Abstract
ToxR, a transmembrane regulatory protein, has been shown to respond to environmental stimuli. To better understand how the aquatic bacterium Vibrio anguillarum, a fish pathogen, responds to environmental signals that may be necessary for survival in the aquatic and fish environment, toxR and toxS from V. anguillarum serotype O1 were cloned. The deduced protein sequences were 59 and 67% identical to the Vibrio cholerae ToxR and ToxS proteins, respectively. Deletion mutations were made in each gene and functional analyses were done. Virulence analyses using a rainbow trout model showed that only the toxR mutant was slightly decreased in virulence, indicating that ToxR is not a major regulator of virulence factors. The toxR mutant but not the toxS mutant was 20% less motile than the wild type. Like many regulatory proteins, ToxR was shown to negatively regulate its own expression. Outer membrane protein (OMP) preparations from both mutants indicated that ToxR and ToxS positively regulate a 38-kDa OMP. The 38-kDa OMP was shown to be a major OMP, which cross-reacted with an antiserum to OmpU, an outer membrane porin from V. cholerae, and which has an amino terminus 75% identical to that of OmpU. ToxR and to a lesser extent ToxS enhanced resistance to bile. Bile in the growth medium increased expression of the 38-kDa OMP but did not affect expression of ToxR. Interestingly, a toxR mutant forms a better biofilm on a glass surface than the wild type, suggesting a new role for ToxR in the response to environmental stimuli.
Vibrio anguillarum is a highly pathogenic bacterium that causes terminal hemorrhagic septicemia in marine fish, resulting in great economic losses within aquaculture (1, 3). The disease vibriosis is associated with high rates of mortality and shares many features with invasive septicemic diseases in humans (1). The infection model of V. anguillarum in salmonid fish has been suggested to be a useful model for studying host-pathogen interactions.
Although the exact mode of infection for V. anguillarum is still unclear, it most likely involves attachment to and colonization of host surfaces, followed by penetration of the tissues. Chemotactic motility has been suggested to aid the entry of V. anguillarum into the fish host (47). Furthermore, adhesion and invasion studies using a Chinook salmon embryo cell line and a set of isogenic motility mutants showed that invasion of but not adhesion to the cell line was significantly decreased in nonmotile or partially motile mutants (45). A smooth-swimming, chemotactic mutant, however, was hyperinvasive.
V. anguillarum utilizes fish skin and intestinal mucus as chemoattractants (48). This mucus should induce smooth swimming, which may aid the entry of V. anguillarum into the fish through the mucus layers. These studies strongly suggest that active motility is required for invasion of the fish host. Once the bacterium has invaded the fish, motility is no longer needed for the progression of vibriosis (37, 45, 47).
Several factors have been suggested to be important in the virulence mechanism of V. anguillarum (for reviews, see references 1 and 3). The well-characterized iron uptake system carried on the virulence plasmid pJM1 is essential for virulence. The siderophore anguibactin is produced, diffuses into the environment, and scavenges for iron which is bound to iron-binding proteins, such as transferrin, lactoferrin, and ferritin, that are found in the serum, secretions, and tissues of the fish, respectively. The siderophore then attaches to a receptor on the surface of the bacterium, leading to transport of iron into the bacterial cells, allowing V. anguillarum to multiply within the host. Lipopolysaccharides are required for virulence and play a role in serum resistance. In addition, although not genetically proven, extracellular products such as hemolysins, lipases, proteases, and a neurotoxic acetylcholinesterase have been suggested to play a role in the pathology of vibriosis.
Many pathogenic bacteria utilize signal transduction systems to regulate virulence gene expression in response to specific stimuli in the external environment (14, 15). In Vibrio cholerae, ToxR regulates multiple virulence genes, and this regulation is influenced by environmental stimuli such as pH, salt, temperature, amino acids, CO2, and cyclic AMP- cyclic AMP receptor protein (for reviews, see references 14 and 60). ToxR resides in the inner membrane and contains an amino-terminal cytoplasmic domain that has homology to DNA-binding domains of response regulators in the two-component family of transcriptional activators (34). ToxR interacts with another transmembrane protein, ToxS, which is thought to stabilize the conformation of ToxR optimal for transcriptional activation (12).
Although ToxR is required for expression of cholera toxin (33), toxin-coregulated pilus (62), and the accessory colonization factor (50), the regulation is indirect, via ToxR's control of expression of an additional activator, ToxT (13). Regulation of ToxT expression is complex and requires additional regulatory proteins, TcpP and TcpH (19). Furthermore, the motility status of the bacterium may be an important signal for the ToxR regulon (17). A ToxR mutant shows an increase in motility, while virulence gene production is decreased, whereas hyperswarmers have a reduction or loss in virulence factor production and nonmotile mutants produce cholera toxin under nonpermissive conditions.
ToxR has recently been found in a number of other Vibrio and Photobacterium species, including three fish pathogens, of which V. anguillarum is one (29, 30, 41, 46, 56, 66). In Vibrio vulnificus, ToxR was shown to positively regulate the expression of the vvh gene, which encodes a highly toxic hemolysin (29), while in Vibrio parahaemolyticus, ToxR regulates the gene encoding the thermostable direct hemolysin, which displays enterotoxic activities in a rabbit ileal loop model (30). In another study, ToxR was shown to aid the resistance of several Vibrio intestinal human pathogens to bile salts, suggesting a role for ToxR in the survival of these Vibrio species within the intestines of the host (53). In contrast to these studies, ToxR from V. anguillarum serotype O2 was shown not to regulate the virulence of the bacterium in an ayu fish model, the production of hemolysins or proteases, or resistance to bile (41). However, ToxR from V. anguillarum did regulate the production of major outer membrane proteins (OMPs), as seen with other Vibrio species (35, 53), and sensitivity to certain β-lactam antibiotics and to the anionic detergent sodium dodecyl sulfate (SDS) (41).
In this study, we have identified both toxR and toxS homologs from an O1 serotype V. anguillarum strain. The ToxR protein was 99% identical to that characterized recently from V. anguillarum (41). As found in the V. anguillarum serotype O2 study, ToxR regulated the production of a major OMP, the amino terminus of which is 75% identical to OmpU of V. cholerae (61), and ToxR is not likely a major regulator of virulence. In contrast to the serotype O2 study, we suggest that ToxR from V. anguillarum serotype O1 is required for enhanced resistance to bile. In fact, bile affected the expression of the major OMP. Unlike a V. cholerae toxR mutant, a V. anguillarum toxR mutant is not hypermotile. In addition, we show that ToxR negatively regulates its own expression and plays a role in biofilm formation.
MATERIALS AND METHODS
Strains, phage, plasmids, and media.
Bacterial strains and plasmids are described in Table 1. Escherichia coli SY327 (λpir) was used for transformation after subcloning fragments into the pDM4 suicide vector. All plasmids to be conjugated into V. anguillarum were transformed into E. coli S17-1 (λpir), which was used as the donor strain. Plasmid transfers from E. coli to V. anguillarum were done as previously described (37). E. coli XL1-Blue was used for bacteriophage lambda infections and for most transformations.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or relevant markers | Reference or source |
|---|---|---|
| E. coli | ||
| SY327 | Δ(lac-pro) argE(Am) rif malA recA56 λpir | 35 |
| S17-1 | thi pro hsdR hsdM+recA RP4-2-Tc::Mu-Km::Tn7 λpir | 59 |
| XL1-Blue | recA1 endA1 gyrA96, thi-1 hsdR17 supE44 relA1 Δ(lac-pro) [F′proAB lacIqlacZΔM15 Tn10(Tetr)] | Stratagene |
| V. anguillarum | ||
| NB10 | Wild type, serotype O1, clinical isolate from the Gulf of Bothnia | 40 |
| SY10 | Deletion of ToxR residues 559-1257 | This study |
| SY11 | Deletion of ToxS residues 1436-1948 | This study |
| Plasmids | ||
| pBluescript | Apr; ColE1 origin | Stratagene |
| pBSToxS-263 | Apr; pBluescript containing 263-bp PCR fragment from toxS (bp 1481-1732) | This study |
| pBSToxS-1 | Apr; pBluescript containing a cloned fragment with toxRS genes | This study |
| pDM4 | Cmr; suicide vector with an R6K origin (pir requiring) and sacBR genes from Bacillus subtilis | 37 |
| pDMToxR1 | Cmr; pDM4 derivative containing toxR bp 321-558 fused to bp 1258-1496 | This study |
| pDMToxS2 | Cmr; pDM4 derivative containing toxS bp 1197-1435 fused to bp 1949-2226 | This study |
| pSup202 | Cmr Tcr Apr; pBR325 derivative (ColE1 origin, RP4 mob+) | 59 |
| pDM8 | Cmr Tcr; pSup202 derivative containing promoterless lacZ gene used for transcriptional gene fusion studies | 10 |
| pDM8-ToxR | Cmr Tcr; pDM8 derivative containing a toxR::lacZ transcriptional gene fusion (residues 297-536) | This study |
| pSup202P | Cmr Tcr; pSup202 derivative (ColE1 origin, RP4 mob+) with a polylinker cloned into PstI within the Apr gene | 36 |
| pToxR | Cmr Tcr; pSup202P derivative containing toxR gene and its promoter (residues 321-1496) | This study |
| pMMB208 | Cmr IncQ lacIqPtac, polylinker from M13mp19 | 38 |
| pMMBToxR | Cmr; pMMB208 derivative containing toxR gene and its promoter (residues 321-1496) | This study |
For complementation analyses, two plasmids were created. The first, pToxR, is a pSup202P derivative and was used in all complementation studies except for the lacZ assays. To create this plasmid, primers ToxR-A (5′-GGACTAGTGTTATTTTCATTCAC-3′) and ToxR-E (5′-CTCGGTACCATAACCAACTACTGA-3′) were used in PCR to amplify a DNA fragment (bp 321 to 1496) carrying the wild-type toxR and its promoter. The fragment was digested with SpeI and KpnI (sites included on the primers) and cloned into the identical sites of pSup202P. The second plasmid, pMMBToxR, was used in lacZ assays because pToxR was not compatible with the pDM8 vector and its derivatives. To create this plasmid, a PCR fragment (bp 321 to 1496) containing the toxR gene and its promoter was obtained by using primers ToxR-A1 (5′-GGGGATCCGTTATTTTCATTCAC-3′) and ToxR-D (5′-CTCGAGCTCATAACCAACTACTGA-3′). Using the BamHI and SacI sites included on the primers, the PCR fragment was cloned into similar restriction sites of the pMMB208 vector. Both plasmids were mobilized into V. anguillarum via conjugation.
E. coli was routinely grown in Luria broth, which contains Bacto-tryptone (10 g/liter), Bacto yeast extract (5 g/liter), and sodium chloride (10 g/liter). For V. anguillarum, Trypticase soy broth medium (TSB) from BBL was used for routine growth, and the cultures were grown with aeration at 24°C. For selection against E. coli after conjugation, two vibrio selective media were used: TCBS agar (Difco Laboratories) and VAM medium (V. anguillarum specific) (2). Biofilm growth medium was minimal M63 salts (58) supplemented with 1% (wt/vol) NaCl, 1.5% (wt/vol) Casamino Acids, 1% (wt/vol) glucose, 1 mM MgSO4, and 10 μg of thiamine per ml.
Antibiotic concentrations for all E. coli strains were ampicillin at 100 μg/ml, tetracycline at 10 μg/ml, kanamycin at 30 μg/ml, and chloramphenicol at 25 μg/ml. Antibiotic concentrations for V. anguillarum in TSB and TCBS were tetracycline at 5 μg/ml and chloramphenicol at 5 μg/ml and, in VAM, chloramphenicol at 1 μg/ml.
DNA techniques and sequencing.
Oligonucleotides were synthesized using Applied Biosystems DNA/RNA synthesizer model 394. Unless otherwise stated, all conditions for the various DNA techniques were as described by Sambrook et al. (57). Reaction conditions for the DNA-modifying enzymes and DNA restriction enzymes were performed as suggested by the manufacturers. Double-stranded DNA sequencing was performed using the dideoxy chain termination method with T7 DNA polymerase (Pharmacia Biotech) and by primer walking in two directions from known regions of DNA sequence. The T7 and T3 primers were used for sequencing fragments in pBluescript (Stratagene).
PCR conditions.
PCR was performed as previously described (31) except that instead of the standard reaction buffer, a buffer containing 1% Thesit was used (51). When a PCR fragment required minimal errors, the high-fidelity Pfu polymerase (Stratagene) was used.
Cloning of the toxRS genes.
Since ToxS has little similarity with other proteins, protein alignment of the V. cholerae and V. parahaemolyticus ToxS proteins (30) was used to design two oligonucleotides, ToxS-1 [5′-GGTGAGCTCAGITGGTT(AG)TA(TC)TGGGG-3′] and ToxS-3 (5′-GGCACTAGTACCTTTCTCTGAAATATTAAT-3′), for use in PCR. ToxS-1 is a degenerate, inosine-containing oligonucleotide that is complementary to V. parahaemolyticus toxS codons for amino acids SWLYWG (30). ToxS-3 is directly complementary to V. parahaemolyticus toxS codons for amino acids INISEKG (30). A 263-bp fragment was amplified from the chromosome of V. anguillarum, purified from 1% agarose using Ultrafree-DA spin columns (Millipore), digested overnight with restriction enzymes SacI and SpeI (sites included on the primers), cloned into similarly digested pBluescript (Stratagene), creating pBSToxS-263, and sequenced. The deduced protein sequence of this fragment was 67% identical to ToxS of V. parahaemolyticus (30).
The 263-bp fragment was used as a probe to screen a previously described (36) genomic library from V. anguillarum in the Lambda Zap bacteriophage (Stratagene). The probe was labeled by random priming using [α-32P]dCTP as described by Sambrook et al. (57). Stringent hybridization and wash conditions were as previously described (37). The pBluescript plasmids containing a chromosomal insert were excised from positive plaques using earlier-described methods (36). One plasmid, pBSToxS-1, was chosen for sequencing of the toxRS genes.
Construction of toxR and toxS mutants.
For functional analyses, toxR and toxS deletion mutant strains were made by allelic exchange as previously described (37). To create the new deletion allele, PCR primers ToxR-A (5′-GGACTAGTGTTATTTTCATTCAC-3′) and ToxR-B (5′-ATGTGATTCATTGTAGATAATCTTCTTATT-3′) were used to create a fragment from bp 321 to 558 and primers ToxR-C (5′-TACAATGAATCACATACTGGAGAA-3′) and ToxR-D (5′-CTCGAGCTCATAACCAACTACTGA-3′) were used to create a fragment from bp 1258 to 1496. These two fragments contained a 15-bp overlap of similar sequence and were used as templates in a second PCR using primers ToxR-A and ToxR-D, creating a fragment that fused residues 321 to 558 to residues 1258 to 1496. Using SpeI and SacI (sites included on primers A and D), the fragment was digested and cloned into the similar sites of pDM4, creating pDMToxR1. After allelic exchange using pDMToxR1, only the last 55 codons of toxR remained, and the strain containing this deletion was called SY10.
A toxS deletion was created in a similar way to that for toxR using primers ToxS-A (5′-GGACTAGTGCCCATCAATCATCC-3′), ToxS-B (5′-ATTACTCGATGGCTAATTTATGCCCTTATT-3′), ToxS-C (5′-TAGCCATCGAGTAATCTGTAACTT-3′), and ToxS-D (5′-CTCGAGCTCTACGTTCCCAAATAC-3′). The second PCR created a fragment that fused residues 1197 to 1435 to residues 1949 to 2216. This fusion removes all codons for the toxS gene. This fragment was cloned as above into pDM4, creating pDMToxS2, which was used in allelic exchange, creating strain SY11. PCR amplification of each region from the chromosome and subsequent DNA sequencing confirmed the toxR deletion in SY10 and the toxS deletion in SY11.
Construction of toxR transcriptional gene fusion.
A transcriptional gene fusion was constructed between the promoter region of toxR and the reporter gene lacZ from E. coli. To create this fusion, a pSup202 derivative, pDM8 (10), which contains the entire lacZ gene and its ribosome-binding site but lacks the lacZ promoter, was used. The toxR promoter region lacking the possible ribosome-binding site (bp 297 to 536 upstream of toxR) was amplified using primers ToxR-βgal-1 (5′-TCCCCCGGGTCATACTAGCTCCAT-3′) and ToxR-βgal-3 (5′-TCCCCCGGGTGTTTAACCGTCCAT-3′). The fragment was digested with SmaI (site included on both primers) and ligated to the unique SmaI site just upstream of lacZ on pDM8, creating pDM8-ToxR. The promoter fusion was sequenced and then conjugated into various V. anguillarum strains.
β-Galactosidase assays.
V. anguillarum cultures were grown overnight at 24°C in TSB containing tetracycline (2 μg/ml). Cell cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in the same medium and further incubated at 24°C with shaking. In some assays, 0.04 and 0.4% bile salts (Difco) were added to the culture after subinoculation. Samples were taken at various time points, and β-galactosidase assays were performed according to Miller (32). Assays were performed in triplicate, using three independent colonies for each culture. After the assay was stopped, the bacterial debris was pelleted, and the A420 was measured for the reaction supernatant. Specific activity was determined as 1,000 × A420 × min−1 × ml−1 × A600−1. The vector, pDM8, without a fused promoter gave a background of approximately 200 Miller units, which were subtracted from the results presented in the figures.
Complementation of the toxR mutation in these assays required the presence of a second plasmid, pMMBToxR. Since double antibiotics in the medium slowed the growth of the complemented strain, only single antibiotic selection was used, and loss of one plasmid during the growth period occurred in approximately 5 to 10% of strains, as measured by growth on the appropriate antibiotic-containing media.
Measurement of resistance to bile salts.
V. anguillarum strains were grown overnight in TSB at 24°C. Cultures were diluted into 0.9% NaCl to 103 cells/ml, and 100 μl was spread onto TSA plates containing various concentrations of bile salts (Difco) or ox bile (Sigma). The same culture dilution was used for all plates so that a comparison could be made between colony counts. After 3 days of incubation at 24°C, single colonies were counted. These measurements were done at least three times.
OMP preparations.
OMP preparations were made essentially according to Filip et al. (16). The OD600 of 5-ml overnight cultures was determined, and equal numbers of cells were pelleted from each culture. The pellet was resuspended in 4.5 ml of distilled water, and the cells were disrupted by sonication on ice (30 cycles of 6-s pulses at 30% output, followed by 4 s off, in a Sonics and Materials Inc. Vibra cell). The cell extracts were centrifuged (5,000 × g for 10 min) to remove whole cells. To solubilize the cytoplasmic membrane, the supernatants were transferred to new tubes, and Sarkosyl (N-lauroyl sarcosine) was added to a final concentration of 2%. This mixture was incubated at room temperature for 30 min. To pellet the outer membranes, the mixture was centrifuged at 121,000 × g for 1 h at 4°C. The pellet, which consists of the outer membranes, was washed once in 4 ml of ice-cold distilled water and centrifuged again at 121,000 × g for 30 min at 4°C. The pellet was resuspended in 100 μl of sample buffer for separation by polyacrylamide gel electrophoresis (PAGE) (28), and 10 μl of each sample was applied to an SDS-12.5% polyacrylamide gel.
Bile induction of OmpU.
The wild-type strain was grown in TSB overnight, subinoculated into TSB containing 0, 0.04, or 0.4% bile salts (Difco), and grown with aeration for 18 h. Each culture was diluted to an OD600 of 1, and the bacterial cells from 1 ml were pelleted. The cell pellet was resuspended in 100 μl of sample buffer for SDS-PAGE (28), and 10 μl was applied to a gel for protein separation.
SDS-PAGE.
Protein separation was done as described by Laemmli (28) by SDS-12.5% PAGE (vertical mini-gel systems from C.B.S. Scientific). Electrophoresis was done for 1.5 h with 20 mA constant current. The gels were fixed and stained with 0.1% Coomassie brilliant blue in 40% methanol-10% acetic acid and then destained in 40% methanol-10% acetic acid.
Western analysis.
For Western analysis, proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Schleicher and Schuell) using a SemiPhor semidry blotter (Hoefer TE 70 series). Enhanced chemiluminescence (ECL) Western blotting was performed according to the manufacturer's instructions (Amersham Life Sciences). The blocking reagent used was 5% skim milk. The primary antibody was a rabbit polyclonal antiserum raised against OmpU from V. cholerae (a gift from James Kaper, University of Maryland School of Medicine) and used at a 1:4,000 dilution.
Motility measurements.
Motility was measured by movement of bacterial cells through TSB containing 0.25% agar. The optical density at 600 nm was determined for overnight cultures of all strains used. Equal amounts of cells were spotted in the center of the plates, and movement away from the center was measured after 24 h of growth at room temperature. This measurement was done three times.
Percent area coverage of a biofilm.
The percent area coverage due to biofilm production was determined essentially as described previously except that fresh medium was not added after 4 h (10). Briefly, the biofilms were grown on glass microscope slides freely suspended in a 400-ml beaker containing 300 ml of biofilm growth medium and 6 ml of an overnight culture diluted to an OD600 of 1. The culture was allowed to grow with stirring at “moderate speed” at room temperature. At various time points after inoculation, a glass slide was removed, and the biofilm was stained immediately with 1 ml of 0.1% acridine orange in potassium phosphate buffer (pH 7.4), washed, and air-dried. All experiments were done in triplicate. A growth curve for each culture was also done by removing culture samples and doing plate counts and OD600 measurements.
Five images were taken of the stained biofilms using a Zeiss Axioplan fluorescent microscope coupled to a charge-coupled device camera. Biofilm formation was quantified by determining the percent area coverage as previously described (10) using the Image Tool software, version 2, from the University of Texas Health Science Center at San Antonio. This was done for all five images taken, and the results were averaged.
Computer analysis.
Database searches were done using the sequence analysis software (11) of the Genetics Computer Group, Inc. (University of Wisconsin).
Fish infections.
Rainbow trout (Oncorhynchus mykiss) weighing approximately 10 to 15 g were infected with V. anguillarum either by intraperitoneal injections or by immersion in seawater containing V. anguillarum as previously described (37). The immersion and intraperitoneal infections were done at least two times. Five fish were infected for each bacterial dilution used. The 50% lethal doses (LD50s) were calculated as described by Reed and Muench (55). The LD50s recorded are an averaged number of all infections for each strain. To aid comparative analysis between strains, the standard deviation of the wild-type LD50 was calculated for both routes of infection. LD50 values were collected from previous studies in our lab and used in determining a standard deviation for the wild-type strain. For infection by immersion, 37 LD50s were used to give a standard deviation of 3.9 × 103 bacteria per ml of seawater. For the intraperitoneal route, 35 LD50s were used, giving a standard deviation of 29 bacterial cells.
Nucleotide sequence accession number.
The complete toxRS DNA sequence has been submitted to GenBank and given accession number AY065624.
RESULTS
Identification and mutagenesis of toxRS genes.
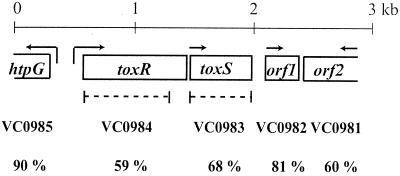
To characterize the toxRS homologs from V. anguillarum serotype O1, degenerate primers were designed from the V. parahaemolyticus toxS DNA sequence because ToxS has fewer domain structures that may have similarity to other proteins (30). Using these primers, a 263-bp PCR fragment was generated that encoded an amino acid sequence with 67% identity to ToxS of V. parahaemolyticus. This PCR fragment was used as a probe to screen a V. anguillarum genomic library for a chromosomal fragment carrying the toxRS homologs. The chromosomal fragment was sequenced, and the genetic organization is shown in Fig. 1.
FIG. 1.
Genetic organization of the toxR DNA locus. Each ORF is indicated by an open box, and the arrows indicate the direction of transcription. The dotted lines below the toxR and toxS genes indicate the deletions made in each gene. The homologous V. cholerae genes from the genomic sequence (20) are indicated below each ORF, along with the percent identity.
Two complete ORFs were identified and named toxR and toxS. The deduced ToxR protein sequence is 99% identical to the newly identified ToxR from V. anguillarum serotype O2 (41), 59% identical to ToxR of V. cholerae and V. parahaemolyticus (30, 33), and 57% identical to ToxR of V. vulnificus (29). The V. anguillarum serotype O1 and O2 ToxR proteins differed in two amino acids located within the variable region (positions 121 and 148). The deduced ToxS sequence is 67, 71, and 65% identical to that of ToxS from V. cholerae, V. parahaemolyticus, and V. vulnificus, respectively (29, 30, 33). ToxS was not completely sequenced in the V. anguillarum O2 serotype study (41).
In addition, one complete and two partial ORFs were also found (Fig. 1). The deduced 92 amino acids of the complete ORF1 are 81% identical to those encoded by the V. cholerae VC0982 gene from chromosome I, and it encodes a possible selenoprotein W-related protein (20). The partial deduced protein sequence of ORF2 is 60% identical to the product of the VC0981 gene from chromosome I of V. cholerae, which encodes a possible serine-threonine phosphatase (20). Upstream of toxR, a partial ORF was also found, and the deduced protein sequence is 90% identical to the product of the VC0985 gene from chromosome I of V. cholerae, which encodes the heat shock protein HtpG. The genetic organization of the five V. anguillarum ORFs is similar to that from the sequenced V. cholerae strain El Tor N16961 (20).
To determine the function of ToxR and ToxS, deletion mutations were made that deleted all but the last 55 codons for toxR and all codons for toxS (Fig. 1). This was done by allelic exchange with a mutated allele carried on a suicide vector as described previously (37). The toxR mutant was called SY10 and the toxS mutant was called SY11. To complement the toxR deletion, the wild-type toxR gene and its promoter were carried on pToxR, which was mobilized into the toxR mutant.
Resistance to bile salts and NaCl.
During isolation of the toxR mutant, we noticed that the toxR mutant had difficulties growing on TCBS selective medium and could not grow on VAM selective medium. The components of these two media were compared for differences that might account for the decreased growth. Since VAM contained 3.5% NaCl and 0.5% bile salts (Difco), whereas TCBS contained 1% NaCl and 0.8% ox bile (Sigma), the sensitivity of the mutant strains to high concentrations of bile and NaCl was tested. To avoid other components in the TCBS and VAM selective media that might also hinder growth, TSA plates containing various concentrations of ox bile (Sigma), bile salts (Difco), and NaCl were used. Equal amounts of bacterial cells were spread onto the bile- or NaCl-containing TSA plates. After 3 days, CFU were counted.
Table 2 shows the results from the bile salts (Difco) media. The wild type was resistant to up to 2% bile salts, after which the CFU decreased by half. The toxR mutant, however, had decreased CFU at the lowest concentration of bile salts tested and was unable to grow in 0.2% bile. For all concentrations of bile, the colony size of the toxR mutant was always much smaller than that of the wild type on a similar medium. Using similar amounts, ox bile (Sigma) had a milder effect on the toxR mutant (data not shown). Colony size was similar to the wild type, CFU were decreased at 0.8%, and growth was inhibited at 2%. For the toxS mutant, both bile salts (Table 2) and ox bile (data not shown) decreased the CFU at the highest concentrations used but never inhibited growth. The NaCl concentration was tested at 0.5, 1, 1.5, 2, 2.5, 3, and 3.5%, and no effect on the number of CFU was seen for the toxR or toxS mutant (data not shown). When the toxR mutation was complemented with the wild-type gene and its promoter, wild-type growth in the presence of all concentrations of bile salts and ox bile was regained. These data suggest that ToxR is required for optimal growth of V. anguillarum in the presence of bile.
TABLE 2.
Effect of bile salts on survival of V. anguillarum
| Genotype | CFU with indicated bile salts (Difco) concn in TSA
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0% | 0.04% | 0.08% | 0.2% | 0.4% | 0.8% | 2% | 4% | |
| Wild type | 90 | 97 | 100 | 127 | 122 | 100 | 117 | 63 |
| ΔtoxS | 130 | 118 | 117 | 107 | 57 | 59 | 24 | 11 |
| ΔtoxR | 65 | 40a | 44a | 0 | 0 | 0 | 0 | 0 |
| ΔtoxR/pSup202P | 60 | 43a | 32a | 13a | 0 | 0 | 0 | 0 |
| ΔtoxR/pToxR | 98 | 97 | 95 | 90 | 87 | 85 | 69 | 50 |
Colony size was greatly reduced compared to the wild type.
Regulation of the major OMP.
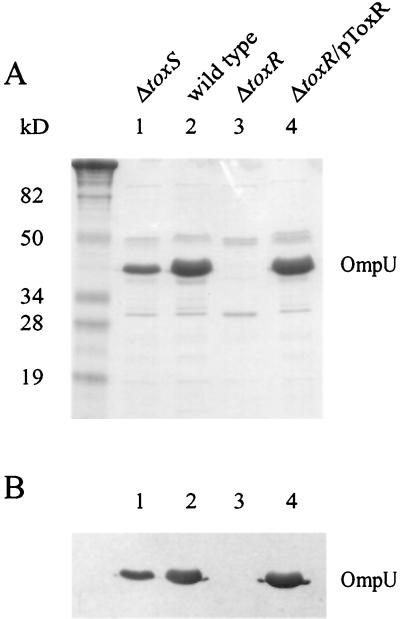
In many Vibrio species, ToxR has been shown to regulate the expression of outer membrane porins (35, 41, 53). In V. anguillarum serotype O2, ToxR was shown to positively regulate a 35-kDa OMP and negatively regulate a 46-kDa OMP (41). OMPs were isolated from the wild-type V. anguillarum serotype O1, the isogenic toxR mutant, and the isogenic toxS mutant and separated on SDS-12.5% PAGE. In Fig. 2A, at least nine OMPs were detected, and one protein approximately 38 kDa in size predominated and appeared to be positively regulated by ToxR. An OMP similar to the 46-kDa OMP of serotype O2 was not detected in any of the strains. The amount of the major 38-kDa OMP was decreased in the toxS mutant and undetectable in the toxR mutant.
FIG. 2.
OMP analyses. OMPs were isolated from the wild type, the toxS mutant, the toxR mutant, and the toxR mutant carrying a plasmid-encoded wild-type toxR gene (pToxR) as described in Materials and Methods. These proteins were applied to an SDS-12.5% PAGE gel and separated. The OMPs were stained with Coomassie blue (A) or transferred to nitrocellulose followed by Western blotting using an antiserum raised against OmpU from V. cholerae (B). Lanes 1, 2, 3, and 4 are labeled and are the same for both A and B. Molecular size markers are included in the leftmost lane, and sizes are indicated.
When the toxR mutation was complemented with the wild-type gene and its promoter, expression of the major OMP returned to wild-type levels. Amino-terminal protein sequencing of the major OMP revealed 25 amino acids (GELYNQDGTSLEMGGRAEARLSLKD) that showed 100% identity with the amino terminus sequence determined for the 35-kDa major OMP of V. anguillarum serotype O2 and 75% identity with OmpU of V. cholerae. Moreover, Western blot analysis using a rabbit polyclonal antiserum against V. cholerae OmpU cross-reacted with the V. anguillarum major OMP (Fig. 2B). These data strongly suggest that this protein is a homolog of OmpU from V. cholerae and that ToxR positively regulates its expression. This major OMP will be called OmpU due to its similarity to OmpU of V. cholerae.
Induced expression of OmpU but not ToxR by bile salts.
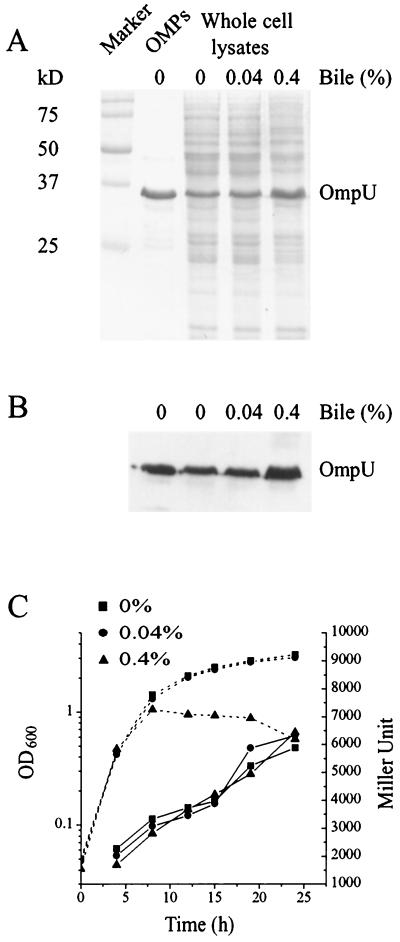
To determine if bile induces the expression of OmpU, as has been shown in other Vibrio species (53), the wild-type V. anguillarum was grown in 0, 0.04, and 0.4% bile salts (Difco), and proteins from whole cells were separated on SDS-12.5% PAGE. Figures 3A and B show that in the wild type, a major protein in the whole-cell lysate that migrates with OmpU and binds antiserum to V. cholerae OmpU is expressed at a higher level in the presence of 0.4% bile salts (Difco).
FIG. 3.
Effect of bile on expression of OmpU and toxR. (A) Production of OmpU in the presence and absence of bile was analyzed using whole-cell lysates of the wild type (NB10). Equal numbers of cells (lanes 3 to 5) from overnight cultures grown in the presence or absence of bile were pelleted, resuspended in loading buffer, applied to an SDS-12.5% PAGE gel, and either stained with Coomassie blue (A) or transferred to nitrocellulose followed by Western blotting using an antiserum raised against OmpU from V. cholerae (B). Lanes 1 to 5 are the same for both A and B. Lane 1, molecular size markers; sizes are indicated at the left. Lane 2, OMPs from the wild-type. Lane 3, wild type grown in the absence of bile. Lane 4, wild type grown in the presence of 0.04% bile. Lane 5, wild type grown in the presence of 0.4% bile. (C) β-Galactosidase activity of the toxR::lacZ transcriptional gene fusion (pDM8-ToxR) was measured in the wild type (NB10) in the presence (0.04 or 0.4%) or absence of bile. Overnight cultures were diluted into fresh TSB medium to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, dotted lines) and β-galactosidase expression (Miller units, solid lines). Miller units for the vector control are between 100 and 200 U, and these were subtracted from the results.
Is this increased expression of OmpU in the presence of bile due to an effect of bile on the expression of ToxR? Since the toxR mutant shows decreased growth in the presence of bile, we could not accurately determine the effect of bile on OmpU expression in this mutant. Instead, a toxR transcriptional gene fusion was made to the E. coli lacZ gene carried on a plasmid (pDM8-ToxR) that was mobilized into the wild-type strain. Figure 3C shows the expression of lacZ from the toxR promoter in the presence of 0, 0.04, and 0.4% bile salts. LacZ expression from the toxR promoter was not affected by bile salts (Difco). These results indicate that bile positively affects the expression of OmpU without affecting the expression of ToxR. However, we cannot rule out that bile may affect the activity of ToxR as a transcriptional factor.
Virulence analyses.
In contrast to V. cholerae, ToxR from V. anguillarum serotype O2 was shown not to be essential for virulence in an ayu fish model (42). We found comparable results when determining the LD50 for both the toxR and toxS mutants of our O1 serotype V. anguillarum. Both the immersion and intraperitoneal infection routes were used. For the immersion route, the LD50s of all three strains were 1.8 × 102, 1.3 × 103, and 2 × 102 bacteria per ml of seawater for the wild type, the toxR mutant (SY10), and the toxS mutant (SY11), respectively. A sevenfold difference in virulence was seen for the toxR mutant; however, the standard deviation of the wild-type LD50 was determined (from numerous previous studies) to be ±3.9 × 103, indicating that this sevenfold difference may not be significant.
For the intraperitoneal route, the LD50s were 92 (standard deviation of ±29 determined from numerous previous studies), 700, and 30 bacteria for the wild type, the toxR mutant (SY10), and the toxS mutant (SY11), respectively. The toxS mutant showed no difference from the wild type, and the toxR mutant showed only a slight increase in both the intraperitoneal route and the immersion route of infection, indicating that ToxR is not likely a major regulator of virulence in V. anguillarum serotype O1 as it is in V. cholerae.
Motility analysis.
In V. cholerae, motility and virulence are thought to be coordinately regulated, and a V. cholerae toxR mutant, which is decreased for virulence, was shown to be hypermotile (17). Since invasion of the fish host by V. anguillarum is enhanced by chemotaxis and motility (37, 45, 47), we wondered if virulence and motility are also coordinately regulated in V. anguillarum. The toxR and toxS mutants were assayed for their ability to migrate away from a point of inoculation in 0.25% agar-TSB. Motility of the toxS mutant was similar to that of the wild type, whereas the toxR mutant was only 79% as motile as the wild type. When the toxR mutation was complemented with the wild-type gene, motility returned to that of the wild type. Thus, ToxR in V. anguillarum does not regulate motility and virulence in the same manner as it does in V. cholerae.
ToxR represses its own production.
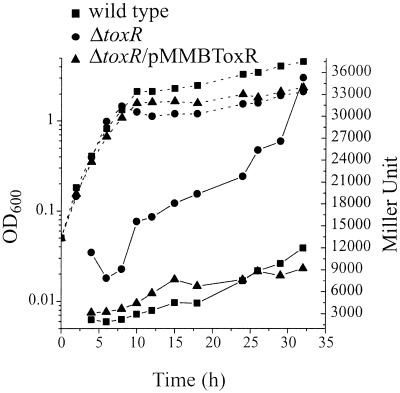
Often, the levels of transcriptional regulators in the bacterial cell are precisely controlled, and as a result, some transcriptional regulators may control their own expression. To test if ToxR is self-regulated, the toxR::lacZ transcriptional gene fusion carried on pDM8-ToxR was mobilized into the wild type and the toxR mutant, and β-galactosidase assays were done throughout the growth curve (Fig. 4). In the toxR mutant, the LacZ activity was three- to sixfold higher throughout growth, indicating that ToxR represses its own expression. When the toxR mutation was complemented (SY10/pMMBToxR) with the wild-type toxR gene and its promoter, LacZ activity returned to wild-type levels.
FIG. 4.
β-Galactosidase activity of toxR::lacZ transcriptional gene fusion in the wild type (NB10), toxR mutant (SY10), and complemented toxR mutant (SY10/pMMBToxR). toxR::lacZ was carried on pDM8-ToxR. Overnight cultures were diluted into fresh TSB medium to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, dotted lines) and β-galactosidase expression (Miller units, solid lines). Miller units for the vector control are between 100 and 200 U, and these were subtracted from the results.
Biofilm formation.
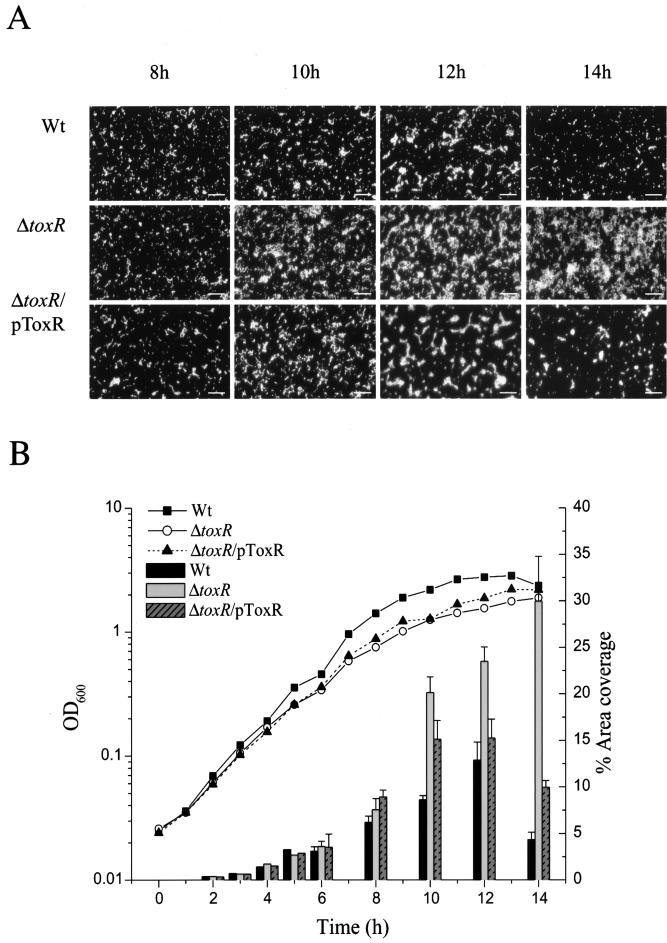
Because the expression of the ToxR regulon in V. cholerae may be an adaptive response to the environment (60) and because aquatic bacteria are normally found attached to surfaces (6), we asked whether ToxR plays a role in initial attachment in biofilm formation in V. anguillarum. The percent area coverage of a biofilm on a glass surface was determined for 2 to 14 h of growth (Fig. 5). The wild type, toxR mutant, and complemented toxR strains showed the same percent area coverage through 8 h of growth. After 8 h, the percent area coverage for the toxR mutant increased three- to fourfold above that of the wild type and maintained this increased level throughout the 14 h. The wild type, on the other hand, never obtained the same level of biofilm coverage as the toxR mutant. In addition, the level of biofilm coverage by the wild type dropped after 12 h. When the toxR mutation was complemented, nearly wild-type levels were obtained. These observations may suggest that the toxR mutant either was more proficient than the wild type at forming a biofilm or was unable to detach from the glass surface as well as the wild type, thus enhancing surface biofilm coverage.
FIG. 5.
Percent area coverage of a biofilm on a glass surface for the wild type (Wt, NB10), the toxR mutant (SY10), and the complemented toxR mutant (SY10/pToxR). (A) Typical view of the progression of biofilm formation for each strain at 8 h, 10 h, 12 h, and 14 h. Slides were stained with acridine orange and then viewed with a fluorescent microscope. Bar, 20 μm. (B) Biofilm formation was quantified by determining the percent area coverage on a glass slide. For each strain, five images were taken at each time point and saved as a computer image. These images were converted into black (background) and white (biofilm) pixels using the Image Tool software, version 2, and the percentage of white pixels is given as the percentage of biofilm covering the glass surface. The data for 2 h, 3 h, 4 h, and 5 h were done as separate experiments from those for 8 h, 10 h, 12 h, and 14 h but are included in the same figure for easy comparison. The data shown are from one experiment. All percent area coverage experiments were done at least three times, and they all gave similar results.
DISCUSSION
In V. cholerae, ToxR plays a major role in the regulation of virulence gene expression (for reviews, see references 14 and 60). Since the discovery of ToxR, the regulation of virulence gene expression in V. cholerae has become much more complex and involves additional regulatory proteins. ToxR, together with a second transmembrane transcriptional activator, TcpP, activate the expression of toxT, which encodes a third cytoplasmically located transcriptional activator (13, 19, 21, 22). ToxT directly activates the expression of over 20 different genes, including the ctx and tcp genes, to make up the ToxR-TcpP regulon (18, 24, 26, 27, 35, 49, 50, 63). This complex regulatory cascade has occurred through horizontal gene transfer via multiple mobile genetic elements. The ctx genes are encoded in a filamentous bacteriophage genome (64), and the toxT and tcp genes, which include TcpP, are found on a pathogenicity island that may also be a filamentous bacteriophage (25, 26).
The virulence genes ctx, tcp, and toxT carried on the bacteriophage of V. cholerae are not likely to be found in V. anguillarum, since the toxR mutant was only slightly less virulent than the wild type in a rainbow trout model. Our data confirm recent data from an ayu fish model suggesting that ToxR of V. anguillarum serotype O2 has no significant role in virulence (41). The slight difference in virulence in the rainbow trout model used in this study may be due to ToxR acting as one of several possible regulators of the virulence factors, and thus only a small effect is seen on virulence. ToxR may not be a major regulator of virulence factors in V. anguillarum. On the other hand, the 20% decrease in motility seen in the toxR mutant compared to the wild type could be the cause of the slight loss in virulence. A partially motile mutant was previously shown to be defective in entry into the fish (37). Interestingly, the effect of the toxR mutation on motility in V. anguillarum is opposite to that seen in V. cholerae. A toxR mutant of V. cholerae is hypermotile, suggesting that ToxR may coordinately regulate motility and virulence gene expression (17). This is not likely to be true for ToxR in V. anguillarum.
ToxR homologs have now been found in several closely related Vibrio-Photobacterium species that are nonpathogenic to humans (41, 46, 56, 66). Instead of evolving to aid colonization of the human intestine, ToxR is suggested to have an ancestral role as a regulator of outer membrane porin production (47). In a ToxT- and TcpP-independent manner, V. cholerae ToxR regulates positively and negatively the outer membrane porins OmpU and OmpT, respectively (5, 9). Furthermore, V. anguillarum, Photobacterium profundum, Vibrio fluvialis, Vibrio mimicus, and V. parahaemolyticus all contain ToxR homologs that regulate expression of OMPs (41, 53, 66). As found in the recent V. anguillarum serotype O2 study (41), ToxR from our V. anguillarum serotype O1 positively regulated the expression of a major OMP of which the amino-terminal amino acids were similar to OmpU of V. cholerae. In addition, cross-reactivity of this major OMP with an antiserum raised against OmpU of V. cholerae provided further evidence that this protein is similar to OmpU of V. cholerae (61), and we have suggested that this protein be called OmpU. In contrast to the V. anguillarum serotype O2 study (41), our data did not show that ToxR regulates the expression of a 46-kDa OMP. In fact, an OMP of this size was not detected in our V. anguillarum strain even in the wild type. One explanation could be that these two vibrios are of different serotypes and that this protein is not found in an O1 serotype strain.
V. anguillarum is suggested to constitute part of the microflora of marine fish (44). When administered orally, V. anguillarum can survive passage through the stomach and can utilize the intestines as a site of attachment, colonization, and proliferation (23, 42, 43). Thus, V. anguillarum should be resistant to bile. Contrary to this assumption, a previous V. anguillarum serotype O2 ToxR study (41) showed that ToxR played no role in bile resistance, suggesting that V. anguillarum is not an enteric bacterium. In contrast to this recent study, our data showed that ToxR is required for enhanced resistance to bile, as it is in other Vibrio species (53).
Little is known about the strain differences between the two V. anguillarum serotypes that would explain this variation in phenotype. However, a possible explanation for the differences in the two V. anguillarum studies is the type of bile that was used. In our studies, we used two types of bile, one from Difco, the exact contents of which we were unable to find out, and another from Sigma that contained sodium salts of taurocholic, glycocholic, desoxycholic, and cholic acids. The Sigma product contained taurocholic acid, which was previously identified as the predominant bile acid found in rainbow trout intestinal mucus (48). Although both types of bile caused a decrease in resistance, the Sigma product showed a milder inhibition of growth, indicating that different combinations of bile acids from different sources can have different effects on inhibition of growth. The bile acids (deoxycholic acid and cholic acid) used in the serotype O2 study were tested as single bile acids and are possibly less effective as a single bile acid than in combination with other bile acids.
Bile may be an important environmental signal that initiates physiological responses in V. anguillarum that are important for survival within the fish host. In support of this possibility, we showed that bile induces an increased expression of OmpU, suggesting that, as in V. cholerae (52, 54), OmpU may be required for enhanced resistance to bile. Contrary to this thought, a toxR mutant, which was decreased in bile resistance and decreased in OmpU protein levels, had approximately wild-type virulence. This effect on virulence may be explained if perhaps something in the intestinal environment signals for increased expression of an additional OMP that compensates for the loss of OmpU and the loss of bile resistance seen under laboratory conditions.
As well as possibly being a part of the microflora of fish, V. anguillarum constitutes part of the normal microflora of the aquatic environment (39, 67). Bacteria in aquatic environments, such as the sea, are normally found attached to surfaces, as opposed to a free-swimming form (6). Thus, adaptation to survival in an aquatic environment may depend on biofilm formation, which may provide an adaptive or survival advantage for aquatic bacteria (7, 8). Biofilm formation has been proposed to be an important factor for the survival of V. cholerae in the aquatic environment (65). As ToxR has been suggested to be a global regulatory protein that responds to environmental signals, attachment to solid surfaces and/or biofilm formation could be a signal to which ToxR responds. In light of these thoughts, we suggest that the V. anguillarum toxR mutant may be altered in its ability to detach from the biofilm or is a better biofilm producer.
The genes required for biofilm formation in V. anguillarum are not known. However, we can make a few predictions as to what types of genes may be involved. First, the toxR mutant is slightly defective in motility, suggesting that motility, which may be regulated by ToxR, either hinders or aids the bacterium in detachment from the biofilm surface. Second, ToxR positively regulates OmpU. Loss of this protein is likely to alter the outer membrane and surface structures or charges of the bacterium. This alteration may lead to surface changes which allow improved surface attachment or cell-cell contact needed for better surface area coverage by the bacterium. Third, ToxR may negatively regulate exopolysaccharide production. In the toxR mutant, exopolysaccharide may be produced in larger amounts, which may either hinder motility and thus detachment of the bacteria or aid production of a thicker biofilm. Fourth, ToxR in P. profundum has been shown to regulate genes involved in the starvation response (4). Similar genes may be needed to signal the bacterium to detach from a biofilm on a surface and to find another nutrient source on which to form a biofilm. Whatever the mechanism, ToxR does seem to play some role in biofilm formation, suggesting a new role for this regulatory protein.
Biofilm formation, OMP expression, and resistance to bile are all physiological responses to environmental signals. A bacterial response to its environment can lead to numerous adaptive physiological changes that are tightly controlled by a number of regulatory proteins or signal molecules. ToxR in V. anguillarum may be part of a regulatory cascade that responds to various environmental signals.
Acknowledgments
Antiserum against OmpU of V. cholerae was kindly provided by James Kaper.
This work was supported by a grant from the Swedish Council for Forestry and Agricultural Research, by a grant from the Carl Tryggers Foundation, Sweden, and by a grant from the Swedish Research Council for Engineering Sciences, which are gratefully acknowledged.
REFERENCES
- 1.Actis, L. A., M. E. Tomalsky, and J. H. Crosa. 1999. Vibriosis, p. 523-557. In P. T. K. Woo and E. W. Bruno (ed.), Fish diseases and disorders, vol. 3: viral, bacterial and fungal infections. Cab International Publishing, Wallingford, United Kingdom [Google Scholar]
- 2.Alsina, M., J. Martínez-Picado, J. Jofre, and A. R. Blanch. 1994. A medium for presumptive identification of Vibrio anguillarum. Appl. Environ. Microbiol. 60:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: disease of farmed and wild fish. Springer and Praxis Publishing Ltd., Chichester, United Kingdom.
- 4.Bidle, K. A., and D. H. Bartlett. 2001. RNA arbitrarily primed PCR survey of genes regulated by ToxR in the deep-sea bacterium Photobacterium profundum strain SS9. J. Bacteriol. 183:1688-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champion, G. A., M. N. Neely, M. A. Brennan, and V. J. DiRita. 1997. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol. Microbiol. 23:323-331. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., K.-J. Cheng, G. G. Geesy, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 10.Croxatto, A., V. J. Chalker, J. Lauritz, J. Jass, A. Hardman, P. Williams, M. Cámara, and D. L. Milton. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184:1617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiRita, V. J., and J. J. Mekalanos. 1991. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64:29-37. [DOI] [PubMed] [Google Scholar]
- 13.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiRita, V. J., N. C. Engleberg, A. Heath, A. Miller, J. A. Crawford, and R. Yu. 2000. Virulence gene regulation inside and outside. Phil. Trans. R. Soc. Lond. B 355:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dziejman, M., and J. J. Mekalanos. 1995. Two-component signal transduction and its role in the expression of bacterial virulence factors, p. 305-317. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 16.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harkey, C. W., K. D. Everiss, and K. M. Peterson. 1995. Isolation and characterization of a Vibrio cholerae gene (tagA) that encodes a ToxR-regulated lipoprotein. Gene 153:81-84. [DOI] [PubMed] [Google Scholar]
- 19.Häse, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins, D. E., E. Nazareno, and V. J. DiRita. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J. Bacteriol. 174:6974-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins, D. E., and V. J. DiRita. 1994. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol. Microbiol. 14:17-29. [DOI] [PubMed] [Google Scholar]
- 23.Horne, M. T., and A. Baxendale. 1983. The adhesion of Vibrio anguillarum to host tissues and its role in pathogenesis. J. Fish Dis. 6:461-471. [Google Scholar]
- 24.Hughes, K. J., K. D. Everiss, D. W. Harkey, and K. M. Peterson. 1994. Identification of a Vibrio cholerae ToxR-activated gene (tagD) that is physically linked to the toxin-coregulated pilus (tcp) gene cluster. Gene 148:97-100. [DOI] [PubMed] [Google Scholar]
- 25.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaolis, D. K., S. Somara, D. R. J. Maneval, J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 27.Kovach, M. E., K. J. Hughes, K. D. Everiss, and K. M. Peterson. 1994. Identification of a ToxR-activated gene, tagE, that lies within the accessory colonization factor gene cluster of Vibrio cholerae 0395. Gene 148:91-95. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. E., S. H. Shin, S. Y. Kim, Y. R. Kim, D. H. Shin, S. S. Chung, Z. H. Lee, J. Y. Lee, K. C. Jeong, S. H. Choi, and J. H. Rhee. 2000. Vibrio vulnificus has the transmembrane transcription activator ToxRS stimulating the expression of the hemolysin gene vvhA. J. Bacteriol. 182:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, Z., K. Kumagai, K. Baba, J. J. Mekalanos, and M. Nishibuchi. 1993. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J. Bacteriol. 175:3844-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGee, K., P. Hörstedt, and D. L. Milton. 1996. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J. Bacteriol. 178:5188-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Miller, V. L., and J. J. Mekalanos. 1984. Synthesis of cholera toxin is positively regulated at the transcriptional level by ToxR. Proc. Natl. Acad. Sci. USA 81:3471-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholerae toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milton, D. L., R. O'Toole, P. Hörstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morales, V. M., A. Bäckman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 39.Muroga, K., M. Iida, H. Matsumoto, and T. Nakai. 1986. Detection of Vibrio anguillarum from waters. Bull. Jpn. Soc. Sci. Fisheries 52:641-647. [Google Scholar]
- 40.Norqvist, A., Å. Hagström, and H. Wolf-Watz. 1989. Protection of rainbow trout against vibriosis and furunculosis by the use of attenuated strains of Vibrio anguillarum. Appl. Environ. Microbiol. 55:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuda, J., T. Nakai, P. S. Chang, T. Oh, T. Nishino, T. Koitabashi, and M. Nishibuchi. 2001. The toxR gene of Vibrio (Listonella) anguillarum controls expression of the major outer membrane proteins but not virulence in a natural host model. Infect. Immun. 69:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsson, J. C., A. Jöborn, A. Westerdahl, L. Blomberg, S. Kjelleberg, and P. L. Conway. 1996. Is turbot, Scophthalmus maximus (L.), intestine a portal of entry for the fish pathogen Vibrio anguillarum? J. Fish Dis. 19:225-234. [Google Scholar]
- 43.Olsson, J. C., A. Jöborn, A Westerdahl, L. Blomberg, S. Kjelleberg, and P. L. Conway. 1998. Survival, persistence, and proliferation of Vibrio anguillarum in juvenile turbot, Scophthalmus maximus (L.), intestine and faeces. J. Fish Dis. 21:1-10. [DOI] [PubMed] [Google Scholar]
- 44.Oppenheimer, C. H. 1962. Marine fish diseases, p. 541-572. In Georg Borgström (ed.), Fish as food, vol. 2. Academic Press, New York, N.Y. [Google Scholar]
- 45.Ormonde, P., P. Hörstedt, R. O'Toole, and D. L. Milton. 2000. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J. Bacteriol. 182:2326-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osorio, C. R., and K. E. Klose. 2000. A region of the transmembrane regulatory protein ToxR that tethers the transcriptional activation domain to the cytoplasmic membrane displays wide divergence among Vibrio species. J. Bacteriol. 182:526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Toole, R., D. L. Milton, and H. Wolf-Watz. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625-637. [DOI] [PubMed] [Google Scholar]
- 48.O'Toole, R., S. Lundberg, S.-Å. Fredriksson, A. Jansson, B. Nilsson, and H. Wolf-Watz. 1999. The chemotactic response of Vibrio anguillarum to fish intestinal mucus is mediated by a combination of multiple mucus components. J. Bacteriol. 181:4308-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsot, C., E. Taxman, and J. J. Mekalanos. 1991. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1641-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson, K. M., and J. J. Mekalanos. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in the intestinal colonization. Infect. Immun. 56:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponce, M. R., and J. L. Micol. 1992. PCR amplification of long DNA fragments. Nucleic Acids Res. 20:623.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Provanzano, D., and K. E. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Provenzano, D., D. A. Schuhmacher, J. L. Barker, and K. E. Klose. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect. Immun. 68:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Provenzano, D., C. M. Lauriano, and K. E. Klose. 2001. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J. Bacteriol. 183:3652-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reed, L. J., and J. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 56.Reich, K. A., and G. K. Schoolnik. 1994. The light organ symbiont Vibrio fischeri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J. Bacteriol. 176:3085-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 58.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions . Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:787-796. [Google Scholar]
- 60.Skorupski, K., and R. K. Taylor. 1997. Control of the ToxR virulenc regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003-1009. [DOI] [PubMed] [Google Scholar]
- 61.Sperandino, V., C. Bailey, J. A. Giron, V. J. DiRita, W. D. Silveirak, A. L. Vettore, and J. B. Kaper. 1996. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect. Immun. 64:5406-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waldor, M. K., and J. J. Mekalanos. 1994. ToxR regulates virulence gene expression in non-01 strains of Vibrio cholerae that cause epidemic cholera. Infect. Immun. 62:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 65.Watnik, P. I., and R. Kolter. 1999. Steps in the development of Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welch, T. J., and D. H. Bartlett. 1998. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol. Microbiol 27:977-985. [DOI] [PubMed] [Google Scholar]
- 67.West, P. A., J. V. Lee, and T. N. Bryant. 1983. A numerical taxonomic study of species of Vibrio isolated from the aquatic environment and birds in Kent, England. J. Appl. Bacteriol. 55:263-282. [DOI] [PubMed] [Google Scholar]