Abstract
We present a complete DNA sequence and metabolic analysis of the dominant oral bacterium Fusobacterium nucleatum. Although not considered a major dental pathogen on its own, this anaerobe facilitates the aggregation and establishment of several other species including the dental pathogens Porphyromonas gingivalis and Bacteroides forsythus. The F. nucleatum strain ATCC 25586 genome was assembled from shotgun sequences and analyzed using the ERGO bioinformatics suite (http://www.integratedgenomics.com). The genome contains 2.17 Mb encoding 2,067 open reading frames, organized on a single circular chromosome with 27% GC content. Despite its taxonomic position among the gram-negative bacteria, several features of its core metabolism are similar to that of gram-positive Clostridium spp., Enterococcus spp., and Lactococcus spp. The genome analysis has revealed several key aspects of the pathways of organic acid, amino acid, carbohydrate, and lipid metabolism. Nine very-high-molecular-weight outer membrane proteins are predicted from the sequence, none of which has been reported in the literature. More than 137 transporters for the uptake of a variety of substrates such as peptides, sugars, metal ions, and cofactors have been identified. Biosynthetic pathways exist for only three amino acids: glutamate, aspartate, and asparagine. The remaining amino acids are imported as such or as di- or oligopeptides that are subsequently degraded in the cytoplasm. A principal source of energy appears to be the fermentation of glutamate to butyrate. Additionally, desulfuration of cysteine and methionine yields ammonia, H2S, methyl mercaptan, and butyrate, which are capable of arresting fibroblast growth, thus preventing wound healing and aiding penetration of the gingival epithelium. The metabolic capabilities of F. nucleatum revealed by its genome are therefore consistent with its specialized niche in the mouth.
Fusobacterium nucleatum is a gram-negative anaerobe, which belongs to the Bacteroidaceae family and is found naturally in the microflora of the mouth in healthy or diseased humans. F. nucleatum is a very long rod with tapering ends and is one of the dominant species of the 500 or more organisms that coexist in the oral cavity (21). Many of the oral flora are commensals but a few are opportunistic pathogens. F. nucleatum can be isolated not only from the mouth but also from infections such as skin ulcers, peritonsillar abscesses, septic arthritis, and endocarditis (5). Several species of Fusobacterium have been isolated and studied, including F. necrophorum (the causative agent of Lemierre's syndrome), F. ulcercans (skin ulcers), F. russi (animal bite infections), and F. varium (eye infections), with F. nucleatum and F. necrophorum considered to be the most pathogenic.
In the initial stages of the periodontal disease process, saccharolytic, aerobic Streptococcus spp. and other bacteria adhere to and colonize the tooth enamel and root surface. This sets the stage for F. nucleatum to coaggregate with these early colonizers and to permit late colonizers, including dental pathogens, to eventually form a biofilm. These complex interactions result in the release of factors that lead to tooth decay. Physical interaction is very specific among various genera in this complex microbial community. Due to the unusual length, adhesive nature, and other cell surface properties of F. nucleatum, periodontal disease-causing bacteria such as Porphyromonas gingivalis, Bacteroides forsythus, Actinobacillus actinomycetemcomitans, Treponema denticola, and Streptococcus spp. aggregate and thrive; hence, F. nucleatum is referred to as a “bridge bacterium” (5). The interaction between F. nucleatum and P. gingivalis has been reported to be very specific (19), mediated by a lactose-binding adhesin (20). The same adhesin protein mediates the binding of F. nucleatum to a variety of eukaryotic cell types including HeLa cells, buccal epithelial cells, macrophages, polymorphonuclear leukocytes, and gingival and periodontal ligaments (46). Helicobacter pylori was also shown to adhere selectively to F. nucleatum (1).
Research on Fusobacterium spp. has focused primarily on species identification, oral ecology, cell-cell communication, extracellular surfaces, amino acid degradation, carbohydrate metabolism, organic acid fermentation, and antibiotic resistance (5, 21). However, many metabolic pathways and their roles in survival in such specialized niches are not known. Genetic experiments have been hampered by the lack of molecular tools, although recently three plasmids, pFN1, pFN2, and pFN3, have been isolated and sequenced from the strain ATCC 10953. A transformation system has been developed for gene manipulation using the pFN1 ori sequence (14).
Although many potential virulence candidates have been described, very few of them have been studied experimentally. In an effort to understand globally its genetic, metabolic, and pathogenic features, we analyzed the genome sequence of F. nucleatum strain ATCC 25586 and present here the results of this analysis. This is the first determination of a Fusobacterium spp. genome sequence.
MATERIALS AND METHODS
Library, sequencing, and assembly.
Three sets of random libraries were constructed from chromosomal DNA from F. nucleatum strain ATCC 25586 (2). Sheared DNA was cloned into plasmid pGEM3 (Promega, Madison, Wis.) containing 2- to 3-kb inserts, in cosmid Lorist 6 carrying 30- to 35-kb inserts, as well as in phage λ DASH (Stratagene, La Jolla, Calif.) carrying 10- to 12-kb inserts. These libraries were maintained in Escherichia coli DH5α (Stratagene). Cosmid end sequences were used as a scaffold for assembly of plasmid contigs. The phage library was useful for sequences unclonable in plasmids. End sequencing was performed on plasmids, cosmids, and phages using Applied Biosystems 3700 DNA sequencers (Perkin-Elmer, Foster City, Calif.). Each of the clones was sequenced from both ends using standard direct and reverse primers. Primes were synthesized at Fidelity Systems, Inc. (Gaithersburg, Md.). About 18,000 sequencing reactions were carried out to give an approximately fivefold genome coverage. In the first step the genome was assembled using Sequencher into 387 contigs. Subsequently, fragments representing the boundaries of repetitive regions were flagged with respect to partial mismatches at the ends of alignments and were independently assembled. The quality of the sequence assembly was checked against the expected insert lengths. More than 1,000 oligonucleotides were designed to walk along appropriate templates for initial gap closure and/or to improve sequence quality. By this procedure the contig number was reduced to 173. Two further rounds of walking reduced the contig number to 15 and finally to a single chromosome.
Bioinformatics.
A combination of CRITICA and an open reading frame (ORF) calling program developed at Integrated Genomics (IG) was used to identify putative ORFs in the sequence. The complete DNA sequence and the predicted ORFs were added into the integrated environment of the ERGO bioinformatics suite for genome annotation and metabolic reconstruction. The ERGO suite now contains 214 partial and complete bacterial genomes.
For a given genome, ERGO employs three steps in annotation. First, sequence similarities are calculated for all the ORFs against the sequences in ERGO using the FASTA algorithm. Based on similarity and other criteria, ORFs are assigned to a preexisiting orthologous gene(s), which automatically assigns a function to this gene (cluster). Second, an exhaustive manual analysis of every ORF is performed using sequence similarity tools such as the motif/pattern databases Pfam, Prosite, Prodom, and COGs (25). At this step, a number of IG proprietary tools that exploit the predictive power of gene context are employed to predict a function in the absence of adequate sequence similarity. These include algorithms for the identification of chromosomal clusters (33), fusion clusters (9), and regulatory ORF clusters. Once functions are assigned to the ORFs, they are connected to corresponding pathway(s). These are a part of the IG collection of over 5,000 biochemical pathways. Each function can be connected to a number of different and/or alternative pathways. Finally, metabolic reconstructions are derived by interconnecting the entire set of pathways to the organism under investigation (41). Once the reconstruction is completed (i.e., when all the known, available pathways are asserted in an organism), it is then possible to identify the missing functions that are expected to occur in the organism but escaped initial identification. This leads to the final step of the annotation process, in which the query becomes the function predicted to be present, and the target is the gene expected to be identified.
Nucleotide sequence accession number.
The genome sequence has been deposited in GenBank under accession no. AE009951. The sequence can be viewed, along with numerous tools for its analysis, at the site http//:www.integratedgenomics.com.
RESULTS AND DISCUSSION
Genome features.
The F. nucleatum strain ATCC 25586 genome consists of a single circular chromosome with 2.17 Mb, about 300 kb fewer than experimentally estimated by pulse-field gel electrophoresis (4). The GC content is 27%, which is lower than that of most bacterial genomes. There are 2,067 predicted ORFs in the genome. Strain ATCC 25586 does not contain any plasmids, although three plasmids have been identified in strain ATCC 10953 (14). The genome statistics are listed in Table 1. About 67% of the ORFs can be assigned functions based on the combination of tools present in the ERGO bioinformatics suite. Nearly one-third (673 ORFs) display sequence similarity to other proteins in the database with no known functions. Based on available genome sequences, about 3.5% (74 ORFs) are unique to F. nucleatum. Nine ORFs have predicted functions based on chromosomal clustering and the presence of protein motifs but do not have orthologs.
TABLE 1.
Characteristics of the F. nucleatum strain ATCC 25586 genome
| Characteristic | Value |
|---|---|
| Genomic features | |
| Number of chromosomes | 1 |
| Total no. of sequence bases | 2,174,500 |
| DNA coding sequence | 1,952,810 |
| %GC content | 27 |
| Plasmids | None in this strain |
| General statistics | |
| Total no. of ORFs | 2,067 |
| ORFs with assigned functions (%) | 1,394 (67) |
| ORFs with functions but no similarities (%) | 2 (0.1) |
| ORFs without assigned functions (%) | 673 (32) |
| ORFs without function, with similaritya (%) | 599 (29) |
| ORFs without assigned function or similarityb (%) | 74 (3.5) |
| ORFs in ortholog clusters (%) | 756 (36) |
| ORFs in paralog clusters (%) | 535 (26) |
| Total no. of paralog clusters | 180 |
| ORFs in chromosomal clusters (%) | 775 (37) |
| ORFs in possible fusion events (%) | 596 (29) |
| ORFs in fusion events as composites (%) | 222 (11) |
| ORFs in fusion events as components (%) | 437 (21) |
Conserved hypotheticals.
Proteins unique to F. nucleatum.
Based on a number of analytical tools from ERGO, nearly 36% (756 ORFs) can be grouped in ortholog clusters and 26% (535 ORFs) can be grouped in paralog clusters. Using the ERGO fusion-finding tool, we identified 596 ORFs that participate in fusion events, 11% (222 ORFs) coding for fusion proteins, and 21% (437 ORFs) coding for components of fusion proteins in other genomes. Sixty-three ORFs are components that form composites in F. nucleatum itself.
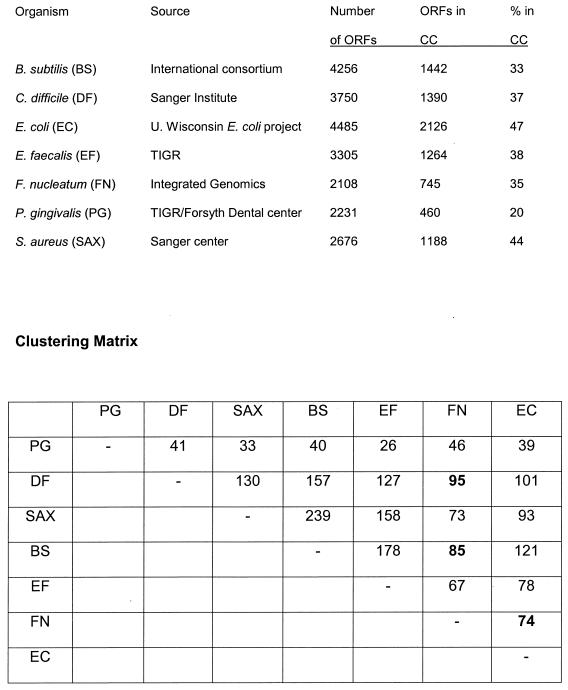
Using the ERGO chromosomal clustering tool, only 35% of the ORFs could be grouped in clusters similar to those of Bacillus subtilis and Clostridium spp., far below the corresponding clustering in E. coli. When compared to the dental pathogen P. gingivalis, only 20% of the ORFs were found to be clustered (Fig. 1). Between F. nucleatum and Clostridium spp., there are 95 chromosomal clusters, 85 between F. nucleatum and B. subtilis, and only 74 between F. nucleatum and E. coli. Based on this cluster analysis, Fusobacterium spp. are evolutionarily closer to gram-positive than to gram-negative bacteria. Based on 16S rRNA sequences, F. nucleatum is placed in the low-GC gram-positive bacterial group, although it has the characteristic cell wall structure of outer membrane, peptidoglycan, and inner membrane that makes it gram negative.
FIG. 1.
Comparisons of chromosomal clustering (CC) of ORFs in F. nucleatum strain ATCC 25586 with B. subtilis, C. difficile, E. coli, E. faecalis, P. gingivalis, and S. aureus EMRSA. Clustering is calculated by searching for pairs of close bidirectional best hits on a run of ORFs on the same strand using the ERGO chromosomal clustering tool. The number of clusters and the comparative matrix are represented below.
There are five rrn loci in F. nucleatum. Two rrn have tandem repeats of 16S, 23S, and 5S and 16S, 23S, phosphopentomutase, and 5S rRNA genes; the fifth locus has only 16S, 23S, and 5S rRNA genes. The 16S rRNA sequence is most similar to that of Lactobacillus lactis strain IL 1403 and Streptococcus pyogenes. None of the five rrn operons includes a tRNA in the cluster.
Transcription and translation.
F. nucleatum components of the transcriptional apparatus, consisting of the ORFs for αββ′β" and ω subunits of RNA polymerase (RNAP), are similar to those of gram-positive polymerase. There are SigA, SigB, SigH (sporulation sigma factor homolog), and an ECF family sigma factor. For transcription termination, there is one ORF that codes for a Rho factor similar to that of Staphylococcus aureus. In addition, there are homologs of NusA and NusG. Three antiterminators, one each for the cel operon, the glycerol uptake operon, and NusB, have also been identified.
Forty-three transcriptional regulators have been identified, of which 12 belong to the TetR and GntR families (6 each), 5 belong to the DoeR family, 8 belong to the LuxR/LysR, MarR, Crp, and MerR families (2 each) and one is an exoenzyme regulatory protein precursor. Six ORFs code for response regulators, one codes for CtrA, one codes for a protein similar to a nitrogen assimilation regulatory protein, and three code for two-component response regulators. We did not find any genes encoding quorum-sensing system proteins such as LuxI or LuxS.
All the typical prokaryotic translation initiation factors, IF-1, IF-2, and IF-3, are present. In addition, there is an ORF for the eukaryotic translation initiation factor EIF-2B subunit 1 (24). Two ORFs for the elongation factor EF-G are present, one similar to that of the gram-negative Thermochromatium tepidum and E. coli and the other similar to that of B. subtilis. There is one ORF each for EF-Tu, EF-Ts, and EF-p (elongation factor for peptide bond synthesis). F. nucleatum encodes only two peptide chain release factors, RF-1 and RF-2. There are 30 large and 20 small ribosomal subunit proteins for the assembly of the ribosome. LSU ribosomal proteins L7AE, L25P, L35P, and L36P and SSU ribosomal proteins S21P, S22P, and S31P are absent. Modifying proteins such as ribosomal protein alanine acetyltransferase and large ribosomal subunit pseudouridine synthase subunits A, B, and D are present.
Forty-seven ORFs code for tRNAs for all 20 amino acids. Interestingly, there are three tRNAs for methionine. Two of these are in a cluster of 11 tRNA genes, and one is in a cluster of 14. They are not similar to one another. Their assignment as Met-tRNA is based on similarity to genes from a gram-negative bacterium and two archaea. All the types of tRNA ligases are present except for Gln-tRNA ligase, although there are two Gln-tRNAs. As in B. subtilis, all the subunits for the Gln-tRNA amidotransferase (GatC, A, and B) are present. This enzyme amidates the misacylated Gln-tRNA charged with Glu to make Gln-tRNAGln. This is the only route to glutamine in F. nucleatum because glutamine synthetase is missing.
DNA repair and replication and cell division.
Most types of DNA repair systems are present in F. nucleatum. Recombination repair is mediated by RecA and is analogous to that of E. coli. In addition, ORFs for proteins such as RecF, RecO, RecR, and single-stranded DNA-binding proteins that are involved in the repair process are present. There are counterparts of the exonucleases SbcC and SbcD that belong to the SMC (structural maintenance of chromosomes) protein family. However, the F. nucleatum SbcC and SbcD proteins are more closely related to the archaeal/eukaryotic RAD50 and MRE11 (RAD32) (42) than they are to E. coli SbcC and SbcD proteins. The F. nucleatum sbcCD genes are located in an operon similar to the archaeal rad50 and mre11 genes. Resolution of homologous recombination intermediates (postsynapsis) occurs via the highly conserved RuvABC complex. There are two copies of the major 5′ to 3′ exonuclease RecJ, one of which (FN2018) is similar to the E. coli RecJ. The N terminus of the second copy of RecJ (FN0374) is similar to E. coli RecJ, while its C terminus is similar to a eukaryotic-like receptor domain.
UV damage is repaired by at least three different pathways, viz., direct damage reversal, nucleotide excision repair, and recombinational repair. F. nucleatum does not have the bacterial PhrB-type photolyase, but it does possess an unusual ORF whose C terminus is homologous to a spore photoproduct lyase that repairs thymine dimers in B. subtilis. In addition, F. nucleatum has a nucleotide excision pathway to repair UV-induced thymine dimers and transcription-coupled errors. Alkylation damage can be repaired by O-6-methylguanine DNA alkyltransferase (EC 2.1.1.63). There is a NAD-dependent DNA ligase similar to one in B. subtilis. The MutS- and MutL-like proteins of the mismatch repair pathway are present. There are several MutS paralogs that belong to the MutS2 protein family. We were unable to identify an ORF for the MutH mismatch endonuclease because mutH, mutL, and mutS are not in an operon. F. nucleatum may use an alternate DNA endonuclease to substitute for MutH. Base excision repair is primarily involved in the removal of modified bases that result from oxidative damage. Although there is not an extensive repertoire of enzymes for base excision repair, there are several DNA glycosylases including uracil DNA glycosylase (ung) to repair deamidated cytosine (FN0100). There are also enzymes to repair oxidatively modified guanine bases (e.g., 8-oxoguanine) such as 8-oxoguanine DNA glycosylase (EC 3.2.2.)/DNA-apurinic or apyrimidinic site (AP) lyase (EC 4.2.99.18) (FN0882), which has both glycosylase and AP lyase domains. Further, this protein is present in archaeal and bacterial ortholog clusters and has strong similarity to Methanococcus jannaschii and Thermotoga maritima enzymes. Orthologs of FN0882 in the archaeal genomes of Ferroplasma acidarmanus, Thermoplasma acidophilum, and Thermoplasma volcanium GSS1 belong to a chromosomal cluster with a superoxide dismutase, another enzyme involved in the quenching of reactive oxygen species that can result in base oxidation damage. Additionally, F. nucleatum possesses a MutT (8-oxo-GTPase) protein that degrades 8-oxo-GTP to the monophosphate. Other F. nucleatum glycosylases also have AP lyase activity, including endonuclease III (NTH) which attacks pyrimidine adducts (other than pyrimidine photodimers) and photoproducts generated as a result of UV or ionizing radiation damage.
There are two 13-mer DnaA-binding boxes that are considered to be the putative origins of replication. The chromosomal replication initiator protein DnaA in most gram-negative bacteria is clustered with the DNA polymerase β chain and the gyrase B subunit protein. In gram-positive bacteria, the gene order is similar except for an unknown ORF between the recF and gyrB genes followed by the gyrA gene. In F. nucleatum, we were unable to find the ORF for the DnaA protein upstream of the recF gene. However, based on the clustering evidence, a hypothetical ORF (637 amino acids [aa]) with no orthologs has been tentatively assigned as the DnaA protein. ORFs for the DnaE primase and RepB protein, also necessary for replication, are found elsewhere.
Two essential ORFs for cell division proteins FtsA and FtsZ are present in an operon. The minCDE operon is distant from the ftsAZ operon, as in most bacteria. The glucose-inhibited cell division gene (gidA) is located five ORFs downstream of the putative dnaA ORF. In addition, there is a gidAB operon located elsewhere on the chromosome, downstream from the potassium uptake operon. Three ORFs for chromosome segregation proteins and two ORFs each for the rod-shape-determining MreB/FtsA and RodA/FtsW protein families have been identified.
F. nucleatum restricts foreign DNA using a type III restriction-modification system. Two ORFs encode methylase-modifying enzymes. Downstream of one of these is a type III deoxyribonuclease. The modifying enzyme is most similar to that found in Pasteurella multocida.
Secretion and Omps.
Fusobacterium spp. appear to secrete very few proteins into the external medium. This bacterium lacks major secretion systems such as type II, type III, and type IV. Components of the sec system that allows proteins to cross the inner membrane, except for the protein translocase subunit SecB, are present. The genes required for protein maturation and release, such as the signal peptidase and the lipoprotein peptidase, are also present. The sec-independent TAT (twin arginine motif translocation) system has not been identified.
Eleven ORFs are assigned as Fusobacterium outer membrane proteins (Fomps). Six are more than 2,000 aa long, of which three (FN2047, FN1893, and FN0387) are identical. Both FN2086 and FN1893 are flanked by insertion (IS) elements. One of these, FN1893, has an IS element only at the C-terminal end. It is believed that the Fomps are involved in pathogenesis. Fomps may be candidates for the production of vaccines. Only one 40-kDa porin, FomA (FN1859), was known prior to the genome sequence (5). However, we have identified several additional ORFs that show similarity to the OmpA-OmpF protein family. A complete list of the predicted outer membrane proteins is given in Table 2. These proteins may stabilize the membrane and/or act as porins. ORF FN0253 is similar to the OmpA protein from B. pertussis, while FN0335 and FN1265 are most similar to the Mesorhizobium loti and Neisseria gonorrhoeae yiaD, respectively, both of which are predicted to be outer membrane lipoproteins. None of the very large predicted Omps was detected on protein gels previously (5). These very large proteins were apparently stuck at the top of the analytical gels.
TABLE 2.
Predicted sizes of the outer membrane proteins of F. nucleatum
| Outer membrane protein type and ORF no. | Predicted size (kDa) |
|---|---|
| Fusobacterium outer membrane proteins (Fomps) | |
| FN2047 | 257 |
| FN1893 | 251 |
| FN0387 | 238 |
| FN1526 | 362 |
| FN2058 | 188 |
| Outer membrane proteins | |
| FN1911 | 77 |
| FN0394 | 28 |
| FN1265 | 21 |
| Outer membrane protein A precursor | |
| FN0253 | 21 |
| Outer membrane porin F precursor | |
| FN0335 | 19 |
| FN1124 | 34 |
| Outer membrane porin P1 precursor | |
| FN1003 | 42 |
| Outer membrane efflux protein | |
| FN0517 | 49 |
| Major outer membrane protein | |
| FN1859 (FomA) | 42 |
Interruptions are due to insertion elements.
Central metabolism.
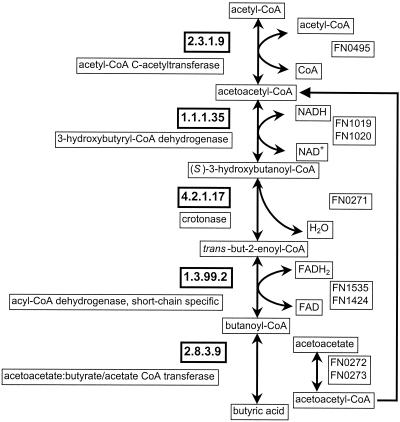
F. nucleatum can obtain carbon and energy from sugars and amino acids. The preferred substrates for energy are amino acids (5), which prevent the metabolism of sugars as long as they are available. Among the sugars, glucose, galactose, fructose, N-acetylglucosamine, N-acetylneuraminate, citrate, and glycerol can be utilized. Galactose and glucose are probably taken in by a galactose/glucose ABC transporter, although a second putative galactose transporter is found clustered with the galactose utilization operon. Galactose is converted to glucose-6-phosphate by the Leloir pathway and feeds into the Embden-Meyerhof pathway. Fructose is taken in by a phosphoenolpyruvate phosphotransferase (PTS) transporter, which is clustered with the ORF for phosphofructokinase that produces fructose-1,6-bisphosphate and feeds into glycolysis. The resulting pyruvate can be fed into formate or lactate fermentation by using the corresponding pyruvate-formate lyase and l- and d-lactate dehydrogenase. Pyruvate can be converted to oxaloacetate by phosphoenolpyruvate carboxykinase and to acetyl coenzyme A (acetyl-CoA) by pyruvate-flavodoxin oxidoreductase. The ORF for the latter is clustered with the ORFs for l-lactate dehydrogenase, phosphate acetyltransferase, and acetate kinase. These enzymes regulate metabolic fluxes to different fermentation products. The ORF for the lactate/acetate transporter is clustered with these genes as well. Acetyl-CoA can be used for anabolic purposes (fatty acid synthesis) or converted to acetate and butyrate. All four genes that encode enzymes for butyrate fermentation are found in the genome, including acetyl-CoA acetyltransferase (FN0495, FipA protein), 3-hydroxybutyryl-CoA dehydrogenase, crotonase, and butyryl-CoA dehydrogenase (Fig. 2). Butyrate can be made in other ways as well. Production of butyrate has several consequences: mouth odor and inhibition of fibroblast proliferation, which prevents healing of wounds in the gum.
FIG. 2.
Metabolic reconstruction of the butyric acid fermentation pathway in F. nucleatum. All the ORFs that code for enzymes involved in the pathway have been identified.
F. nucleatum should be able to utilize ethanolamine as a carbon and nitrogen source, due to the presence of the eut operon. In E. coli and Salmonella enterica serovar Typhimurium, two tandem operons encode the eutSPQTDMNEJG and eutHABCLK genes, followed by an AraC-type transcriptional regulator, EutR. However, the eut operon in F. nucleatum begins with eutS followed by three unrelated ORFs and then eutABCLMKETNHPQR. The ORF for eutD and eutJ are in a different distant cluster. Such a cluster organization is similar in Clostridium difficile and Enterococcus faecalis. The EutB and EutC subunits form the cobalamin-dependent ethanolamine ammonia lyase, which catalyzes the conversion of ethanolamine to acetaldehyde and NH3. The EutA protein is a chaperonin, which prevents the inhibition of lyase by cyanocobalamin B12. The eutE gene codes for an acetaldehyde dehydrogenase (EC 1.2.1.10) and eutT for a cobalamine adenosyl transferase (EC 2.5.1.17). Proteins EutK, L, M, N, and S are homologs of the shell proteins of the carboxysome of S. enterica serovar Typhimurium (18, 39, 43). Growth of F. nucleatum on ethanolamine has not been shown.
F. nucleatum can also obtain energy by degradation of small peptides. It can grow on glutamate, histidine, serine, and lysine as energy sources (38). There is a Xaa-His dipeptidase homolog (FN0278) similar to that found in Streptococcus pyogenes (EC 4.3.13.3) and YtjP of B. subtilis to convert carnosine (N-β-alanyl-l-histidine) to l-histidine and β-alanine. In addition, there are three ORFs (FN1804, FN1408, and FN1277), each of which can also function as an aminoacyl-histidine dipeptidase or carnosinase. Of these, one is clustered with an ORF that encodes a transporter, an integral membrane protein, and histidine ammonia lyase. Arginine can be degraded by the bifunctional ornithine decarboxylase (EC 4.1.1.17)/arginase (EC 3.5.3.1) enzyme (FN0501) which yields ornithine and urea. Urea is degraded to ammonia and CO2.
The ORF for glutamate synthase was not found, although a NAD-specific glutamate dehydrogenase (GluD), which converts 2-oxoglutarate and ammonia to l-glutamate, is present. The ORFs that encode both small and large subunits of carbamoyl-phosphate synthase are present. These compounds are important for pyrimidine synthesis. The enzyme glutamate racemase, EC 5.1.1.3, which converts l-glutamate to d-glutamate, is present. All ORFs (murC, murD, murE, and murF) that encode enzymes for the conversion of d-glutamate to UDP-MurNac pentapeptide are present, as are other enzymes in the peptidoglycan synthetic pathway such as GlmU, MurA, and MurB.
Proline synthesis from l-glutamate and l-alanine is absent. l-alanine can be synthesized from l-cysteine, resulting in the release of sulfide and ammonia, by the NifS protein. l-cysteine might be obtained from o-acetyl-serine by cysteine synthase, but the enzyme for the first step of this pathway, serine-O-acetyltransferase, is not present. Thus cysteine must be imported or made from methionine. Similarly, l-serine might be synthesized from 3-phosphoglycerate using phosphoglycerate dehydrogenase and phosphoserine phosphatase. However, the enzyme phosphoserine aminotransferase, required to transfer the amino group to serine, is absent, and therefore serine must be imported also.
All the genes for MetB, MetC, and MetH are present. Cystathionine β lyase (MetB) produces l-cystathionine from O-succinyl-l-homoserine, and MetC further converts l-cystathionine to l-homocysteine. Finally, 5-methyl-THF-homo-cysteine methyltransferase (MetH) converts homocysteine to methionine.
Since the lysine biosynthetic pathway is absent, F. nucleatum is unable to synthesize meso-diaminopimelate, an important component of peptidoglycan. Instead of meso-diaminopimelic acid (meso-DAP), the peptidoglycan layer contains the unusual amino acid meso-lanthionine. meso-Lanthionine is synthesized as such and then incorporated into peptidoglycan pentapeptide (11). Though the pathway for biosynthesis of meso-lanthionine is not known, Richaud et al. (36) were able to construct an E. coli strain with meso-lanthionine replacing meso-diaminopimelate. This was achieved by overexpression of cystathionine γ-synthase (MetB), which is known to produce l-lanthionine from cysteine, and knockout of the meso-DAP biosynthetic genes and cystathionine beta-lyase (MetC), since the latter enzyme destroys cystathionine and meso-lanthionine. The epimerase DapF was shown to convert l-lanthionine to meso-lanthionine, but this enzyme is missing from F. nucleatum, so the mechanism for conversion of the l-lanthionine to meso-lanthionine remains unknown.
The ORFs that encode enzymes for chorismate synthesis from erythrose 4-phosphate are present, and anthranilate can be synthesized from chorismate by anthranilate synthase, but other genes in the pathway to tryptophan are absent. Three ORFs that code for chorismate mutase, one of them a bifunctional enzyme with phospho-2-dehydro-3-deoxyheptonate aldolase and chorismate mutase activities, can produce prephenate. Further, although aspartate aminotransferases and tyrosine aminotransferase to make tyrosine from prephenate are present, an ORF for the prephenate dehydrogenase has not been identified. Thus, F. nucleatum cannot make tryptophan, tyrosine, or phenlyalanine. Two ORFs encode aspartate aminotransferases, which convert oxaloacetate to aspartate, and another ORF codes for the ammonia ligase to make asparagine from aspartate. There are no ORFs for the conversion of homoserine or threonine from aspartate or isoleucine from threonine. Although Kim et al. (16) have reported that F. nucleatum strain K-60 is capable of quercitrin cleavage to quercitin and l-rhamnose, we did not identify an ORF for the l-rhamnosidase in the genome. There are no ORFs for enzymes for aromatic compound degradation.
Glutamate fermentation via the 2-oxoglutarate pathway begins with deamidation of glutamate by NAD(−) glutamate dehydrogenase (FN1020), followed by the reduction of 2-oxoglutarate by 2-hydroxyglutarate dehydrogenase. An ORF (FN1383) homologous to a 2-hydroxglutarate dehydrogenase was found clustered with glutamate dehydrogenase with a molecular weight similar to that found in Micrococcus aerogenes (26). The 2-butenoyl-CoA is then degraded by the glutaconyl-CoA pathway. All the enzymes including a sodium ion pump glutaconyl-CoA decarboxylase are present in F. nucleatum (3). The butenoyl-CoA can be further converted to butyrate via the butyrate fermentation pathway. The glutamate fermentation pathway is the main source of butyrate during bacterial growth on amino acid-based chemically defined medium (38). We did not find any enzymes for 4-aminobutyrate fermentation, consistent with the experimental observation of Gharbia and Shah (12). None of the enzymes of the mesaconate pathway was detected (12). Glutamate mutase (methylaspartate mutase) is present, but methylaspartate ammonia-lyase is not found in the genome. As mentioned above, butyrate may contribute to the pathogenicity of the F. nucleatum consortia.
Amino acid degradation.
The oral microbial community includes several bacteria that are capable of releasing proteases that break down protein released by dead bacteria and left over from food. F. nucleatum also releases proteases and can convert histidine and glutamine to glutamate, which can then enter the glutamate fermentation pathway. In the case of histidine, the first three enzymes and the reactions are similar to those of B. subtilis. The pathways diverge after N-formimino-l-glutamate. In B. subtilis, this is hydrolyzed to glutamate and formamide, but in F. nucleatum the formimino group is transferred to tetrahydrofolate (THF) and subsequently to 5,10-methyl-THF by a cyclodeaminase. The latter two steps are similar to the eukaryotic pathway. There are two chromosomal clusters for histidine utilization, one cluster including ORFs for a permease, imidazolonepropionase, cyclodeaminase, histidine lyase, glutamate formiminotransferase, and histidine dipeptidase. The second cluster consists of the hutHU genes adjacent to a sodium:histidine symporter.
l-Lysine is fermented by F. nucleatum, and a putative lysine utilization operon has been identified. The pathway of lysine degradation is similar to that of Clostridium. The first step of the reaction is the formation of l-β-lysine from l-lysine by l-lysine 2,3-aminomutase, and F. nucleatum contains a homolog of the l-lysine 2,3-aminomutase from Clostridium subterminale. The second step is conversion of l-β-lysine to 3,5-diaminohexanoic acid by l-β-lysine 5,6-aminomutase. A homolog of the d-lysine 5,6-aminomutase of Clostridium sticklandii, which also acts on l-β-lysine, is located proximal to the ORF encoding the 2,3-aminomutase (6).
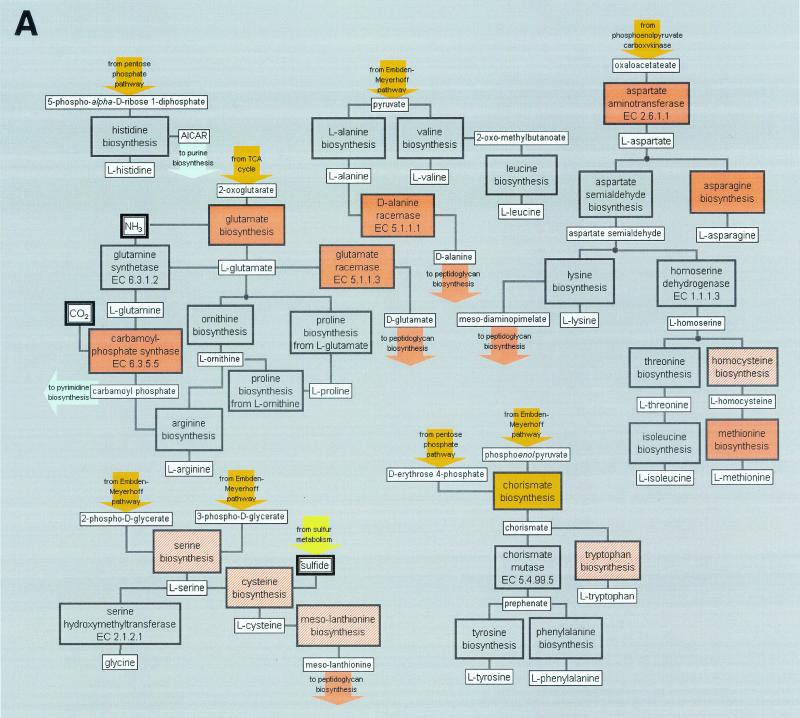
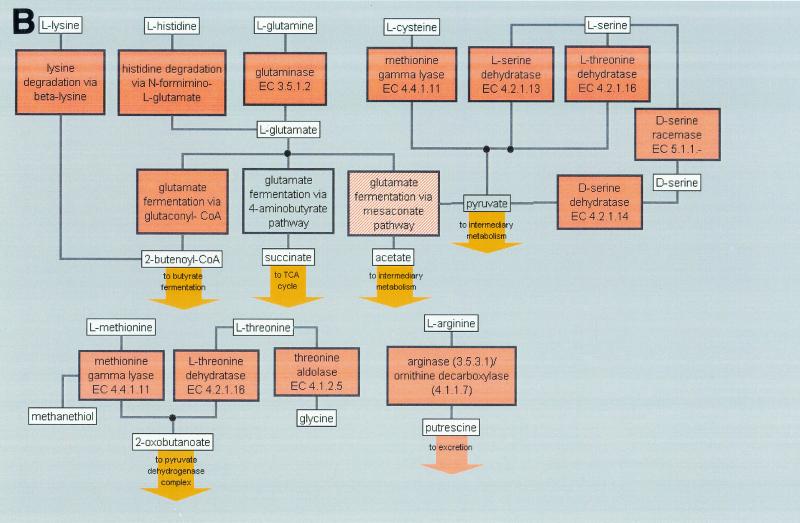
F. nucleatum is known to produce volatile sulfur compounds from the degradation of cysteine and methionine (34). A cysteine desulfurase (NifS), which converts cysteine to alanine and sulfide, is present. Enzymes that convert methionine to ammonia, 2-ketobutyrate and methyl mercaptan as well as γ-lyase that degrades methionine are found, similar to those of Trichomonas vaginalis. Alternatively, cysteine can also be converted to cystathionine by cystathionine β synthase and γ-lyase to form ammonia, pyruvate, and homocysteine. Homocysteine can be further desulfurated or methylated to form methionine. There are two ORFs with similarity to cystathionine metabolic enzymes proximal to each other but on complementary strands, as well as an ORF that codes for a homocysteine methyltransferase. Based on the genome analysis, an overall scheme of amino acid synthesis and degradation pathways is depicted in Fig. 3A and B, respectively.
FIG. 3.
An overview of amino acid synthesis (A) and degradation (B) in F. nucleatum. Orange, brick red, or gold boxes indicate that the pathway is present. A gray box indicates a missing pathway, and the striped box indicates that a few candidates of the pathway are present.
Peptides are probably the second most important source for carbon, nitrogen, and energy in F. nucleatum. Very little is known about the proteins involved in peptide transport as well as degradation. It is not known whether the peptides are broken down outside the cell and imported as amino acids or taken up intact and then degraded. Using PSORT analysis, we predict that a majority of the peptidases are localized in the cytosol. We have identified 17 cytosolic peptidases similar to previously characterized proteins, including both broad- and narrow-specificity peptidases. Because there are numerous ORFs for probable peptide transporters, we believe that most peptides are imported and then degraded, but this remains to be demonstrated experimentally.
Two ORFs identified as γ-glutamyltranspeptidase involved in glutathione metabolism have been identified. Based on PSORT analysis, one of them is periplasmic and the other is membrane bound. A periplasmic γ-glutamyl-transpeptidase, characterized by Mineyama and Saito (29), is similar to one from Helicobacter pylori that is involved in virulence (7, 28).
Nucleotide metabolism.
All the ORFs for de novo IMP synthesis appear to be present. The synthetic genes are organized in an operon interrupted by two hypothetical ORFs, unlike other gram-negative bacteria. One of the hypothetical proteins is homologous to a methyltransferase, and the other is unknown. Genes for UMP synthesis are also organized in an operon interrupted by a conserved hypothetical ORF. ORFs that code for the enzymes of UMP biosynthesis are closely related to the gram-positive Enterococcus spp. and Staphylococcus spp. orthologs. Nucleoside monophosphate kinases for all types of nucleotides are present. As in Mycoplasma spp. and Clostridium spp., the nucleoside diphosphate kinase (NdK) is absent, and a polyphosphate kinase substitutes for the Ndk function, as demonstrated for Pseudomonas aeruginosa (15, 22), or by a glycolytic kinase as in Mycoplasma spp. The deoxyribonucleotides can be synthesized under both aerobic and anaerobic conditions by ribonucleoside-diphosphate and ribonucleoside-triphosphate reductases.
Enzymes necessary for the purine and pyrimidine salvage pathway are also present. The purine salvage enzymes and uracil phosphoribosyltransferase are highly homologous to the corresponding enzymes of gram-positive bacteria. TMP is formed by thymidylate synthase from dUMP, providing the only interconversion pathway between pyrimidine nucleotides. In addition, there are four ORFs for the xanthine/uracil permease family of proteins involved in the transport of free bases. Thus, based on the genome analysis we conclude that F. nucleatum can utilize exogenous bases and nucleosides.
There are primarily two methods for phosphorous transport: a sodium-dependent phosphate transporter (FN0276) and a phosphonate transporter. The phn operon (phosphonate) encodes a phosphonate-binding periplasmic protein, transport protein PhnC, and a permease protein (PhnE). None of the phosphonate lyase or other phosphonate degradation enzymes is present, suggesting that phosphonates can be transported but not utilized further.
Polyphosphate kinase is absent, but there is an exopolyphosphatase, which is able to degrade polyphosphates. The exopolyphosphatase is fused with 3-dehydroquinate synthase (EC 4.6.13). The lack of phosphate transporters and a phosphate storage system suggests that Fusobacterium spp. use alternate sources for their phosphorus requirements, such as phospholipids.
THF in synthesized in a nine-step reaction as in most bacteria. THF is the carrier involved in many one-carbon transfer reactions (27). For example, in amino acid synthesis, the cobalamin-dependent methionine synthase (MetH) utilizes 5-methyltetrahydrofolate. However, an ORF for the methylenetetrahydrofolate reductase has not been identified. ORFs for 10-formyltetrahydrofolate synthase and methenyltetrahydrofolate cyclohydrolase, both of which can synthesize 10-formyl-THF, are present. The enzymes in purine metabolism that utilize 10-formyl-THF, namely, glycinamide ribonucleotide transformylase and 5-aminoimidazole-4-carboxamide ribonucleotide transformylase have also been found. The ORF for the methylenetetrahydrofolate dehydrogenase that interconverts 5,10-methylenetetrahydrofolate and 5,10-methenyltetrahydrofolate is also present.
Lipid and LPS metabolism.
F. nucleatum uses the nonmevalonate pathway for isoprenoid biosynthesis. All the fab enzymes necessary for fatty acid synthesis have been found except for the fatty acid desaturase (fabA homolog), suggesting that the bacterium is not capable of synthesizing unsaturated fatty acids. The enoyl-ACP reductase is a homolog of Streptococcus pneumoniae FabK. ORFs for the 2,4-dienoyl-CoA reductase and 3,2-trans-enoyl-CoA isomerase that are necessary for the utilization of unsaturated fatty acids are absent. We have identified the ORFs for the enzymes for phospholipid synthesis. F. nucleatum can synthesize phosphatidylglycerol, cardiolipin, phosphatidylserine, and phosphatidylethanolamine. However, ORFs for the first acyltransferase responsible for phosphatidate synthesis and glycerol-3-phosphate acyltransferase (PlsB) have not been found, as is also the case for gram-positive Bacillus spp. and Streptococcus spp.
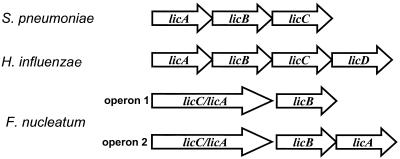
Genes encoding enzymes necessary for lipidA synthesis such as lpxA, lpxC, lpxD, lpxB, lpxK, and kdtA are present. However, ORFs for enzymes similar to E. coli lpxL (lauroyl transferase), lpxM (myristoyl transferase), and lpxP (palmitoyl transferase), which participate in the formation of acyloxyacyl chains in lipopolysaccharide (LPS), were not found. We predict that F. nucleatum is able to decorate its LPS with choline due to the presence of the lic1 operon, which is involved in phosphocholine decoration of LPS in Haemophilus influenzae and teichoic and lipoteichoic acids in S. pneumoniae. Attachment of phosphocholine to cell wall components has been linked to virulence in S. pneumoniae (17) and phase variation in H. influenzae (45). There are no ORFs for a choline-binding protein (40).
The organization of the lic1 operon in several bacteria is shown in Fig. 4. The lic1 operon in H. influenzae consists of a choline kinase (licA), a putative choline transporter (licB), a phosphocholine cytidylyltransferase (licC), and a presumable phosphotransferase (licD) that catalyzes the attachment of phosphorylcholine to the carbohydrate, releasing CMP. The corresponding operon in S. pneumoniae has a similar structure, but it lacks the licD gene, which is located on the opposite strand five ORFs upstream of licA. F. nucleatum has two loci that resemble the lic1 operon found in S. pneumoniae and H. influenzae. The first one consists of ORFs FN0110 and FN0111, and the second includes ORFs FN1670 and FN1669. Interestingly, two of these are distinct fusion proteins (FN0110 and FN1670), which are predicted to exhibit both choline kinase and phosphocholine cytidylyltransferase activities. Moreover, the second operon has a third gene whose product is homologous to LicA. In addition, there is a LicD homolog encoded nine ORFs upstream of FN0111. Based on these findings, we believe that the LPS of F. nucleatum structurally resembles the lipo-oligosaccharide of H. influenzae.
FIG. 4.
Comparison of the lic1 operon of F. nucleatum with that of S. pneumoniae and H. influenzae. F. nucleatum has two operons that resemble the lic1 loci in the other bacteria. Operon 1 consists of two ORFs (FN0110 and FN0111), while operon 2 consists of three ORFs (FN1668, FN1669, and FN1670). The F. nucleatum choline kinase and phosphocholine cytidylyl-transferase genes are fused and presumably form two bifunctional enzymes. One of these operons might have arisen by gene duplication in F. nucleatum more recently than the gene fusion event.
Coenzyme and cofactor metabolism.
F. nucleatum has genes that code for enzymes involved in synthesis of coenzymes and cofactors such as biotin, CoA, thiamin, protoporphyrin IX, siroheme, cobalamin, NAD, NADP, riboflavin, flavin mononucleotide, flavin adenine dinucleotide, dihydrofolate, tetrahydrofolate, tetrahydrofolyl-polyglutamate, tetrahydrobiopterin, dihydrobiopterin, and 2Fe-2S and 4Fe-4S clusters. The first step in biotin synthesis, which involves the methyltransferase BioC protein that converts pimelic acid to biotin, is present, but the carboxyhexanoate-CoA ligase is absent. In enterobacteria, BioC acts synergistically with the BioH protein. In most other proteobacteria, the gene that codes for BioH is absent. In F. nucleatum, BioC is a part of an operon that includes a hypothetical ORF weakly similar to BioH.
The synthesis of CoA begins with pantothenate, enabled by the presence of an ORF for a sodium:pantothenate transporter. No ORFs for molybdenum-containing cofactor or Mo-utilizing enzymes such as dimethyl sulfoxide reductase, dissimilatory nitrate reductase, formate dehydrogenase, xanthine oxidase, or aldehyde oxidase were found.
Genes related to lipoate synthesis and transporters are absent; most anaerobes lack them. There are also no ORFs for the synthesis of ubiquinone and menaquinone. Of the genes necessary for pyridoxine synthesis (pdxA, pdxJ, serC, pdxH, pdxK) in gram-negative bacteria, only the ORF for PdxA (pyridoxal phosphate biosynthetic protein) is present in the genome.
Respiration and proton transfer.
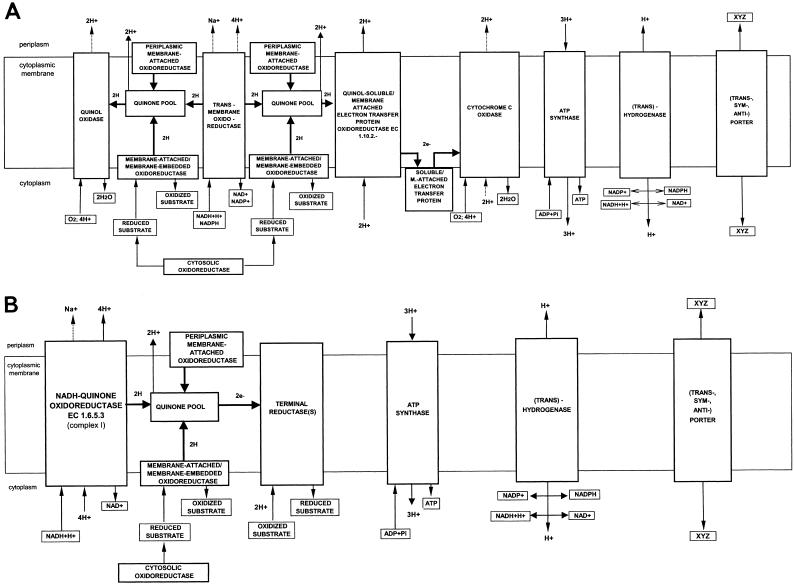
F. nucleatum should be capable of both aerobic and anaerobic respiration. Although there are genes that encode glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12), NAD (P)+-dependent glycerol-3-phosphate dehydrogenase (EC 1.1.1.94) and glycerol-3-phosphate dehydrogenase (EC 1.1.99.5), and d-lactate dehydrogenase (EC 1.1.2.4), it lacks many of the key proteins and protein complexes known to be important in aerobic complexes. There are no terminal cytochrome or quinol oxidases or complexes I, II, and III of the aerobic respiratory chain. In such situations, the bacterium might use an NADH oxidase (EC 1.6.99.3) for growth under normal oxygen concentrations. NADH oxidase represents an alternative way to directly reduce oxygen to water. F. nucleatum cannot accommodate oxidative stress due to the absence of superoxide dismutases or catalases, and therefore NADH oxidases have to play a major role in removing peroxides. Several proteins have been identified as putative NADH dehydrogenases. These are unlikely to be quinone-related proteins, as we could not identify ORFs for the synthesis of quinones or menaquinones. Thus, proteins that might be involved in electron transfer during respiration would have to be flavin dependent (Fig. 5A).
FIG. 5.
(A) Representation of the aerobic respiration pathway and proton flow in F. nucleatum. (B) Representation of the anaerobic respiration pathway in F. nucleatum.
Under anaerobic conditions, arsenate reductase and d- and l-lactate dehydrogenases (EC 1.1.1.28 and EC 1.1.1.27, respectively) serve as electron donors. There is an ORF for the flavoprotein subunit of fumarate reductase complex, but ORFs for iron-sulfur, cytochrome b55x, or hydrophobic anchor protein subunits are absent. Several proteins contribute to the proton gradient, including a proton:sodium-glutamate symporter, a sodium:proton antiporter, a V-type H+-translocating ATP synthase (EC 3.6.1.34), and a Na+-transporting ATP synthase (EC 3.6.3.15) (Fig. 5B).
Transport systems.
About 6% of the ORFs in the genome are dedicated to transport of a variety of compounds by primary and secondary transport systems. These transporters are energized by either ATP, sodium, or proton gradients. There are 36 complete ABC transporter operons and four additional substrate-binding proteins; only one belongs to the major facilitator super family transporter class. The predominant substrates for ABC transporters appear to be oligopeptides and iron compounds. There are ABC transporters for other metal ions such as cobalt, nickel, manganese, zinc, and copper. In contrast, there is a single ABC transporter for sugars and two for amino acids. Three PTS transporters and four P-type ATPases are found.
The transmembrane sodium gradient appears to be as important for transport as the proton gradient. Most of the amino acid transporters are sodium dependent. In prokaryotes, an ABC transporter and/or a proton-dependent inorganic phosphate transporter commonly takes up phosphate, but in F. nucleatum there is only a single phosphate transporter and it is homologous to a eukaryotic sodium-dependent phosphate transporter. There are two potassium uptake systems; one is a sodium symporter, and the other is a proton symporter. Ten predicted sodium:proton antiporters were found, nine of which are homologs of the NhaC protein family, similar to that found in Bacillus firmus. F. nucleatum uses these antiporters to balance ion gradients and to adjust to the pH changes in the oral environment.
There are transporters for all of the essential ions except for sulfate and molybdate. The sulfur requirement may be substituted by the uptake of sulfur-containing amino acids. F. nucleatum strain ATCC 25586 has been reported to take up low levels of glycine and all l-amino acids except leucine and valine (12). We have identified transporters for all the l-amino acids except for asparagine, methionine, and phenylalanine. Of these, phenlyalanine cannot be made internally. Absence of a transporter means that phenylalanine enters as an oligo- or dipeptide or that phenlyalanine is transported by the one ABC transporter that could not be assigned on the basis of similarity. There also appears to be a d-serine transporter homologous to the DsdX protein found in E. coli. Although Gharbia and Shah (12) did not find leucine or valine transporters, there are several potential branched chain amino acid transporters that could also transport leucine or valine. One of the amino acid ABC transporters is similar to the P. aeruginosa branched chain amino acid transporter, and in addition there are two proteins homologous to the branched chain amino acid carrier protein BrnQ of P. aeruginosa.
Two sets of genes encode the TonB, ExbB, and ExbD proteins that are involved in chelated-metal uptake. There are five TonB-dependent outer membrane receptors, which are probably involved in heme and/or siderophore uptake. Three of the receptors are adjacent to ABC transporters involved in chelated iron uptake. There is also a ferric iron ABC transporter, located in tandem with an ORF for an iron-sulfur protein.
F. nucleatum is known to take up fructose, glucose, and galactose. These sugars are either used for biosynthesis or stored intracellularly as a glucose polymer, as long as the preferred energy-providing substrates, amino acids, are present. A non-PTS transporter has been demonstrated to import glucose and galactose, similar to the E. coli glucose/galactose ABC transporter. Fructose is taken up by a PTS system. We have also identified a number of other carbohydrate transporters for substrates such as glycerol and citrate that are not known to be utilized by F. nucleatum. The glycerol channel protein GlpF, which imports glycerol, and other secondary transporters for citrate and N-acetylneuraminate have been identified. Two additional PTS systems are present, of which one is homologous to an N-acetylglucosamine PTS system, and the other is similar to a mannose transporter. However, the true substrates of these two systems are not known.
Several ORFs for amino acid transporters and carbohydrate transporters are linked to catabolic enzymes, facilitating their annotation. This has resulted in expansion of the substrate range for several transporter families. The proposed tryptophan and tyrosine transporters are members of the sodium:neurotransmitter family. Two of the sodium:alanine/glycine transporter family members are proposed to take up cysteine and glutamine, respectively. The putative histidine transporter is not homologous to any characterized transporter, but the homolog in this organism is clustered with histidine catabolism genes, as in Bacillus halodurans. The N-acetylneuraminate transporter is a binding-protein-dependent secondary transporter of the TRAP-T family. The TRAP-T transporter usually consists of a binding protein and two membrane proteins. The F. nucleatum N-acetylneuraminate transporter contains only one membrane protein, since the two transmembrane subunits are fused together. These are clustered with N-acetylneuraminate catabolic genes in F. nucleatum, Vibrio cholerae, and other pathogens. The LysE homolog is adjacent to the lysine catabolic genes. This suggests that Fusobacterium spp. take up lysine by a LysE family protein, and the LysE family may not function exclusively in amino acid efflux.
Motility, signal transduction, stress.
None of the species of Fusobacterium is motile. No genes encoding ORFs for either motility or chemotaxis were identified. However, there is one ORF whose predicted amino acid sequence is similar to that of the response regulator CheY (FN0076) linked in tandem to a sensory transduction protein kinase (FN0077), as in Enterococcus spp. A pair of sensor histidine kinases and the two-component response regulators similar to those of Clostridium spp. are also present. These appear to be the only two-component signal transduction systems in F. nucleatum, although there are seven single histidine kinase genes and three single response regulator genes.
The ORFs for the heat shock proteins are clustered, as in gram-positive bacteria, beginning with the transcriptional repressor hrcA gene, and grpE, dnaK, and dnaJ genes. An ORF for an alkyltransferase is found between dnaK and dnaJ only in F. nucleatum. Both GroEL and GroES chaperonins are present. In addition, there is an ORF that codes for the heat shock protein 15, present only in gram-positive bacteria, as well as an ORF for Hsp90 protein. The heat shock sigma factor rpoH gene is absent. Only one ORF for the cold shock protein CspB has been identified.
Antibiotic resistance.
There are a number of multidrug efflux transporters that appear to be primarily driven by sodium. There are five ORFs for acriflavin resistance protein, including an acriflavin resistance protein precursor A and two copies of acriflavin resistance proteins B and D, which can protect the bacterium from hydrophobic inhibitors. Goldstein et al. (13) have shown that erythromycin (and other macrolides) are ineffective against several species of F. nucleatum due to the macrolide efflux protein (FN1168). The presence of two genes for 5-nitroimidazole resistance is an unexpected finding, as several studies have indicated that Fusobacterium spp. are sensitive to nitroimidazoles, especially metronidazole (35). Resistance to β-lactams is due to the presence ofβ-lactamase (FN1375), which is similar to that of Clostridium spp. (10, 32). There is a second β-lactamase similar to the archaeal lactam utilization protein, LamB, which is involved in utilization of pyrrolidinone. An ORF for chloramphenicol resistance is also found in the genome.
There is an ORF that shows homology to the phenazine synthesis protein PhzF. Because the phzF gene is normally clustered with other phenazine synthetic enzymes and such a cluster is absent in Fusobacterium spp., it is unlikely that phenazine is synthesized. Alternatively, this ORF might code for a 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase. Similarly, there is an ORF (FN0388) that encodes tetracenomycin polyketide O-methyltransferase, although the rest of the polyketide synthetase cluster is absent.
Virulence.
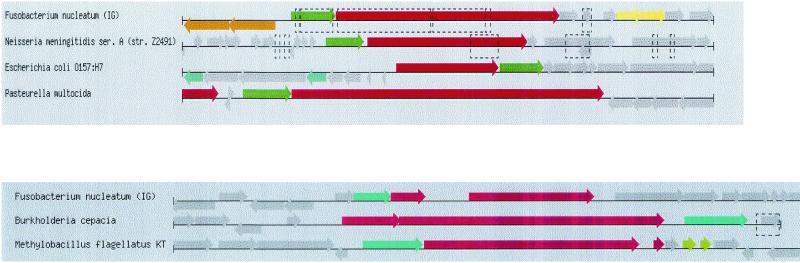
Although Fusobacterium is not a major pathogen per se, there are a number of ORFs that code for putative virulence factors. An ORF similar to the one encoding integral membrane protein MviN (mouse virulence) of Salmonella spp. is present (23). An ORF for a virulence-associated protein I has also been identified. Two copies (FN1817 and FN0132) of the hemolysin precursor protein (304 kDa) and activator protein precursor have been identified; both are on a transposable element. The ORF FN1817 is similar in sequence and organization to the hemolysin from Neisseria meningitidis and E. coli O157:H7 (Fig. 6, upper panel). The third hemolysin (FN1885 and FN0132 are truncated) is similar to ORFs FN1516 and FN2031 and clusters with the hemolysin of Burkholderia cepacia (Fig. 6, lower panel). Finally, there is an ORF on a transposable element for a hemolysin III protein similar to a hemolysin of S. aureus.
FIG. 6.
(Upper panel) Clustering of hemolysin genes (red arrow) of F. nucleatum with those of N. meningitidis, E. coli O157:H7, and P. multocida. Except in the case of E. coli O157:H7, the ORF for hemolysin activator (green arrow) is upstream of the hemolysin gene. The broken lines indicate the presence of insertion elements. The“pinned region” algorithm was used to describe the pattern organization. The yellow, brown, and gray arrows represent ORFs and operons that are not conserved in this hemolysin cluster. (Lower panel) Clustering of a second hemolysin gene (red arrow) of F. nucleatum with those of B. cepacia and Methylobacillus flagellatus. This hemolysin gene in F. nucleatum is truncated.
Two ORFs with similarity to filamentous hemagglutinin, an adhesin homologue of Bordetella pertussis and a surface antigen D15 homolog of H. influenzae have been identified. In addition, there is an ORF for a protein similar to Omp of V. cholerae and a VacJ lipoprotein of Shigella flexneri. The functions of these proteins are unknown in F. nucleatum, but VacJ protein from Shigella spp. has been shown to be involved in virulence (44). There is a large family of related Omps (over 2,000 aa long) in F. nucleatum, with weak similarity to adhesins of various pathogens. The functions of these are not known. A major virulence factor, a leukotoxin (LktA) of F. necrophorum (30), is absent from F. nucleatum.
The LPS synthesis genes are organized in an operon consisting of nine ORFs. The first ORF is similar to a polysaccharide deacetylase, followed by a β-1,4-glucosyltransferase, heptosyltransferase-I, heptosyltransferase-II, and two core LPS synthesis proteins. An ORF for RecA protein, RecA interacting protein RecX, and the O-sialoglycoprotein endopeptidase that cleaves sialated glycoproteins are clustered. In addition, there are ORFs for lipo-oligosaccharide synthesis and an ORF for teichoic acid biosynthesis similar to that found in gram-positive bacteria. F. nucleatum strain FDC 364 produces an immunosuppressive factor FipA protein (8), which is an acetyl-CoA acetyltransferase that is normally not secreted in bacteria.
Insertion sequences and transposons.
There are 73 assignments of possible IS elements. Two are large: FINSEL51 (4,736 bp) and FINSEL24 (3,525 bp). There are 11 with sizes varying between 1,000 and 2,000 bp, 19 between 500 and 1,000 bp, and 41 less than 500 bp in length. The smallest is 179 bp long. These elements can also be grouped into seven classes based on sequence similarity. Interestingly, GMP synthase and DNA helicase are flanked by an active IS element (FINSEL8, 9, 10, and 11) that has in it a transposase and integrase. Downstream from this IS element cluster is an outer membrane protein gene. Similarly, the nickel transport/uptake operon and the pyrophosphate synthesis ribADF operon are flanked by a transposable element (IS 12, 13, 14, and 15). Other genes that have IS elements are hemolysin precursor, hemolysin activator precursor protein, hemolysin III, cold shock protein, serine protease, glycerol phosphate diester phosphodiesterase, and a restriction/modification gene.
In general, the transposases are similar to those of the gram-positive bacteria. There are 41 ORFs assigned with transposase function. Major transposases include a group of six (493 aa) that are related to S. aureus transposases and a group of five (391 aa) related to a Lactobacillus spp. family of transposases. Six additional ORFs that are 207 aa long are similar to Mycoplasma spp. transposases. Finally, there is a smaller family (below 170 aa) of six transposases similar to those found in Streptococcus equi. The remaining ORFs are remnants of earlier transposition events.
Conclusion.
Although a large number of bacteria coexist in the mouth, larynx, and upper respiratory tract in humans, they all have evolved to localize and form a microbial community with complex physical and biochemical interactions. Several questions remain unanswered as to such localization and interdependence. Genome sequence analysis has revealed answers to some of these questions; for example, the absence of genes that encode choline-binding proteins in F. nucleatum limits its ecological niche to the oral cavity, unlike S. pneumoniae or H. influenzae, which can colonize the naso-pharynx. Since F. nucleatum is an oral resident, it has perhaps lost the leukotoxin gene as it rarely encounters leukocytes, unlike F. necrophorum.
Because several essential amino acid synthetic pathways are missing, F. nucleatum requires an extensive repertoire for amino acid transport. It also has a di- and oligo-peptide uptake transport system to utilize the peptides that are freely available from the proteolysis of food or of dead bacteria. Genome data have revealed several adhesin proteins that might interact with dental pathogens, although only one has been experimentally shown to interact with P. gingivalis and various human cell types.
The novel amino acid meso-lanthionine in the peptidoglycan might provide a specific target for anti-Fusobacterium drugs. Further work is needed to validate the pathway proposed for the synthesis of l-lanthionine and to discover the epimerase needed for its conversion to the substrate for peptidoglycan synthesis. The genome also showed that there are many outer membrane proteins of very high molecular weight, at least six that were previously unsuspected. These might be useful as immunogens. Finally, the genome sequence has laid a foundation for microarrray and proteomics experiments. The results from such experiments should provide potential drug targets for treatment and control of periodontal diseases.
Acknowledgments
V.K. and I.A. contributed equally towards this work.
We thank the members of the Integrated Genomics sequence and assembly groups. We also thank Karl Reich for critically reading the manuscript and providing valuable suggestions.
This work was supported by SBIR grant R44 GM61431 from the NIH.
REFERENCES
- 1.Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1998. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol. Immunol. 13:51-54. [DOI] [PubMed] [Google Scholar]
- 2.Baker, H. A., J. M. Khan, and L. Hedrick. 1982. Pathway of lysine degradation in Fusobacterium nucleatum. J. Bacteriol. 152:201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatrix, B., K. Bendrat, S. Rospert, and W. Buckel. 1990. The biotin-dependent sodium ion pump glutaconyl-CoA decarboxylase from Fusobacterium nucleatum (subsp. nucleatum). Comparison with the glutaconyl-CoA decarboxylases from Gram-positive bacteria. Arch. Microbiol. 154:362-369. [DOI] [PubMed] [Google Scholar]
- 4.Bolstad, A. I. 1994. Sizing of the Fusobacterium nucleatum genome by pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 123:145-152. [DOI] [PubMed] [Google Scholar]
- 5.Bolstad, A. I., H. B. Jensen, and V. Bakken. 1996. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin. Microbiol. Rev. 9:55-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, C. H., and P. A. Frey. 2000. Cloning, sequencing, heterologous expression, purification, and characterization of adenosylcobalamin-dependent D-lysine 5,6-aminomutase from Clostridium sticklandii. J. Biol. Chem. 275:106-114. [DOI] [PubMed] [Google Scholar]
- 7.Chevalier, C., J. M. Thiberge, R. L. Ferrero, and A. Labigne. 1999. Essential role of Helicobacter pylori γ-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol. Microbiol. 31:1359-1372. [DOI] [PubMed] [Google Scholar]
- 8.Demuth, D. R., R. Savary, E. Golub, and B. J. Shenker. 1996. Identification and analysis of fipA, a Fusobacterium nucleatum immunosuppressive factor gene. Infect. Immun. 64:1335-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, A. J., I. Iliopoulos, N. C. Kyrpides, and C. A. Ouzounis. 1999. Protein interaction maps for complete genomes based on gene fusion events. Nature 402:86-90. [DOI] [PubMed] [Google Scholar]
- 10.Fosse, T., I. Madinier, C. Hitzig, and Y. Charbit. 1999. Prevalence of β-lactamase-producing strains among 149 anaerobic gram-negative rods isolated from periodontal pockets. Oral Microbiol. Immunol. 14:352-357. [DOI] [PubMed] [Google Scholar]
- 11.Fredricksen, A., E. N. Vasstrand, and H. B. Jensen. 1991. Peptidoglycan precursor from Fusobacterium nucleatum contains lanthionine. J. Bacteriol. 173:900-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gharbia, S. E., and H. N. Shah. 1991. Pathways of glutamate catabolism among Fusobacterium species. J. Gen. Microbiol. 137:1201-1206. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein, E. J., D. M. Citron, C. V. Merriam, Y. Warren, and K. Tyrrell. 1999. Activities of telithromycin (HMR 3647, RU 66647) compared to those of erythromycin, azithromycin, clarithromycin, roxithromycin, and other antimicrobial agents against unusual anaerobes. Antimicrob. Agents Chemother. 43:2801-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haake, S. K., S. C. Yoder, G. Attarian, and K. Podkaminer. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J. Bacteriol. 182:1176-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishige, K., and T. Noguchi. 2001. Polyphosphate: AMP phosphotransferase and polyphosphate: ADP transferase activities of Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 281:821-826. [DOI] [PubMed] [Google Scholar]
- 16.Kim, D. H., S. Y. Kim, S. Y. Park, and M. J. Han. 1999. Metabolism of quercitrin by human intestinal bacteria and its relation to some biological activities. Biol. Pharm. Bull. 22:749-751. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. O., and J. N. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368-377. [DOI] [PubMed] [Google Scholar]
- 18.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander, P. E., R. N. Andersen, and L. V. Holdeman. 1985. Coaggregration of oral Bacteriodes with other bacteria: central role in coaggregation bridges and competitions. Infect. Immun. 3:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolenbrander, P. E., and R. N. Andersen. 1989. Inhibition of coaggregration between Fusobacterium nucleatum and Porphyromonas gingivalis by lactose and related sugars. Infect. Immun. 10:3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 23.Kutsukake, K., T. Okada, T. Yokoseki, and T. Iino. 1994. Sequence analysis of the flgA gene and its adjacent region in Salmonella typhimurium, and identification of another flagellar gene, flgN, Gene 1:49-54. [DOI] [PubMed] [Google Scholar]
- 24.Kyrpides, N. C., and C. Woese. 1998. Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B alpha-beta-delta subunit families. Proc. Natl. Acad. Sci. USA 7:3726-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyrpides, N. C., C. A. Ouzounis, I. Iliopoulos, V. Vonstein, and R. Overbeek. 2000. Analysis of the Thermotoga maritima genome combining a variety of sequence similarity and genome context tools. Nucleic Acids Res. 28:4573-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerud, R. F., and H. R. Whitely. 1971. Purification and properties of α-ketoglutarate reductase from Micrococcus aerogenes. J. Bacteriol. 106:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maden, B. E. H. 2000. Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carriers in C1 metabolism. Biochem. J. 350:609-629. [PMC free article] [PubMed] [Google Scholar]
- 28.McGovern, K. J., T. G. Blanchard, J. A. Gutierrez, S. J. Czinn, S. Krakowka, and P. Youngman. 2001. γ-Glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect. Immun. 69:4168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mineyama, R., and K. Saito. 1997. Some properties of γ-glutamyl transpeptidase from Fusobacterium nucleatum. Microbios 90:187-200. [PubMed] [Google Scholar]
- 30.Narayanan, S. K., T. S. Nagaraja, M. M. Chengappa, and G. C. Stewart. 2001. Cloning, sequence, and expression of the leukotoxin gene from Fusobacterium necrophorum. Infect. Immun. 9:5447-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noack, J., G. Dongowski, L. Hartmann, and M. Blaut. 2000. The human gut bacteria Bacteroides thetaiotaomicron and Fusobacterium varium produce putrescine and spermidine in cecum of pectin-fed gnotobiotic rats. J. Nutr. 5:1225-1231. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto, A. C., E. Gaetti-Jardim, Jr., S. Cai, and M. J. Avila-Campos. 2000. Influence of antimicrobial sub-inhibitory concentrations on hemolytic activity and bacteriocin-like substances in oral Fusobacterium nucleatum. New Microbiol. 23:137-142. [PubMed] [Google Scholar]
- 33.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA 96:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pianotti, R., S. Lachette, and S. Dills. 1986. Desulfuration of cysteine and methionine by Fusobacterium nucleatum. J. Dent. Res. 65:913-917. [DOI] [PubMed] [Google Scholar]
- 35.Poulet, P. P., D. Duffaut, and J. P. Lodter. 1999. Metronidazole susceptibility testing of anaerobic bacteria associated with periodontal disease. J. Clin. Periodontol. 26:261-263. [DOI] [PubMed] [Google Scholar]
- 36.Richaud, C., D. Mengin-Lecreulx, S. Pochet, E. J. Johnson, G. N. Cohen, and P. Marlier. 1993. Directed evolution of biosynthetic pathways. Recruitment of cysteine thioethers for constructing the cell wall of Escherichia coli. J. Biol. Chem. 268:26827-26835. [PubMed] [Google Scholar]
- 37.Rock, C. O., R. J. Heath, H. W. Park, and S. Jackowski. 2001. The licC gene of Streptococcus pneumoniae encodes a CTP:phosphocholine cytidylyltransferase. J. Bacteriol. 183:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers, A. H., J. Chen, P. S. Zilm, and N. J. Gully. 1998. The behavior of Fusobacterium nucleatum chemostat-grown in glucose and amino acid-based chemically defined media. Anaerobe 4:111-116. [DOI] [PubMed] [Google Scholar]
- 39.Roof, D. M., and J. R. Roth. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171:3316-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 41.Selkov, E., R. Overbeek, Y. Kogan, L. Chu, V. Vonstein, D. Holmes, S. Silver, R. Haselkorn, and M. Fonstein. 2000. Functional analysis of gapped microbial genomes: amino acid metabolism of Thiobacillus ferrooxidans. Proc. Natl. Acad. Sci. USA 97:3509-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharples, G. J., and D. R. Leach. 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17:1215-1217. [DOI] [PubMed] [Google Scholar]
- 43.Stojiljkovic, I., A. J. Baumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, T., T. Murai, I. Fukuda, T. Tobe, M. Yoshikawa, and C. Sasakawa. 1994. Identification and characterization of a chromosomal virulence gene,vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11:31-41. [DOI] [PubMed] [Google Scholar]
- 45.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65:943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss, E. I., B. Shaniztki, M. Dotan, N. Ganeshkumar, P. E. Kolenbrander, and Z. Metzger. 2000. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol. Immunol. 15:371-377. [DOI] [PubMed] [Google Scholar]