Abstract
Bacteroides fragilis, a component of the normal intestinal flora, is an obligate anaerobe capable of long-term survival in the presence of air. Survival is attributed to an elaborate oxidative stress response that controls the induction of more than 28 peptides, but there is limited knowledge concerning the identities of these peptides. In this report, RNA fingerprinting by arbitrarily primed PCR identified five new genes whose expression increased following exposure to O2. Nucleotide sequence analysis of the cloned genes indicated that they encoded an outer membrane protein, an aspartate decarboxylase, an efflux pump, heat shock protein HtpG, and an NrdA ortholog constituting the large subunit of a class Ia ribonucleotide reductase (RRase). Attention was focused on the nrdA gene since class I RRases are obligate aerobic enzymes catalyzing the reduction of ribonucleoside 5′-diphosphates by a mechanism that requires molecular oxygen for activity. Sequence analysis of the nrd locus showed that two genes, nrdA and nrdB, are located in the same orientation in a 4.5-kb region. Northern hybridization and primer extension experiments confirmed induction of the genes by O2 and suggested they are an operon. The B. fragilis nrdA and nrdB genes were overexpressed in Escherichia coli, and CDP reductase assays confirmed that they encoded an active enzyme. The enzyme activity was inhibited by hydroxyurea, and ATP was shown to be a positive effector of CDP reductase activity, while dATP was an inhibitor, indicating that the enzyme was a class Ia RRase. A nrdA mutant was viable under anaerobic conditions but had decreased survival following exposure to O2, and it could not rapidly resume growth after O2 treatment. The results presented indicate that during aerobic conditions B. fragilis NrdAB may have a role in maintaining deoxyribonucleotide pools for DNA repair and growth recovery.
Compared to other Bacteroides species, Bacteroides fragilis is a minor component of the indigenous intestinal tract microflora, yet it is the most commonly isolated anaerobic pathogen found in clinical samples (9). Several virulence factors may contribute to the success of B. fragilis as an opportunistic pathogen, including the complex capsular polysaccharide matrix and a neuraminidase (5, 10, 40). In addition, this anaerobe is extremely aerotolerant, capable of surviving more than 48 h of exposure to O2, and it is thought that this may play some role in the initiation or persistence of infection (32). Further, it may be important for the indigenous anaerobes to withstand oxidative stress during the process of transmission between hosts and for the initial steps in colonization of the intestinal tract. Regardless of the benefits, the physiological and genetic basis for this aerotolerance is not well understood but it has become clear that the organism mounts an elaborate response, which is required for survival. This oxidative stress response (OSR) is complicated, in part because anaerobes do not replicate in the presence of air, and accordingly more than 28 peptides are induced by O2 (or H2O2) but an even greater number are repressed, all in a rapid, coordinated fashion (27).
Several strategies have been used to identify components of the B. fragilis OSR, and most have focused on the identification of genes or gene products that are up-regulated by O2 or H2O2. Most of the genes identified to date are involved in detoxification or protection from oxygen radicals. In B. fragilis, catalase (katB), superoxide dismutase (sod), alkyl hydroperoxide reductase (ahpCF), and Dps (nonspecific DNA binding protein; dps) have been demonstrated to increase dramatically during exposure to oxidative stress (12, 13, 26, 28, 31). For KatB, Dps, and AhpCF, mutational studies have shown that each plays a role in protection of the organism, but interestingly none of these mutants has an exceptionally severe oxidative stress survival defect. This suggests that there is considerable redundancy built into the protective component of the OSR, but much less is known concerning the repair or adaptive aspects of the response.
Coordinated expression of the OSR genes is in part mediated by the redox-sensitive regulator OxyR, which controls katB, ahpCF, and dps in B. fragilis as well as several additional OSR loci 26; C. D. Herren, unpublished data). The OxyR-regulated genes are induced equally by either H2O2 or O2, but the actual effector molecule(s) is not yet known. Thus there is a significant relationship among the genes induced by molecular oxygen and those induced by H2O2. In fact, 23 of 28 induced peptides were identical in cultures treated with either of these agents, indicating considerable overlap in the response (27). This stands in contrast to the limited overlap in Escherichia coli, where there are separate superoxide and peroxide responses, each of which is controlled by an independent transcriptional regulator (4, 11). The OxyR regulon in E. coli is a peroxide-specific response, which has been shown to include AhpCF, KatG, Dps, Fur, TrxC, and GorA (38). Recent results using microarray technology have identified new members of the E. coli OxyR regulon, and there are now more than 30 genes that are thought to be part of this regulon (41). Among the newly identified OxyR-controlled proteins are several whose function is involved in macromolecule repair.
To identify novel gene products important for oxidative stress that are not directly involved in detoxification reactions, we used RNA fingerprinting by random arbitrarily primed PCR (RAP-PCR) to screen for unique mRNA sequences that were induced during exposure of B. fragilis to atmospheric O2. Here we make the novel observation that one of the induced genes, nrdA, encodes the large subunit for a class I, aerobic-type ribonucleotide reductase (RRase). RRase enzymes catalyze the synthesis of deoxyribonucleotides required for DNA synthesis in all living cells, and the presence of an obligate aerobic-class enzyme in an anaerobe was unexpected. We demonstrated that the B. fragilis nrdAB genes were in an operon induced by O2 and that the genes encoded an active RRase. Our results led us to speculate that NrdAB could supply the deoxyribonucleotides needed to repair oxidatively damaged DNA.
MATERIALS AND METHODS
Strains and growth conditions.
E. coli strain DH10B (F− mcrA Δ[mrr-hsd RMS-mcrBC] φ80d lacZ ΔlacX74 endA1 recA1 deoR Δ[ara, leu]7697 araD139 galU galK nupG rpsL λ−) was used for routine analyses, and E. coli strain BL21(DE3) (F− ompT hsdSB[rB− mB−] gal dcm) was used for expression of recombinant NrdA and NrdB. E. coli was grown aerobically in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) or spectinomycin (40 μg/ml).
B. fragilis 638R (rifampin resistant [24]) and its isogenic derivatives, IB263 [oxyR(Con) (26, 30)], IB267 (nrdA; see below), and IB271 (638ΩermF [26]) were grown anaerobically in the complex medium BHIS (34) or a chemically defined medium (27). To induce oxidative stress, cultures were grown anaerobically at 37°C to mid-logarithmic phase (2 × 108 per ml) and then split. One half was shaken (250 rpm) in air at 37°C in a conical flask (10:1 culture-to-flask ratio) for 1 h, while the remaining (anaerobic) half was immediately harvested by centrifugation at 4°C.
DNA manipulation and sequence analysis.
Routine DNA analyses were performed according to standard protocols (33), and B. fragilis chromosomal DNA was isolated from 5-ml cultures as described previously (34). For Southern hybridizations probes were generated by random oligonucleotide priming (Pharmacia LKB, Piscataway, N.J.) with [32P]dCTP.
Nucleotide sequences were determined by automated DNA sequencing (Molecular Biology Resource Facility, University of Tennessee, Knoxville) using a primer walking strategy. Nucleotide and protein sequences were compiled and analyzed using the University of Wisconsin Genetics Computer Group (GCG) DNA sequence analysis software (6). Parsimony analysis was generated with the PHYLIP phylogeny inference package (version 3.5) (8) from an alignment of peptide sequences derived using the GCG Pileup program.
Northern hybridizations and primer extension analysis.
RNA isolation using the hot phenol method and Northern blot analysis using formaldehyde gels have been described previously (29). 32P-labeled probes were generated as described above or by incorporation of [α-32P]dCTP during PCR amplification. Primer extension mapping of transcriptional start sites was carried out essentially as previously described using 50 μg of total RNA (29). Primers were end labeled with [γ-32P]dATP, and reaction products were electrophoresed on 6% polyacrylamide gels containing urea.
RAP-PCR analysis.
The general method of Sokolov and Prockop (36) was used for the generation and analysis of the RAP-PCR gels. Briefly, a random pool of cDNAs was generated with 400 U of Superscript II RNaseH reverse transcriptase (Life Technologies, Gaithersburg, Md.) from total RNA (5, 50, or 100 μg) primed with 100 ng of fully degenerate hexamers. Next, pairs of arbitrarily derived 10-mer oligonucleotide primers (Genosys, The Woodlands, Tex.) were used to amplify aliquots of the cDNA with the Stoffel fragment of Taq polymerase (Perkin-Elmer Corp., Norwalk, Conn.) in the manufacturer's buffer with the addition of 50 μCi of [α-32P]dCTP under the following amplification conditions: 45 cycles at 94°C for 1 min, 35°C for 1 min, and 72°C for 1 min, followed by 1 cycle of 72°C for 10 min. The resulting products were electrophoresed on nondenaturing polyacrylamide gels buffered with 1× TTE (88 mM Tris-HCl, 88 mM taurine, 0.5 mM EDTA). Following electrophoresis the gel was exposed to film overnight. Autoradiographs then were aligned over the gel, putative RAPs were excised and boiled in 100 μl of 20 mM Tris (pH 8.0) for 5 min to elute the DNA, and then 5 μl was amplified with Taq polymerase by using the same pair of arbitrary primers.
Cloning of RAPs and cognate genes.
RAPs were purified from agarose gels and ligated into the SmaI site of B. fragilis suicide vector pFD516 (spectinomycin [aad9] and erythromycin [ermF] resistance [35]). The plasmid inserts then were sequenced, and where relevant they were transferred to B. fragilis 638R by triparental mating (14), resulting in single-crossover insertions into the chromosome. These crossover events in the transconjugants were confirmed by Southern hybridization. The nrdA mutant strain, IB267, was derived from the single-crossover insertion of RAP osr4 cloned into pFD516.
To clone RAP-associated genes, chromosomal DNA from the B. fragilis transconjugants was digested with SmaI (no intact sites present in the plasmid), purified, precipitated with ethanol, and then suspended in 25 μl of Tris-EDTA buffer. Approximately 300 ng of the digested DNA was self-ligated with T4 DNA ligase in a 20-μl volume and electroporated into E. coli DH10B. Spectinomycin-resistant clones were examined for pFD516 containing the RAP fragment and flanking chromosomal DNA.
Viable-count assays and recovery curves.
Viable-count and recovery curve assays used IB267 (nrdA) and control strain IB271. Both strains contain a single copy of the ermF gene in the chromosome to account for any possible growth defects caused by the erythromycin resistance phenotype. For control strain IB271 the suicide vector pFD516 was inserted in the chromosome in an irrelevant site (bglA gene) (26, 30). Cultures were grown to about 2 × 108 cells/ml in either BHIS or chemically defined media and then removed from the anaerobic chamber and shaken (250 rpm) in air at 37°C as described above. Viable counts were taken immediately prior to oxidative stress and at 24-h intervals for up to 96 h (27).
For recovery curves IB267 (nrdA) and control IB271 were exposed to O2 in BHIS broth and, after 48 h of exposure to O2, approximately 107 CFU (as determined from previous viable count assays) were harvested by centrifugation. The supernatants were removed, and the pellets were transferred to the anaerobic chamber, where they were suspended in 1 ml of reduced BHIS broth. This was then used to inoculate 9 ml of reduced BHIS, and the A550 of the culture was monitored every hour until growth was observed.
Expression of NrdA and NrdB in pET16b.
The nrdA and nrdB open reading frames (ORFs) were amplified by PCR individually from the chromosome of B. fragilis 638R using Hi-Fidelity Taq polymerase (Life Technologies). The amplified products were cloned into pGem-T. The nrdA and nrdB ORFs were isolated from the resulting plasmids by digestion with SalI and NdeI and ligated into the XhoI and NdeI sites of pET16b (Novagen, Darmstadt, Germany) in frame with the codons for the six-His tag. The nrdA and nrdB constructs were confirmed by nucleotide sequencing. Both pET16b:nrdA and pET16b:nrdB were moved into permissive host E. coli BL21(DE3), which expresses T7 polymerase. A time course of induction with IPTG was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and used to confirm expression of the two proteins.
RRase assays.
Cell extracts from E. coli BL21(DE3) containing pET16b, pET16b:nrdA, or pET16b:nrdB were prepared from mid-logarithmic-phase cultures induced with IPTG for 2 h. Cultures were harvested, and the pellet was washed once in ice-cold 20 mM Tris-HCl (pH 7.0)-1 mM dithiothreitol. Following resuspension in the same buffer, cells were lysed by sonication and cell debris was removed by centrifugation.
CDP reductase activity was assayed as described by Steeper and Steuart (37). The reaction mixtures contained 20 mM sodium phosphate buffer, 6 mM magnesium acetate, 9 mM dithiothreitol, 1.2 mM ATP (unless stated otherwise), and 10 μM [2-14C]CDP (Moravek Biochemicals, Brea, Calif.) in a final volume of 100 μl. The reaction was started by the addition of cell extract, and the reaction mixture was incubated at 37°C for 30 min. The reaction was stopped by boiling, and then the reactants and products were dephosphorylated with snake venom phosphodiesterase I (Sigma-Aldrich Co., St. Louis, Mo.) for 1.5 h at 37°C. The reactants and products were separated on Dowex-1-borate columns equilibrated with deionized water, which retained the ribonucleosides. The column was washed with 4 ml of deionized water, and the concentration of deoxyribonucleoside was determined by liquid scintillation counting. One unit of CDP reductase is the amount of enzyme that produces 1 pmol of dCDP per min at 37°C. Specific activity is expressed as units per milligram of crude extract.
Nucleotide sequence accession numbers.
The GenBank accession numbers obtained during this study are as follows: RAP products osr1 to osr5, AF055866 to AF055870, respectively; complete genes nrdAB, AY043208; htpG, AF404759; asdA, AF404820.
RESULTS
Identification of oxygen-induced genes.
Based largely on the identification of several abundant proteins in constitutively active oxyR(Con) strain IB263, previous work has recognized several highly expressed OSR genes in B. fragilis (30). These genes, such as katB, ahpC, dps, and sod, generally encode detoxification and protective enzymes. To obtain a more balanced picture of the global OSR, RAP-PCR was used to enrich for genes expressed during O2 exposure. Although there are a number of strategies available for the identification of differentially expressed genes in procaryotes, many depend on a well-developed genetic system (e.g., transposon fusions are not available in B. fragilis), but RAP-PCR can be applied to any organism from which RNA can be reliably isolated.
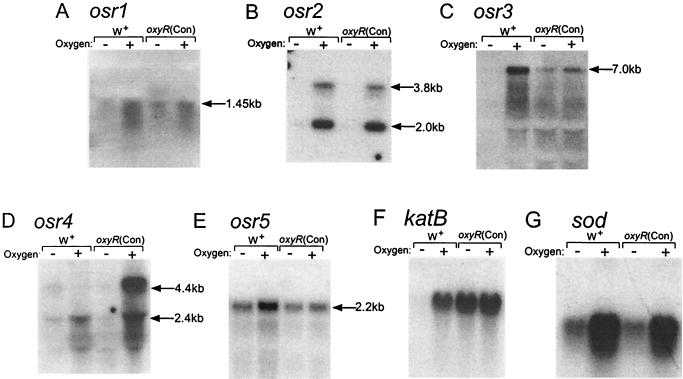
Complex RNA fingerprints from cultures of the oxyR(Con) mutant and wild-type strain 638R with and without O2 were generated with pairs of arbitrary 10-mer oligonucleotide primers following 45 amplification cycles. The patterns were compared, and unique bands found in both of the O2-exposed cultures were reamplified using the same pairs of primers. The products generated (RAPs) were cloned and then used as probes in Northern hybridization experiments to confirm their induction by O2. Of the 55 RAPs examined, five proved to be up-regulated by O2 and were designated osr1 to -5 (for OSR). As shown in Fig. 1, the Northern analysis revealed the presence of three expression classes among the osr genes compared to patterns seen with katB and sod. The katB gene, regulated by OxyR, is representative of the peroxide response and shows constitutive expression, under anaerobic or aerobic conditions, in the oxyR(Con) mutant, whereas sod was highly up-regulated by O2 in an OxyR-independent manner. The sod class was represented by osr1, osr2, and osr4, and osr3 was the only RAP that seemed to fit in the oxyR-controlled group, being constitutively expressed in the oxyR(Con) mutant. One gene, osr5, seemed not to fit well in either of the established classes, as it was increased in the parent strain during exposure to O2 but not in the oxyR(Con) mutant.
FIG. 1.
Autoradiographs of Northern blots probed with O2-induced RAPs. Mid-log-phase cultures of wild-type strain 638R (W+) and strain IB263 [oxyR(Con)] were grown anaerobically (−) or stressed by aerobic incubation for 1 h (+). (A to E) Probing with the RAP products; (F) probing with an internal katB fragment; (G) probing with an internal sod fragment. All lanes contain 50 μg of total RNA except for those in panel A, which contain 80 μg/lane. Approximate sizes of the hybridizing transcripts in kilobases are shown and were extrapolated from a molecular weight ladder in an adjacent lane.
Sequence analysis of oxygen-induced RAPs.
The lengths of the RAPs were consistently short, ranging from 93 to 240 bp, and this precluded accurate identification of the cognate genes by standard GenBank database searches. Thus, the complete ORFs corresponding to the RAPs were cloned and sequenced. Using the complete nucleotide sequence, a diverse set of genes was identified (Table 1). Osr1 is related to a family of cation efflux pumps, originally identified as CzcD, which was implicated in metal resistance (Co, Zn, and Cd) (22). Osr2, which appeared to be the most tightly controlled by exposure to O2, was related most closely to an aspartate decarboxylase encoded by asdA from Alcaligenes faecalis (3). Homology to AsdA was present over the entire length of the molecule, but there also was a classic pyridoxal phosphate binding domain present in the central region of each of these proteins, which has very high similarity to those of aminotransferase proteins. Gene osr3 was transcribed as part of a large operon, nearly 7 kb. The putative gene product had homology to the Sus family of outer membrane proteins from the closely related organism Bacteroides thetaiotaomicron (25) and to RagA, the major cell surface protein in Porphyromonas gingivalis (2).
TABLE 1.
Sequence analysis of oxygen-induced RAPsa
| RAP | Size (bp) | Corresponding gene; size (bp) | Closest homolog; % identityc | Putative function |
|---|---|---|---|---|
| osr1 | 93 | czcDb; 909 | Caulobacter crescentus; 46 | Cation efflux pump |
| osr2 | 226 | asdA; 1,647 | A. faecalis; 46 | Aspartate decarboxylase |
| osr3 | 181 | susCb; 3,006 | B. thetaiotaomicron; 47 | Outer membrane protein; starch uptake |
| osr4 | 240 | nrdA; 2,520 | Treponema pallidum; 60 | RRase |
| osr5 | 143 | htpG; 2,046 | P. gingivalis; 60 | Heat shock protein |
For accession numbers of RAPs and genes nrdAB, htpG, and asdA, see Materials and Methods.
The gene was deduced from the Sanger Center database for B. fragilis ATCC 25285.
The percent identity at the amino acid level.
Heat shock protein HtpG from P. gingivalis was found to be about 60% identical to the putative product encoded by osr5 (19). This is a large protein of 681 residues that is in the family of hsp90s found ubiquitously in procaryotes and eukaryotes and generally thought to be molecular chaperones (23). The last O2-induced RAP was located in the ORF for the ribonucleotide diphosphate reductase gene, nrdA. NrdA is the large subunit of the class Ia RRase, and the ORF product had 60% identity to Treponema pallidum NrdA, which was identified by genome sequence analysis. NrdA was particularly interesting in that the class Ia enzymes function solely under aerobic conditions, and, in fact, NrdAB enzymes utilize molecular oxygen as part of the catalytic mechanism for radical generation (17). In E. coli the NrdAB RRase is essential for DNA replication during aerobic growth, and, since B. fragilis does not replicate during exposure to O2, it was necessary to determine the role of the nrdA locus in this anaerobe.
Characterization of the nrdAB locus.
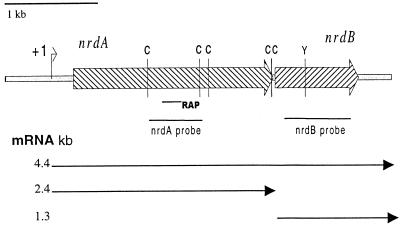
Nucleotide sequence analysis of the nrdA locus revealed two ORFs, which are separated by 28 bp (Fig. 2). The first ORF was 2,520 bp in length and encoded a large protein (839 amino acids) with a predicted molecular mass of 95.8 kDa. The second ORF was 1,050 bp in length and encoded a protein with a predicted molecular mass of 41.3 kDa. Sequence analysis of the two ORFs indicated that they encoded the large (nrdA) and small (nrdB) subunits of a class Ia RRase. This was confirmed by alignments of the peptide sequences of the B. fragilis NrdA with orthologs from other species (data not shown). As summarized in Fig. 2, detailed analysis of the peptide sequence showed that the five conserved cysteine residues, characteristic of the large subunits of class Ia enzymes, are present in the B. fragilis ortholog. In addition, analysis of the NrdB subunit revealed the conserved tyrosyl residue which is responsible for the formation of the oxygen-linked free radical.
FIG. 2.
Genetic map of the nrdAB locus. Hatched arrows, nrdAB ORFs; C, conserved NrdA cysteines; Y, conserved NrdB tyrosine; solid lines, DNA fragments used as probes in Northern hybridizations and location of RAP osr4; +1, transcriptional start site of nrdA. Approximate sizes of the mRNAs are shown by the arrows. All of the features are drawn to scale.
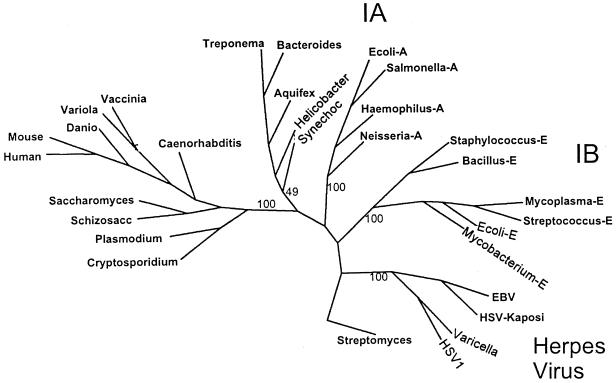
Parsimony analysis of NrdA orthologs strengthened the argument that the B. fragilis enzyme belongs to the class Ia subgroup of enzymes (Fig. 3). The Bacteroides enzyme was located in a cluster of class Ia enzymes that included those of several other genera representing early procaryote lineages. This cluster was distinct from class Ia enzymes of other eubacteria and eukaryotes, but it clearly was not within the herpesvirus group or in the group of class Ib enzymes. Consistent with this finding, the sequence ITKRNG (amino acids 3 to 8), located in the amino terminus of the large subunit, is highly similar to the motif VXKRDG, which is found in class Ia and class III enzymes but not the class Ib enzymes (17). Some weakness in the bootstrap values for the Treponema/Bacteroides group was due primarily to the Synechocystis sp. enzyme which occasionally aligned with the major eubacterial class Ia enzymes.
FIG. 3.
Unrooted phylogenetic tree showing the relationship of B. fragilis NrdA to representative orthologs of class I RRase large subunits. A 148-amino-acid internal region of the peptides was aligned using the PILEUP program from the Wisconsin GCG software package. The parsimony program (Protpars) was used to generate the consensus tree from 100 bootstrap analyses, but not all species used in the alignment are shown on the tree for the sake of clarity. The bootstrap values for the major branches of the tree are shown and the Streptomyces clavuligerus class II enzyme (labeled Streptomyces) was used as the outgroup. Abbreviations: Mycoplasma-E, Mycoplasma genitalium; Streptococcus-E, Streptococcus dysgalactiae; Mycobacterium-E, Mycobacterium tuberculosis; Ecoli-E, E. coli nrdE, Salmonella-A, S. enterica serovar Typhimurium nrdA; Bacillus-E, Bacillus subtilis; Haemophilus-A, Haemophilus influenzae; Neisseria-A, Neisseria meningitidis; Ecoli-A, E. coli nrdA, mouse, Mus musculus; human, Homo sapiens; Danio, Danio rerio; vaccinia, vaccinia virus; variola, variola virus; Caenorhabditis, Caenorhabditis elegans; Schizosacc, Schizosaccharomyces pombe; Saccharomyces, Saccharomyces cerevisiae; Plasmodium, Plasmodium falciparum; Cryptosporidium, Cryptosporidium parvum; Synechoc, Synechocystis spp.; Bacteroides, Bacteroides fragilis; Aquifex, Aquifex aeolicus; Helicobacter, Helicobacter pylori; Treponema, Treponema pallidum, HSV-Kaposi, Kaposi's sarcoma-associated herpesvirus; EBV, Epstein-Barr virus; varicella, varicella-zoster virus; HSV1, human herpes simplex virus type 1.
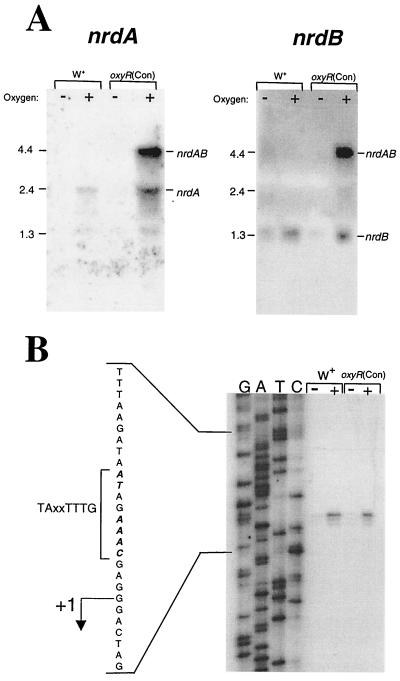
Expression of the nrdAB operon.
As shown above, when RAP osr4 was used as a probe in Northern blots, a 2.4-kb mRNA was observed in cultures induced by O2. In addition, a larger transcript (∼4.4 kb) was observed when the oxyR(Con) mutant was exposed to oxidative stress. Based on the sequence analysis of the nrdA locus it seemed likely that nrdA and nrdB formed a bicistronic operon. Northern hybridizations probed with internal regions of both nrdA and nrdB supported this idea (Fig. 4A). The nrdA-specific probe hybridized to a 2.4-kb mRNA in oxidatively stressed cultures and a second, larger mRNA of about 4.4 kb in strain IB263 exposed to O2. The nrdB-specific probe also hybridized to the 4.4-kb mRNA in stressed cultures of the oxyR(Con) mutant and a smaller 1.35-kb mRNA in both strain 638R and the mutant. In both cases, results with the gene-specific probes corresponded to the predicted size of nrdA or nrdB, implying that both genes may contain their own promoters, but, as clearly seen in IB263, nrdAB also are expressed as an operon, as indicated by the 4.4-kb mRNA (summarized in Fig. 2). It is not known why the 4.4-kb message was seen only in IB263, but recent work from our laboratory indicates that constitutively active OxyR [oxyR(Con)] can substantially increase mRNA stability (E. R. Rocha and C. J. Smith, unpublished data).
FIG. 4.
Transcriptional control of the nrdAB locus. Analysis of anaerobic (−) and oxidatively stressed (+) RNA from strain 638R (W+) and the oxyR(Con) mutant, IB263. Oxidative stress was induced by shaking in air at 37°C for 1 h, and each lane contains 50 μg of total RNA. (A) Northern hybridizations probed with either an nrdA- or nrdB-specific probe. Approximate sizes of the hybridizing transcripts were extrapolated from a molecular weight ladder in an adjacent lane. (B) Primer extension analysis of B. fragilis nrdA with an oligonucleotide primer located at bp 849 to 873c within the nrdA coding sequence. The transcriptional start site of nrdA is indicated (+1), and the −7 consensus promoter motif is shown.
Primer extension analysis was used to map the location of the nrdAB promoters. The nrdA promoter was mapped 271 bp upstream from the translational start site (Fig. 4B). This transcriptional start site was positioned just three nucleotides downstream from consensus B. fragilis −7 transcription promoter sequence TAXXTTTG (1) and was clearly induced by oxidative stress in strains 638R and IB263. The same transcription start site appeared to be used during both induced and uninduced conditions and in either the wild type or the oxyR(Con) mutant.
Several attempts to map the transcriptional start site of nrdB were not successful. Analysis of the nucleotide sequence upstream of the nrdB translational start site also did not reveal any identity to the consensus B. fragilis RNA polymerase promoter. Therefore it is not possible to determine at this time whether the mRNA product seen with Northern hybridization using an nrdB-specific probe was derived from a nrdB-specific promoter or whether it was the result of mRNA processing.
Activity of NrdA and NrdB.
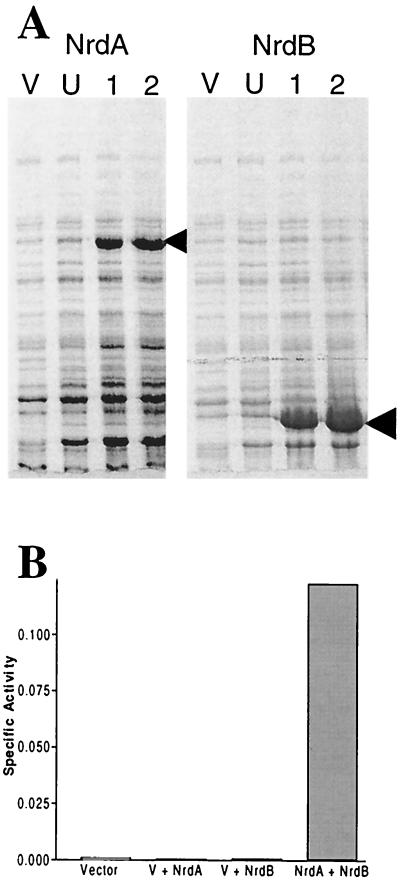
To demonstrate that B. fragilis nrdA and nrdB encode a functional RRase, the genes were cloned individually into E. coli expression vector pET16b. Previous attempts at cloning the genes together in E. coli had indicated that they were lethal (data not shown). Both pET16b:nrdA and pET16b:nrdB produced IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible proteins in E. coli BL21(DE3) (Fig. 5A). The ability of the crude cell extracts from both E. coli strains to reduce CDP was tested. Only when crude extracts from strains containing pET16b:nrdA were mixed with those containing pET16b:nrdB was significant RRase activity observed (Fig. 5B). The highest CDP reductase activity was observed when the concentration of crude cell extract from cells containing nrdB was twice that from cells containing nrdA. This ratio was used to show that CDP reductase activity was linear with respect to increasing concentrations of enzyme (data not shown) and for all subsequent studies. At comparable protein concentrations, no or very low activity was observed in the vector control containing just pET16b and in controls with extracts from cells with either pET16b:nrdA or pET16b:nrdB alone (Fig. 5B). This ruled out the possibility that the activity was derived from E. coli and clearly demonstrated that B. fragilis genes nrdA and nrdB encode an active RRase. In contrast, preliminary studies with oxidatively stressed B. fragilis 638R crude cell extracts and extracts precipitated with ammonium sulfate failed to produce any RRase activity. It is possible that the enzyme was not sufficiently induced or was unstable under the conditions tested.
FIG. 5.
Expression and activity of recombinant B. fragilis RRase. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of strains containing pET16b:nrdA and pET16b:nrdB following induction with IPTG. Cultures were grown with shaking at 37°C in Luria-Bertani broth to an A550 of 0.5 and then induced by the addition of 1 mM IPTG. Lane V, pET16b vector control; lane U, samples taken from the uninduced culture; lanes 1 and 2, samples following IPTG induction for 1 and 2 h, respectively. Arrowheads, major induced products. (B) Measurement of CDP reductase (RRase) activity in cell extracts of E. coli BL21(DE3) containing the vector alone pET16b (25 μg), recombinant B. fragilis NrdA (60 μg) plus the vector (V), recombinant NrdB (60 μg) plus the vector, or NrdA plus NrdB. Each reaction mixture contained positive effector ATP (1.2 mM).
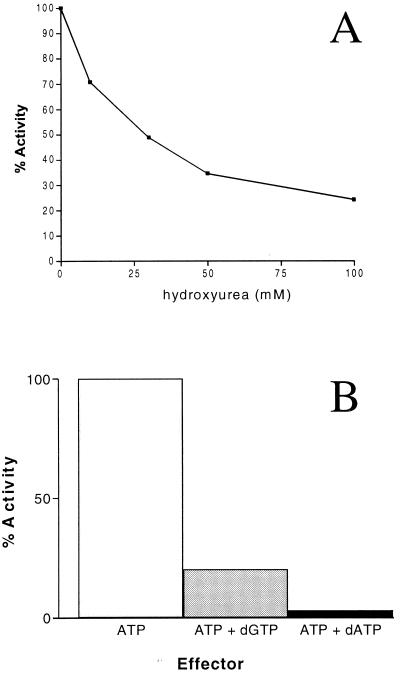
Radical-scavenging molecule hydroxyurea inhibits class I enzymes but not class III RRases. This inhibition is the result of hydroxyurea quenching the tyrosyl-free radical in the small subunit of class I enzymes; in contrast, in class III enzymes the free radical is generated through S-adenosylcobalamin and is resistant to hydroxyurea (7). It can clearly be seen that increasing concentrations of hydroxyurea correlate with a decreased CDP reductase activity (Fig. 6A). This is consistent with the presence of the conserved tyrosine residue in the primary peptide sequence of the B. fragilis NrdB (Fig. 2; GenBank accession no. AY043208).
FIG. 6.
The effect of inhibitors and allosteric effectors on RRase activity. For all reactions 180 μg of total crude cell extract was used (60 μg of NrdA, 120 μg of NrdB), and the results are given as percentages of the activity with positive effector ATP (1 mM) alone, which corresponded to 5.6 U. (A) Effect of increasing concentrations of hydroxyurea on the reduction of CDP by B. fragilis RRase. The results are given as percentages of the activity in the absence of hydroxyurea. (B) Effect of purine deoxynucleoside triphosphates (1 mM) on the activity of B. fragilis RRase in the presence of positive effector ATP (1 mM).
To examine the allosteric regulation of the B. fragilis RRase with respect to the substrate CDP, the abilities of different effector compounds to stimulate activity were examined. As would be expected of a class Ia enzyme, the B. fragilis RRase had the highest level of activity when ATP was present (data not shown). In class Ia enzymes, the reduction of all four ribonucleoside diphosphates is typically inhibited by dATP, and CDP reduction is also inhibited by dGTP. This is not observed in class Ib enzymes, in which dATP is a positive effector of enzyme activity (17). In the presence of positive effector ATP, the B. fragilis enzyme was strongly inhibited by dATP and to a lesser extent dGTP (Fig. 6B), thus confirming that the B. fragilis enzyme belongs to the class Ia subgroup. However it was interesting that relatively high levels of activity were seen in the absence of any effector (data not shown).
Role of nrdAB in oxidative stress.
Experiments described above clearly showed that nrdA and nrdB are induced relatively quickly following oxidative stress (1 to 2 h) but not significantly during anaerobic growth. To determine if NrdAB was required for normal anaerobic growth, a nrdA mutant was constructed. As shown in Fig. 7A, the mutant grew at the same rate as the wild-type control under anaerobic conditions. During the course of another study, we obtained evidence for a B. fragilis class III (anaerobic enzyme) RRase that could be used during anaerobic growth. We fortuitously cloned a segment of the B. fragilis nrdD gene which has significant homology to several NrdD sequences, including those of Haemophilus influenzae (34% identity) and E. coli (32%). The entire gene has now been identified in the B. fragilis genome sequencing project and is located in an apparent operon with nrdG. Northern hybridizations with the nrdD gene segment showed that the gene was constitutively expressed during anaerobic growth, and this suggests that it encodes the major RRase used for normal growth (data not shown).
FIG. 7.
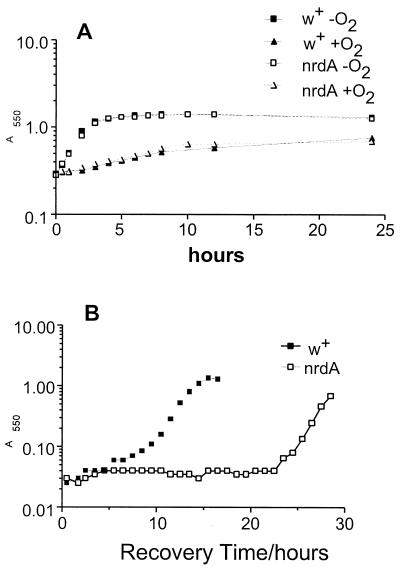
Effect of nrdA mutation on growth of B. fragilis. (A) Growth curves of strains IB267 (nrdA) and IB271 (control, W+) during exposure to O2. At time zero, mid-logarithmic-phase cultures were exposed to O2 by being shaken in air at 37°C or were maintained under anaerobic conditions in stoppered tubes in the anaerobic chamber. Growth was monitored by measurement of A550 at the specified intervals. (B) Recovery curve of B. fragilis IB267 and IB271 following exposure to O2 for 48 h in BHIS. At time zero, approximately 107 viable, stressed bacteria were added to 10 ml of prereduced BHIS. The culture was incubated anaerobically at 37°C, and the A550 was monitored at 1-h intervals until growth was observed. The data presented are representative of the results from three independent experiments.
NrdAB may function to maintain deoxynucleoside triphosphate (dNTP) pools within the cell immediately following oxidative stress, allowing the bacterium to complete any rounds of replication that had previously been initiated. However this hypothesis was not supported by growth curve experiments in which the nrdA mutant was shifted to an aerobic atmosphere (Fig. 7A). In this case there was no difference in the time taken for either the mutant or the control strain (IB271) to complete the last replication cycle following growth inhibition by oxidative stress.
During prolonged exposure to O2 the intracellular deoxyribonucleotide pools are probably depleted due to the repair of damaged DNA. Therefore the presence of an RRase that can function in the presence of O2 may allow B. fragilis to maintain deoxynucleotide pools and thus the integrity of the chromosome. One approach used to test this was to examine the survival of a nrdA mutant following exposure to O2. Viable-count assays showed that nrdA enhanced survival approximately sixfold following a 72-h exposure in complex medium (BHIS), and this effect was somewhat more pronounced in experiments using in a chemically defined medium (data not shown).
Another manifestation of low-deoxynucleotide pools may be a negative impact on the ability of the organisms to resume growth following extended exposure to O2. That is, anaerobes lacking an aerobic-type RRase would not be able to synthesize deoxyribonucleotides under aerobic conditions and this would limit their ability to repair DNA damage and begin synthesis of new chromatin rapidly when anaerobic conditions were reestablished. To test this hypothesis, the ability of a nrdA mutant to resume growth following prolonged O2 exposure was tested (Fig. 7B). Cultures of the mutant and the control were exposed to O2 for 48 h and then adjusted to the same level of viable cells (107 per ml) and returned to the anaerobic chamber. In every case examined, the nrdA mutant took longer to resume growth than the control strain of B. fragilis.
DISCUSSION
The OSR of anaerobic bacteria is an elaborate response mounted to protect the organisms from a lethal stress. In contrast to that of facultative organisms, where most of the research has focused, the OSR of anaerobes takes on added complexity as there is an immediate shift in metabolism to a stationary-phase-like state, where replication ceases and physiology enters into a mode designed to protect macromolecules and prepare for the resumption of growth or long-term survival. Recent studies with anaerobes have resulted in the identification of new defense mechanisms such as the superoxide reductase enzymes first found in Pyrococcus furiosus (16), and analogous systems have now been found in other anaerobic organisms (20). Similarly, in B. fragilis a large 5.6-kb locus containing several putative membrane proteins of unknown function was shown to be required for survival in the presence of air (39). In the present report, we have identified five new genes that are induced upon exposure of B. fragilis to O2. Foremost of these was nrdA, which will be discussed below, but the others also may provide some insight into the challenges facing anaerobes as they are exposed to lethal oxidative stress. The aspartate decarboxylase gene, asdA (osr2), was strongly induced and may have a role in recycling resources. The product of this reaction, β-alanine, enters into the pantothenate biosynthetic pathway, ultimately ending up in the major cellular cofactors coenzyme A and the acyl carrier protein. Another strongly induced gene (osr3) encodes a putative outer membrane protein with similarity to a proteins involved in starch uptake in B. thetaiotaomicron and to a major temperature-modulated cell surface protein in P. gingivalis, RagA, of unknown function (2, 25). These outer membrane proteins appear to be unique to Bacteroides and related organisms, but within this phylogenetic group they are widespread and may have diverse functions. Others such as cation efflux pump CzcD (osr1) and heat shock protein HtpG (osr5) were not as highly induced, but it is interesting that both of these proteins have been implicated in stress responses in other organisms (15, 22).
Three expression classes were observed for the OSR RAPs, and only one of the RAPs, osr3, seemed to be in the OxyR regulon. The most common expression class was similar to the sod response, which is not OxyR controlled but which is nonetheless highly up-regulated in the presence of molecular O2 (but not H2O2, data not shown). The finding of these additional expression classes supports the idea that the response of B. fragilis to O2 is a complicated physiological adaptation that induces multiple regulons (27). A regulator for the sod expression class has not yet been identified, and there currently are no obvious candidates since a search of the B. fragilis genome database did not find any obvious soxR homologs. It may be that the Sox system is restricted to organisms closely related to the Enterobacteriaceae since this system has not been found in other organisms whereas OxyR seems to be more widespread (38). Future studies will be needed to determine the mechanism(s) that controls the OSR genes in this regulon.
A major focus of this paper was the identification of an aerobic class RRase in an anaerobic organism. There are three major classes of RRase enzymes, which have different sensitivities and requirements for O2 (17). Class I reductases require oxygen and therefore are unable to function in its absence, class II enzymes are unaffected by O2 and function in both its presence and absence, and class III enzymes only function in the absence of O2. Evidence presented here shows that the anaerobe B. fragilis has at least two RRase enzymes, a class I (NrdAB) enzyme that is induced during oxidative stress and a class III enzyme (NrdDG) that is transcribed under anaerobic conditions and that is presumably produced during normal growth. The limited biochemical properties of NrdAB examined here were consistent with a class I enzyme. Most importantly, enzyme sensitivity to hydroxyurea (Fig. 6A) is consistent with the O2-mediated radical generation mechanism of class I enzymes but not the class III mechanism (7). In addition, dATP inhibition (Fig. 6B) is a feature of class Ia but not class Ib, and this also is consistent with the sequence homology (17). While this is the first example of an active class I enzyme in an anaerobe, there are several cases where facultative organisms and obligate aerobes have homologs of all three enzymes. For example, the facultative anaerobes E. coli and Salmonella enterica serovar Typhimurium both encode three RRases (17). The class Ia and class III enzymes are utilized for growth in the presence and absence of O2, respectively. Recent work has shown that, among its other roles, the class Ib enzyme (NrdEF) may be important during oxidative stress (21) and could function as NrdAB does in B. fragilis. Regulatory studies with oxyR or soxRS mutants suggest that these do not control E. coli nrdEF expression, so at the present time the mechanism(s) of induction are not clear. The aerobe Pseudomonas aeruginosa also encodes all three classes of enzymes (18). Interestingly, both the class I and class II enzymes are expressed during aerobic conditions but the class III enzyme was not expressed under any of the conditions tested. The remarkable redundancy of RRase enzymes in procaryotes is a testament to their essential roles throughout the growth phase and during times of environmental stress, and the B. fragilis NrdAB described here nicely fits this model.
There are several possible functions for this enzyme in an obligate anaerobe. First, the enzyme may help to maintain intracellular dNTP pools during extended periods of oxidative stress to allow DNA repair and thus extend the viability of the organism. In addition, maintenance of the dNTP pools may allow the bacterium to recover from the effects of oxidative stress at a faster rate than obligate anaerobes that do not contain an aerobically functioning enzyme and that have to synthesize their dNTP pools. Although the dNTP pool sizes were not measured, experiments showed that in complex media a nrdA mutation has an effect on the viability of B. fragilis after about 48 h of exposure to O2 (data not shown) and that there was a severe effect on the ability of B. fragilis to resume growth when it was returned to anaerobic conditions (Fig. 7B). These effects were more pronounced in chemically defined media than complex media, presumably since complex media contain bases and other complex molecules which could bypass the need for de novo synthesis. Clearly there is a need to examine dNTP pool sizes during a variety of growth conditions in order to elucidate the precise roles of the different RRases. A second function for the NrdAB enzyme may be to enable growth of B. fragilis during microaerophilic conditions. The ability of NrdA to benefit B. fragilis during microaerophilic conditions was not tested in this study, but the organism is capable of growth at greatly reduced O2 tensions (<2%). Interestingly, the Pseudomonas class Ia enzyme was maximally expressed during growth under microaerophilic conditions, suggesting that these enzymes have the capacity to function under these conditions.
The class I, oxygen-requiring RRase enzymes are thought to have evolved in response to the appearance of an oxygen atmosphere (17). Thus, the existence of these in an obligate anaerobe poses an interesting evolutionary question. That is, did the organisms obtain this enzyme by horizontal transfer or has it been a part of the genome during the course of evolution? The G+C content of the nrdAB operon was 46%, which is marginally higher than that of the genome as a whole, but more importantly the codon usage was nearly identical to that in genes in our Bacteroides codon usage database (35). These data suggest that the genes are not recent acquisitions. Only through continued study of organisms with multiple RRase enzymes will it be possible to discern their natural history and perhaps the origin of nrdAB in Bacteroides.
Acknowledgments
We thank J. G. Cory for help with the RRase assays and M. McClelland for helpful discussions on RAP-PCR strategies. We also acknowledge the help of C. D. Herren for the cloning of the nrdD gene and E. Pesci for critically reading the manuscript.
This work was supported by Public Health Service grant AI40588 to C.J.S. from the National Institutes of Health.
REFERENCES
- 1.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 2.Bonass, W. A., P. D. Marsh, R. S. Percival, J. Aduse-Opoku, S. A. Hanley, D. A. Devine, and M. A. Curtis. 2000. Identification of ragAB as a temperature-regulated operon of Porphyromonas gingivalis W50 using differential display of randomly primed RNA. Infect. Immun. 68:4012-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. C., T. L. Chou, and C. Y. Lee. 2000. Cloning, expression and characterization of l-aspartate-decarboxylase gene from Alcaligenes faecalis CCRC 11585. J. Ind. Microbiol. Biotechnol. 25:132-140. [Google Scholar]
- 4.Christman, M. F., R. W. Morgan, F. S. Jacobson, and B. N. Ames. 1985. Positive control of a regulon for defense against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753-762. [DOI] [PubMed] [Google Scholar]
- 5.Coyne, M. J., W. Kalka-Moll, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2000. Bacteroides fragilis NCTC9343 produces at least three distinct capsular polysaccharides: cloning, characterization, and reassignment of polysaccharide B and C biosynthesis loci. Infect. Immun. 68:6176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrenberg, A., and P. Reichard. 1972. Electron spin resonance of the iron-containing protein B2 from ribonucleotide reductase. J. Biol. Chem. 247:3485-3488. [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1989. PHYLIP--Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 9.Finegold, S. M., and W. L. George. 1989. Anaerobic infections in humans. Academic Press, San Diego, Calif.
- 10.Godoy, V. G., M. M. Dallas, T. A. Russo, and M. H. Malamy. 1993. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect. Immun. 61:4415-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory, E. M. 1985. Characterization of the O2-induced manganese-containing superoxide dismutase from Bacteroides fragilis. Arch. Biochem. Biophys. 238:83-89. [DOI] [PubMed] [Google Scholar]
- 13.Gregory, E. M., J. B. Kowalski, and L. V. Holdeman. 1977. Production and some properties of catalase and superoxide dismutase from the anaerobe Bacteroides distasonis. J. Bacteriol. 129:1298-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiney, D. G., P. Hasegawa, and C. E. Davis. 1984. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc. Natl. Acad. Sci. USA 81:7203-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 16.Jenney, F. E., Jr., M. F. Verhagen, X. Cui, and M. W. Adams. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306-309. [DOI] [PubMed] [Google Scholar]
- 17.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 18.Jordan, A., E. Torrents, I. Sala, U. Hellman, I. Gibert, and P. Reichard. 1999. Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 181:3974-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopatin, D. E., A. Combs, D. G. Sweier, J. C. Fenno, and S. Dhamija. 2000. Characterization of heat-inducible expression and cloning of HtpG (Hsp90 homologue) of Porphyromonas gingivalis. Infect. Immun. 68:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lumppio, H. L., N. V. Shenvi, A. O. Summers, G. Voordouw, and D. M. Kurtz, Jr. 2001. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183:101-108. (Erratum, 183:2970.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monje-Casas, F., J. Jurado, M. J. Prieto-Alamo, A. Holmgren, and C. Pueyo. 2001. Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. J. Biol. Chem. 276:18031-18037. [DOI] [PubMed] [Google Scholar]
- 22.Nies, D. H. 1992. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 174:8102-8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panaretou, B., C. Prodromou, S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1998. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17:4829-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Privitera, G., A. Dublanchet, and M. Sebald. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 139:97-101. [DOI] [PubMed] [Google Scholar]
- 25.Reeves, A. R., G. R. Wang, and A. A. Salyers. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha, E. R., G. Owens, Jr., and C. J. Smith. 2000. The redox-sensitive transcriptional regulator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocha, E. R., T. Selby, J. P. Coleman, and C. J. Smith. 1996. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J. Bacteriol. 178:6895-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocha, E. R., and C. J. Smith. 1995. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J. Bacteriol. 177:3111-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocha, E. R., and C. J. Smith. 1997. Regulation of Bacteroides fragilis katB mRNA by oxidative stress and carbon limitation. J. Bacteriol. 179:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocha, E. R., and C. J. Smith. 1998. Characterization of a peroxide-resistant mutant of the anaerobic bacterium Bacteroides fragilis. J. Bacteriol. 180:5906-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rocha, E. R., and C. J. Smith. 1999. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 181:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolfe, R. D., D. J. Hentges, J. T. Barrett, and B. J. Campbell. 1977. Oxygen tolerance of human intestinal anaerobes. Am. J. Clin. Nutr. 30:1762-1769. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Smith, C. J., and D. R. Callihan. 1992. Analysis of rRNA restriction fragment length polymorphisms from Bacteroides spp. and Bacteroides fragilis isolates associated with diarrhea in humans and animals. J. Clin. Microbiol. 30:806-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, C. J., L. A. Rollins, and A. C. Parker. 1995. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid 34:211-222. [DOI] [PubMed] [Google Scholar]
- 36.Sokolov, B. P., and D. J. Prockop. 1994. A rapid and simple PCR-based method for isolation of cDNAs from differentially expressed genes. Nucleic Acids Res. 22:4009-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steeper, J. R., and C. D. Steuart. 1970. A rapid assay for CDP reductase activity in mammalian cell extracts. Anal. Biochem. 34:123-130. [DOI] [PubMed] [Google Scholar]
- 38.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 39.Tang, Y. P., M. M. Dallas, and M. H. Malamy. 1999. Characterization of the Batl (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol. Microbiol. 32:139-149. [DOI] [PubMed] [Google Scholar]
- 40.Tzianabos, A. O., A. B. Onderdonk, B. Rosner, R. L. Cisneros, and D. L. Kasper. 1993. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 262:416-419. [DOI] [PubMed] [Google Scholar]
- 41.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]