Abstract
Salmonella enterica serovar Typhimurium is resistant to the action of bile salts, and resistance to bile is enhanced in strains in which the PhoP-PhoQ (PhoPQ) two-component regulatory system has been activated. To identify genes necessary for bile resistance, MudJ transposon mutagenesis was performed on a strain containing a phoP mutation that results in constitutive expression of PhoP-activated genes. After screening >10,000 mutants for the loss of growth on Luria-Bertani broth-bile plates, 14 bile-sensitive mutants were identified. Of these 14 mutants, 3 were found to retain the bile sensitivity phenotype upon P22 transduction, to possess wild-type growth characteristics, and to contain a smooth lipopolysaccharide. Southern hybridization experiments showed that all three strains contained unique insertions. DNA sequencing of the transposon-chromosomal-DNA fusion junctions of these strains showed all to be linked to the putative Salmonella orf1-tolQRA operon, with insertions in tolQ, orf1, and a gene upstream of the orf1-tolQRA operon not previously associated with Tol function (orfX). Through the use of transcriptional fusions, none of the putative tol (or tol-associated) genes were shown to be regulated by PhoPQ, bile, or the RcsC-RcsB two-component system; however, all of the genes (orfX, orf1, tolQRA) are predicted to be cotranscribed. This is the first identification of Salmonella serovar Typhimurium Tol homologs and the first demonstration of their role in bile resistance in this organism. In addition, the observed regulation, operon arrangement, and phenotypes associated with these tol genes demonstrate significant differences from their Escherichia coli homologs.
Bile salts are detergent-like compounds that aid in the digestion and dispersion of dietary fats in the intestine. As detergent-like compounds, they are also bactericidal. Enteric organisms such as Salmonella enterica serovar Typhimurium are resistant to the action of bile salts at concentrations even above that found in the small intestine. Salmonella spp. can also enter into a carrier state in which the bacteria reside in the gallbladder, the storage site for bile. To survive in this microenvironment, mechanisms are required for resistance to high concentrations of bile.
Although research into bacterial mechanisms of bile resistance has been performed, relatively little is known about this resistance at the molecular level. Resistance mediated by efflux pumps which actively remove bile from the cell has been studied thoroughly (24, 30, 32). This system removes bile by active efflux from the cell. For Vibrio cholerae, it has been shown that resistance to bile involves the regulator ToxR and the ToxR-regulated porins OmpU and OmpT (27). In addition, bile can modulate the activity of the AraC-like activator ToxT in V. cholerae (28).
Recently, we have demonstrated that Salmonella serovar Typhimurium can resist the action of extremely high concentrations of bile (>60%) and that this high-level resistance requires the virulence regulators PhoP and PhoQ (PhoPQ) (33). PhoPQ is a two-component regulatory system that is not only necessary for resistance to bile but is also required for mouse and human virulence (9, 18, 20) and the expression of several virulence phenotypes and factors (1-3, 8-10, 16, 20, 22, 25). None of the 22 previously identified PhoPQ-activated or -repressed loci tested were responsible for this PhoPQ-regulated bile resistance phenotype (33).
The present study was undertaken to attempt to identify genes necessary for bile resistance through the mutagenesis of a high-level-resistance strain (PhoP constitutive [PhoPc]). Saturating MudJ mutagenesis identified three unique insertions closely linked to tol or tol-associated genes. The analysis of these genes demonstrated that the tolQRA region of Salmonella serovar Typhimurium has differential levels of expression, means of regulation, and, likely, functions from its corresponding region in Escherichia coli.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and reagents.
Bacterial strains used in this study are shown in Table 1. Cultures were grown overnight at 37°C with aeration in Luria-Bertani (LB) broth or in microtiter plates as described below. When necessary, media were supplemented with chloramphenicol (25 μg/ml), ampicillin (50 μg/ml), or kanamycin (45 μg/ml).
TABLE 1.
Salmonella serovar Typhimurium strains used
| Strain | Genotype and/or phenotype | Source or reference |
|---|---|---|
| JSG206 | PhoP− (phoP::Tn10d-cam) | 20 |
| JSG208 | PhoPc (pho24) | 21 |
| JSG210 | Wild-type 14028s | ATCCa |
| JSG212 | hsdLT hsdSA hsdSB galE, rough strain (LB5010) | 5 |
| JSG642 | TT10288 MudJ donor, hisD9953::MudJ hisA9944::MudI | 20 |
| JSG1168 | rcsC::luc | This study |
| JSG1287 | orfX::luc | This study |
| JSG1328 | tolR::luc | This study |
| JSG854 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG855 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG856 | JSG208 orf1::MudJ, bile sensitive | This study |
| JSG857 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG858 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG861 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG862 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG871 | JSG208 MudJ insertion, bile sensitive, rough LPS | This study |
| JSG872 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG873 | JSG208 orfX::MudJ, bile sensitive | This study |
| JSG878 | JSG208 MudJ insertion, bile sensitive | This study |
| JSG879 | JSG208 tolQ::MudJ, bile sensitive | This study |
| JSG880 | JSG208 MudJ insertion, bile sensitive, rough LPS | This study |
| JSG881 | JSG208 MudJ insertion, bile sensitive, rough LPS | This study |
| JSG1584 | JSG856 MudJ insertion in JSG210 | This study |
| JSG1585 | JSG873 MudJ insertion in JSG210 | This study |
| JSG1586 | JSG879 MudJ insertion in JSG210 | This study |
ATCC, American Type Culture Collection, Manassas, Va.
Conjugated and unconjugated bile salts were purchased from Sigma (St. Louis, Mo.). Bile used in this work is labeled “sodium choleate” but is a crude ox bile extract which contains salts of taurocholic, glycocholic, deoxycholic, and cholic acids.
Transposon mutagenesis and detection of bile-sensitive strains.
Bile-sensitive Salmonella serovar Typhimurium strains were created by MudJ transposon mutagenesis as previously described (14). Briefly, the MudJ transposon from strain JSG642 (TT10288) was introduced into Salmonella serovar typhimurium PhoPc by P22HT int phage transduction. PhoPc strains containing MudJ transposons were selected on LB agar-kanamycin plates. Individual colonies were screened for a bile sensitivity phenotype by replica plating onto plates containing LB agar plus 10% bile and were screened for loss of growth on these bile-containing plates. Colonies were reconfirmed as being bile sensitive after single-colony isolation and after P22-mediated transduction of the region containing the MudJ insertion into a fresh PhoPc background.
Standard MIC assays of resistance.
Bacterial cells were challenged with an unconjugated bile acid (deoxycholic acid) or conjugated bile acids (glycocholic, taurocholic, and glycochenodeoxycholic acids), and MICs for them were determined under nonaerated conditions. Stationary-phase cultures were diluted such that samples of 2 × 103 to 5 × 103 CFU/ml were subjected to various concentrations of bile acids in polypropylene microtiter plates (Costar Corp., Cambridge, Mass.). The plates were incubated overnight at 37°C, and the wells of the plate were visually analyzed to determine MICs.
Southern blot analysis of bile-sensitive strains.
Chromosomal DNAs were digested, electrophoresed on a 1% agarose slab gel, and transferred to nylon membranes as previously described (13). Membranes were hybridized with digoxigenin-labeled probes, and detection was achieved by chemiluminescence. The probe was specific to the aph (kanamycin) gene of the MudJ transposon and was generated by PCR with primers JG217 and JG218, whose sequences are shown in Table 2. All digoxigenin labeling and detection were carried out by using reagents purchased from Boehringer Mannheim (Indianapolis, Ind.).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| JG31 | 5"-AAACTGCAGTTAGCGCAATTCAAAAAG-3" |
| JG45 | 5"-GCTCTAGAGGGAGAAGAGATGATGCGCGTACTGGTTGTAG-3" |
| JG134 | 5"-GGAATTCATGACGATGTCCAGACGC-3" |
| JG135 | 5"-GGGGTACCGGTTAAAGGTGTTGGTCG-3" |
| JG136 | 5"-GGAATTCGACGCGCAAATCCATG-3" |
| JG137 | 5"-GGGGTACCGTTCTTCGCCGACAACCC-3" |
| JG217 | 5"-CGCATGATTGAACAAGATGG-3" |
| JG218 | 5"-CCCGCTCAGAAGAACTC-3" |
| JG49 | 5"-CTAATCCCATCAGATCCC G-3" |
| JG340 | 5"-GGAATTCGCGTCGCGTGCGATGCGC-3" |
| JG341 | 5"-GGGGTACCGGCTGCGTCATTAAGCC-3" |
| JG269 | 5"-GGAATTCGACCACCCGATTGTACTG-3" |
| JG270 | 5"-GGGGTACCCGCTCTCTTTTGGCGACAG-3" |
| JG259 | 5"-GGAATTCATTTATGCGGTAATGGAC-3" |
| JG260 | 5"-GGGGTACCTGGATTATGCCTGGCAAAC-3" |
SDS-PAGE analysis of LPS.
As previously described by Hitchcock and Brown (17), bacterial cells in stationary phase were lysed in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (0.125 M Tris [pH 6.75], 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.1% bromophenol blue) and incubated with proteinase K (10 mg/ml) for 1 h at 60°C. Lipopolysaccharide (LPS) samples were separated by SDS-PAGE (15% polyacrylamide) at 10 mA overnight and detected by silver staining.
Examination of bile-sensitive strains for MudJ insertions in phoPQ, marAB, or acrB.
A combination of Southern blot analysis, PCR, and P22-mediated transduction was used with bile-sensitive strains to confirm the absence of MudJ insertions within the phoPQ, marAB, and acrB loci. Southern blot analysis was performed as described above. Probes were specific to the phoP, marAB, and acrB genes and were produced by PCR with primers JG31 and JG45 (phoP), JG134 and JG135 (marAB), and JG136 and JG137 (acrB). All primer sequences are listed in Table 2. PCR was used to amplify the regions of interest from chromosomal DNAs of bile-sensitive strains, and the molecular weights of the products were compared to those of the parental strain. P22-mediated transduction was used to examine linkages between known markers in or near the phoPQ, marAB, and acrB loci and the MudJ insertions of the bile-sensitive strains.
Transcription assays.
P22 phage-mediated transduction was used to transfer the MudJ insertions of bile-sensitive strains (in a PhoP-constitutive background) to PhoP-null, RscB-null, or wild-type strains. A comparison of the levels of MudJ-encoded β-galactosidase activity among the various strains was accomplished as previously described (13). For bile regulation assays, strains were grown to log phase in LB broth or LB broth with 2.5% bile, washed twice, and assayed for β-galactosidase activity. Gene fusions to firefly luciferase were created by PCR amplification of a region whose 3" end was within the target gene (tolR, JG340 plus JG341; rcsC, JG269 plus JG270; orfX, JG259 plus JG260). The PCR fragment was ligated to the suicide plasmid pGPL01, which, upon being mated and recombined with the chromosome, produces a transcriptional fusion to the luc gene (15). The tolR::luc and orfX::luc chromosomal fusions were moved by P22 transduction into various backgrounds. The level of firefly luciferase activity was determined as previously described (15).
Cloning and DNA sequencing of MudJ-chromosomal-DNA fusion junctions.
Chromosomal DNAs were digested with SalI or BamHI and ligated to the low-copy-number vector pWSK29 (11). SalI and BamHI cut at the left end of the MudJ transposon but outside of the kanamycin resistance gene. Transformants were selected on LB broth plates containing kanamycin and ampicillin. The MudJ-chromosomal-DNA junction was sequenced with primer JG49, which binds to the left end of the MudJ transposon. Sequencing was accomplished with an ABI sequencer as previously described (14).
RESULTS
Mutagenesis of a PhoPc strain identifies loci necessary for resistance to bile.
Previously, we demonstrated that a strain constitutively expressing PhoP-activated genes exhibited high-level bile resistance (MIC, >60%; minimum bactericidal concentration, >60%) (33). Therefore, this high-level bile-resistant strain was mutagenized with the MudJ transposon to identify genes necessary for resistance to bile. As well as resulting in a gene disruption, the MudJ transposon can create active lacZ fusions if inserted properly into the target gene. The PhoPc strain could grow on LB agar plates containing 10% bile, while a PhoP− strain could not grow on plates containing bile concentrations of >8%. Approximately 11,000 colonies from 11 different mutagenesis experiments were replica plated onto 10% bile plates. Of these, we identified 14 mutants unable to grow on the plates containing 10% bile.
Determination of unique bile-sensitive mutants.
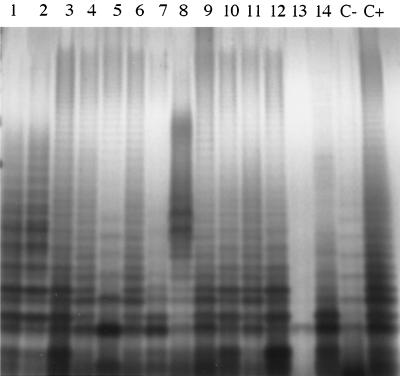
It has been demonstrated that mutants lacking LPS with a complete O side chain (rough mutants) are more sensitive to detergents and other compounds, including bile salts (26). Therefore, we examined our 14 mutants for a rough LPS phenotype by SDS-PAGE analysis of purified LPS and by cross-streaking strains against a lytic P22 phage (O antigen is the P22 phage receptor). As seen in Fig. 1, the typical ladder appearance of the O side chain was absent in strains JSG871, JSG880, and JSG881 (unlike in the PhoPc parental strain and LB5010, a galE mutant and rough strain). The cross-streaking experiments with a lytic P22 phage confirmed the SDS-PAGE analysis, and therefore, strains JSG871, JSG880, and JSG881 were eliminated from further analysis. In addition, two other strains (JSG855 and JSG857) were also eliminated at that time from further analysis because of poor growth, even in rich media.
FIG. 1.
Analysis of LPS in MudJ-mutagenized bile-sensitive strains. Samples were prepared from stationary-phase cultures, electrophoresed on an SDS-15% PAGE gel, and detected by silver staining. Lanes: 1, JSG854; 2, JSG855; 3, JSG856; 4, JSG857; 5, JSG858; 6, JSG861; 7, JSG862; 8, JSG871; 9, JSG872; 10, JSG872; 11, JSG878; 12, JSG879; 13, JSG880; 14, JSG881; C−, LB5010; C+, Salmonella serovar Typhimurium PhoPc. The strains are designated smooth (presence of the typical O-antigen ladder, as seen for the parental Salmonella serovar Typhimurium PhoPc strain) (lane C+) or rough (absence of the typical O-antigen ladder, as seen for the galE mutant LB5010) (lane C−). Rough strains (JSG871, JSG880, and JSG881) were subsequently discarded from further characterization of the bile sensitivity phenotype.
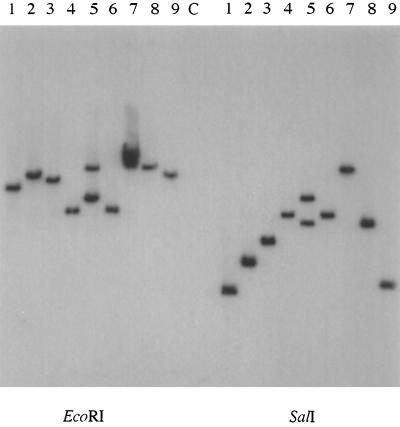
To examine the uniqueness of the remaining nine bile-sensitive mutants, Southern blot hybridization experiments were conducted by using a MudJ-specific probe. Figure 2 shows that with two different enzymes to digest the chromosomal DNAs, these insertions showed unique hybridization patterns, except possibly in strains JSG861 and JSG872. The hybridization patterns in those two strains looked slightly different, and both strains were retained for further analysis (noted differences in the MICs for them are discussed below). Strain JSG862 possessed two MudJ insertions, with one having a hybridization pattern identical to that of strain JSG879. Therefore, because the insertion likely responsible for the bile sensitivity phenotype in JSG862 was not unique, this strain was not further characterized.
FIG. 2.
Differentiation of unique bile-sensitive strains by Southern blot analysis. DNAs were digested with EcoRI or SalI (each cuts once within MudJ) as indicated below the gel, and blots were probed with DNA specific for the aph (kanamycin) gene of the transposon. Lanes: 1, JSG854; 2, JSG856; 3, JSG858; 4, JSG861; 5, JSG862; 6, JSG872; 7, JSG873; 8, JSG879; 9, JSG878; C, Salmonella serovar Typhimurium 14028s. The strains in lanes 5 and 8 appear to be similar, with that in lanes 5 exhibiting multiple insertions. Lanes 5 (JSG862) were not considered to contain a unique strain due to its similarity to the strain with the single insertion in lanes 8 (JSG879), an insertion which is alone responsible for the bile sensitivity phenotype. Although the strains in lanes 4 (JSG861) and 6 (JSG872) show similar banding patterns, both were still considered unique due to marked differences in the MICs for them.
As a final determination of the validity of the bile sensitivity phenotype, strains were transduced with phage P22 back into JSG208 (PhoPc). Of the eight strains tested, five lost the bile sensitivity phenotype upon back transduction. As the Southern blot experiments discussed above showed that these strains contained only one MudJ insertion, it was concluded that a spontaneous mutation not associated with the transposon insertion had resulted in the bile sensitivity phenotype.
It is possible that the mutations resulting in the bile sensitivity phenotype are in loci known to affect bile resistance, such as phoPQ, acr, and mar (19, 30). The acr and mar loci encode or are involved in the expression of efflux pumps, which have been implicated in E. coli bile resistance (32). Therefore, Southern hybridization experiments, in association with PCR analysis, were conducted to screen for insertions in these loci. These experiments demonstrated that all strains had banding patterns similar to that of the parental PhoPc strain, indicating that none of the three remaining insertions (JSG856, JSG873, and JSG879) were located in these obvious gene candidates (data not shown).
MIC analysis of bile-sensitive strains.
MIC analysis was performed with the bile-sensitive mutants (including those of strains JSG854, JSG858, JSG861, JSG872, and JSG878, which were not linked to the transposon and for which data are not included in Table 3) to further confirm bile sensitivity and to extend the observed phenotype to individual bile components and other detergents. Several individual bile salts and nonionic (Triton X-100) and ionic (SDS) detergents were tested. While all strains showed bile sensitivity (1.3- to 8-fold), the mutants showed various sensitivities to the other tested detergents and bile salts (Table 3). None of the mutants had sensitivity patterns or MIC levels similar to those of the PhoP− strain, suggesting that the identified bile-sensitive genes were not solely responsible for the observed PhoP-mediated bile resistance phenotype and that they may not be regulated by PhoPQ. Furthermore, the JSG856, JSG873, and JSG879 insertions in a wild-type background also showed bile sensitivity, demonstrating an effect even in the absence of PhoP activation.
TABLE 3.
Bile salt and detergent MICs for identified bile-sensitive strains
| Strain | MIC (%) ofa:
|
||||||
|---|---|---|---|---|---|---|---|
| Bile | DC | GCDC | TC | GC | SDS | Triton X-100 | |
| JSG208 (PhoPc) | 24 | 5 | 20 | 20 | 20 | 14.5 | 14.5 |
| JSG206 (PhoP−) | 6 | 0.3 | 0.3 | 10 | 10 | 14.5 | 14.5 |
| JSG210 | 15 | ND | ND | ND | ND | 14.5 | 14.5 |
| JSG856 | 6 | 2.5 | 0.6 | 2.5 | 20 | 3.6 | 14.5 |
| JSG873 | 6 | 2.5 | 10 | 2.5 | 2.5 | 0.03 | 14.5 |
| JSG879 | 3 | 0.6 | 20 | 2.5 | 1.25 | 0.03 | 14.5 |
| JSG1584 | 7.5 | ND | ND | ND | ND | 0.25 | 14.5 |
| JSG1585 | 3 | ND | ND | ND | ND | 0.06 | 14.5 |
| JSG1586 | 3 | ND | ND | ND | ND | 0.06 | 14.5 |
DC, deoxycholate; GCDC, glycochenodeoxycholate; TC, taurocholate; GC, glycocholate; ND, not determined.
Cloning and sequencing of MudJ fusion junctions.
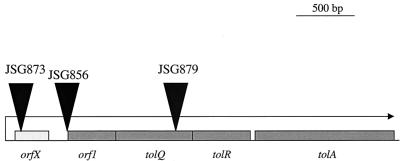
The DNA sequence upstream of the MudJ insertion site was determined for each of the three unique bile-sensitive strains. The sequence was obtained from clones containing the transposon-chromosomal-DNA fusion junctions, which were selected by screening for those clones containing the Kanr gene of the transposon. The sequencing revealed that all three insertions were located in different regions of the Salmonella serovar Typhimurium tol gene cluster. These genes encode important membrane colicin transport and phage receptor proteins, which also appear to play a major role in membrane integrity. One insertion was shown to be located in tolQ (JSG879), one was in orf1 (JSG856), and one was in an open reading frame (ORF) upstream of the orf1-tolQRA operon (JSG873) (Fig. 3). The insertion in tolQ was located near the 3" end of the gene. Based on genome sequence information, Salmonella serovar Typhimurium tolQ shows >86% DNA identity (>94% at the protein level) to E. coli tolQ. The JSG856 MudJ insertion is located in an untranslated region 22 bp upstream from the orf1 ATG start codon. This gene product is also very similar to its E. coli homolog, sharing >92% identity at the amino acid level. The third insertion is located within a small ORF (orfX) located immediately upstream of the orf1-tolQRA operon. This gene is predicted to produce a 9.0-kDa protein that has not previously been assigned a Tol function. Of the Salmonella serovar Typhimurium tol cluster gene products, OrfX shares the least identity with its E. coli counterpart (82%).
FIG. 3.
Chromosomal map of the orf1-tolQRA chromosomal region showing the locations of MudJ transposon insertions. Solid triangles above the map show the locations of the MudJ insertions that mapped to this region.
Analysis of bile resistance gene regulation and operon arrangement.
Because a PhoPc strain which exhibits enhanced bile resistance compared to the level of the wild-type strain was used as a target of MudJ mutagenesis, it was possible that the identified bile resistance loci were PhoPQ regulated. To determine if the identified loci were regulated by PhoPQ, the MudJ insertions were transduced into a strain with a PhoP− background and β-galactosidase activities in strains with PhoPc and PhoP− backgrounds were compared. For orfX, which contained a MudJ insertion in the wrong orientation to produce an active lacZ fusion product, a chromosomal gene fusion to firefly luciferase was created. The levels of β-galactosidase and luciferase activity did not significantly vary in any of the mutants between these two backgrounds, indicating that the identified genes were not regulated by PhoPQ (data not shown).
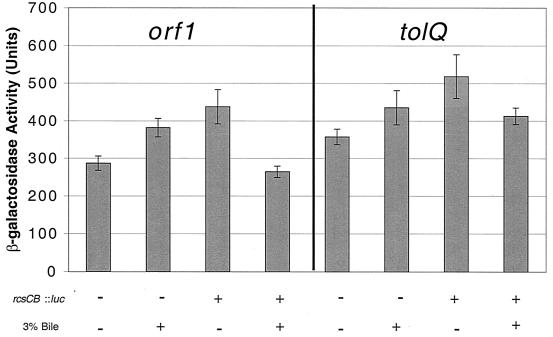
The tolQRA genes in E. coli are regulated by the RcsCB two-component system (6). This can be observed genetically with transcriptional fusions. An rcsCB mutation was constructed by the recombination of a suicide plasmid within this locus. The rcsC::pGPL01 mutation was moved by P22-mediated transduction into strains containing transcriptional fusions to orf1 and tolQ. Transcription assays showed no differences between the fusions in the rcsC::pGPL01 background and those in the wild-type background (Fig. 4). Therefore, the RcsCB system does not appear to regulate the transcription of the tol genes in Salmonella serovar Typhimurium.
FIG. 4.
Effects of bile and RcsCB on the transcription of orf1 and tolQ. Strains were grown in the presence (+) or absence (−) of a functional RcsCB two-component system. Transcription was also measured after growth in the presence (+) or absence (−) of 3% bile.
The three unique bile-sensitive strains were also examined for regulation by environmental bile. The MudJ fusions were transduced into a wild-type background, grown in the presence or absence of 2.5% bile, and assayed for β-galactosidase activity. As with the assays for PhoPQ regulation, none of these fusions were activated or repressed in the presence of bile (data for orf1 and tolQ are shown in Fig. 4).
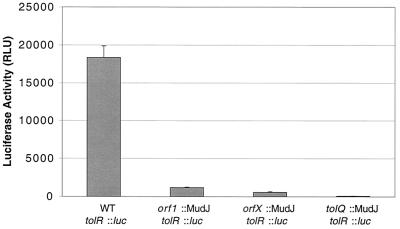
The analysis of the transcription of the tol cluster of E. coli has produced conflicting results, but recent data suggest that orf1 is cotranscribed with the tolQRA operon (23, 34). However, no data exist concerning orfX and the potential of its operonic arrangement with the rest of the tol cluster other than the suggestion that a Rho-independent terminator might exist 3" of orfX in E. coli (23). To determine the transcriptional units within this cluster, the effect of transposon insertions in orfX, orf1, and tolQ on a chromosomal tolR::luciferase reporter was measured. These results demonstrate that the entire tol cluster, including orfX and orf1, is likely cotranscribed (Fig. 5), as tolR transcription in orfX, orf1, and tolQ transposon mutants is reduced 16-, 32-, and 294-fold, respectively. These data suggest that, as predicted for E. coli, orf1 and tolQRA are cotranscribed, but the data contradict the prediction that orfX would not be cotranscribed with these genes.
FIG. 5.
Effects of insertion mutations in upstream genes on the transcription of tolR. Mutations in orf1, orfX, and tolQ were individually placed on the chromosome upstream of a tolR::luc reporter. The level of luciferase activity (measured in relative light units [RLU]) was determined for each strain, which demonstrated the cotranscription of the entire orfX-orf1-tolQRA cluster.
DISCUSSION
As enteric pathogens, Salmonella spp. are resistant to the action of bile. One to three percent of individuals infected with Salmonella typhi develop a chronic, asymptomatic infection most often located in the gallbladder. In this environment, a greater resistance to bile than that expressed in the intestine would be necessary, and it is of interest to define those genes necessary for high-level resistance to bile. Because of this interest, a strain with high-level bile resistance (constitutively expressing the PhoPQ two-component system) was used as a target for mutagenesis in this study.
MudJ mutants were replica plated onto LB agar plates with 10% bile and screened for those unable to grow. Fourteen strains were identified as bile sensitive. Of these, three grew very poorly and three were shown by SDS-PAGE and P22 phage cross-streaking to be rough and as such were eliminated from further analysis. It has been previously shown that E. coli LPS mutants, especially those that are deep rough and missing core heptose residues, possess increased sensitivity to bile salts (26).
Southern blot analysis was also performed to determine the uniqueness of the insertions in these mutants, a process which eliminated one strain from further consideration. Finally, backcrosses of MudJ from the mutant strains into PhoPc strains were used to confirm that the bile sensitivity phenotype was associated with the transposon. Three of the eight remaining insertions retained their phenotype upon transduction back into JSG208 (PhoPc), suggesting that in the four other strains, a mutation not linked to the transposon resulted in bile sensitivity.
Upon cloning and sequencing the MudJ-chromosomal-DNA fusion junctions from JSG856, JSG873, and JSG879, all were found to be located in the tolQRA chromosomal region. Most of the tol genes in E. coli and other organisms are found in two linked operons consisting of orf1-tolQRA and tolB-pal-orf2. Salmonella homologs of this tol cluster had not previously been described. The genetic organization of the tol cluster appears to be fairly well conserved, as diverse organisms such as Pseudomonas spp. and E. coli have similar organizations of these genes (7, 29, 31). The tol genes are responsible for the transport of colicin A into the cell and for the uptake of filamentous phage DNA (4, 31, 35). Strains containing a mutation in tolQ are therefore colicin A and filamentous phage resistant and have increased sensitivity to some antibiotics and detergents, including bile salts. It is thought that the increased detergent sensitivity is due to a loss of membrane integrity, but the molecular details of this sensitivity are unclear.
The function of the orf1 product is not known, and it has been reported that a transposon insertion in orf1 does not possess the tol phenotype (31). This is surprising, as this transposon insertion should be polar on downstream genes known to elicit the tol phenotype, and such an operon arrangement (orf1-tolQRA) has been described for E. coli. Our data suggest that orf1 is both cotranscribed with tolQRA and involved in the Tol phenotype of bile salt resistance.
While an E. coli tolA mutant could not be constructed, a tolQ mutant with a polar effect on tolA has been described to grow more slowly and have reduced 07 LPS production (12). Reduced O-antigen or LPS production does not appear to be a phenotype of the Salmonella serovar Typhimurium tol mutants, as all produced full-length LPSs comparable to that of the parental strain. In addition, the tol mutants possessed no growth defects in comparison to the parental strain (data not shown).
Interestingly, the MudJ insertion in JSG873 was located in an ORF (orfX) directly upstream of the orf1-tolQRA operon. This gene has not been assigned a Tol function. Although this gene is transcribed in the same direction as orf1 and tolQRA, it was not predicted to be cotranscribed, since a gap of 150 bp exists between upstream orfX and orf1 and a putative Rho-independent terminator exists 3" of the orfX gene. However, the data presented here demonstrate that a transposon insertion in orfX is polar on the transcription of the downstream tol genes, suggesting that orfX is cotranscribed with downstream orf1 and tolQRA. Also consistent with our findings is the possibility that the orfX product affects the transcription of orf1 and tolQRA. However, we feel that this scenario is unlikely, as OrfX is not similar in structure or sequence to transcriptional activators. Furthermore, given the fact that the interruption of this gene results in a bile sensitivity phenotype, we propose that orfX is a member of the tol cluster in Salmonella serovar Typhimurium (and likely in E. coli).
In E. coli, the RcsCB two-component system, which regulates capsule synthesis and tolQRA, activates cps to produce a protective capsule in response to perceived membrane damage, such as the loss of tol gene function (6). This also requires growth at low temperatures (20 to 30°C), such that the growth of an E. coli tol mutant at 20 to 30°C produces a mucoid colony. The fact that this phenotype was not apparent in the Salmonella serovar Typhimurium tol mutants (data not shown) suggests that the RcsCB system of Salmonella serovar Typhimurium does not sense membrane damage due to Tol loss, the cps cluster is not regulated by RcsCB in Salmonella serovar Typhimurium, or the entire repertoire of cps genes is not present in Salmonella serovar Typhimurium. In addition, analysis of tolQ transcription in an rcsCB mutant showed that it was unaffected compared to tolQ transcription in a wild-type background; thus, our data suggest that RcsCB may not regulate transcription of the tolQRA cluster or mucoidy in response to membrane damage (e.g., tol mutations) in Salmonella serovar Typhimurium.
Examination of the transcriptional activities of the orfX, orf1, and tolQ genes in PhoPc and PhoP− backgrounds showed no significant differences. Furthermore, regulation of the β-galactosidase activities of the mutant strains grown in the presence or absence of bile also showed no differences. These data suggest that these loci are not part of the PhoP regulon or part of the bile resistance pathway that can be upregulated by encountering bile in the medium.
Although the number of mutants screened was conservative, at a >90% level of confidence (approximate) of hitting all genes in the genome, we were surprised by the fact that no efflux pump-associated mutations (mar or acr) or mutations in phoPQ were identified. This may suggest that our mutagenesis was not complete enough; however, three independent insertions were found in the orf1-tolQRA region. Numerous potential explanations exist for these findings, including the presence of MudJ hotspots or the selection of certain mutations based on the methodology of mutant screening. Regardless, the fact that screening at a >90% confidence level uncovered no PhoP-regulated genes necessary for the bile resistance phenotype may suggest that the phenotype is not controlled by a single PhoP-regulated gene. Further studies using two-dimensional gels to identify bile-regulated protein spots and comparing these spots to known PhoP-regulated proteins may help to identify novel PhoP-regulated and non-PhoP-regulated genes involved in bile resistance.
Acknowledgments
We thank other members of the Gunn laboratory for their suggestions and comments on the manuscript.
This work was supported by the San Antonio Area Foundation.
REFERENCES
- 1.Alpuche-Aranda, C. M., E. P. Berthiaume, B. Mock, J. A. Swanson, and S. I. Miller. 1995. Spacious phagosome formation within mouse macrophages correlates with Salmonella serotype pathogenicity and host susceptibility. Infect. Immun. 63:4456-4462. [DOI] [PMC free article] [PubMed]
- 2.Alpuche-Aranda, C. M., E. L. Racoosin, J. A. Swanson, and S. I. Miller. 1994. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J. Exp. Med. 179:601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourdineaud, J.-P., S. P. Howard, and C. Lazdunski. 1989. Localization and assembly into the Escherichia coli envelope of a protein required for entry of colicin A. J. Bacteriol. 171:2458-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullas, L. R., and J.-I. Ryu. 1983. Salmonella typhimurium LT2 strains which are r− m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clavel, T., J. C. Lazzaroni, A. Vianney, and R. Portalier. 1996. Expression of the tolQRA genes of Escherichia coli K-12 is controlled by the RcsC sensor protein involved in capsule synthesis. Mol. Microbiol. 19:19-25. [DOI] [PubMed] [Google Scholar]
- 7.Dennis, J. J., E. R. Lafontaine, and P. A. Sokol. 1996. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J. Bacteriol. 178:7059-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 9.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, J. W., and H. K. Hall. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, R., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 12.Gaspar, J. A., J. A. Thomas, C. L. Marolda, and M. A. Valvano. 2000. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38:262-275. [DOI] [PubMed] [Google Scholar]
- 13.Gunn, J. S., C. M. Alpuche-Aranda, W. P. Loomis, W. J. Belden, and S. I. Miller. 1995. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177:5040-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 15.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, L., K. Lim, J. S. Gunn, B. Bainbridge, R. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 17.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohmann, E. L., C. A. Oletta, and S. I. Miller. 1995. Evaluation of phoP/phoQ deleted aroA-deleted live oral S. typhi vaccine in humans. Vaccine 14:19-24. [DOI] [PubMed] [Google Scholar]
- 19.Lacroix, F. J., A. Cloeckaert, O. Grepinet, C. Pinault, M. Y. Popoff, H. Waxin, and P. Pardon. 1996. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol. Lett. 135:161-167. [DOI] [PubMed] [Google Scholar]
- 20.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, S. I., and J. J. Mekalanos. 1990. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, S. I., W. S. Pulkkinen, M. E. Selsted, and J. J. Mekalanos. 1990. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium. Infect. Immun. 58:3706-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller, M. M., and R. E. Webster. 1997. Characterization of the tol-pal and cyd region of Escherichia coli K-12: transcript analysis and identification of two new proteins encoded by the cyd operon. J. Bacteriol. 179:2077-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169-181. [DOI] [PubMed] [Google Scholar]
- 26.Picken, R. N., and I. R. Beacham. 1977. Bacteriophage-resistant mutants of Escherichia coli K12. Location of receptors within the lipopolysaccharide. J. Gen. Microbiol. 102:305-318. [DOI] [PubMed] [Google Scholar]
- 27.Provenzano, D., and K. E. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuhmacher, D. A., and K. E. Klose. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 181:1508-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen, K., D. J. Sikkema, and T. F. Murphy. 1996. Isolation and characterization of the Haemophilus influenzae tolQ, tolR, tolA and tolB genes. Gene 178:75-81. [DOI] [PubMed] [Google Scholar]
- 30.Sulavik, M. C., M. Dazer, and P. F. Miller. 1997. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 179:1857-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, T.-P., and R. E. Webster. 1987. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J. Bacteriol. 169:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vianney, A., M. M. Muller, T. Clavel, J. C. Lazzaroni, R. Portalier, and R. E. Webster. 1996. Characterization of the tol-pal region of Escherichia coli K-12: translational control of tolR expression by TolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J. Bacteriol. 178:4031-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster, R. E. 1991. The tol gene products and the import of macromolecules into Escherichia coli. Mol. Microbiol. 5:1005-1011. [DOI] [PubMed] [Google Scholar]