Abstract
Termination of replication forks at the natural termini of the rDNA of Saccharomyces cerevisiae is controlled in a sequence-specific and polar mode by the interaction of the Fob1p replication terminator protein with the tandem Ter sites located in the nontranscribed spacers. Here we show, by both 2D gel analyses and chromatin immunoprecipitations (ChIP), that there exists a second level of global control mediated by the intra-S-phase checkpoint protein complex of Tof1p and Csm3p that protect stalled forks at Ter sites against the activity of the Rrm3p helicase (“sweepase”). The sweepase tends to release arrested forks presumably by the transient displacement of the Ter-bound Fob1p. Consistent with this mechanism, very few replication forks were arrested at the natural replication termini in the absence of the two checkpoint proteins. In the absence of the Rrm3p helicase, there was a slight enhancement of fork arrest at the Ter sites. Simultaneous deletions of the TOF1 (or CSM3), and the RRM3 genes restored fork arrest by removing both the fork-releasing and fork-protection activities. Other genes such as MRC1, WSS1, and PSY2 that are also involved in the MRC1 checkpoint pathway were not involved in this global control. This observation suggests that Tof1p–Csm3p function differently from MRC1 and the other above-mentioned genes. This mechanism is not restricted to the natural Ter sites but was also observed at fork arrest caused by the meeting of a replication fork with transcription approaching from the opposite direction.
Keywords: protein–protein interaction, replication terminus, terminator protein
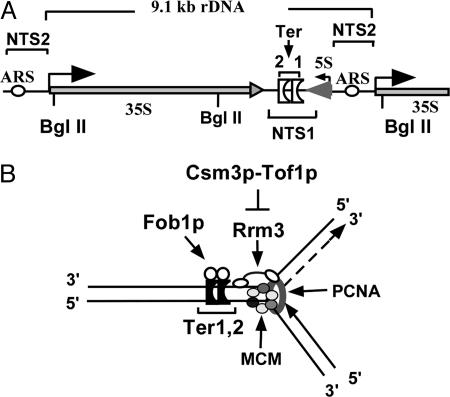
Site-specific replication termini (Ter sites), also called replication fork barriers, are present in many prokaryotic chromosomes and at certain specific regions of eukaryotic chromosomes, such as the nontranscribed spacers of rDNA of Saccharomyces cerevisiae (Fig. 1A; refs. 1–3). Binding of the cognate replication terminator proteins to the Ter sites causes programmed fork arrest that has special physiological functions (4). The terminator proteins of prokaryotes antagonize the activity of the replicative hexameric helicases in a polar mode (5–7), not only by binding to the Ter DNA but also by protein–protein contact with the replicative helicase (8, 9). Several aspects of the mechanism of fork arrest have been reviewed (4, 10).
Fig. 1.
Diagrammatic representation of a single repeat of the rDNA array of S. cerevisiae and a model of a terminated replication fork. (A) The repeated sequence present in the rDNA showing the 35S region transcribed by RNA polymerase I, the 5S region transcribed by RNA polymerase III, nontranscribed spacer I (NTS1) that contains tandem Ter sites and the nontranscribed spacer II (NTS2) that contains an origin of bidirectional replication (ARS). (B) A model of a terminated fork showing the locations of the two Ter sites that are bound to Fob1p, the putative replicative helicase MCM2–7p, the proliferating cell nuclear antigen sliding clamp (lavender ring), the Tof1p–Csm3p protein complex that either directly or indirectly antagonizes the Rrm3p helicase that progresses 5′ to 3′ on DNA and tends to release the terminated forks probably by transiently displacing Fob1p.
Eukaryotic replication termini are located in the nontranscribed spacers of rDNA of both budding and fission yeast (2, 3, 11, 12). A protein called Fob1p is necessary for fork arrest at the tandem Ter sites present at the nontranscribed spacers of S. cerevisiae (13, 14). A number of Ter-binding proteins have been discovered in Schizosaccharomyces pombe to date that bind to the replication termini located near the mating type locus and at the nontranscribed spacers of rDNA (15–19). Whether these proteins, like their analogues in prokaryotes, terminate replication by antagonizing the replicative helicase is not known at this time.
In addition to the terminus-binding protein, two checkpoint proteins called swi1 and swi3 of S. pombe are required for stable fork arrest (15, 17). These proteins also serve to stabilize replication fork arrest caused by the depletion of the dNTP pool by hydroxyurea (20, 21). The TOF1 and CSM3 genes of S. cerevisiae are the known homologs of swi1 and swi3, respectively, of S. pombe (22, 23). The two budding yeast proteins interact with each other and act at the intra-S-phase checkpoint pathway to stabilize stalled forks by maintaining the replisome at the arrested sites (24–28). In the absence of Tof1p and Mrc1p, a signal transducer of the intra-S-phase checkpoint pathway (29, 30), the replisome dissociates from the stalled forks and apparently translocates away from its original location at the pause site due to possible uncoupling of the DNA polymerase and the replicative helicase (26).
Unimpeded progression of the replication fork during normal DNA replication requires the activity of the helicase called Rrm3p that unwinds DNA in a 5′ to 3′ direction, and the deletion of the RRM3 gene causes fork arrest at numerous locations, some of which have been mapped to the binding sites of nonhistone proteins (31–33). In the absence of Rrm3p, fork arrest at the natural Ter sites of S. cerevisiae is enhanced by a factor of ≈2, suggesting thereby that Rrm3p causes a minority of the forks to escape the fork barrier at the sites even in the WT cells (34).
This work was prompted by our attempts to answer the following question. Besides the binding of the terminator proteins to the yeast Ter sites that arrest forks, which other proteins are involved in the regulation of fork passage past the Ter sites of S. cerevisiae? Keeping in mind that Tof1p and Csm3p proteins are homologous to the Swi1p and Swi3p of S. pombe, respectively (17, 21), we examined whether these proteins are also involved in fork arrest at the natural Ter sites and at the points of collision between transcription and DNA replication (35) in budding yeast. We report here that the answer to the above-mentioned questions is in the affirmative. However, the central point of this paper is that it illuminates a mechanism of action of the Tof1p–Csm3p checkpoint protein complex that promotes fork arrest at Ter sites and elsewhere by antagonizing the action of the Rrm3p helicase. Unrestrained Rrm3p activity tends to the release of forks arrested at the Ter sites during normal DNA replication that is unchallenged by genotoxic stress. The data revealed that, unlike Rrm3p, the Sgs1p and Srs2p helicases do not significantly contribute to fork release. The recently uncovered mechanism appears to serve as a second level of global control that, along with the Fob1p-Ter DNA interaction, controls the frequency of fork passage through natural Ter sites.
Results
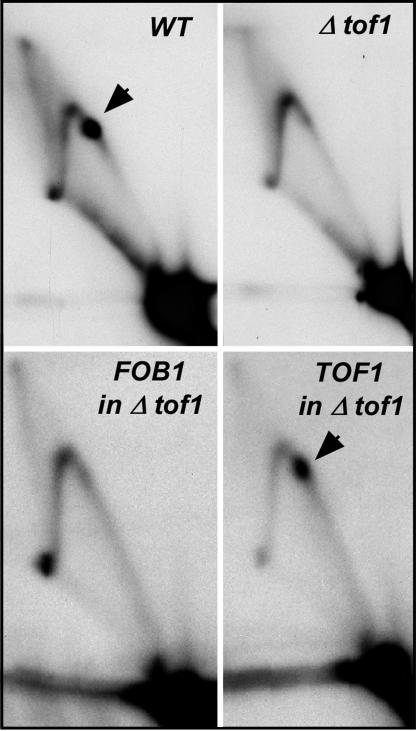
Tof1p and Csm3p Are Needed for Stable Fork Arrest at the Ter Sites. Fig. 1 A presents a physical map of the region of the replication termini present in the rDNA of S. cerevisiae. A model of the mechanism of fork arrest at Ter is presented in Fig. 1B. The major goal of the present work was to test some of the predictions of this model. We wished to answer the following question pertaining to the model. Besides Fob1p that binds to the Ter sites of S. cerevisiae (13, 14) and causes polar fork arrest, are there other proteins that regulate the number of forks that pass through the replication terminus and, if so, what is their mechanism of action? Keeping in mind the findings that swi1 and swi3 of S. pombe are involved in fork arrests at the natural Ter sites at the mating type locus and at the three Ter sites of rDNA called Ter1–3 (15, 17), we wished to determine whether the homologous genes, namely TOF1 and CSM3 of S. cerevisiae, might also be performing similar functions and if so, try to determine their mechanism of action. We examined the effects of deletions Δtof1and Δcsm3 on fork progression at the Ter sites by 2D gel electrophoresis, as contrasted with the isogenic WT strain (Fig. 2). The results showed that Δtof1 cells had a marked reduction of the termination spot in comparison with that of the WT cells. The Ter spot was not restored by complementation of the Δtof1 cells by FOB1, but positive complementation was achieved by plasmid-borne TOF1, thus proving the absence of Tof1p rather than a cryptic mutation in Fob1p caused the failure to arrest the forks at the Ter sites (Fig. 2). Similar experiments were performed with Δcsm3 cells and are shown in Fig. 7, which is published as supporting information on the PNAS web site. Tof1p and Csm3p are known to exist as an equimolar complex in vivo and appear to work together as a complex (28).
Fig. 2.
Autoradiograms of 2D gels showing the role of Tof1p on fork arrest at Ter1 and -2. The Ter spots in WT yeast (arrow) are almost completely abolished in Δtof1 cells. There was failure to complement (and thereby restore the Ter spot) by a plasmid expressing Fob1p in Δtof1 cells and complementation of Δtof1 cells by plasmid-borne WT TOF1.
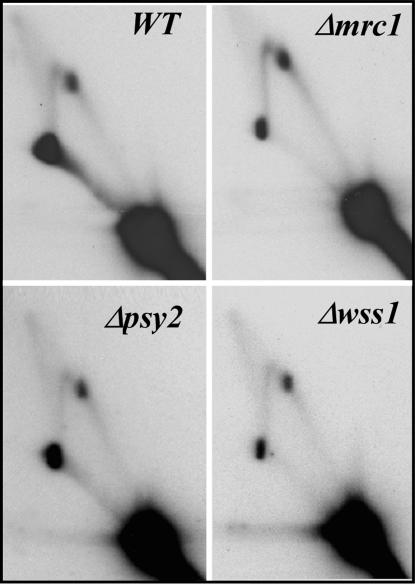
According to a published report (26), Tof1p and Mrc1p were shown to be present at the moving replication forks. Because of this close association between the two proteins, we checked to see whether deletion of MRC1 (29, 30) and other genes of the TOF1-MRC1 pathway, called PSY2 and WSS1 (36), also resulted in a similar loss of termination activity at the Ter sites. We observed that Δmrc1, Δwss1, and Δpsy2 cells showed persistent Ter spots of similar intensity (Fig. 3), suggesting that TOF1/CSM3 have a different function with regard to fork arrest than other proteins such as MRC1, PSY2, and WSS1 of the intra-S-phase checkpoint pathway. Because RAD9 has been suggested to play a role during DNA damage protection in the absence of Tof1p or Mrc1p (24, 25), we examined the effect of Δrad9 on termination activity in the rDNA and found no difference from that of the WT cells (not shown). It should be noted that RAD9 is known to interact functionally with RRM3 (37), and the latter protein, as will be shown later, plays a key role in preventing fork arrest at Ter.
Fig. 3.
Autoradiogram of 2D gels of replication intermediates of rDNA in Δmrc1, Δwss1, Δpsy2 strains showing that deletions of these genes had no effect on replication termination (all of the samples showed distinct termination spots).
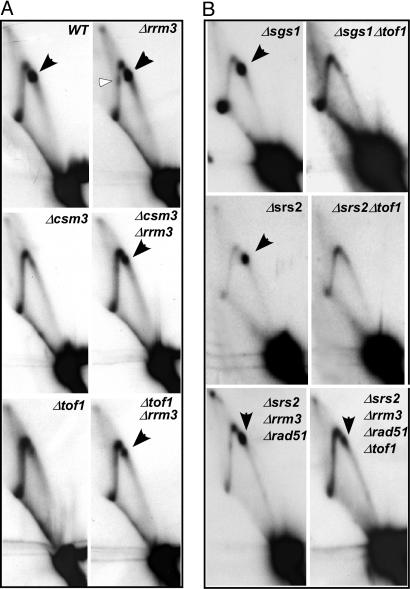
Tof1p and Csm3p Protect Stalled Forks Against the Fork-Releasing Activity of Rrm3p Helicase. The Rrm3p helicase is known to be required for normal fork progression through rDNA, because in its absence, there is enhanced replication fork arrest at the Ter site and fork pausing at numerous other regions of the chromosome, including the telomeric segment (31, 38). The Ter spot in the Δrrm3 cells was either similar in intensity to that of the WT cells (Fig. 4A) or enhanced slightly in some 2D gels, thus suggesting that even in normal cells containing Tof1p–Csm3p, the Rrm3p helicase appears to be acting as a persistent “sweepase,” probably by transiently displacing a fraction of the Terbound Fob1p to reduce fork arrest at these sites (see ref. 34). Because the Δtof1/Δcsm3 cells, as contrasted with the WT cells, showed substantial abolition of fork arrest at the rDNA Ter sites, we wished to test whether the activity of Tof1p–Csm3p counteracted that of Rrm3p.
Fig. 4.
Autoradiograms of 2D gels showing termination activity in WT strain and its derivatives. (A) The Ter spot is slightly enhanced (≈2-fold) in Δrrm3 cells in comparison with the WT but is mostly missing in Δtof1 and Δcsm3 cells. The Δtof1Δrrm3 and Δcsm3Δrrm3 double deletions at least partially restored the Ter spot. Black arrowheads show the Ter spot(s). The unfilled arrowhead indicates fork stalling on the descending part of the arc in Δrrm3 cells. (B) In Δsgs1 cells, a ≈2-fold enhancement of Ter spot over that of the WT was observed. Simultaneous deletions of both sgs1 and tof1 (Δsgs1Δtof1) caused disappearance of the Ter spot; removal of srs2 (Δsrs2) did not reduce fork arrest at Ter; double deletions, Δsrs2 Δtof1 did not cause restoration of fork arrest not unlike in the Δsgs1Δtof1 cells; Δsrs2Δrrm3 Δrad51 (Δrad51 was included to overcome synthetic lethality of Δsrs2Δrrm3) did not affect the frequency of fork arrest, whereas the quadruple deletion that also contained Δtof1 shows reduced fork arrest.
In designing experiments to test this hypothesis, we reasoned that the double deletions Δtof1Δrrm3 or Δcsm3Δrrm3 should restore fork arrest at Ter because of simultaneous removal of both the helicase that promotes fork escape from Ter and the helicase antagonists that oppose fork release. We carried out 2D gel analysis of replication intermediates of chromosomal rDNA in Δtof1Δrrm3, Δcsm3Δrrm3 double deletion strains and compared and contrasted the gel patterns with those obtained from the isogenic WT and the Δtof1, Δcsm3, and Δrrm3 single deletion strains. As shown in Fig. 4A, Δrrm3 cells retained a prominent Ter spot and in addition showed fork stalling at the descending part of the arc in comparison with the WT strain (Fig. 4A; compare WT and Δrrm3). As expected, the fork arrest was mostly abolished in the Δtof1 and Δcsm3 cells. In contrast, both the Δtof1 Δrrm3 and Δcsm3Δrrm3 double deletion cells showed a partial restoration of the termination spot that was ≈30–40% of the WT levels. Scanning the gels with a phosphorimager and then calculating the ratios of the intensity of the Ter spot over that of the integrated intensity of the corresponding replication arc provided the above-mentioned quantifications.
We then asked why the fork arrest in Δcsm3Δrrm3 and Δtof1Δrrm3 cells was incomplete in comparison with that of WT cells. Could it be, we asked, due to the actions of other helicases such as the RecQ helicase Sgs1p (39, 40) or the helicase Srs2p (41) that might also be promoting fork escape from the Ter sites by acting at rDNA? To address this question, we examined fork arrest at Ter sites in the Δsgs1, Δsgs1Δtof1, Δsrs2, Δsrs2Δtof1, Δsrs2Δsgs1 (SGS1ts) (not shown), Δsrs2Δsgs1 Δtof1 (not shown), Δsrs2Δrrm3Δrad51 [removal of rad51 relieved the synthetic lethality caused by removal of both the helicases (42)], and in the quadruple deletion Δsrs2Δ rrm3Δ rad51Δtof1 (Fig. 4B). The data are consistent with the conclusion that removal of Sgs1p and Srs2p singly or in various combinations with Rrm3p did not significantly enhance the restoration of the termination spot when Tof1p was simultaneously removed. Therefore, perhaps some hitherto unidentified activity reduced the recovery of the termination spot in Δtof1/Δcsm3 and Δrrm3 double deletion strains. Identification of the gene(s) responsible for this effect should be a topic of future work.
Complementation of a Δcsm3Δrrm3 strain with the WT RRM3 and mutant forms lacking the N-terminal 55 and 230 amino acids showed that the N-terminal part, including the putative proliferating cell nuclear antigen interaction domain, was necessary for fork release from Ter sites (see Fig. 8, which is published as supporting information on the PNAS web site).
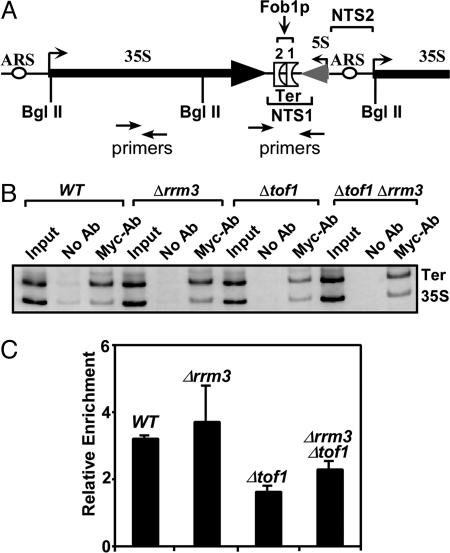
ChIP Analyses Confirmed 2D Gel Data. We wished to confirm the conclusion that was drawn exclusively from the 2D gel analyses by an independent technique. Using a strain containing myc-tagged MCM7p (43), we performed ChIP analyses to measure the frequency of fork arrest at Ter, as revealed by the extent of Ter DNA that was associated with the MCM7p, a protein that is an integral component of the putative MCM2-7 replicative helicase that travels with the replication fork (44). The cells were synchronized by treatment with the α factor, then released from cell cycle arrest at G1 and, following various time periods after release, the endogenous chromosome-bound MCM7p was crosslinked to DNA in vivo with formaldehyde (HCHO), and the chromatin extracted, sonicated, and immunoprecipitated with anti-myc antibodies. Under our experimental conditions, samples obtained after 30′ following release yielded the most consistent results. After reversing the crosslinks, the immunoprecipitated DNA was amplified by PCR with Ter-specific and control primer pairs as described (44), excepting that in some cases, in addition to the unlabeled primers, 5′end-labeled primers were also used for amplification by PCR (Fig. 5A) and the products resolved in 6% polyacrylamide gels (Fig. 5B). Three independent sets (with three replicates each) of experiments were performed. The ratios of the intensities of the Ter bands and the corresponding control bands were measured with an image scanner and quantified by using the imagequant (Bio-Rad) software (Fig. 5C). The ratio was taken as a measure of the relative frequency of fork arrest at Ter (in cells of the various genetic backgrounds stated above) in comparison with a control site(s) located on the 35S DNA. The data (Fig. 5 B and C) showed, as expected, that there was a net arrest of forks at the Ter site in the WT cells and enhanced fork arrest in the Δrrm3 cells but reduced arrest in the Δtof1 cells. In contrast, as expected, the Δtof1Δrrm3 doubly deleted cells showed higher levels of accumulation of forks in comparison with the Δtof1 cells. It should be noted that, under these experimental conditions, the restoration of the Ter spot in the Δtof1Δrrm3 double deletions was somewhat higher than the data obtained from the analyses of the 2D gels (Fig. 4). Thus, the ChIP analysis data were at least qualitatively consistent with the conclusion drawn from the 2D gel analysis.
Fig. 5.
ChIP analysis of fork arrest at the Ter sites in different genotypes. (A) Diagram showing the location of the primer pairs used to amplify Ter-specific and control DNA fragments that were crosslinked to myc-tagged minichromosome maintenance protein (MCM7p) and immunoprecipitated by anti-myc antibodies. (B) A representative photograph of the of the PCR products resolved in a 6% polyacrylamide gel. (C) Quantification of the ratios of the (from three independent sets of experiments) Ter fragment over that of the control fragment. Intensities of the DNA bands were calculated by using an image scanner and the imagequant software of ethidium bromide-stained gels. Computing the ratio of intensity of Ter band over that of the control band and normalizing that by multiplying with the inverse of the corresponding ratio of the input DNA gave the relative enrichment index. The primer pairs used are shown in Supporting Text.
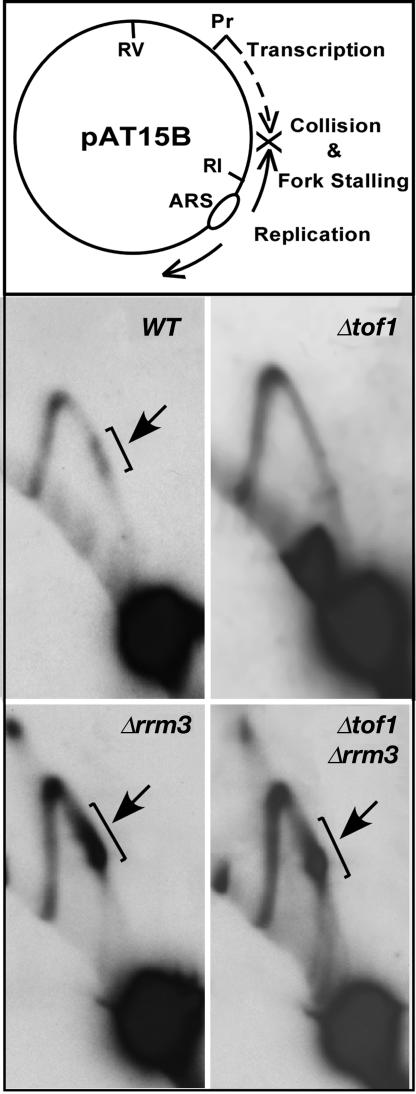
The Tof1p–Csm3p Complex Also Acts Against Rrm3p to Preserve Transcription-Dependent Fork Stalling. We wished to determine whether Tof1p and Csm3p also protected stalled forks at other regions of the chromosome by counteracting Rrm3p. In budding yeast, encounters between transcription initiated from a promoter and extended by RNA polymerase III and a replication fork moving toward it from the opposite direction causes polar pausing of forks (35). We made use of this observation to investigate whether Tof1p and Csm3p also counteracted the possible impact of Rrm3p on transcription-induced fork stalling in a plasmid context. We followed replication fork movement in the plasmid pAT15B (35) in the WT cells and in cells containing the following deletions: Δrrm3, Δtof1, and Δtof1Δrrm3. The plasmid replication intermediates of pAT15B prepared from these strains were analyzed by 2D gel electrophoresis. The plasmid construct is shown in Fig. 6 Top. As shown in Fig. 6 Bottom, the WT cells showed two closely spaced regions of fork arrest (Fig. 6, WT, arrow). In contrast, the pause was almost completely eliminated in the Δtof1 cells (Fig. 6). We then analyzed the collision-induced fork arrest in Δrrm3 and Δrrm3Δtof1 double deletion strains and discovered that (Fig. 6 Bottom), whereas fork stalling was almost completely absent in the Δtof1 cells, the Δrrm3 cells showed a major increase in the arrested forks in comparison with the WT and Δtof1 cells. In the Δrrm3Δtof1 cells, the intensity of the pause sites was much higher than that of the WT cells and, of course, that of the Δtof1 cells. The results showed that the protective action of Tof1p and Csm3p also extended to fork pausing outside the chromosomal rDNA, during normal replication.
Fig. 6.
Effects of Tof1p and Rrm3p on forks stalled by encounter between a RNA polymerase III transcription complex and a fork coming from the opposite direction. (Top) Diagram showing the structure of the plasmid pAT15B. (Middle and Bottom) Autoradiograms of 2D gels showing that encounter between transcription emanating from a tRNA promoter (by RNA pol III) and the fork moving counterclockwise from the ARS generate two elongated spots in WT cells (see parentheses and arrow); the spots are either missing or greatly attenuated in the Δtof1 and significantly enhanced in the Δrrm3 cells. Double deletion of the two genes (see Δrrm3Δtof1) restored the forks to a level significantly higher than that of the WT cells.
Discussion
In this paper, we have shown, both by 2D gel analysis of fork progression and by ChIP analyses, that the Tof1p–Csm3p protein complex of S. cerevisiae is needed for stable fork arrest at the natural Ter sites of rDNA and at RNA polymerase III transcription-induced fork arrest caused by the encounter of a replication fork with transcription approaching from the opposite direction. We have shown further that the checkpoint protein complex appears to function by protecting terminated or stalled forks from the fork-dislodging activity of the Rrm3p helicase either directly or indirectly, thereby preventing fork escape from the Ter and other sites. Rrm3p is known to remove nonhistone proteins from in front of a replication fork (34, 38), similar to the action of the dda helicase of phage T4 (45). Thus, a subset of intra-S-phase checkpoint proteins appears to control fork passage through Ter sites and other barriers, such as those created by transcribing RNA polymerase III, during normal DNA replication.
How does the Tof1p–Csm3p complex restrain unfettered Rrm3p activity? Is the inhibition direct or indirect? The precise answer to this question will have to await the purification of the relevant proteins and in vitro biochemical work. However, regarding a mechanism, the following consideration should be kept in mind. The Rrm3p helicase acts as a powerful “sweepase” that removes nonhistone proteins bound to several regions of the yeast chromosome (31, 33, 38). In fact, the observation that the termination spot in 2D gels is enhanced by a factor of ≈2 in Δrrm3 cells in comparison with the WT cells suggests that, even in the presence of functional Tof1p and Csm3p, a certain amount of Ter-bound Fob1p is being transiently displaced by Rrm3p. We have proposed a simple model to explain the control of fork arrest (Fig. 1B). Rrm3p is recruited to a replication fork by proliferating cell nuclear antigen (46) and probably moves with the replisome until it encounters a bound Fob1p (or other DNA-bound proteins) and transiently displaces it from the Ter site, much in the way that the dda helicase of phage T4 displaces RNA polymerase from DNA (45). An indirect mechanism could conceivably be that, without Tof1p–Csm3p, the replisome would be in a more open conformation that would allow less-restricted Rrm3p activity, thereby causing unfettered fork release from the Ter sites. Alternatively, the Tof1p–Csm3p complex may be directly inhibiting the helicase activity. Development of an in vitro system for investigating fork arrest and purification of several proteins, such as Tof1p, Csm3p, Rrm3p, and the minichromosome maintenance (MCM) proteins and investigations of their activities in vitro in fork arrest will be needed in the future to distinguish between the alternative mechanisms mentioned above.
Does Rrm3p really remove the Ter-bound Fob1p and lead to detectable vacant Ter sites in vivo? We have taken a step in this direction by asking whether Rrm3p actually displaced Fob1p from Ter sites by performing normal PCR and quantitative PCR in α factor arrest-induced synchronized populations of the WT and the isogenic Δtof1 cells to measure Fob1p-bound Ter DNA by ChIP. We found no detectable difference between the two, i.e., the frequency of occupation of Ter sites by Fob1p was indistinguishable between the two types of cells (Fig. 9, which is published as supporting information on the PNAS web site). Perhaps this is not surprising, because the helicase could have transiently displaced the Fob1p from DNA and the detection of this displacement might require the presence of an excess of a Fob1p-binding trap consisting of an excess of Ter DNA (47). An experiment using an excess of Fob1p-binding trap DNA could be done in vitro in the future to look for possible eviction of Fob1p from the Ter by the Rrm3p helicase.
The Tof1p and Csm3p proteins are homologous not only with swi1 and swi3 of S. pombe but also with the mammalian “Timeless” (48) and the Timeless-interacting protein, TIPIN (49), respectively. It is interesting to note that the timeless gene of worms is involved in chromosome cohesion (50). A single homologue of the Rrm3/Pif1p family of helicases has been found in the distantly related fission yeast (51) and in mammalian cells (31). In view of the apparent conservation of the key proteins, the mechanism of control of fork arrest reported in this work could be evolutionarily conserved.
While this work was being reviewed, two other groups also reported that Tof1p–Csm3p, but not Mrc1p, are needed for stable fork arrest at the Ter sites of yeast (52, 53). However, we wish to point out that, unlike the above-mentioned reports, this paper identifies Rrm3p helicase as at least one of the major activities against which the Tof1p–Csm3p checkpoint proteins, but not Mrc1p, Wss1p, and Psy2p, protect terminated or stalled replication forks during normal DNA replication, thus moving closer to providing a mechanism of fork protection and release at the natural Ter sites. We have shown further that two other helicases, namely Sgs1p and Srs2p, do not appear to promote significant removal of arrested forks from Ter.
Experimental Procedures
Strains and Plasmids. Yeast strains BY4742 (MAT α his3-1 leu2 lys2 ura3), EMY74.7 (Mat a his3-Δ1 leu2-3, –112 trp1D ura3-52), and RDKY3615 (Mat a ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 yel069::URA3) and their derivatives were used in all experiments of 2D gel analysis. YB511 W303 MATa dbf2-1b MCM4(T7)-His-3 MCM6(HA3)-LEU2 MCM7(Myc7)-URA3 that was used for the ChIP analysis was a gift from Bruce Stillman (43). The plasmid pAT15B (35), shown in Fig. 6 and used in experiments on RNA polymerase III transcription-induced replication fork arrest, was a gift from Carol Newlon (35).
2D Gel Electrophoresis. Replication intermediates for 2D gel analysis were prepared and analyzed as described [refs. 54 and 55, and as modified (ref. 13)].
ChIP. ChIP analysis was performed exactly as described (44). The sequences of the oligonucleotide primer pairs used to amplify the immunoprecipitated Ter, and the control (segments of the 35S DNA) DNA fragments have been presented in Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Drs. Virginia Zakian (Princeton University, Princeton), Carol Newlon (University of Medicine and Dentistry, Newark, NJ), Mark Longtine (Oklahoma State University, Stillwater, OK), Richard Kolodner (University of California, San Diego), Phil Hieter (University of British Columbia, Vancouver), Steve Bell (Massachusetts Institute of Technology, Cambridge, MA), Louise and Satya Prakash (University of Texas Medical Branch, Galveston, TX), and Bruce Stillman (Cold Spring Harbor Laboratory, Plainview, NY) for many gifts of strains and plasmids. We acknowledge the technical support of the DNA sequencing core facility of Medical University of South Carolina. This work was supported by grants from the National Institute of Allergy and Infectious Diseases and the National Institute of General Medical Sciences of the National Institutes of Health (to D.B.).
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: ChIP, chromatin immunoprecipitation.
References
- 1.Brewer, B. J. & Fangman, W. L. (1988) Cell 55, 637–643. [DOI] [PubMed] [Google Scholar]
- 2.Ward, T. R., Hoang, M. L., Prusty, R., Lau, C. K., Keil, R. L., Fangman, W. L. & Brewer, B. J. (2000) Mol. Cell. Biol. 20, 4948–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linskens, M. H. K. & Huberman, J. A. (1988) Mol. Cell. Biol. 8, 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastia, D. & Mohanty, B. K. (1996) in DNA Replication in Eukaryotic Cells, ed. DePamphilis, M. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 177–215.
- 5.Khatri, G. S., MacAllister, T., Sista, P. R. & Bastia, D. (1989) Cell 59, 667–674. [DOI] [PubMed] [Google Scholar]
- 6.Lee, E. H., Kornberg, A., Hidaka, M., Kobayashi, T. & Horiuchi, T. (1989) Proc. Natl. Acad. Sci. USA 86, 9104–9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaul, S., Mohanty, B. K., Sahoo, T., Patel, I., Khan, S. A. & Bastia, D. (1994) Proc. Natl. Acad. Sci. USA 91, 11143–11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulugu, S., Potnis, A., Shamsuzzaman, Taylor, J., Alexander, K. & Bastia, D. (2001) Proc. Natl. Acad. Sci. USA 98, 9569–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manna, A. C., Pai, K. S., Bussiere, D. E., Davies, C., White, S. W. & Bastia, D. (1996) Cell 87, 881–891. [DOI] [PubMed] [Google Scholar]
- 10.Rothstein, R., Michel, B. & Gangloff, S. (2000) Genes Dev. 14, 1–10. [PubMed] [Google Scholar]
- 11.Brewer, B. J., Lockshon, D. & Fangman, W. (1992) Cell 71, 267–271. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez, J. A., Kim, S. M. & Huberman, J. A. (1998) Exp. Cell Res. 238, 220–230. [DOI] [PubMed] [Google Scholar]
- 13.Mohanty, B. K. & Bastia, D. (2004) J. Biol. Chem. 279, 1932–1941. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi, T. (2003) Mol. Cell. Biol. 23, 9178–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalgaard, J. Z. & Klar, A. J. (2000) Cell 102, 745–751. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Gorostiaga, A., Lopez-Estrano, C., Krimer, D. B., Schvartzman, J. B. & Hernandez, P. (2004) Mol. Cell. Biol. 24, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krings, G. & Bastia, D. (2004) Proc. Natl. Acad. Sci. USA 101, 14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krings, G. & Bastia, D. (2005) J. Biol. Chem. 280, 39135–39142. [DOI] [PubMed] [Google Scholar]
- 19.Mejia-Ramirez, E., Sanchez-Gorostiaga, A., Krimer, D. B., Schvartzman, J. B. & Hernandez, P. (2005) Mol. Cell. Biol. 25, 8755–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noguchi, E., Noguchi, C., Du, L. L. & Russell, P. (2003) Mol. Cell. Biol. 23, 7861–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguchi, E., Noguchi, C., McDonald, W. H., Yates, J. R., III, & Russell, P. (2004) Mol. Cell. Biol. 24, 8342–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren, C. D., Eckley, D. M., Lee, M. S., Hanna, J. S., Hughes, A., Peyser, B., Jie, C., Irizarry, R. & Spencer, F. A. (2004) Mol. Biol. Cell 15, 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer, M. L., Pot, I., Chang, M., Xu, H., Aneliunas, V., Kwok, T., Newitt, R., Aebersold, R., Boone, C., Brown, G. W. & Hieter, P. (2004) Mol. Biol. Cell 15, 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foss, E. J. (2001) Genetics 157, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foss, E. J. (2000) Cold Spring Harb. Symp. Quant. Biol. 65, 347–351. [DOI] [PubMed] [Google Scholar]
- 26.Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K. & Shirahige, K. (2003) Nature 424, 1078–1083. [DOI] [PubMed] [Google Scholar]
- 27.Zegerman, P. & Diffley, J. F. (2003) Nat. Struct. Biol. 10, 778–779. [DOI] [PubMed] [Google Scholar]
- 28.Mayer, M. L., Pot, I., Chang, M., Xu, H., Aneliunas, V., Kwok, T., Newitt, R., Aebersold, R., Boone, C., Brown, G. W. & Hieter, P. (2004) Mol. Biol. Cell 15, 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcasabas, A. A., Osborn, A. J., Bachant, J., Hu, F., Werler, P. J., Bousset, K., Furuya, K., Diffley, J. F., Carr, A. M. & Elledge, S. J. (2001) Nat. Cell Biol. 3, 958–965. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, K. & Russell, P. (2001) Nat. Cell Biol. 3, 966–972. [DOI] [PubMed] [Google Scholar]
- 31.Ivessa, A. S., Zhou, J. Q., Schulz, V. P., Monson, E. K. & Zakian, V. A. (2002) Genes Dev. 16, 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres, J. Z., Bessler, J. B. & Zakian, V. A. (2004) Genes Dev. 18, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres, J. Z., Schnakenberg, S. L. & Zakian, V. A. (2004) Mol. Cell. Biol. 24, 3198–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivessa, A. S., Zhou, J. Q. & Zakian, V. A. (2000) Cell 100, 479–489. [DOI] [PubMed] [Google Scholar]
- 35.Deshpande, A. M. & Newlon, C. S. (1996) Science 272, 1030–1033. [DOI] [PubMed] [Google Scholar]
- 36.O'Neill, B. M., Hanway, D., Winzeler, E. A. & Romesberg, F. E. (2004) Nucleic Acids Res. 32, 6519–6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hazbun, T. R., Malmstrom, L., Anderson, S., Graczyk, B. J., Fox, B., Riffle, M., Sundin, B. A., Aranda, J. D., McDonald, W. H., Chiu, C. H., et al. (2003) Mol. Cell 12, 1353–1365. [DOI] [PubMed] [Google Scholar]
- 38.Ivessa, A. S., Lenzmeier, B. A., Bessler, J. B., Goudsouzian, L. K., Schnakenberg, S. L. & Zakian, V. A. (2003) Mol. Cell 12, 1525–1536. [DOI] [PubMed] [Google Scholar]
- 39.Weitao, T., Budd, M. & Campbell, J. L. (2003) Mutat. Res. 532, 157–172. [DOI] [PubMed] [Google Scholar]
- 40.Versini, G., Comet, I., Wu, M., Hoopes, L., Schwob, E. & Pasero, P. (2003) EMBO J. 22, 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfander, B., Moldovan, G. L., Sacher, M., Hoege, C. & Jentsch, S. (2005) Nature 436, 428–433. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, K. H. & Kolodner, R. D. (2004) Mol. Cell. Biol. 24, 3213–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinreich, M., Liang, C. & Stillman, B. (1999) Proc. Natl. Acad. Sci. USA 96, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aparicio, O. M., Weinstein, D. M. & Bell, S. P. (1997) Cell 91, 59–69. [DOI] [PubMed] [Google Scholar]
- 45.Bedinger, P., Hochstrasser, M., Jongeneel, C. V. & Alberts, B. M. (1983) Cell 34, 115–123. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, K. H., Derry, K. L. & Kolodner, R. D. (2002) J. Biol. Chem. 277, 45331–45337. [DOI] [PubMed] [Google Scholar]
- 47.Mohanty, B. K., Sahoo, T. & Bastia, D. (1998) J. Biol. Chem. 273, 3051–3059. [DOI] [PubMed] [Google Scholar]
- 48.Zylka, M. J., Shearman, L. P., Levine, J. D., Jin, X., Weaver, D. R. & Reppert, S. M. (1998) Neuron 21, 1115–1122. [DOI] [PubMed] [Google Scholar]
- 49.Gotter, A. L. (2003) J. Mol. Biol. 331, 167–176. [DOI] [PubMed] [Google Scholar]
- 50.Chan, R. C., Chan, A., Jeon, M., Wu, T. F., Pasqualone, D., Rougvie, A. E. & Meyer, B. J. (2003) Nature 423, 1002–1009. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, J. Q., Qi, H., Schulz, V. P., Mateyak, M. K., Monson, E. K. & Zakian, V. A. (2002) Mol. Biol. Cell 13, 2180–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calzada, A., Hodgson, B., Kanemaki, M., Bueno, A. & Labib, K. (2005) Genes Dev. 19, 1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tourriere, H., Versini, G., Cordon-Preciado, V., Alabert, C. & Pasero, P. (2005) Mol. Cell 19, 699–706. [DOI] [PubMed] [Google Scholar]
- 54.Huberman, J. A., Spotila, L. D., Nawotka, K. A., el-Assouli, S. M. & Davis, L. R. (1987) Cell 51, 473–481. [DOI] [PubMed] [Google Scholar]
- 55.Brewer, B. J. & Fangman, W. (1987) Cell 51, 463–471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.