Abstract
We report here the first use of directed mutagenesis in Methanosarcina acetivorans C2A. The method employs homologous recombination-mediated gene replacement and was used to construct a variety of proline auxotrophs with mutations in the proABC locus. Each mutation was also complemented in trans with autonomously replicating Methanosarcina-Escherichia plasmid shuttle vectors.
Studies of methane-producing archaea have been hampered by a lack of genetic tools; however, in recent years this has been changing rapidly. Useful genetic tools have now been developed for organisms in two genera, Methanococcus and Methanosarcina. These tools include selectable markers, transformation methods, plasmids, reporter genes, and a variety of mutagenesis protocols (reviewed in references 13 and 23). Despite these advances, serious deficiencies in the genetic tools available for many methanoarchaea are still evident. In particular, robust protocols for directed mutagenesis of Methanosarcina species, which are the most metabolically diverse of the methanoarchaea, are still lacking, while plasmid-based complementation of mutations has yet to be reported for any methanogen.
Directed mutagenesis of Methanosarcina species is clearly possible. Transformation and integration of foreign DNA into the chromosome via homologous recombination have been demonstrated for Methanosarcina mazei S-6 (6). In that study, the first to demonstrate the transformation of any Methanosarcina species, a selectable marker inserted into the grpE-dnaK intergenic region was recombined onto the M. mazei chromosome after chemical transformation. The process was, however, inefficient, requiring 50 μg of DNA to produce transformed cultures. More recently, a very efficient liposome-mediated transformation method for Methanosarcina species was developed. This method allows isolation of up to 108 transformants per μg of DNA in Methanosarcina acetivorans C2A. The availability of this high-frequency transformation method led us to attempt directed mutagenesis in this species. To examine the factors involved in this type of experiment and to verify that the process of homologous recombination occurs in a reliable and predictable manner, we decided to mutagenize genes with known functions and predictable phenotypes. For this purpose, we chose to examine genes involved in the biosynthesis of proline.
Cloning the proline biosynthetic genes of M. acetivorans.
Previous studies demonstrated the feasibility of cloning proline biosynthetic genes from methanoarchaea by complementation of Escherichia coli mutants (10). Therefore, we used the same method to clone the proC gene from each of the three Methanosarcina species frequently used in our laboratory. Chromosomal DNAs, isolated as previously described (3) from M. acetivorans C2A, Methanosarcina thermophila TM1, and Methanosarcina barkeri Fusaro, were used to construct genomic libraries in the single-copy, dual-cos vector pWM357 (Table 1). The libraries contain ca. 330,000, 14,000, and 50,000 independent clones, respectively, and were prepared essentially as described previously (2). Numerous cosmids carrying the pro locus from each organism were subsequently obtained as plasmids that allow growth of the E. coli proC mutant XS72 on glucose-MOPS (morpholinepropanesulfonic acid) agar (19) without added proline (Pro+ phenotype).
TABLE 1.
Microorganisms and plasmids used in the study
| Species and strain or plasmid | Relevant characteristic(s) or description and/or constructionb | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | 8 |
| DH5α/λpir | λpir/φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | 18 |
| DH10B | φ80dlacZαM15/araD139 Δ(ara leu)7697 ΔlacX74 galU galK rpsL deoR endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | 8 |
| BW10244 | proC::Tn5-132 | 15 |
| BW21116 | Δ(lacZYA-argF)U169 creC510 hsdR514 uidA(ΔMluI)::pir+ | 14 |
| XS72a | Δ(lacZYA-argF)U169 creC510 hsdR514 uidA(ΔMluI)::pir+proC::Tn5-132 | This study |
| GM119 | dam3 dcm9 metB1 galK2 galT27 lacY1 tsx-78 supE44 thi-1 mel1 tonA31 | 21 |
| Salmonella enterica serovar Typhimurium | ||
| MST951 | proA41 | 11 |
| MST952 | proB8 | 11 |
| MST953 | proC90 | 11 |
| TR5877 | hsdL6 hsdSA29 ilv-452 metA22 trpC2 metE551 xly-404 fla-66 rpsL120 H1-b H2-e,n,x (Fels2)−nml | 20 |
| M. acetivorans C2A | Wild type (DSM2834) | Lab stock |
| M. thermophila TM1 | Wild type (DSM1825) | Lab stock |
| M. barkeri Fusaro | Wild type (DSM804) | Lab stock |
| Plasmid | ||
| pBace3.6 | Cmr, oriF vector | 7 |
| Supercos1 | Dual cos vector | Stratagene; La Joya, Calif. |
| pBluescript KS(+) | Apr cloning vector | Stratagene; La Joya, Calif. |
| pSL1180 | Apr cloning vector | 4 |
| pPB35 | Par AprMethanosarcina-Escherichia shuttle vector | 3 |
| pWM321 | Pur AprMethanosarcina-Escherichia shuttle vector | 16 |
| pJK5 | pac-ori-aph cassette plasmid | 24 |
| pBEND2 | Apr symmetrical polylinker vector | 12 |
| pPB12 | Par AprMethanosarcina/Escherichia shuttle vector | 3 |
| pWM353 | Eco47III deletion of Supercos1 | This study |
| pWM357 | oriF, dual cos vector; XhoI-cut (added restriction sites shown in bold) dual-cos PCR fragment from pWM353 using primers CGCGCGCTCGAGCCTATAAAAATAGGCGTATCACGAGG and CGCGCGCTCGAGTTGAAGGCTCTCAAGGGCATCGGTCG ligated to a 6.3-kbp SalI fragment from pBace3.6 | This study |
| pJK201 | Pro+ clone from M. acetivorans in pWM367 | This study |
| pAW1 | proB-complementing subclone; Sau3AI partial of pJK201 in BamHI-cut pBluescript KS(+) | This study |
| pAW2 | proB-complementing subclone; Sau3AI partial of pJK201 in BamHI-cut pBluescript KS(+) | This study |
| pAW3 | proA-complementing subclone; Sau3AI partial of pJK201 in BamHI-cut pBluescript KS(+) | This study |
| pAW4 | proA-complementing subclone; Sau3AI partial of pJK201 in BamHI-cut pBluescript KS(+) | This study |
| pCK1 | proC-complementing subclone; Sau3AI partial of pJK201 BamHI cut in pSL1180 | This study |
| pCK2 | proC-complementing subclone; Sau3AI partial of pJK201 BamHI cut in pSL1180 | This study |
| pJK68 | M. acetivorans proABC subclone; 4,611-bp NdeI of pJK201 into NdeI-cut pSL1180 | This study |
| pJK69 | M. acetivorans proABC subclone; 4,611-bp NdeI of pJK201 into NdeI-cut pSL1180 (orientation opposite to that of pJK68) | This study |
| pJK72 | M. acetivorans proABC subclone; NotI-to-AvrII insert from pJK68 into NotI- and AvrII-cut pWM321 | This study |
| pPB36 | M. acetivorans proABC subclone; BamHI-to-MluI insert from pJK72 into BamHI- and MluI-cut pPB35 | This study |
| pJK74 | M. acetivorans proAB subclone; KpnI deletion of pJK72 | This study |
| pJK75 | M. acetivorans proA subclone; BclI deletion of pJK72 | This study |
| pJK76 | M. acetivorans proA subclone; BclI deletion of pPB36 | This study |
| pWM403 | Apr vector with unique PvuII site; PvuII deletion of pBluescript KS(+) | This study |
| pWM404 | PvuII-cut PCR product of EZ::TN <KM-1> (Epicentre, Madison, Wis.) with the primer GAATTCCAGCTGTCTCTTATACACATCTCAACC into PvuII-cut pWM403 | This study |
| pWM407 | 2.9-kbp KpnI-to-XbaI pac-ori-aph cassette of pJK5 into KpnI-XbaI-deleted pWM404 | This study |
| pTn5-407 | Tn5-407 vector; circularization of the PvuII fragment from pWM407 | This study |
| pJK70 | Tn5-ileS12 vector; the XbaI-SphI fragment from pPB12 was cloned into XbaI-SphI-digested pWM404 | This study |
| pJK71 | Tn5-ileS12 vector; the XbaI-SphI fragment from pPB12 was cloned into the XbaI-SphI-digested pWM405 | This study |
| pJK61 | pac-ori-aph cassette plasmid with symmetrical flanking sites; XbaI fragment of pJK5 into XbaI-cut pBend2 | This study |
| pCK11 | BamHI fragment of pCK1 into BamHI-cut pBluescript KS(+) | This study |
| pCK12 | BamHI fragment of pCK1 into BamHI-cut pBluescript KS(+) (orientation opposite to that of pCK11) | This study |
| pCK14 | proC1::pac-ori-aph plasmid; BamHI cassette from pJK61 into BglII-cut pCK12 | This study |
| pCK16 | proC2::pac-ori-aph plasmid; BamHI cassette from pJK61 into BglII-cut pCK11 | This study |
| pJK68- proA1::Tn5-407 | In vitro transposon mutagenesis of pJK68 with Tn5-407c | This study |
| pJK68- proA2::Tn5-407 | In vitro transposon mutagenesis of pJK68 with Tn5-407c | This study |
| pJK68- proA3::Tn5-ileS12 | In vitro transposon mutagenesis of pJK68 with Tn5-ileS12c | This study |
| pJK68- proA4::Tn5-ileS12 | In vitro transposon mutagenesis of pJK68 with Tn5-ileS12c | This study |
| pJK68- proB1::Tn5-407 | In vitro transposon mutagenesis of pJK68 with Tn5-407c | This study |
| pJK68- proB2::Tn5-ileS12 | In vitro transposon mutagenesis of pJK68 with Tn5-ileS12c | This study |
| pJK68- proB3::Tn5-ileS12 | In vitro transposon mutagenesis of pJK68 with Tn5-ileS12c | This study |
| pJK68- proC3::ileS12 | In vitro transposon mutagenesis of pJK68 with Tn5-ileS12c | This study |
| pJK68- proC4::ileS12 | In vitro transposon mutagenesis of pJK68 with Tn5-ileS12c | This study |
The proC::Tn5-132 allele of BW10244 was moved into BW21116 by P1 kc transduction to construct XS72. Tetracycline-resistant transductants were selected on TYE-tetracycline agar (22) and screened for Pro− phenotype on glucose-MOPS agar with and without proline.
Standard methods were used throughout for isolation and manipulation of plasmid DNA from E. coli (1). E. coli strains DH5α and DH10B were used as the hosts for most cloning experiments; DH5α/λpir was used as the host for cloning experiments involving pir-dependent replicons. Plasmids used for constructions involving BclI were isolated from E. coli GM119. Plasmids to be transformed into Salmonella serovar Typhimurium that were originally isolated from E. coli were usually passed through Salmonella serovar Typhimurium TR5877 first, to avoid problems associated with host restriction systems.
In vitro mutagenesis was performed using EZ::TN transposase (Epicentre) as recommended. Transposon Tn5-407 as used in these reactions was prepared by linearization of pTn5-407 with PvuII; transposon Tn5-ileS12 was prepared by purification of the 6.8-kbp FspI fragment from either pJK70 or pJK71. Mutagenized plasmids were recovered from Ampr and Kanr transformants of DH10B. The orientation and insertion site of each mutation were verified by DNA sequencing and are shown in Fig. 1. The primers used for determining the DNA sequences of transposon insertion junctions were pJK5forI (5"-GTATATTACGAATAGGGCG-3") and pJK5revI (5"-AGCTGCTGGTGAAAGAGAC-3") for Tn5-407 and EZTNkanrev (5"-GGTACCGAGCTCGAATTCAT-3") and ileS12rev (5"-CGAGCAGGTGATGAAAAGG-3") for Tn5-ileS12.
Two cosmids from each organism were moved into an isogenic set of Salmonella enterica serovar Typhimurium proA, proB, and proC mutants. Surprisingly, all of the cosmids complemented not only the proC mutation but also the proA and proB mutations, suggesting that each carries functional homologs of all three proline biosynthetic genes or a gene(s) that allows synthesis of proline via an alternate route. To address this issue, we carried out a series of subcloning and complementation experiments using one of the cosmids (pJK201) isolated from M. acetivorans. Random subclones of pJK201 were generated as described in Table 1. Pro+ subclones were then isolated based on their ability to complement the proA mutant Salmonella serovar Typhimurium MST951(pAW3 and pAW4), the proB mutant Salmonella serovar Typhimurium MST952(pAW1 and pAW2), or the proC mutant E. coli BW10244(pCK1 and pCK2). In all cases, the subclones isolated by complementation of a particular pro mutation failed to complement the other two pro mutations, strongly supporting the conclusion that proline biosynthesis in M. acetivorans occurs via the same three-step pathway used by Salmonella (5). The fact that similar clones were isolated from M. barkeri and M. thermophila suggests that the same is true in these species as well.
DNA sequence analysis of the M. acetivorans proABC locus.
The DNA sequences of the inserts in each of the pJK201 subclones were determined by automated dye terminator sequencing at the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois. Upon analysis, these sequences could be compiled into a single continuous DNA fragment of 5,139 bp containing three large open reading frames, which, based on sequence analysis, were designated proA, proB, and proC (Fig. 1). The predicted protein products of the proA, proB, and proC genes show very significant homology to known ProA (glutamate semialdehyde dehydrogenase), ProB (glutamate kinase), and ProC (1-pyrroline-5-carboxylate reductase) proteins, with between 30 and 50% identities to the top 20 BLAST matches for all three proteins (see below). The results of these homology searches are in complete agreement with the complementation data presented above. Accordingly, the only complete gene found on the proA-complementing plasmids is proA, the only complete gene found on the proB-complementing plasmids is proB, and the only complete gene on the proC-complementing plasmids is proC (Fig. 1). Thus, in combination, these data provide strong support for the function of the genes in the M. acetivorans proABC locus, i.e., the biosynthesis of proline via the standard pathway found in most organisms.
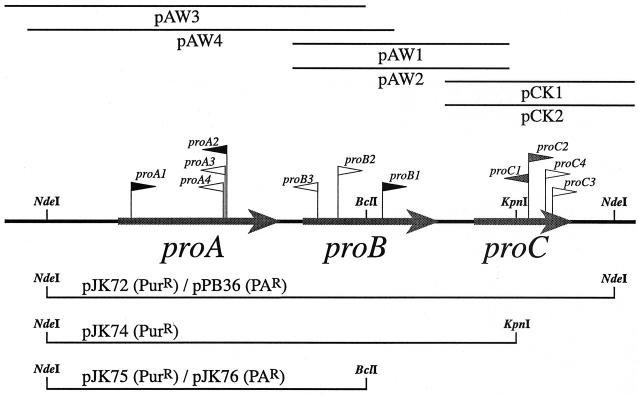
FIG. 1.
The M. acetivorans C2A proABC locus. The physical structure of a 5,139-bp fragment of the M. acetivorans chromosome is shown in the center (heavy black line), with the positions of the proA, proB, and proC genes shown as gray arrows. The regions covered by the six plasmids whose DNA sequences were determined are shown as thin black lines at the top, with the names of the respective plasmids shown below each line. The position and relative orientation of each in vitro-constructed insertion mutation are indicated by a flag on the center line. The black flags represent Tn5-407 insertions, the white flags represent Tn5-ileS12 insertions, and the gray flags represent insertions of the pac-ori-aph cassette. The regions contained in several plasmid shuttle vectors used for complementation studies are shown as thin black lines at the bottom. The names of the respective plasmids and antibiotic resistance markers are shown above each line. The endonuclease restriction sites used for construction of these shuttle vectors are indicated.
Although the findings regarding the function of M. acetivorans genes in the standard proline biosynthesis pathway are not surprising per se, they are at odds with the situation observed for other archaea. Recent data suggest that proline is synthesized from ornithine in several methanoarchaea (9). Moreover, examination of the 10 complete archaeal genome sequences presently available failed to identify complete sets of genes required for the standard biochemical pathway (searches were performed in September 2001). Thus, recognizable ProA homologs were not observed (based on a BLAST E value of <0.04 when M. acetivorans ProA is used as the query sequence) in any of the completed archaeal genome sequences, which include those of Aeropyrum pernix K1, Archaeoglobus fulgidus, Halobacterium sp. strain NRC-1, Methanobacterium thermoautotrophicum, Methanococcus jannaschii, Pyrococcus abyssi, Pyrococcus horikoshii, Sulfolobus acidocaldarius, Thermoplasma acidophilum, and Thermoplasma volcanium. In contrast, sequence comparisons of M. acetivorans ProA with dozens of other ProA proteins produced highly significant scores (the top 50 matches produced E values of <4 × 10−44); these proteins included many with biochemically and genetically demonstrated function. Using M. acetivorans ProB as the query sequence, a possible ProB homolog was identified in Methanobacterium thermoautotrophicum (E value of 3 × 10−4) but not in the remaining archaeal genomes (based on E values of <0.01). When compared with the top 50 known and putative ProB proteins, M. acetivorans ProB produced scores of <7 × 10−20. In contrast, easily recognizable ProC proteins were found in 5 of the 10 available archaeal genomes. Thus, it appears that recognizable ProC homologs are more common, but not universal, among archaea. As described above, a proC gene was previously identified in Methanobrevibacterium smithii by genetic complementation (10), but this gene is not among the closest homologs to M. acetivorans proC. In sum, it is clear that the evolution of proline biosynthesis in Methanosarcina species is dramatically different than in the other members of the domain Archaea for which data are available; they are the only known archaea to possess the standard pathway for proline biosynthesis.
Directed mutagenesis of M. acetivorans proabc by homologous recombination-mediated allele replacement.
A variety of pro mutations were constructed on plasmids for subsequent recombination onto the M. acetivorans chromosome as described in Table 1. These gene disruption mutations consist of those with insertions of either the pac gene (in the form of the pac-ori-aph cassette or Tn5-407) or the ileS12 gene (in the form of Tn5-ileS12), which encode resistance to puromycin or pseudomonic acid, respectively, in Methanosarcina species (3, 16). Both transposons and the pac-ori-aph cassette also encode resistance to kanamycin in E. coli to simplify the plasmid constructions. The sites and orientations of each insertion mutation are shown in Fig. 1.
Each of the plasmid-borne pro alleles was transferred onto the M. acetivorans chromosome by simple homologous recombination after liposome-mediated transformation (3, 16) with linear DNA. For these experiments, the plasmids were linearized with restriction enzymes that cut only the plasmid backbone, as close to the insert as possible. Recombinants carrying Tn5-407 or pac-ori-aph cassette insertions were selected as puromycin-resistant (Purr) clones; recombinants carrying Tn5-ileS12 insertions were selected as pseudomonic acid-resistant (PAr) clones. In both cases, selection for recombinants was carried out in medium containing 50 mM proline to allow growth of the expected auxotrophic mutants. (There is no available evidence to suggest that proline is transported by M. acetivorans. We used this relatively high proline concentration in the hope of overcoming potential transport defects.) In these experiments, transformation with ca. 2 μg of linearized plasmid DNA yielded from a few hundred to a few thousand antibiotic-resistant colonies. After purification of the recombinant clones by streaking for single colonies on solid-medium plates, 80 to 100% of these transformants (20 were tested for each mutation) required proline for growth (Pro− phenotype). All 11 of the pro mutations yielded Pro− transformants.
Verification of the chromosomal structure of the M. acetivorans Pro−. mutants.
Because allele replacement has not previously been employed in M. acetivorans, we examined the physical structure of each mutant chromosome in detail by using DNA hybridization. DNA hybridizations were performed essentially as described previously (17); however, a digoxigenin-labeled probe (pJK68) and chemiluminescence detection (DIG High Prime and CDP-Star, respectively; Roche Molecular Biochemicals, Indianapolis, Ind.) were used for visualization of hybridizing bands in place of 31P-labeled probes. For all 11 M. acetivorans Pro− mutants, there was complete agreement between the sizes of the hybridizing bands predicted from the known DNA sequences and those obtained experimentally (data not shown). Thus, the mutant strains that we constructed were indistinguishable from those that would result from simple homologous recombination, and this finding provides strong support for the occurrence of this genetic process in M. acetivorans C2A.
Plasmid-based complementation of M. acetivorans proABC mutants.
A series of Methanosarcina-E. coli shuttle vectors have been reported (3, 16); however, these have never been used for complementation studies because Methanosarcina mutants were not previously available. To examine whether plasmid-borne alleles could complement genomic mutations, each of the PAr Pro− mutants was transformed with a puromycin resistance plasmid, pJK72, carrying the entire proABC locus. The shuttle vector pWM321 (with no insert) was used as a negative control. In a similar way, each of the Purr Pro− mutants was transformed with the pseudomonic acid resistance proABC plasmid, pPB36, by using the shuttle vector pPB35 as a control (Table 2). All of the strains transformed by the proABC plasmids regained the Pro+ phenotype, indicating that each chromosomal mutation was complemented by the plasmid-borne, wild-type allele. Strains transformed by the vector controls remained Pro−. Because these mutant strains are recombination proficient, it is possible that restoration of the Pro+ phenotype occurred by marker rescue (i.e., recombinational repair) rather than by true complementation. However, we do not believe this was the case. Selection for both antibiotics was maintained throughout the experiment. Therefore, the antibiotic resistance marker on the chromosome was not removed by recombination unless a reciprocal recombination event exchanged the markers on the plasmid and the chromosome. In each case, we were able to recover the intact original plasmid from the complemented strain, indicating that no such event had occurred (data not shown).
TABLE 2.
Plasmid complementation of M. acetivorans proABC mutationsa
| Mutation | Complementation with: | |||
|---|---|---|---|---|
| Vector only | proABC | proAB | proA | |
| pPB35 | pPB36 | None | pJK76 | |
| proA1::Tn5-407 | − | + | NDb | + |
| proA2::Tn5-407 | − | + | ND | + |
| proB1::Tn5-407 | − | + | ND | ND |
| proC2::pac-ori-aph | − | + | ND | ND |
| proC1::aph-ori-pac | − | + | ND | ND |
| pWM321 | pJK72 | pJK74 | pJK75 | |
| proA3::Tn5-ileS12 | − | + | + | + |
| proA4::Tn5-ileS12 | − | + | + | + |
| proB2::Tn5-ileS12 | − | + | + | ND |
| proB3::Tn5-ileS12 | − | + | + | ND |
| proC3::Tn5-ileS12 | − | + | ND | ND |
| proC4::Tn5-ileS12 | − | + | ND | ND |
Complementation was scored according to ability to grow on HA-MA medium with puromycin and pseudomonic acid but without proline. Plasmids used are described in the text and shown in Fig. 1.
ND, not done.
Although all the mutations tested resulted in proline auxotrophy, it is not clear from mutant analysis alone whether proA and proB have a role in proline biosynthesis. Given the potential operon structure of the proABC locus, it was formally possible (although perhaps unlikely) that only proC was required for proline biosynthesis and that the effects of the proA and proB mutations were via polarity on proC. Accordingly, we transformed the proA::Tn5-ileS12 mutants with puromycin resistance plasmids carrying only proA (to check for polarity on both proB and proC) or proAB (to check for polarity on proC). In a similar way, the proB::Tn5-ileS12 mutants were transformed with a puromycin resistance plasmid carrying only proAB to check for polarity on proC (Table 2). In all cases, the plasmids were able to complement the transposon insertion mutations, indicating that these Tn5-ileS12 mutations were not polar on the downstream genes and that proA and proB are required for proline biosynthesis in M. acetivorans. Similar experiments involving complementation of Tn5-407 insertions by pseudomonic resistance plasmids yielded identical results (Table 2).
As discussed above, the ability to complement chromosomal mutations by plasmid-borne alleles was critical in defining the functions of proA and proB. To our knowledge, this is the first example of plasmid complementation in a methanogenic archaeon. The ability to use plasmids in this way will greatly expand our ability to perform genetic analysis in Methanosarcina species. Most importantly, plasmid complementation will allow verification that a particular phenotype is caused by a mutation in a specific gene (namely, the one carried on the complementing plasmid). However, innumerable genetic techniques require the use of replicating plasmids. Among these are the identification of essential genes and regulatory proteins and the introduction of foreign genes, or genes carrying defined mutations, into strain backgrounds with mutated alleles on the chromosome.
The use of plasmids for complementation of mutations created by the insertion of selectable markers requires, of course, multiple selectable markers. Currently, only two such markers, the ileS12 gene for resistance to pseudomonic acid (3) and the pac cassette for resistance to puromycin (16), are available for use in Methanosarcina. Our demonstration that plasmids carrying proline genes can complement proline auxotrophic mutations suggests the possibility of developing a third selectable marker from these genes. This would require a suitable proline auxotroph as a host strain, preferably one that did not already carry an antibiotic insertion in the chromosome. Experiments designed to develop such a system are presently under way in our laboratory.
These new techniques for directed mutagenesis and plasmid complementation, along with the recently presented method for random transposon-based mutagenesis (24), now complete a set of basic tools for genetic analysis of Methanosarcina species. These tools, in combination with the metabolic diversity of members of this genus (relative to other methanoarchaea), make Methanosarcina species very attractive model organisms for the study of methanogenesis and of archaeal biology in general.
Nucleotide sequence accession number.
The sequence for the M. acetivorans C2A proABC locus has been deposited in GenBank under accession number AF305580.
Acknowledgments
We thank Stanley Maloy, Charles Miller, and Barry Wanner for strains used in this study; Xun Shi, April Stanley, and Brian Matlock for technical assistance; and Ralph Wolfe and Stanley Maloy for useful advice.
This work was supported by grant MCB-987459 from the National Science Foundation and by a Searle Scholars Award from the Chicago Community Trust to W.W.M.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology, vol. 1 and 2. John Wiley & Sons, New York, N.Y.
- 2.Bates, P. 1987. Double cos site vectors: simplified cosmid cloning. Methods Enzymol. 153:82-94. [DOI] [PubMed] [Google Scholar]
- 3.Boccazzi, P., J. K. Zhang, and W. W. Metcalf. 2000. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri Fusaro. J. Bacteriol. 182:2611-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius, J. 1989. Superpolylinkers in cloning and expression vectors. DNA 8:759-777. [DOI] [PubMed] [Google Scholar]
- 5.Csonka, L. N., and A. Baich. 1983. Proline biosynthesis, p. 35-51. In K. M. Herrmann and R. L. Somerville (ed.), Amino acids: biosynthesis and genetic regulation. Addison-Wesley, Reading, Mass.
- 6.Demacario, E. C., M. Guerrini, C. B. Dugan, and A. J. L. Macario. 1996. Integration of foreign DNA in an intergenic region of the archaeon Methanosarcina mazei without effect on transcription of adjacent genes. J. Mol. Biol. 262:12-20. [DOI] [PubMed] [Google Scholar]
- 7.Frengen, E., D. Weichenhan, B. Zhao, K. Osoegawa, M. van Geel, and P. J. de Jong. 1999. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics 58:250-253. [DOI] [PubMed] [Google Scholar]
- 8.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graupner, M., and R. H. White. 2001. Methanococcus jannaschii generates l-proline by cyclization of l-ornithine. J. Bacteriol. 183:5203-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton, P. T., and J. N. Reeve. 1985. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol. Gen. Genet. 200:47-59. [DOI] [PubMed] [Google Scholar]
- 11.Itikawa, H., and M. Demerec. 1968. Salmonella typhimurium proline mutants. J. Bacteriol. 95:1189-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, J., C. Zwieb, C. Wu, and S. Adhya. 1989. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene 85:15-23. [DOI] [PubMed] [Google Scholar]
- 13.Metcalf, W. W. 1999. Genetic analysis in the domain Archaea, p. 277-326. In M. Smith and L. Sockett (ed.), Methods in microbiology: genetic methods for diverse prokaryotes, vol. 29. Academic Press, London, England.
- 14.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ α for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 15.Metcalf, W. W., P. M. Steed, and B. L. Wanner. 1990. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J. Bacteriol. 172:3191-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metcalf, W. W., J. K. Zhang, X. Shi, and R. S. Wolfe. 1996. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J. Bacteriol. 178:5797-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palva, E. T., P. Liljestrom, and S. Harayama. 1981. Cosmid cloning and transposon mutagenesis in Salmonella typhimurium using phage lambda vehicles. Mol. Gen. Genet. 181:153-157. [DOI] [PubMed] [Google Scholar]
- 21.Wanner, B. L. 1986. Bacterial alkaline phosphatase clonal variation in some Escherichia coli K-12 phoR mutant strains. J. Bacteriol. 168:1366-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 23.Whitman, W. B., D. L. Tumbula, J. P. Yu, and W. Kim. 1997. Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors 6:37-46. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, J. K., M. A. Pritchett, D. J. Lampe, H. M. Robertson, and W. W. Metcalf. 2000. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. USA 97:9665-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]