Abstract
Selenium can provoke contrasting effects on living organisms. It is an essential trace element, and low concentrations have beneficial effects, such as the reduction of the incidence of cancer. However, higher concentrations of selenium salts can be toxic and mutagenic. The bases for both toxicity and protection are not clearly understood. To provide insights into these mechanisms, we analyzed the proteomic response of Escherichia coli cells to selenate and selenite treatment under aerobic conditions. We identified 23 proteins induced by both oxides and ca. 20 proteins specifically induced by each oxide. A striking result was the selenite induction of 8 enzymes with antioxidant properties, particularly the manganese and iron superoxide dismutases (SodA and SodB). The selenium inductions of sodA and sodB were controlled by the transcriptional regulators SoxRS and Fur, respectively. Strains with decreased superoxide dismutase activities were severely impaired in selenium oxide tolerance. Pretreatment with a sublethal selenite concentration triggered an adaptive response dependent upon SoxRS, conferring increased selenite tolerance. Altogether, our data indicate that superoxide dismutase activity is essential for the cellular defense against selenium salts, suggesting that superoxide production is a major mechanism of selenium toxicity under aerobic conditions.
At high concentrations, selenium salts are toxic compounds (40, 44). They are mutagenic in prokaryotes (30, 41) and have been reported to cause several types of diseases (37). However, selenium is an essential trace element for many living organisms from bacteria to mammals (28, 40). Small amounts of selenium are required to synthesize the amino acid selenocysteine present in a few proteins, such as formate dehydrogenases in Escherichia coli or glutathione peroxidases and thioredoxin reductases in higher eucaryotes (reviewed in reference 45). Another benefit of dietary selenium in mammals is to prevent chemicals from inducing tumors (14, 40, 41), but the mechanism of this prevention is not clearly understood. Selenium has been shown to inhibit the intracellular JNK/SAPK signaling and p38MAPK cascades (32) and some transcription factors (13, 16, 43). Some of these inhibitions occur through a thiol redox mechanism (31, 32), but it is not known whether this mechanism is involved in the anticarcinogenic properties of this element.
Selenium is naturally occurring as selenate (SeO42−) and selenite (SeO32−) in the environment (40). These inorganic oxidized forms, particularly selenite, are toxic. In the biogeochemical cycle of selenium, various redox reactions are produced by microorganisms (24, 46): in E. coli, selenate and selenite are detoxified through their reduction into the elemental selenium (Se0) or metabolized to volatile hydrogen selenide (HSe−), which can be incorporated into selenocysteine. In vitro studies (51) have shown that the reduction of selenite involves reactions with sulfhydryl groups of thiol-containing molecules such as glutathione, leading to the production of intermediate metabolites selenodiglutathione (GS-Se-SG), glutathioselenol (GS-SeH), and hydrogen selenide (HSe−) and finally to elemental selenium (10, 15). Certain reactions of this pathway produce hydrogen peroxide (H2O2) and superoxide (O2· −) (17, 39), which can cause damage to cell membranes and DNA (for a review, see reference 49). Thus, it has been suggested that selenite toxicity is due to oxidative stress (33, 39), while selenate has toxic effects only after being reduced to selenite or selenol (-SeH) (51). Consistent with this hypothesis, Kramer and Ames (17) showed that the oxyR1 mutant of Salmonella enterica serovar Typhimurium, in which a general defense against oxidative stress is constitutively expressed, is hyperresistant to selenite. However, other in vivo evidences for this hypothesis are lacking. Furthermore, in prospect of the potential use of selenium in cancer therapy or prevention, it is important to understand its toxicity mechanism since the difference between the nutritional level of selenium and its toxic level for human health is quite narrow.
In the present work, we used two-dimensional (2D) gel electrophoresis to identify proteins induced by selenite and selenate in E. coli. The identity of these proteins provides new insights into the mechanism of selenium toxicity and the cellular protection against this compound under aerobic conditions. In particular, our data strongly suggest that the toxicity of selenite is mostly due to the formation of superoxide radicals.
MATERIALS AND METHODS
Materials, bacterial strains, and culture conditions.
Sodium selenite, sodium selenate, and methyl-viologen (paraquat) were purchased from Sigma-Aldrich, and hydrogen peroxide was obtained from Fluka. 32P-labeled nucleotide and [35S]methionine were purchased from Amersham.
Bacterial strains used in this study were derivatives of E. coli K-12 strain and are listed in Table 1. ΔoxyR::kan (from GSO9, a gift from G. Storz), Δahp::kan (from DSA103, a gift from R. Hayward), Δhns-1001::Tn5 seq-1 (21), katE::Tn10 (UM120 [23]), katG::Tn10 (UM202 [23]), and ΔgshA::kan (WP748 [36]) mutations were introduced by P1 transductions as previously described (6). To construct katE::Tn10 katG::Tn10, the katG::Tn10 mutation was introduced into MG1655 katE::Tn10 by cotransduction with rha::Tn5 and kanamycin-resistant (Kmr) transductants were screened for their inability to produce bubbles when treated with hydrogen peroxide. The strain was further transduced for rha+ Kms and a transductant selected for the inability to bubble in the presence of hydrogen peroxide. Cultures were grown aerobically at 37°C in Luria-Bertani (LB) medium or M9 glucose medium containing 10 μg of thiamine/ml in Erlenmeyer flasks in a rotary shaker. When appropriate, the medium was supplemented with chloramphenicol (25 μg/ml), tetracycline (20 μg/ml), ampicillin (100 μg/ml), or kanamycin (50 μg/ml).
TABLE 1.
E. coli bacterial strains used in this study
| Bacterial strain | Relevant genotype or characteristicsa | Source or reference |
|---|---|---|
| MC4100 | F−araD139 Δ(lacIPOZYA-argF)U169 rpsL thi | Laboratory collection |
| GC4468 | F− ΔlacU169 rpsL Smr | 5 |
| QC772 | GC4468 Φ(sodA-lacZ)49 Cmr Lac+ | 5 |
| QC773 | GC4468 Φ(sodB-kan)1-Δ2 Kmr | 5 |
| QC1726 | GC4468 ΔsodA3, sodB::MudPR3, Cmr | 48 |
| QC6019 | GC4468 gshA20::kan Kmr | This work |
| QC6020 | GC4468 ΔsodA3, sodB::MudPR3, gshA20::Tn10 Cmr Kmr | This work |
| BW829 | GC4468 Δsox8::cat Smr Cmr | 50 |
| QC1732 | GC4468 fur::kan | 7 |
| MG1655 | wt | Genetic Center |
| QC2413 | MG1655 ΔoxyR::kan Kmr | This work |
| QC2809 | MG1655 Δahp::kan Kmr | This work |
| QC2476 | MG1655 katE::Tn10 katG::Tn10 Tetr | This work |
| QC2816 | QC2476 Δahp::kan Tetr Kmr | This work |
| QC2736 | MG1655 Δhns-1001::Tn5 seq-1 | 21 |
| DHB4 | F′ lac-pro lacI /Δ(ara-leu)7697 araD139 ΔlacX74 galE galK rpsL phoR Δ(phoA)PvuII ΔmalF3 thi | 36 |
| AD494 | DHB4 trxB::kan Kmr | 36 |
| WP840 | DHB4 gor522…mini-Tn10 Tetr | 36 |
| UC5710 | arg56 nad113 araD81 Δ(uvrB-bio) Δ(ogt-fnr)1 | 35 |
| UC844 | UC5710ΔtrxA | 35 |
| UC1369 | UC5710ΔtrxA gshA | 35 |
Cmr, chloramphenicol resistance; Smr, streptomycin resistance; Tetr, tetracycline resistance; Kmr, kanamycin resistance; wt, wild type.
Labeling and 2D gel electrophoresis, spot identification, and quantification.
Labeling experiments were performed in M9 medium at 37°C. Exponentially growing cells (2 ml) were treated with selenate or selenite (2 mM) for 30 or 120 min and then labeled with 200 μCi of l-[35S]methionine/ml. After 20 min, cells were harvested by centrifugation for 5 min at 3,000 × g and washed twice in water at 4°C, and the pellet was frozen at −180°C. Proteins were extracted and separated on a 2D gel as described previously (25) on a Millipore Investigator apparatus. After electrophoresis, the gels were stained with Coomassie brilliant blue R-250, dried, and processed for autoradiography by standard procedures.
Spots of interest were excised from 2D gels, washed, and then reswollen with a porcine trypsin solution. The digests were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (42), providing a list of peptide masses (5 to 14, depending on the spot). This peptide mass fingerprint was then submitted to an appropriated software database (MS-FIT or PROFOUND), and proteins were identified with a minimum of 80% matching fragments.
The spots on the radioactive gels were recorded by PhosphorImager technology (Molecular Dynamics) and analyzed with a 2D gel analysis software (MelanieII; Bio-Rad). The spot intensities were obtained in pixel units and normalized to the total radioactivity of the gel. The selenium oxide stimulation index was calculated as the ratio of spot intensity between the selenium oxide and standard conditions. The standard deviation of the analysis ranged from 20 to 25%.
Adaptation experiments.
Overnight cultures were used to inoculate 5 ml of glucose M9 medium to an initial optical density at 600 nm (OD600) of 0.05. When the suspension had reached an OD600 of 0.2, 30 μM H2O2, 100 μM paraquat, 500 μM SeO42−, or 250 μM SeO32− was added. In some pretreatment experiments, 100 μg of chloramphenicol/ml was also added. After 60 min, cells were challenged with 5 mM H2O2 or 25 mM SeO32−. After 0, 30, 60, 120, and 360 min, aliquots of the cell culture were withdrawn, diluted in minimal medium, and spread on nutrient broth plates. Plates were incubated at 37°C and, after 20 h, the CFU were counted.
Sensitivity assays.
Patch assays were performed as follows: 5-μl aliquots containing ca. 103 cells of an overnight culture were spotted on rich LB medium containing SeO42− or SeO32− at the indicated concentrations. Plates were monitored after 2 days of incubation at 37°C.
Preparation of crude extracts and enzymatic activities.
After exposure to stress conditions, cells were harvested by centrifugation for 20 min at 20,000 × g (4°C) and were resuspended in ice-cold 50 mM (pH 7.8) phosphate buffer, 1 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride]. Cells were disrupted by using a French press (1.4 × 108 Pa). Unbroken cells were removed by centrifugation at 17,000 × g (4°C) for 10 min. The soluble and membrane fractions of cell extracts were separated by ultracentrifugation for 1 h at 150,000 × g (4°C). The protein concentration was determined by the method of Bradford (4) with bovine serum albumin as the calibrating standard. Catalase activity was determined by a spectrophotometric method previously described (2). Total superoxide dismutase (SOD) activity in extracts was assayed by the method described by McCord and Fridovich (27). One unit of enzyme activity was defined as the amount of enzyme required to cause 50% inhibition in the rate of reduction of ferricytochrome c under the conditions of the assay. Detection of SOD in nondenaturing 8% polyacrylamide gels was carried out by an in situ staining procedure according to the riboflavin-nitroblue tetrazolium method described by Beauchamp and Fridovich (1). This method is based on the production of superoxide by photochemical reaction with riboflavin. Superoxide reacts with nitroblue tetrazolium to form formazan blue. The dismutation of superoxide by SOD in gel prevents the coloration. The enzymatic activity values are means of three independent determinations.
Northern blot analysis.
Exponentially growing cells (OD600 of 0.35) were treated with 2 mM SeO42− or SeO32− and collected after 0, 10, 20, or 30 min. Total cell RNA was prepared by the hot phenol protocol (38). For each condition tested, 10-μg RNA samples were separated by electrophoresis in 0.8% agarose-3.7% formaldehyde gels and transferred onto nylon membrane (Roche Molecular Biochemicals). The probes used were obtained by PCR and were labeled with the Megaprime DNA labeling systems (Amersham) with [α-32P]dCTP. Blots were hybridized overnight at 65°C in 0.5 M NaPO4-5% sodium dodecyl sulfate-10 mM EDTA with the indicated specific DNA probe. Signals were detected by autoradiography and PhosphorImager technology (Molecular Dynamics) for quantification. The radioactivity associated with the hybridized RNA bands was normalized to the amount of rRNA present in each lane to correct for differences in sample loading.
RESULTS
Adaptive response to selenium salts.
In a preliminary experiment, we tested E. coli tolerance to selenium salts by spreading cells on LB plates containing increasing concentrations of selenite (SeO32−) or selenate (SeO42−). A significant decrease in the frequency of colony formation was observed with selenite concentrations of >5 mM. This frequency dropped to 4% with 8 mM selenite. In contrast, selenate concentrations as high as 400 mM had no effect on E. coli growth (data not shown).
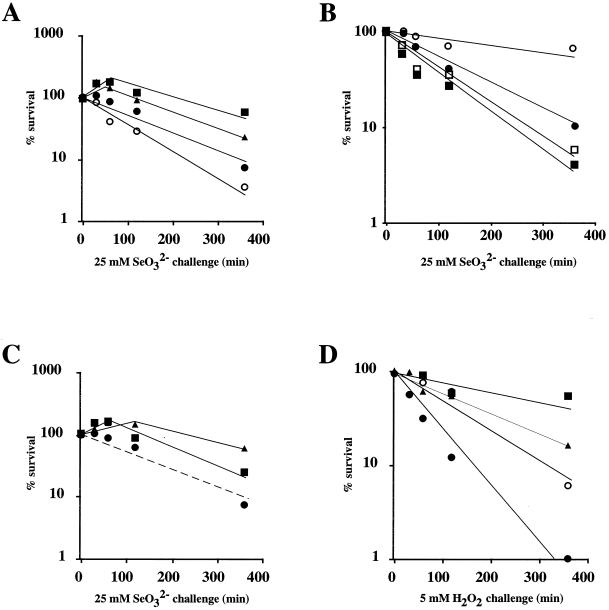
The survival after selenite exposure was tested by adding 25 mM selenite to exponentially growing cultures. The survival decreased to 10% after 6 h (Fig. 1A). When cultures were pretreated 1 h before the challenge with sublethal concentrations of selenate or selenite (500 or 250 μM, respectively), a significant protection against selenite killing was observed (Fig. 1A), suggesting an adaptive response. When chloramphenicol was added (100 μg/ml) with the selenite pretreatment, the protective effect of the pretreatment was abolished, indicating that this protection requires the synthesis of a new set of proteins. Pretreatment with paraquat and H2O2, which induces a protective response against oxidative stress, also protected from selenite killing (Fig. 1C). Likewise, selenate or selenite pretreatment protected cells from H2O2 (Fig. 1D) or paraquat killing (data not shown). These results, which are consistent with previous data obtained with Salmonella serovar Typhimurium (17), indicate that oxidative stress is a significant factor in selenate or selenite toxicity and that antioxidant defenses are probably induced by selenate and selenite treatment.
FIG. 1.
Adaptive response of E. coli to selenite and H2O2 stresses. Exponentially growing cells (E. coli MC4100, GC4468, or BW829) were pretreated or not with 30 μM H2O2, 100 μM paraquat, 500 μM SeO42−, or 250 μM SeO32− in the presence or absence of 100 μg of chloramphenicol/ml. After 60 min of treatment, 25 mM selenite (A, B, and C) or 5 mM H2O2 (D) was added to the cultures. At intervals, samples were diluted and plated onto LB agar to monitor cell viability. The data are mean values from at least three experiments, with essentially similar results. (A) MC4100 strain. Symbols: •, not pretreated; ▪, pretreated SeO32−; ▴, pretreated SeO42−; ○, pretreated chloramphenicol plus SeO32−. (B) GC4468 strain (symbols: •, not pretreated; ○, pretreated SeO32−) and BW829 strain (symbols: ▪, not pretreated; □, pretreated SeO32−). (C) MC4100 strain. Symbols: •, not pretreated; ▪, pretreated H2O2; ▴, pretreated paraquat. (D) MC4100 strain. Symbols: •, not pretreated; ▪, pretreated H2O2; ▴, pretreated SeO32−; ○, pretreated SeO42−.
To analyze whether the induced resistance to selenium by selenite pretreatment was due to induction of the regulons involved in the defense against superoxide and/or hydrogen peroxide, the experiment was repeated in ΔsoxRS and ΔoxyR mutants. Induced resistance was suppressed in ΔsoxRS (Fig. 1B) but was maintained in ΔoxyR (data not shown), indicating that the induction of defense against superoxide stress was responsible for the increased protection against selenite.
Induction of genes encoding antioxidant enzymes.
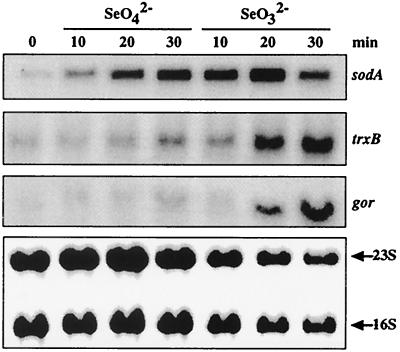
We thus analyzed whether some selected genes known to be induced by various oxidative stresses were also induced by selenate or selenite treatment. The genes chosen for this analysis were sodA, gor, and trxB, encoding, respectively, the manganese SOD, the glutathione reductase, and the thioredoxin reductase. The former gene is induced by superoxide, whereas hydrogen peroxide induces expression of the last two genes (reviewed in reference 47). Cells were treated with 2 mM selenate or selenite. This concentration was arbitrarily chosen within a range of concentrations which does not affect cell survival. After 10 to 30 min, total mRNA was prepared and submitted to Northern blot analysis with sodA, trxB, and gor probes. These genes were strongly induced by selenite with induction factors of >5 after 20 to 30 min of treatment (Fig. 2). Selenate treatment gave less-intense inductions (by a factor of ∼2). Note that with each treatment, sodA presented the more pronounced and rapid induction. These results indicate that several genes of the oxidative stress stimulon are induced and that the induction of the oxidative stress responses is a significant part of the cellular response to selenium salts.
FIG. 2.
Transcriptional induction of antioxidant genes in response to selenate and selenite. Total cellular RNA was isolated from cells treated with SeO42− (2 mM) or SeO32− (2 mM) for 0, 10, 20, and 30 min. The Northern blots were hybridized with sodA, trxB, or gor radiolabeled DNA probes. 16S and 23S rRNA were used for loading calibration.
Proteomic response to selenium salts.
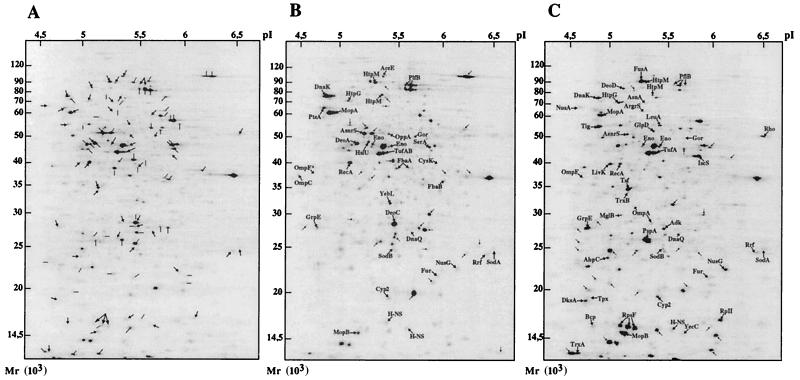
The above mRNA analysis was limited to three selected genes and did not provide data concerning the synthesis of the corresponding enzymes. We thus undertook a large-scale proteome analysis based on 2D gel electrophoresis to examine the modification of protein expression in response to selenium salts. Exponentially growing cells were untreated or treated with 2 mM selenite or selenate for 30 min, pulse-labeled with [35S]methionine, and analyzed by comparative 2D gel electrophoresis (Fig. 3). The changes in spot intensity between untreated and treated cells were quantified by PhosphorImager and software analysis (see Materials and Methods). The analysis focused on the protein spots showing a variation of expression higher than a factor of 1.7. This threshold was selected as being sufficiently high to limit the number of spots to analyze and to be certain that the induction is significant. According to this criterion, more than 100 protein spots were induced after selenite or selenate treatment, and more than 60 other proteins were significantly repressed. Proteins induced by the selenium salts were extracted and analyzed by MALDI-TOF mass spectrometry (see Materials and Methods). Among 85 protein spots analyzed by mass spectrometry, 70 spots were identified, corresponding to 58 distinct gene products. The identified proteins could be sorted in functional groups: enzymes with antioxidant properties, heat shock proteins, components of the transcription machinery, enzymes involved in protein synthesis, carbohydrate and amino acid metabolism enzymes, proteins involved in the transport of small molecules, and other unclassified proteins or proteins with unknown function (Table 2).The proteome analysis was also performed 2 h after selenate or selenite treatment (Table 2). The pattern of induced proteins and of their expression levels was similar to the one obtained at 30 min, indicating that the inductions were not transient. Among 46 proteins induced by selenite and 37 proteins induced by selenate, 23 were induced by both oxides. The others were specific to either selenite or selenate. Note, for example, the specific induction by selenate of both fructose bisphosphate aldolases FbaA and FbaB. Conversely, proteins such as PspA, DksA, or IscS were strongly induced by selenite but not by selenate.
FIG. 3.
Comparative 2D gel electrophoresis analyses of total E. coli proteins expressed in response to selenium oxide treatment. Autoradiograms of 2D gels performed with total E. coli extracts from [35S] methionine-labeled cells as described in Materials and Methods are shown. The extracts were prepared from control untreated cells (A), from cells exposed to SeO42− (2 mM) for 30 min (B), and from cells exposed to SeO32− (2 mM) for 30 min (C). Proteins whose synthesis rate is stimulated upon SeO42− or SeO32− exposure were identified by mass spectrometry and are indicated on the map. Protein spots induced but not characterized are also indicated by an arrow. Proteins repressed by SeO42− or SeO32− are indicated by a black bar in panel A.
TABLE 2.
Identification of proteins induced by selenate and selenite
| Protein source or type | Protein name | Stimulation index after exposure to:
|
|||||
|---|---|---|---|---|---|---|---|
| Selenate (2 mM) at:
|
Selenite (2 mM) at:
|
||||||
| 30 min | 120 min | 30 min | 120 min | ||||
| Antioxidant protein | |||||||
| Glutathione reductase | Gor | 1.4 | 2 | 2 | 3.3 | ||
| Iron superoxide dismutase | SodB | 1.9 | 2 | 3 | >3 | ||
| Manganese superoxide dismutase | SodA | 1.4 | 2.8 | 1.9 | 3.4 | ||
| Alkyl hydroperoxide reductase C22 protein | AhpC | NIa | 1.5 | >2 | >2 | ||
| Protein with similarity to AhpC | Bcp | NI | NI | 2.6 | 2.8 | ||
| Thiol peroxidase | Tpx | NI | NI | 1.8 | 2.2 | ||
| Thioredoxin reductase | TrxB | NI | NI | 2.1 | 4 | ||
| Thioredoxin | TrxA | NI | 1.5 | 3.4 | 4.1 | ||
| Heat shock protein, chaperone, and protease | |||||||
| Heat shock protein Hsp70 | DnaK | 4.4 | 3.4 | 1.5 | 1.5 | ||
| Hsp70 cofactor | GrpE | 2.7 | 2.1 | 4.5 | 3.5 | ||
| GroEL protein-Hsp | MopA | 3.2 | 2.2 | 1.9 | 2 | ||
| GroES protein-Hsp | MopB | 1.6 | 2.5 | 4.1 | 3.7 | ||
| Heat shock protein HslU | HslU | 1.4 | 2.1 | NI | NI | ||
| Peptidyl-prolyl cis-trans isomerase B | Cyp2 | 1.9 | 1.7 | 2.2 | 2.1 | ||
| Heat shock protein C62.5 | HtpG | 2.3 | 2.2 | 2.6 | 4.2 | ||
| ATP-dependent Clp proteinase regulatory chain B | HtpM | 3.2 | 2.9 | 2.3 | 2.1 | ||
| Trigger factor (protein export) | Tig | NI | NI | 2.6 | 2.7 | ||
| Carbohydrate metabolism | |||||||
| Formate acetyltransferase I | PlfB | 4.8 | 6.5 | 5.5 | 3.6 | ||
| Fructose bisphosphate aldolase class I | FbaB | 2.1 | 2 | NI | NI | ||
| Fructose bisphosphate aldolase class II | FbaA | 5.4 | 7 | NI | NI | ||
| Aerobic glycerol-3-phosphate dehydrogenase | GlpD | NI | NI | 1.6 | 1.9 | ||
| Pyruvate dehydrogenase E1 component | AceE | 1.9 | 1.8 | NI | NI | ||
| Phosphoenolpyruvate-protein phosphotransferase | PtsA | 1.5 | 1.9 | NI | NI | ||
| Enolase | Eno | 3.4 | 5.7 | 1.7 | 1.7 | ||
| Protein translation | |||||||
| Elongation factor EF-G | FusA | NI | NI | 1.6 | 1.9 | ||
| Elongation factor EF-Ts | Tsf | NI | NI | 2.4 | 2.3 | ||
| Protein chain elongation factor EF-Tu | TufAB | 1.7 | 2.6 | 2.3 | 1.8 | ||
| 30S ribosomal protein S6 | RpsF | NI | 2.1 | 3.9 | 3.3 | ||
| Ribosome recycling factor | Rrf | 1.4 | 2.9 | 1.7 | NI | ||
| Amino acid metabolism | |||||||
| Aspartate-ammonia ligase | AsnA | NI | NI | 1.7 | 2.3 | ||
| Asparaginyl-tRNA synthetase | AsnrS | 1.9 | 2 | 1.7 | 1.7 | ||
| Arginyl-tRNA synthetase | ArgrS | NI | NI | 1.5 | 1.9 | ||
| 2-Isopropylmalate synthase | LeuA | NI | NI | 3.2 | 2.4 | ||
| Cysteine synthase A | CysK | 1.8 | 2.2 | NI | NI | ||
| d-3-Phosphoglycerate dehydrogenase | SerA | 2 | 2.9 | NI | NI | ||
| Transcription | |||||||
| DNA-binding protein H-NS | H-NS | 2 | 2 | 1.7 | 2 | ||
| Ferric uptake regulation protein | Fur | 1.8 | 1.5 | >2 | >2 | ||
| DNA polymerase III, epsilon chain | DnaQ | 1.9 | 1.8 | 1.9 | 2 | ||
| Homologue to N-terminal domain of RNA polymerase β | RpII | NI | NI | 4.1 | 3.3 | ||
| Component in transcription anti-terminaiton | NusG | 2.7 | 4.3 | 3.1 | 2.6 | ||
| Transcription termination factor Rho | Rho | NI | NI | 1.5 | 1.7 | ||
| Component in transcription | NusA | NI | NI | 2.3 | 1.8 | ||
| Nucleotide and deoxyribonucleotide catabolism | |||||||
| Purine nucleoside phosphorylase | DeoD | NI | 1.6 | NI | 1.8 | ||
| Deoxyribose-phosphate aldolase | DeoC | 1.6 | 1.7 | NI | NI | ||
| Thymidine phosphorylase | DeoA | 2.1 | 1.8 | NI | NI | ||
| Transport of small molecules | |||||||
| Outer membrane protein A | OmpA | NI | NI | 2.7 | 1.5 | ||
| Outer membrane protein C | OmpC | 2.6 | 2.4 | NI | NI | ||
| Outer membrane protein F (matrix protein) | OpmF | 3.8 | 3.2 | 4.1 | 4 | ||
| Leucine specific binding protein | LivK | NI | NI | 2.5 | 2.6 | ||
| d-Galactose-binding periplasmic protein precursor | MglB | NI | NI | 2 | 1.9 | ||
| Amino acid ABC transporter binding protein | YecC | NI | NI | 3.2 | 2.2 | ||
| Periplasmic oligopeptide-binding protein | OppA | 1.8 | 2.1 | NI | 2 | ||
| Unclassified | |||||||
| Adenylate kinase | Adk | NI | NI | 2.9 | 2.1 | ||
| Phage shock protein A | PspA | NI | NI | 7.3 | 8 | ||
| NifS protein homolog | IscS | NI | NI | 4.3 | 4.3 | ||
| RecA protein | RecA | 2.1 | 2.6 | NI | >2 | ||
| DnaK suppressor protein | DksA | NI | NI | 9.2 | 5.4 | ||
| YebL protein | YebL | 3 | 2.2 | NI | NI | ||
NI, not induced.
Antioxidant enzymatic activities are increased.
As expected, numerous enzymes with antioxidant properties have been found induced by selenate or selenite treatment (Table 2). These included enzymes involved in the degradation of superoxide anion (SodA and SodB) and proteins involved in protection against hydrogen peroxide (TrxA, TrxB, and Tpx) which are supposed to be the two main reactive oxygen species produced during selenate or selenite metabolization (17, 39). The catalases could not be analyzed because the corresponding spots have not been identified on our 2D maps. We thus directly measured the catalase activity (Table 3) and found that it was augmented nearly twofold after selenate treatment and more than fourfold after selenite challenge. The total SOD activitiy was also significantly increased in response to both treatments (Table 3). The respective activities of manganese and iron SODs (MnSOD and FeSOD) were also measured (Table 4). We found that SodA and SodB activities were increased by selenium salt treatment, a finding consistent with the induction level of these enzymes measured on 2D gels (Table 2). These results suggest that, for most of the proteins of the selenate or selenite stimulon, the increase in enzyme activity correlates with the increase in enzyme expression.
TABLE 3.
Total SOD and catalase activities in E. coli MC4100 crude extracts after selenium oxide treatment
| Enzyme assayeda | Sp act (U/mg) ± SD of:
|
|||||
|---|---|---|---|---|---|---|
| Control at 0 min | SeO42− at:
|
SeO32− at:
|
||||
| 30 min | 120 min | 30 min | 120 min | |||
| Total SOD | 14.7 ± 1.5 | 22.2 ± 2 | 32.3 ± 3 | 35.6 ± 4 | 62.3 ± 6 | |
| Total catalase | 10 ± 1.5 | 17.3 ± 2.6 | 19.2 ± 3 | 38.5 ± 6 | 44.8 ± 7 | |
Cells (crude extracts) were treated with 2 mM SeO42− or SeO32− for 30 or 120 min. All assays were performed on exponential-phase cells and monitored by the spectrophotometry direct method as described in Materials and Methods.
TABLE 4.
MnSOD and FeSOD activities in extracts of wild-type and mutant strains
| Strain | SOD | Mean SOD activity ± SDa of:
|
||
|---|---|---|---|---|
| Control | SeO42− | SeO32− | ||
| GC4468 (reference) | MnSOD | 4.8 ± 0.5 | 7.2 ± 0.7 | 13.9 ± 1.5 |
| FeSOD | 5.5 ± 0.6 | 9.6 ± 1 | 12.7 ± 1.5 | |
| QC772 (ΔsodA) | FeSOD | 6.9 ± 0.7 | 11.4 ± 1.5 | 12.1 ± 1.5 |
| QC773 (ΔsodB) | MnSOD | 5.8 ± 0.6 | 8.1 ± 0.8 | 12.6 ± 1.5 |
Cells were treated with 2 mM SeO42− or SeO32− for 30 min. The SOD activities (MnSOD, SodA; FeSOD, SodB) are expressed in arbitrary units and are estimated from the gel densitometer tracings of nondenaturing gels.
sod mutants are hypersensitive to selenium salts.
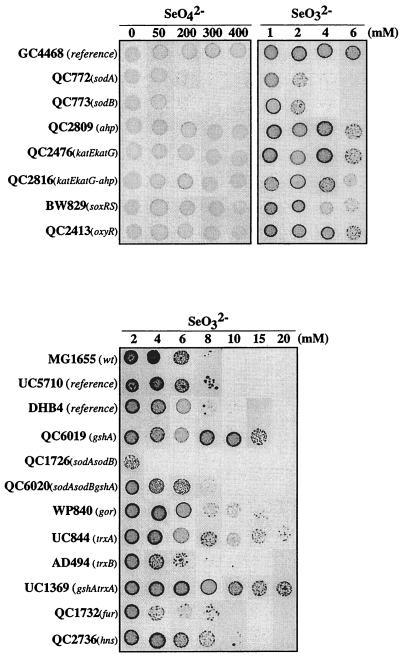
We sought to identify enzymatic activities important in selenite and/or selenate tolerance. As oxidative stress is supposed to play a role in selenate or selenite toxicity (reference 39 and the present study), we focused on antioxidant activities and particularly enzymes shown to be induced in response to selenium salt treatment. We analyzed the tolerance of a series of strains carrying a deletion in different antioxidant genes (Fig. 4). Strains lacking either sodA or sodB were hypersensitive to both selenite and selenate. Moreover, the sodA sodB double mutant was markedly more sensitive to selenite. However, the selenate sensitivity of the double mutant was not increased compared to that of the single mutants. When these mutants were transformed with a plasmid overexpressing sodA, they recovered a wild-type tolerance phenotype (data not shown). In contrast, deletion of either katEkatG, ahp, trxA, trxB, gor, or gshA did not resulted in selenite or selenate sensitivity. On the contrary, ΔgshA, ΔtrxA, and ΔgshAΔtrxA strains were significantly more resistant than the reference strains (Fig. 4). Interestingly, the gshA sodA sodB triple mutant was markedly more resistant to selenite than was the sodA sodB mutant (Fig. 4), indicating that the absence of glutathione relieves the hypersensitivity of strains lacking SODs.
FIG. 4.
Selenium salt tolerance of wild-type and mutant strains. Mutant strains altered in antioxidant functions were compared to parental strains (GC4468, MG1655, UC5710, and DHB4) by patch assay for their ability to grow on solid LB medium containing SeO42− or SeO32− at the indicated concentrations. For each strain, 5 μl of an overnight culture (103 cells) was spotted onto plates.
SoxRS and Fur are regulators of the selenium response.
The sodA sodB double mutant was the most sensitive strain to selenium salts, indicating that SOD activities play a particular role in selenium resistance. Since both SOD activities were found to be induced upon selenium salt treatment, we attempted to determine by which mechanisms selenium triggers these inductions. sodA expression has been shown to be controlled by SoxRS in response to oxidative stress (6, 12). On the contrary, sodB is not induced by oxidative stress (9) but is induced in iron sufficiency via Fur and is repressed via H-NS in iron deficiency (7). Interestingly, both Fur and H-NS were among the proteins induced by the selenium salts and identified on 2D gels (Table 2). We then analyzed the induction of SodA and SodB activities in response to selenite for the three strains devoid of the transcriptional regulators SoxRS, Fur, or H-NS (Table 5). We found that the induction of SodA activity is defective in the ΔsoxRS strain and that the induction of SodB activity is defective in the Δfur strain. The strain with a deletion of hns showed normal inductions of manganese and iron SOD activities.
TABLE 5.
MnSOD and FeSOD induction factors by a 30-min selenite treatment in regulatory mutant strains
| Strain | SOD activitya of:
|
|
|---|---|---|
| MnSOD | FeSOD | |
| GC4468 (reference) | 2.9 | 2.3 |
| BW829 (ΔsoxRS) | 1.1 | 2.5 |
| QC1732 (Δfur) | 2.5 | 1 |
| MG1655 (reference) | 2.6 | 2.1 |
| QC2413 (ΔoxyR) | 2.5 | 2 |
| QC2736 (Δhns) | 3.0 | 2.6 |
Cells were treated with 2 mM SeO32−. The standard error for the induction factor values was ca. 20%.
The strains lacking the different regulators were also tested for selenite tolerance. As expected, the ΔsoxRS and Δfur strains defective in full SOD induction were hypersensitive to selenite (Fig. 4).
DISCUSSION
To gain insights into the biological effects of selenium salts and to identify activities relevant to its toxic and protective effects, we analyzed the proteomic response of E. coli cells to selenate and selenite treatment under aerobic conditions. Among the 23 proteins induced by both oxides, antioxidant enzymes and particularly the manganese and iron SODs (SodA and SodB) were evidenced. SOD activity was essential for selenium tolerance, suggesting that superoxide production is a major mechanism of selenium toxicity.
Selenium treatment causes in vivo superoxide production.
In most microorganisms, selenium salts are detoxified through their reduction into the elemental selenium (Se0) or metabolized to volatile hydrogen selenide (HSe−), which can be incorporated into selenocysteine. In vitro analyses have shown that some of the oxido-reductive reactions of selenium metabolism produce hydrogen peroxide (H2O2) and superoxide (O2· −) (17, 39), suggesting that selenite toxicity is due to oxidative stress. Consistent with that hypothesis, we have found that eight enzymes with antioxidant properties are induced by selenite treatment and particularly enzymes involved in superoxide degradation (SodA and SodB) and in hydrogen peroxide degradation (TrxA, TrxB, Tpx, and catalase). Both SOD and catalase activities were increased in response to selenium salts. However, only SOD and not catalase and peroxidase was essential for selenium tolerance. Although soxRS can be activated by signals other than superoxide (22, 26, 52, 53), its activation by selenium oxides suggests that superoxide is generated during the selenium reduction process. Furthermore, the induced resistance after sublethal selenite treatment was dependent on SoxRS, the regulator of the global response to superoxide, but not on OxyR, the global response regulator to hydrogen peroxide. Altogether, these results strongly suggest that the in vivo production of superoxide and not hydrogen peroxide is responsible for the toxicity of selenium salts. This conclusion is consistent with the in vitro data of Kramer and Ames (17) showing that H2O2 is predominantly formed at a low thiol concentration and O2· − is mainly produced at a high thiol concentration. In E. coli, the intracellular concentration of glutathione measured in the millimolar range (11), and the presence of other sulfhydryls of proteins probably favor superoxide production.
Although these data strengthen the notion that superoxide production is a major mechanism of selenium oxide toxicity, other mechanisms of selenium salt toxicity exist certainly. In particular, under anaerobic conditions in which superoxide cannot be produced, selenite is also toxic, although at higher concentrations than under aerobic condition (data not shown). Anaerobic mechanism of toxicity remains to be characterized.
Importance of cellular sulfhydryls in selenite toxicity.
According to in vitro studies (10, 15), the cellular O2· − production is the result of reactions between selenite and cellular sulfhydryl compounds such as reduced glutathione or reduced cysteine residues of proteins. Consistent with this model, E. coli and a Salmonella serovar Typhimurium strain devoid of glutathione (ΔgshA) were hyper-resistant to selenite (17; the present study). The fact that the sensitivity of the sodA sodB double mutant was relieved by the ΔgshA mutation also argues that glutathione plays an important role in the production of superoxide. Moreover, we observed the hyper-resistance of the ΔtrxA strain, suggesting that thioredoxin also participates in the toxicity mechanism. In support of this hypothesis, selenite and GS-Se-SG have been shown to be efficient oxidants of E. coli thioredoxin (3, 18).
Induction of IscS, a selenocysteine lyase.
Selenate metabolization is supposed to produce selenite in vivo (51). It was then surprising that some proteins such as PspA, DksA, or IscS are strongly induced by selenite but not by selenate. Our interpretation is that the intracellular selenite concentration resulting from selenate uptake and metabolization is probably lower than the intracellular concentration of selenite when cells are exposed to this compound. Such a limiting step in the selenate uptake or metabolization to selenite would also account for the low toxicity of selenate compared to selenite.
Among the proteins induced by selenite but not by selenate, we identified IscS, one of the three E. coli NifS homologs. These enzymes have been shown to be involved in different aspects of sulfur metabolism: the repair of the Fe-S cluster, the desulfurization of cysteine, and sulfur transfer in the biosynthesis of thiamine, NAD, and thionucleosides (20). A major effect of superoxide is iron sulfur cluster damage (49) and, in this context, the induction of IscS would be a logical response for repairing this damage. Interestingly, these enzymes have also a selenocysteine lyase activity catalyzing the decomposition of selenocysteine to alanine and elemental selenium Se0 (29). It also functions as a selenide delivery protein in the biosynthesis of selenophosphate and Se-tRNAs (19). Thus, the function of IscS is not clear in the response to selenium salts but could be a part of the cellular response to increase the production of Se0, the inactive form of selenium, and to trap the highly toxic and reactive coumpond HSe−.
Activation of SodA and SodB production by selenium salts.
SodA and SodB production and activity are significantly increased after selenium oxide treatment. sodA is a member of the soxRS regulon which is activated by superoxide. We found that the increase of SodA in response to selenium salt treatment is soxRS dependent. More surprising was the Fur-dependent increase of SodB. The mechanism of activation of sodB expression is still unknown and presumably is mediated by another regulatory protein, itself regulated by Fur (7, 8). Since no other Fur-regulated protein was identified on the 2D gel, the selenium effect on sodB expression might rather be mediated via this putative intermediate regulator.
Remarkably, the selenium oxide treatment is the only condition described thus far that triggers the induction of both SODs. Indeed, oxidative stress generated by paraquat or hydrogen peroxide treatment induces sodA but not sodB (34, 53). Conversely, sodB is highly expressed when iron is present in excess, a condition that decreases sodA expression (7, 49). The coordinated induction of both enzymes when cells are treated by selenium oxides is probably necessary to maximize the cellular SOD activity for the defense against these compounds.
Acknowledgments
M.B. was supported by a Contrat de Formation par la Recherche from CEA.
We thank Carmen Pueyo and Arne Holmgren for the gifts of mutant strains. We thank Francis Biville and Evelyne Turlin for help with analysis of the 2D gels and Jacques Coves, Benoit Pinson, Daniel Spector, Michel Toledano, and André Sentenac for critical reading of the manuscript.
REFERENCES
- 1.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 2.Beers, R. F., and I. W. Sizer. 1952. A spectrophotometric method for measuring breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133-140. [PubMed] [Google Scholar]
- 3.Bjornstedt, M., S. Kumar, and A. Holmgren. 1992. Selenodiglutathione is a highly efficient oxidant of reduced thioredoxin and a substrate for mammalian thioredoxin reductase. J. Biol. Chem. 267:8030-8034. [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compan, I., and D. Touati. 1993. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J. Bacteriol. 175:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrac, S., and D. Touati. 2002. Fur-mediated transcriptional and post-transcriptional regulation of FeSOD expression in Escherichia coli. Microbiology 148:147-156. [DOI] [PubMed]
- 9.Fee, J. A. 1991. Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol. Microbiol. 5:2599-2610. [DOI] [PubMed] [Google Scholar]
- 10.Ganther, H. E. 1968. Formation by the reaction of thiols with selenious acid. Biochemistry 7:2898-2905. [DOI] [PubMed] [Google Scholar]
- 11.Gérard-Monnier, D., and J. Chaudière. 1996. Metabolism and antioxidant function of glutathione. Pathol. Biol. 44:77-85. [PubMed] [Google Scholar]
- 12.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handel, M. L., C. K. Watts, A. deFazio, R. O. Day, and R. L. Sutherland. 1995. Inhibition of AP-1 binding and transcription by gold and selenium involving conserved cysteine residues in Jun and Fos. Proc. Natl. Acad. Sci. USA 92:4497-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ip, C. 1998. Lessons from basic research in selenium and cancer prevention. J. Nutr. 128:1845-1854. [DOI] [PubMed] [Google Scholar]
- 15.Kice, J. L., T. W. S. Lee, and S.-T. Pan. 1980. Mechanism of the reaction of thiols with selenite. J. Am. Chem. Soc. 18:102-113. [Google Scholar]
- 16.Kim, I. Y., and T. C. Stadtman. 1997. Inhibition of NF-κB DNA binding and nitric oxide induction in human T cells and lung adenocarcinoma cells by selenite treatment. Proc. Natl. Acad. Sci. USA 94:12904-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer, G. F., and B. N. Ames. 1988. Mechanisms of mutagenicity and toxicity of sodium selenite (Na2SeO3) in Salmonella typhimurium. Mutat. Res. 201:169-180. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., M. Bjornstedt, and A. Holmgren. 1992. Selenite is a substrate for calf thymus thioredoxin reductase and thioredoxin and elicits a large non-stoichiometric oxidation of NADPH in the presence of oxygen. Eur. J. Biochem. 207:435-439. [DOI] [PubMed] [Google Scholar]
- 19.Lacourcière, G. M., H. Mihara, T. Kurihara, N. Esaki, and T. C. Stadtman. 2000. Escherichia coli NifS-like proteins provide selenium in the pathway for the biosynthesis of selenophosphate. J. Biol. Chem. 275:23769-23773. [DOI] [PubMed] [Google Scholar]
- 20.Lauhon, C. T., and R. Kambampati. 2000. The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 275:20096-20103. [DOI] [PubMed] [Google Scholar]
- 21.Laurent-Winter, C., S. Ngo, A. Danchin, and P. Bertin. 1997. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response-identification of targets by two-dimensional electrophoresis. Eur. J. Biochem. 244:767-773. [DOI] [PubMed] [Google Scholar]
- 22.Liochev, S. I., L. Benov, D. Touati, and I. Fridovich. 1999. Induction of the soxRS regulon of Escherichia coli by superoxide. J. Biol. Chem. 274:9479-9481. [DOI] [PubMed] [Google Scholar]
- 23.Loewen, P. C., J. Switala, and B. L. Triggs Raine. 1985. Catalases HPI and HPII in Escherichia coli are induced independently. Arch. Biochem. Biophys. 243:144-149. [DOI] [PubMed] [Google Scholar]
- 24.Macy, J. M., S. Rech, G. Auling, M. Dorsch, E. Stackenbrandt, and L. I. Sly. 1993. Thauera selenatis gen. nov., sp. nov., a member of the beta subclass of Proteobacteria with a novel type of anaerobic respiration. Int. J. Syst. Bacteriol. 43:135-142. [DOI] [PubMed] [Google Scholar]
- 25.Maillet, I., G. Lagniel, M. Perrot, H. Boucherie, and J. Labarre. 1996. Rapid identification of yeast proteins on two-dimensional gels. J. Biol. Chem. 271:10263-10270. [DOI] [PubMed] [Google Scholar]
- 26.Manchado, M., C. Michan, and C. Pueyo. 2000. Hydrogen peroxide activates the soxRS regulon in vivo. J. Bacteriol. 182:6842-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 28.McKeehan, W. L., W. G. Hamilton, and R. G. Ham. 1976. Selenium is an essential trace nutrient for growth of WI-38 diploid human fibroblasts. Proc. Natl. Acad. Sci. USA 73:2023-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihara, H., T. Kurihara, T. Watanabe, T. Yoshimura, and N. Esaki. 2000. cDNA cloning, purification, and characterization of mouse liver selenocysteine lyase. Candidate for selenium delivery protein in selenoprotein synthesis. J. Biol. Chem. 275:6195-6200. [DOI] [PubMed] [Google Scholar]
- 30.Noda, M., T. Takano, and H. Sakurai. 1979. Mutagenic activity of selenium compounds. Mutat. Res. 66:175-179. [DOI] [PubMed] [Google Scholar]
- 31.Park, H. S., S. H. Huh, Y. Kim, J. Shim, S. H. Lee, I. S. Park, Y. K. Jung, I. Y. Kim, and E. J. Choi. 2000. Selenite negatively regulates caspase-3 through a redox mechanism. J. Biol. Chem. 275:8487-8491. [DOI] [PubMed] [Google Scholar]
- 32.Park, H. S., E. Park, M. S. Kim, K. Ahn, I. Y. Kim, and E. J. Choi. 2000. Selenite inhibits the c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) through a thiol redox mechanism. J. Biol. Chem. 275:2527-2531. [DOI] [PubMed] [Google Scholar]
- 33.Pinson, B., I. Sagot, and B. Daignan-Fornier. 2000. Identification of genes affecting selenite toxicity and resistance in Saccharomyces cerevisiae. Mol. Microbiol. 36:679-687. [DOI] [PubMed] [Google Scholar]
- 34.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prieto-Alamo, M. J., J. Jurado, R. Gallardo-Madueno, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 36.Prinz, W. A., F. Åslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272:15661-15667. [DOI] [PubMed] [Google Scholar]
- 37.Rayman, M. P. 2000. The importance of selenium to human health. Lancet 356:233-241. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seko, Y., and N. Imura. 1997. Active oxygen generation as a possible mechanism of selenium toxicity. Biomed. Environ. Sci. 10:333-339. [PubMed] [Google Scholar]
- 40.Shamberger, R. J. 1983. The biochemistry of selenium. Plenum Press, Inc., New York, N.Y.
- 41.Shamberger, R. J. 1985. The genotoxicity of selenium. Mutat. Res. 154:29-48. [DOI] [PubMed] [Google Scholar]
- 42.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 43.Spyrou, G., M. Bjornstedt, S. Kumar, and A. Holmgren. 1995. AP-1 DNA-binding activity is inhibited by selenite and selenodiglutathione. FEBS Lett. 368:59-63. [DOI] [PubMed] [Google Scholar]
- 44.Stadtman, T. C. 1974. Selenium biochemistry. Science 183:915-922. [DOI] [PubMed] [Google Scholar]
- 45.Stadtman, T. C. 1996. Selenocysteine. Annu. Rev. Biochem. 65:83-100. [DOI] [PubMed] [Google Scholar]
- 46.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of arsenic and selenium. FEMS Microbiol. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 47.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 48.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 50.Tsaneva, I. R., and B. Weiss. 1990. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 172:4197-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1998. Selenium metabolism in Escherichia coli. Biometals 11:223-227. [DOI] [PubMed] [Google Scholar]
- 52.Wu, J., and B. Weiss. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]