] [PubMed] [Google Scholar]
- 26.de Rudder, K. E., I. M. Lopez-Lara, and O. Geiger. 2000. Inactivation of the gene for phospholipid N-methyltransferase in Sinorhizobium meliloti: phosphatidylcholine is required for normal growth. Mol. Microbiol. 37:763–772. [DOI] [PubMed] [Google Scholar]
- 27.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270. [DOI] [PubMed] [Google Scholar]
- 29.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. [DOI] [PubMed] [Google Scholar]
- 30.Fraser, G. M., J. C. Bennett, and C. Hughes. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569–580. [DOI] [PubMed] [Google Scholar]
- 31.Georgellis, D., O. Kwon, and E. C. Lin. 1999. Amplification of signaling activity of the arc two-component system of Escherichia coli by anaerobic metabolites. An in vitro study with different protein modules. J. Biol. Chem. 274:35950–35954. [DOI] [PubMed] [Google Scholar]
- 32.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the Arc two-component system of bacteria. Science 292:2314–2316. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein, S. F., K. F. Buttle, and N. W. Charon. 1996. Structural analysis of the Leptospiraceae and Borrelia burgdorferi by high-voltage electron microscopy. J. Bacteriol. 178:6539–6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Pasayo, R., and E. Martínez-Romero. 2000. Multiresistance genes of Rhizobium etli CFN42. Mol. Plant-Microbe Interact. 13:572–577. [DOI] [PubMed] [Google Scholar]
- 35.Hamblin, P. A., B. A. Maguire, R. N. Grishanin, and J. P. Armitage. 1997. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol. Microbiol. 26:1083–1096. [DOI] [PubMed] [Google Scholar]
- 36.Hauri, D. C., and J. Ross. 1995. A model of excitation and adaptation in bacterial chemotaxis. Biophys. J. 68:708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol. Microbiol. 31:361–371. [DOI] [PubMed] [Google Scholar]
- 38.Huang, X., and J. D. Helmann. 1998. Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search. J. Mol. Biol. 279:165–173. [DOI] [PubMed] [Google Scholar]
- 39.Jasuja, R., J. Keyoung, G. P. Reid, D. R. Trentham, and S. Khan. 1999. Chemotactic responses of Escherichia coli to small jumps of photoreleased L-aspartate. Biophys. J. 76:1706–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelsbak, L., and L. Søgaard-Andersen. 1999. The cell surface-associated intercellular C-signal induces behavioral changes in individual Myxococcus xanthus cells during fruiting body morphogenesis. Proc. Natl. Acad. Sci. USA 96:5031–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jelsbak, L., and L. Søgaard-Andersen. 2000. Pattern formation: fruiting body morphogenesis in Myxococcus xanthus. Curr. Opin. Microbiol. 3:637–642. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan, H. B., and L. A. Plamann. 1996. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol. Lett. 139:89–95. [DOI] [PubMed] [Google Scholar]
- 43.Kato, J., T. Nakamura, A. Kuroda, and H. Ohtake. 1999. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 63:155–161. [DOI] [PubMed] [Google Scholar]
- 44.Kim, K. K., H. Yokota, and S.-H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787–792. [DOI] [PubMed] [Google Scholar]
- 45.Kirby, J. R., C. J. Kristich, M. M. Saulmon, M. A. Zimmer, L. F. Garrity, I. B. Zhulin, and G. W. Ordal. 2001. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol. Microbiol. 42:573–586. [DOI] [PubMed] [Google Scholar]
- 46.Ko, M., and C. Park. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303:371–382. [DOI] [PubMed] [Google Scholar]
- 47.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55. [DOI] [PubMed] [Google Scholar]
- 48.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946–950. [Google Scholar]
- 49.Kwon, O., D. Georgellis, A. S. Lynch, D. Boyd, and E. C. Lin. 2000. The ArcB sensor kinase of Escherichia coli: genetic exploration of the transmembrane region. J. Bacteriol. 182:2960–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, S.-Y., H. S. Cho, J. G. Pelton, D. Yan, R. K. Henderson, D. S. King, L.-S. Huang, S. Kustu, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 8:52–56. [DOI] [PubMed] [Google Scholar]
- 51.Lewis, R. J., S. Krzywda, J. A. Brannigan, J. P. Turkenburg, K. Muchova, E. J. Dodson, I. Barak, and A. J. Wilkinson. 2000. The trans-activation domain of the sporulation response regulator Spo0A revealed by X-ray crystallography. Mol. Microbiol. 38:198–212. [DOI] [PubMed] [Google Scholar]
- 52.Li, C., A. Motaleb, M. Sal, S. F. Goldstein, and N. W. Charon. 2000. Spirochete periplasmic flagella and motility. J. Mol. Microbiol. Biotechnol. 2:345–354. [PubMed] [Google Scholar]
- 53.Liu, J. D., and J. S. Parkinson. 1991. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J. Bacteriol. 173:4941–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, Y., M. Levit, R. Lurz, M. G. Surette, and J. B. Stock. 1997. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 16:7231–7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717–1723. [DOI] [PubMed] [Google Scholar]
- 56.Magariyama, Y., S. Sugiyama, K. Muramota, Y. Maekawa, I. Kawagishi, Y. Imae, and S. Kudo. 1994. Very fast flagellar rotation. Nature 371:752. [DOI] [PubMed] [Google Scholar]
- 57.Martin, M. E., and Y. V. Brun. 2000. Coordinating development with the cell cycle in Caulobacter. Curr. Opin. Microbiol. 3:589–595. [DOI] [PubMed] [Google Scholar]
- 58.McEvoy, M. M., D. R. Muhandiram, L. E. Kay, and F. W. Dahlquist. 1996. Structure and dynamics of a CheY-binding domain of the chemotaxis kinase CheA determined by nuclear magnetic resonance spectroscopy. Biochemistry 35:5633–5640. [DOI] [PubMed] [Google Scholar]
- 59.McNamara, B. P., and A. J. Wolfe. 1997. Coexpression of the long and short forms of CheA, the chemotaxis histidine kinase, by members of the family Enterobacteriaceae. J. Bacteriol. 179:1813–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505–524. [DOI] [PubMed] [Google Scholar]
- 61.Milburn, M. V., G. G. Privé, D. L. Milligan, W. G. Scott, J. Yeh, J. Jancarik, D. E. Koshland, Jr., and S.-H. Kim. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342–1347. [DOI] [PubMed] [Google Scholar]
- 62.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059–1066. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell, J. G., L. Pearson, A. Bonazinga, S. Dillon, H. Khouri, and R. Paxinos. 1995. Long lag times and high velocities in the motility of natural assemblages of marine bacteria. Appl. Environ. Microbiol. 61:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311–1320. [DOI] [PubMed] [Google Scholar]
- 65.Morton-Firth, C. J., T. S. Shimizu, and D. Bray. 1999. A free-energy-based stochastic simulation of the Tar receptor complex. J. Mol. Biol. 286:1059–1074. [DOI] [PubMed] [Google Scholar]
- 66.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ninfa, E. G., A. Stock, S. Mowbray, and J. Stock. 1991. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 266:9764–9770. [PubMed] [Google Scholar]
- 68.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740. [DOI] [PubMed] [Google Scholar]
- 69.Paul, S., X. Zhang, and F. M. Hulett. 2001. Two ResD-controlled promoters regulate ctaA expression in Bacillus subtilis. J. Bacteriol. 183:3237–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel ς54- and ς28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595–1609. [DOI] [PubMed] [Google Scholar]
- 71.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodrigue, A., Y. Quentin, A. Lazdunski, V. Mejean, and M. Foglino. 2000. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 8:498–504. [DOI] [PubMed] [Google Scholar]
- 73.Rosario, M. M. L., and G. W. Ordal. 1996. CheC and CheD interact to regulate methylation of Bacillus subtilis methyl-accepting chemotaxis proteins. Mol. Microbiol. 21:511–518. [DOI] [PubMed] [Google Scholar]
- 74.Rosenblueth, M., M. F. Hynes, and E. Martínez-Romero. 1998. Rhizobium tropici teu genes involved in specific uptake of Phaseolus vulgaris bean-exudate compounds. Mol. Gen. Genet. 258:587–598. [DOI] [PubMed] [Google Scholar]
- 75.Samatey, F. A., K. Imada, S. Nagashima, F. Vonderviszt, T. Kumasaka, M. Yamamoto, and K. Namba. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410:331–337. [DOI] [PubMed] [Google Scholar]
- 76.Schuster, M., R. E. Silversmith, and R. B. Bourret. 2001. Conformational coupling in the chemotaxis response regulator CheY. Proc. Natl. Acad. Sci. USA 98:6003–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu, T. S., N. Le Novère, M. D. Levin, A. J. Beavil, B. J. Sutton, and D. Bray. 2000. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2:792–796. [DOI] [PubMed] [Google Scholar]
- 79.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740–751. [DOI] [PubMed] [Google Scholar]
- 81.Sourjik, V., and R. Schmitt. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327–2335. [DOI] [PubMed] [Google Scholar]
- 82.Spiro, P. A., J. S. Parkinson, and H. G. Othmer. 1997. A model of excitation and adaptation in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 94:7263–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spormann, A. M. 1999. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 63:621–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215. [DOI] [PubMed] [Google Scholar]
- 85.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959–964. [DOI] [PubMed] [Google Scholar]
- 86.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143–1146. [DOI] [PubMed] [Google Scholar]
- 87.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tcherpakov, M., E. Ben-Jacob, and D. L. Gutnick. 1999. Paenibacillus dendritiformis sp. nov., proposal for a new pattern-forming species and its localization within a phylogenetic cluster. Int. J. Syst. Bacteriol. 49:239–246. [DOI] [PubMed] [Google Scholar]
- 89.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsai, J.-W., and M. R. K. Alley. 2001. The proteolysis of the Caulobacter McpA chemoreceptor is cell-cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 183:5001–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang, L., C. Fabret, K. Kanamaru, K. Stephenson, V. Dartois, M. Perego, and J. A. Hoch. 2001. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J. Bacteriol. 183:2795–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, W., R. Kim, J. Jancarik, H. Yokota, and S. Kim. 2001. Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 Å resolution. Structure 9:65–72. [DOI] [PubMed] [Google Scholar]
- 93.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu, J., N. Ohta, J. L. Zhao, and A. Newton. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. USA 96:13068–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu, D., C. Yang, and H. B. Kaplan. 1998. Myxococcus xanthus sasN encodes a regulator that prevents developmental gene expression during growth. J. Bacteriol. 180:6215–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan, D., H. S. Cho, C. A. Hastings, M. M. Igo, S. Y. Lee, J. G. Pelton, V. Stewart, D. E. Wemmer, and S. Kustu. 1999. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. USA 96:14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang, C., and H. B. Kaplan. 1997. Myxococcus xanthus sasS encodes a sensor HK required for early developmental gene expression. J. Bacteriol. 179:7759–7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang, Z., Y. Geng, D. Xu, H. B. Kaplan, and W. Shi. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123–1130. [DOI] [PubMed] [Google Scholar]
- 99.Yang, Z. M., X. Y. Ma, L. M. Tong, H. B. Kaplan, L. J. Shimkets, and W. Y. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yi, T.-M., Y. Huang, M. I. Simon, and J. Doyle. 2000. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. USA 97:4619–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yonekura, K., S. Maki, D. G. Morgan, D. J. DeRosier, F. Vonderviszt, K. Imada, and K. Namba. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148–2152. [DOI] [PubMed] [Google Scholar]
- 102.Zhou, H., D. F. Lowry, R. V. Swanson, M. I. Simon, and F. W. Dahlquist. 1995. NMR studies of the phosphotransfer domain of the histidine kinase CheA from Escherichia coli: assignments, secondary structure, general fold, and backbone dynamics. Biochemistry 34:13858–13870. [DOI] [PubMed] [Google Scholar]
- 103.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhulin, I. B. 2001. The superfamily of microbial chemotaxis transducers: from physiology to genomics and back. Adv. Microbial Physiol. 45:157–198. [DOI] [PubMed] [Google Scholar]
- 105.Zimmer, M. A., J. Tiu, M. A. Collins, and G. W. Ordal. 2000. Selective methylation changes on the Bacillus subtilis chemotaxis receptor McpB promote adaptation. J. Biol. Chem. 275:24264–24272. [DOI] [PubMed] [Google Scholar]