Abstract
The discovery that two distinct enzyme catalysts, purine hydroxylase (PH) and xanthine dehydrogenase (XDH), are required for the overall conversion of hypoxanthine to uric acid by Clostridium purinolyticum was unexpected. In this reaction sequence, hypoxanthine is hydroxylated to xanthine by PH and then xanthine is hydroxylated to uric acid by XDH. PH and XDH, which contain a labile selenium cofactor in addition to a molybdenum cofactor, flavin adenine dinucleotide, and FeS centers, were purified and partially characterized as reported previously. In the present study, the activities of these two enzymes were measured in cells grown in media containing various concentrations of selenite, molybdate, and various purine substrates. The levels of PH protein in extracts were determined by immunoblot assay. The amount of PH protein, as well as the specific activities of PH and XDH, increased when either selenite or molybdate was added to the culture medium. PH levels were highest in the cells cultured in the presence of either adenine or purine. XDH activity increased dramatically in cells grown with either xanthine or uric acid. The apparent increases in protein levels and activities of PH and XDH in response to selenium, molybdenum, and purine substrates demonstrate that these enzymes are tightly regulated in response to these nutrients.
The family of enzymes known as the molybdenum hydroxylases have been the focus of intense investigation for many decades. These enzymes are distinguished from other molybdenum-containing oxidoreductases by the utilization of H2O as the source of oxygen for the hydroxylation reaction. The best-studied enzyme of this family is bovine xanthine oxidoreductase (XOR), primarily because of its relative abundance and ease of purification. XOR is responsible for the conversion of hypoxanthine to xanthine and xanthine to uric acid in the purine degradative pathway in mammals and certain bacteria (12, 15). In addition to a molybdenum cofactor, XOR contains two 2Fe-2S clusters and flavin adenine dinucleotide (FAD) (15). A few molybdenum hydroxylases, characterized from clostridial species, also contain selenium in the form of a labile cofactor (4, 5, 16, 17). These include nicotinic acid hydroxylase (NAH) and xanthine dehydrogenase (XDH) from Clostridium barkeri (4, 16) as well as XDH from Clostridium purinolyticum (17), Clostridium acidi-urici (20), and Clostridium cylindrosporum (20). A new enzyme, purine hydroxylase (PH) from C. purinolyticum, has been added to this small family of selenium-dependent molybdenum hydroxylases (17). In contrast to the previously studied purine degradation systems in both bacteria and mammals, two enzymes are required for hydroxylation of hypoxanthine to uric acid in C. purinolyticum, and both are selenium-dependent molybdoenzymes. Although not yet well defined, this labile selenium cofactor is required for catalytic activity of all of the above-mentioned clostridial enzymes. Based on electron paramagnetic resonance (EPR) studies, selenium is in close proximity to the molybdenum atom at the active site of NAH (8), but the interactions between molybdenum and selenium in PH and XDH from C. purinolyticum are presently under investigation.
Besides the labile selenium cofactor-containing enzymes, there are several selenium-dependent enzymes in mammals and bacteria that contain selenocysteine, which is encoded by a UGA codon in the mRNA. The pathway for the carefully regulated incorporation of selenocysteine into the appropriate position of the peptide backbone has been well-defined in certain bacteria, especially Escherichia coli (1). Investigations of formate dehydrogenase H (FDHH) from E. coli led to the discovery of the gene products required for selenocysteine incorporation into selenoenzymes as described by August Böck and his associates (1). The crystal structure of E. coli FDHH has been solved (2), thus confirming the precise nature and coordination of selenium present in the polypeptide. In all of these cases, the presence of selenium in a nonlabile moiety of the translated protein has been predicted by DNA sequence analysis of the gene encoding the selenoenzyme and in many cases was confirmed by Edman degradative peptide analyses. Although the presence of a TGA codon can be found in the genome databases of many bacteria and mammals, the presence of labile selenium cofactors such as those in PH and XDH from C. purinolyticum cannot be detected in silico. It is possible that labile selenium cofactors also may exist in certain eukaryotic enzymes.
In recent years, significant advances have been made in the study of the regulation of molybdoenzymes at the transcriptional level in bacteria. In E. coli, the molybdate regulatory protein ModE, initially described as the repressor of the modABC operon encoding a high-affinity molybdate transporter (9), has also been shown to positively regulate transcription of the narXL operon (involved in nitrate-dependent up-regulation of nitrate reductase [18]) and the dmsABC operon (encoding DMSO reductase [13]) and to increase the expression of the hyc operon (encoding the FDHH-linked hydrogenase 3 isoenzyme [18]). This increase in transcription of these genes, all encoding either molybdoenzymes or proteins involved in the biosynthesis or regulation of molybdoenzyme systems, is controlled by the level of molybdate in the cytoplasm (13, 18). These genes are also tightly regulated by the level of oxygen in the medium and, with the exception of the dmsABC operon, the presence of the substrate for each enzyme (3, 10, 13, 19). Given this response at the transcriptional level to the need for molybdenum, it is possible that the members of the family of selenium-dependent molybdenum hydroxylases in clostridia may also be regulated in response to molybdenum levels, purine substrates, and selenium availability. In this report, levels of PH and XDH from C. purinolyticum were investigated with respect to the presence of various purines as well as in response to limitation of selenium and molybdenum in the culture medium.
Growth of C. purinolyticum.
C. purinolyticum cells were cultured in a minimal medium as previously described (17). Selenium was added to the culture medium as selenite at 1 μM, except where indicated in the figure legend. Purines were added to the culture medium at a final concentration of 12 mM after heating with the slow addition of base (as needed) to ensure solubility. Cultures were grown in volumetric flasks as previously described (17) overnight at 37°C. Optical density (OD) measurements of cultures at 600 nm were determined immediately prior to harvesting using a Cary 100 Bio UV-visible spectrophotometer. Various purines were the highest grade available and were purchased from Sigma (St. Louis, Mo.). All other chemicals and components of the media were of the highest grade available.
Extract preparation.
Cells were harvested by centrifugation at 9,500 × g, washed once with buffer A (0.1 M Tricine [pH 8.0], 0.1 mM phenylmethylsulfonyl fluoride, 0.5 mM EDTA), sedimented again, and resuspended in buffer A. Rupture of cells was accomplished by sonication using a Branson 450 Sonifier (Danbury, Conn.). Cell extracts were clarified by centrifugation at 18,500 × g for 15 min at 4°C. Protein concentration in this cell extract was determined by dye-binding assay (Bio-Rad protein assay; Hercules, Calif.).
Enzyme assays.
XDH activity was determined by following the xanthine-dependent reduction of potassium ferricyanide (FeCN), which was monitored by measuring the decrease in absorbance at 420 nm (1,020 M−1 cm−1) in 0.1 M sodium phosphate buffer, pH 8.0. The rate of reduction of FeCN (0.5 mM) by cell extract was followed until a steady-state baseline was attained, and then 1 mM xanthine was added. The initial linear rate of xanthine-dependent reduction of FeCN was determined and corrected for the baseline reduction. These assays were performed under aerobic conditions since no change in the rate of reaction was observed when assays were performed anaerobically.
Purine hydroxylase activity was determined by following the hypoxanthine-dependent reduction of NADP to NADPH at 340 nm (6,220 M−1 cm−1) in 0.1 M sodium phosphate buffer, pH 7.75, under either an argon or a nitrogen atmosphere. The reaction mixture contained 1 mM NADP and 1 mM hypoxanthine, and the initial rate of reduction of NADP was determined after the addition of substrate. Details of the reduction of NADP by PH are to be described elsewhere in an in-depth biochemical analysis of PH. Purified PH exhibits a maximum activity of 154 μmol min−1 (mg of protein)−1 under these conditions. All assays were performed at 25°C.
Immunoblot analysis.
Rabbit polyclonal antibodies to native purified PH were obtained from Rockland, Inc. (Gilbertsville, Pa.). Polyclonal antibodies to PH were partially purified from rabbit serum using a protein G immunoaffinity column (Pierce, Rockford, Ill.). Cell extracts and purified PH were separated under denaturing and reducing conditions essentially as previously described (11) using sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) gels (Invitrogen, Carlsbad, Calif.). Proteins were transferred to a polyvinylidene diflouride membrane, blocked with Tris-buffered saline-Tween (TBS-Tween; Biosource International, Camarillo, Calif.) containing 2% bovine serum albumin for 1 h at 25°C, and incubated with primary antibody overnight at 4°C. After being washed with TBS-Tween buffer, the membrane was incubated with goat anti-rabbit immunoglobulin G (IgG) labeled with alkaline phosphatase (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) at 25°C for 1 h. After being washed in TBS-Tween, the blot was developed using a 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT) phosphatase substrate system (Kirkegaard & Perry Laboratories).
PH expression in response to purines.
In initial studies of PH and XDH from C. purinolyticum, it was determined that PH is responsible for the hydroxylation of hypoxanthine to xanthine but is unable to hydroxylate xanthine (17). XDH, also isolated from the same organism, was efficient in the hydroxylation of xanthine to uric acid. The fact that C. purinolyticum requires adenine for growth (6) even in the presence of other purines indicates that adenine is required for some critical metabolic function. Hypoxanthine alone cannot replace adenine in the medium for growth of the organism (data not shown). Whether this need for adenine for growth is related to the utilization of a two-enzyme pathway for purine catabolism in C. purinolyticum is not yet known. PH will reduce NADP but XDH does not reduce pyridine nucleotides, and the electron acceptor (with the exception of dyes) for XDH is not known. Because of these differences in electron acceptor and substrate specificity, activities of either enzyme can be measured in the presence of its substrate and the appropriate acceptor without concern for the contribution by the other.
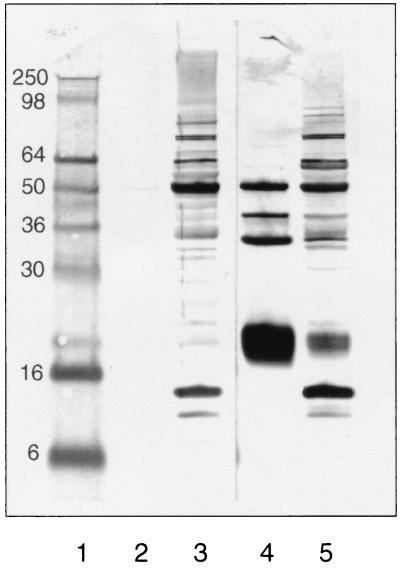
In addition to PH and XDH activities, PH protein levels were determined using polyclonal antibodies generated in rabbits. Figure 1 demonstrates that although there are a number of antibodies present in the preimmune serum of the rabbit which react with proteins other than PH, each of the four subunits of PH present in the cell extract is reactive with the immune serum (lanes 3 and 5). The smallest subunit is the only one of the four subunits to transfer quantitatively to the polyvinylidene difluoride membrane (data not shown); therefore, this subunit was selected as the best indicator of the level of PH in the cell extracts determined by Western blot analysis.
FIG. 1.
Immunoblot analysis of PH using rabbit polyclonal antibodies. Lane 1, SDS-PAGE protein marker, molecular masses are indicated at left (in kilodaltons); lanes 2 and 4, 0.5 μg of purified PH; lanes 3 and 5, 25 μg of cell extract from C. purinolyticum. Lanes 1 through 3 were probed with preimmune rabbit serum, and lanes 4 and 5 were probed with immune serum after injection with native PH enzyme.
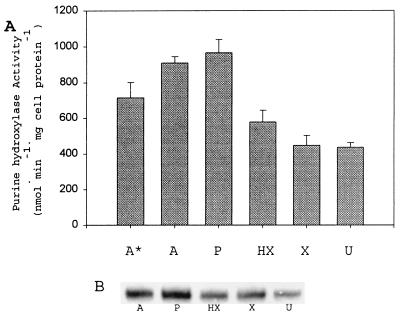
C. purinolyticum cells were cultured using either adenine, purine, hypoxanthine, xanthine, or uric acid as the added substrate (12 mM) in the medium. Because adenine is required for growth (6), it was added to all cultures at a final concentration of 1 mM, with ammonium sulfate (2.3 mM) also being supplied as the primary nitrogen source. As a control, C. purinolyticum was cultured in the presence of only 1 mM adenine. The final OD of this control culture was only (mean ± standard deviation [SD]) 0.32 ± 0.02 at 600 nm, whereas the typical OD is between 1.7 and 2.0. Clearly, as shown in Fig. 2A, the level of PH activity in cell extracts was higher when the cells were grown on either adenine, purine, or hypoxanthine than when grown on the more oxidized purines (Fig. 2A). The basal level of PH activity in cells cultured in 12 mM xanthine and uric acid is most likely due to the adenine also present (needed for growth) in the medium and in fact is slightly lower than the activity of cells grown with only 1 mM adenine. The presence of high levels of oxidized purines in the culture medium caused a slight decrease in the PH activity compared to the adenine (1 mM)-grown cells. Figure 2B demonstrates that the level of PH protein present correlates well with the specific activity seen in Fig. 2A. It is possible that hypoxanthine may be a required effector for the activation of transcription of the genes encoding PH or, alternatively, may be involved in the posttranslational regulation of PH levels. This possibility is difficult to test since so far no conditions have been found for the growth of this organism in the absence of adenine in the medium, but it is clear from the above data that PH activity is induced by adenine, purine, and hypoxanthine.
FIG. 2.
PH activity response to purine present in culture medium. (A) PH specific activities from cell extracts of C. purinolyticum cultured on different purines (12 mM) except where indicated. Activities given represent the mean derived from at least three independent cultures, and error bars indicate 1 SD from the mean. A*, adenine-grown cell extract (1 mM); A, adenine-grown cell extract; P, purine-grown cell extract; HX, hypoxanthine-grown cell extract; X, xanthine-grown cell extract; U, uric acid-grown cell extract. (B) Western analysis of the small subunit of PH from an extract of cells grown on each purine at a 12 mM concentration in the medium. A, adenine-grown cell extract (10 μg); P, purine-grown cell extract (10 μg); HX, hypoxanthine-grown cell extract (10 μg); X, xanthine-grown cell extract (10 μg); U, uric acid-grown cell extract (10 μg).
XDH is expressed at significant levels only in the presence of more oxidized purines.
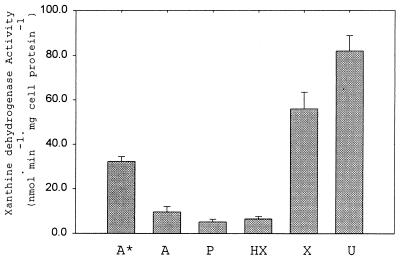
Figure 3 illustrates the dramatic increase in XDH activity when cells are cultured in the presence of either xanthine or uric acid. Only about one-tenth of the activity was detected when either adenine, purine, or hypoxanthine was supplied as a substrate and added at a concentration of 12 mM. The level of XDH activity in cells cultured only with 1 mM adenine was slightly higher than that in cells cultured in the presence of excess adenine. In the absence of excess adenine (or purine or hypoxanthine) in the medium, the level of XDH expression was higher, which suggests that these purines have an inhibitory effect on XDH expression. There was a significant up-regulation of XDH in response to high levels of oxidized purines in the culture medium. It is possible that the genes encoding the three subunits of XDH are regulated positively in response to either xanthine or uric acid at the transcriptional level and the presence of either in the medium may act as a gratuitous inducer of the expression of XDH, whereas the physiologically relevant inducer may be only xanthine. Rabinowitz and Barker reported previously (14) that XDH in a related purinolytic species, C. cylindrosporum, was reversible in vivo, and this might explain the increase in XDH activity in the presence of uric acid in the medium. Other possibilities, such as the failure to insert a cofactor into the apoenzyme when the cells were cultured in the presence of adenine or hypoxanthine or degradation of XDH in the absence of significant amounts of xanthine or uric acid, cannot be ruled out. Although a significant intracellular pool of xanthine should be generated in the cytoplasm by the action of PH even when cells are cultured on adenine or hypoxanthine, the level of XDH activity is not as high when cells are cultured in the presence of hypoxanthine.
FIG. 3.
XDH activity response to the purine source. XDH-specific activities from cell extracts of C. purinolyticum were cultured on different purines (12 mM) except where indicated. Activities given represent the mean from at least three independent cultures, and error bars indicate 1 SD from the mean. A*, adenine-grown cell extract (1 mM); A, adenine-grown cell extract; P, purine-grown cell extract; HX, hypoxanthine-grown cell extract; X, xanthine-grown cell extract; U, uric acid-grown cell extract.
Level of selenium in growth medium significantly affects specific activity of PH.
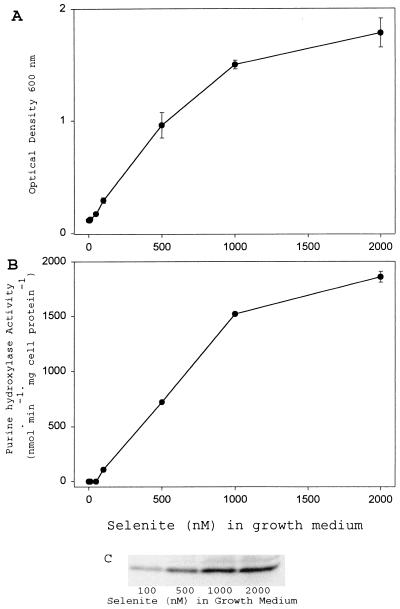
As was shown earlier by Dürre and Andreesen (5), selenium supplementation is required for the growth of C. purinolyticum cells in a defined medium, and a positive response of the level of XDH activity in the cell extracts to the presence of increased levels of selenium was observed (data not shown). Since it now has been established that both PH and XDH contain selenium and require selenium for activity, the effects of various concentrations of selenite in the culture medium on the level of PH in cells grown on hypoxanthine as substrate were examined (Fig. 4).
FIG. 4.
PH activity and protein level responses to availability of selenium in culture medium. (A) Final ODs at 600 nm of cultures grown with various concentrations of selenite are plotted. Densities given represent the mean from at least three independent cultures, and error bars indicate 1 SD from the mean. (B) PH specific activities from cell extracts of C. purinolyticum cells cultured on various concentrations of selenite as indicated. Activities given represent the mean from at least three independent cultures, and error bars indicate 1 SD from the mean. (C) Western analysis of small subunit of PH of a representative extract of cells grown in various concentrations of selenite. The concentration of selenite in the growth medium is indicated below each lane; 10 μg of cell extract was loaded in each lane.
As observed earlier, growth of C. purinolyticum cells was limited by low selenium levels (Fig. 4A). A low visible turbidity in all cultures is presumed to be due to carryover of selenium (0.3 nM) from the inoculum. In these selenium-limited cultures, no detectable PH activity was present (Fig. 4B). Growth increased as the level of selenite added to the medium increased above 100 nM (Fig. 4A). In accordance with this increase in growth, PH catalytic activity also increased (Fig. 4B), as did the level of PH protein detected by antibody (Fig. 4C).
C. purinolyticum also contains a selenium-dependent glycine reductase (GR) which is key to the growth of the organism under these culture conditions (7). Thus, limitation of growth by low selenium availability would be due not only to a lack of active PH or XDH enzyme but also to a lack of GR. It is not yet known whether the cell, under limiting selenium conditions, would preferentially incorporate selenium into selenocysteine-containing enzymes, such as glycine reductase, or into the labile selenium cofactor-containing molybdoenzymes. A hierarchy for selenium incorporation has been demonstrated in other systems. The order of preference of selenium incorporation will be an interesting question for future studies when more is known about the labile selenium cofactor bound to PH and XDH as well as the mode of incorporation of this labile selenium moiety.
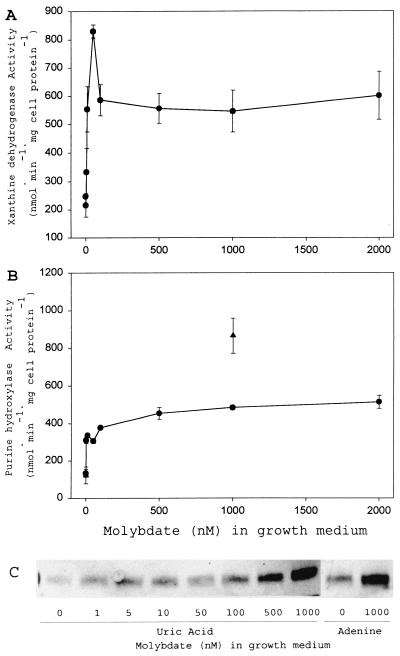
PH also responds positively to level of molybdate in culture medium.
Omission of molybdate supplementation from the minimal culture medium only slightly reduced the growth yield of C. purinolyticum with either uric acid or adenine as substrate (data not shown). The specific activities of both XDH and PH were significantly lower in the absence of added molybdate (Fig. 5A and B), indicating that residual molybdenum in the culture medium is low enough to be limiting for either the maximal expression of the subunits of PH and XDH or the insertion of Mo cofactor into PH and XDH. This residual activity of both enzymes may explain why there was not a significant decrease in the growth yield of the organism, as seen with selenium limitation.
FIG. 5.
Responses of PH and XDH activities to the availability of molybdenum in the culture medium. (A) XDH specific activities in extracts of C. purinolyticum cells cultured on various concentrations of molybdate as indicated. (B) PH specific activities in extracts of C. purinolyticum cells cultured on various concentrations of molybdate as indicated. Activities given represent the mean derived from at least three independent cultures, and error bars indicate 1 SD from the mean. Cells were cultured with uric acid (•) and adenine (▴). (C) Western analysis of small subunit of PH from a representative extract of cells grown in various concentrations of molybdate. The concentration of molybdate in the growth medium is indicated below each lane, and 10 μg of cell extract was added to each lane. The purines added to the growth medium are also indicated.
Although significant amounts of PH and XDH were present in cells in the absence of molybdate supplementation, the catalytic activities of both enzymes responded positively to increased molybdate in the medium (Fig. 5A and B). An apparent maximal level of XDH activity in extracts was reached after the addition of 100 nM molybdate (Fig. 5A). The level of PH activity in cell extracts also responded in a similar fashion when cells were cultured in the presence of molybdate at 100 nM or greater (Fig. 5B). The amount of PH protein detected by antibody increased as the level of molybdate supplementation increased, whether the cells were cultured in the presence of uric acid or adenine (Fig. 5C). From the immunoblot analysis (Fig. 5C), it appears that in the presence of saturating molybdenum concentrations (above 200 nM) there was still an increase in the amount of PH protein produced without a concomitant increase in PH activity. This would suggest that the activity of PH may be limited by the synthesis of one of the various cofactors required (Mo cofactor, labile Se cofactor, Fe-S clusters, FAD).
Nonetheless, these increases in activity of both PH and XDH and in the levels of PH protein in the cell in response to molybdenum indicate that the expression of the genes encoding these enzymes is sensitive to the level of molybdate in the cytoplasm, as has been seen with other molybdoenzymes (18). The protein(s) involved in the molybdate-dependent up-regulation of PH and XDH in C. purinolyticum might prove to be similar to that found in E. coli, in which the molybdate response regulator ModE has been shown to be required for maximal expression of molybdenum-dependent enzymes (13, 18).
In C. purinolyticum, which was initially isolated as an adenine fermenter, a two-enzyme system is responsible for the conversion of hypoxanthine to uric acid. Although NADP is utilized in vitro by PH and is the likely candidate as the physiological electron acceptor in vivo, the only known electron acceptors for XDH are artificial dyes. The significance of a two-enzyme system for purine metabolism in C. purinolyticum cannot be fully appreciated until the identity of the final electron acceptor is uncovered experimentally. Nonetheless, regulation of the levels of active XDH and PH in response to the presence of selenium, molybdenum, and the purine substrates appears complex and probably reflects this organism's need to tightly regulate the amounts and activity of these enzymes. Determination of whether this regulation involves systems consisting of transcriptional activators or posttranscriptional signals to regulate either mRNA or protein levels or both will be a fruitful area of future investigation. Such studies may shed light on the regulation of selenoenzymes and molybdoenzymes in other biological systems.
Acknowledgments
I thank Thressa Stadtman for her guidance and support during the research and the preparation of the manuscript.
REFERENCES
- 1.Böck, A., K. Forchhammer, J. Heider, W. Leinfelder, G. Sawers, and B. F. Veprek. 1991. Selenocysteine: the 21st amino acid. Mol. Microbiol. 5:515-520. [DOI] [PubMed] [Google Scholar]
- 2.Boyington, J. C., V. N. Gladyshev, S. V. Khangulov, T. C. Stadtman, and P. D. Sun. 1997. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275:1305-1308. [DOI] [PubMed] [Google Scholar]
- 3.Cotter, P. A., and R. P. Gunsalus. 1989. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J. Bacteriol. 171:3817-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dilworth, G. L. 1982. Properties of the selenium-containing moiety of nicotinic acid hydroxylase from Clostridium barkeri. Arch. Biochem. Biophys. 219:30-38. [DOI] [PubMed] [Google Scholar]
- 5.Dürre, P., and J. R. Andreesen. 1982. Selenium-dependent growth and glycine fermentation by Clostridium purinolyticum. J. Gen. Microbiol. 128:1457-1466. [DOI] [PubMed] [Google Scholar]
- 6.Dürre, P., W. Andersch, and J. R. Andreesen. 1981. Isolation and characterization of an adenine-utilizing, anaerobic sporeformer, Clostridium purinolyticum sp. nov. Int. J. Syst. Bacteriol. 31:184-194. [Google Scholar]
- 7.Garcia, G. E., and T. C. Stadtman. 1991. Selenoprotein A component of the glycine reductase complex from Clostridium purinolyticum: nucleotide sequence of the gene shows that selenocysteine is encoded by UGA. J. Bacteriol. 173:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gladyshev, V. N., S. V. Khangulov, and T. C. Stadtman. 1994. Nicotinic acid hydroxylase from Clostridium barkeri: electron paramagnetic resonance studies show that selenium is coordinated with molybdenum in the catalytically active selenium-dependent enzyme. Proc. Natl. Acad. Sci. USA 91:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grunden, A. M., W. T. Self, M. Villain, J. E. Blalock, and K. T. Shanmugam. 1999. An analysis of the binding of repressor protein ModE to modABCD (molybdate transport) operator/promoter DNA of Escherichia coli. J. Biol. Chem. 274:24308-24315. [DOI] [PubMed] [Google Scholar]
- 10.Hopper, S., M. Babst, V. Schlensog, H. M. Fischer, H. Hennecke, and A. Böck. 1994. Regulated expression in vitro of genes coding for formate hydrogenlyase components of Escherichia coli. J. Biol. Chem. 269:19597-19604. [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Leimkuhler, S., M. Kern, P. S. Solomon, A. G. McEwan, G. Schwarz, R. R. Mendel, and W. Klipp. 1998. Xanthine dehydrogenase from the phototrophic purple bacterium Rhodobacter capsulatus is more similar to its eukaryotic counterparts than to prokaryotic molybdenum enzymes. Mol. Microbiol. 27:853-869. [DOI] [PubMed] [Google Scholar]
- 13.McNicholas, P. M., R. C. Chiang, and R. P. Gunsalus. 1998. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol. Microbiol. 27:197-208. [DOI] [PubMed] [Google Scholar]
- 14.Rabinowitz, J. C., and H. A. Barker. 1955. Purine fermentation by Clostridium cylindrosporum. J. Biol. Chem. 218:147-160. [PubMed] [Google Scholar]
- 15.Sanders, S. A., and V. Massey. 1999. The thermodynamics of xanthine oxidoreductase catalysis. Antioxid. Redox Signal. 1:371-379. [DOI] [PubMed] [Google Scholar]
- 16.Schrader, T., A. Rienhofer, and J. R. Andreesen. 1999. Selenium-containing xanthine dehydrogenase from Eubacterium barkeri. Eur. J. Biochem. 264:862-871. [DOI] [PubMed] [Google Scholar]
- 17.Self, W. T., and T. C. Stadtman. 2000. Selenium-dependent metabolism of purines: a selenium-dependent purine hydroxylase and xanthine dehydrogenase were purified from Clostridium purinolyticum and characterized. Proc. Natl. Acad. Sci. USA 97:7208-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Self, W. T., A. M. Grunden, A. Hasona, and K. T. Shanmugam. 1999. Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology 145:41-55. [DOI] [PubMed] [Google Scholar]
- 19.Stewart, V. 1982. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151:1320-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner, R., and J. R. Andreesen. 1979. Selenium requirement for active xanthine dehydrogenase from Clostridium acidiurici and Clostridium cylindrosporum. Arch. Microbiol. 121:255-260. [DOI] [PubMed] [Google Scholar]