Abstract
The involvement of two primary alcohol dehydrogenases, BDH and BOH, in butane utilization in Pseudomonas butanovora (ATCC 43655) was demonstrated. The genes coding for BOH and BDH were isolated and characterized. The deduced amino acid sequence of BOH suggests a 67-kDa alcohol dehydrogenase containing pyrroloquinoline quinone (PQQ) as cofactor and in the periplasm (29-residue leader sequence). The deduced amino acid sequence of BDH is consistent with a 70.9-kDa, soluble, periplasmic (37-residue leader sequence) alcohol dehydrogenase containing PQQ and heme c as cofactors. BOH and BDH mRNAs were induced whenever the cell's 1-butanol oxidation activity was induced. When induced with butane, the gene for BOH was expressed earlier than the gene for BDH. Insertional disruption of bdh or boh affected adversely, but did not eliminate, butane utilization by P. butanovora. The P. butanovora mutant with both genes boh and bdh inactivated was unable to grow on butane or 1-butanol. These cells, when grown in citrate and incubated in butane, developed butane oxidation capability and accumulated 1-butanol. The enzyme activity of BOH was characterized in cell extracts of the P. butanovora strain with bdh disrupted. Unlike BDH, BOH oxidized 2-butanol. The results support the involvement of two distinct NAD+-independent, PQQ-containing alcohol dehydrogenases, BOH (a quinoprotein) and BDH (a quinohemoprotein), in the butane oxidation pathway of P. butanovora.
Pseudomonas butanovora (ATCC 43655) is an aerobic gram-negative proteobacterium closely related to the genera Thauera and Azoarcus as shown by analysis of its 16S rRNA (1). This organism has been classified in the genus Pseudomonas based on its morphology, physiology, and biochemistry (39, 40). P. butanovora was isolated from activated sludge from an oil-refining company for the purpose of generating biomass from n-alkanes (39, 40). P. butanovora can derive energy for growth from C2 to C9 n-alkanes and any of their oxidation products as well as from a variety of other carbon sources (39, 40). Butane-grown P. butanovora can oxidize some chlorinated hydrocarbons by cometabolism through the action of a monooxygenase (18) and thus may have applications in bioremediation schemes.
The pathway for the oxidation of butane in P. butanovora proceeds primarily from butane to 1-butanol, to butyraldehyde, to butyrate (2), and then probably to the β-oxidation pathway of fatty acid oxidation. As in other alkane utilizers (3, 27, 36), in P. butanovora the oxidation of the alkane (butane) is initiated by the action of a monooxygenase (19). Each intermediate in the pathway accumulated in the presence of appropriate inhibitors, supported cell growth, and stimulated O2 consumption (2). The presence of a terminal butane oxidation pathway (i.e., production of 1-butanol) in P. butanovora was indicated (2). Although butane-grown cells consumed 2-butanol, 2-butanol production (indicative of a subterminal oxidation pathway) was not demonstrated, even in the presence of appropriate inhibitors of 2-butanol consumption. For P. butanovora four different alcohol dehydrogenases (ADHs) with different specificities towards primary and secondary alcohols were identified on native gels stained for activity (45). Among these ADHs, 1-butanol dehydrogenase (BDH) was characterized biochemically (45). BDH enzyme activity was detected in butane- and 1-butanol-grown cells but not lactate-grown cells. BDH is a soluble, periplasmic, type II NAD+-independent quinohemoprotein that contains 1.0 mol of pyrroloquinoline quinone (PQQ) and 0.25 mol of ratio heme c as prosthetic groups and exists as a monomer with an apparent molecular mass of 67 kDa (45).
The liquid-alkane metabolisms of other gram-negative proteobacteria such as Pseudomonas oleovorans and Pseudomonas putida (6, 10, 11) and Acinetobacter sp. (14, 23, 28, 29) have been studied. From these studies and from our research with the gaseous alkane utilizer P. butanovora, several differences among the enzymes involved in the metabolism of alkanes are starting to emerge. First, the essential enzymes in the utilization of the alkane differ in cellular location among these proteobacteria and P. butanovora. For example, the oxidation of octane in P. oleovorans and Acinetobacter sp. strain ADP1 proceeds through membrane-bound monooxygenases. In P. butanovora the oxidation of butane proceeds via a soluble alkane monooxygenase (D. Arp Laboratory, unpublished results). Second, the oxidation of the resulting alcohol in n-alkane metabolism proceeds via diverse enzymes depending on the bacterium. In P. oleovorans there is an inducible ADH which is a flavin-containing enzyme (43), while for P. butanovora BDH, an inducible PQQ- and heme c-containing ADH, has been described (45). Constitutive ADH activity was not detected in P. butanovora (33), while Acinetobacter sp. strain HO1-N has at least one constitutive ADH activity (35). Third, these alkane-utilizing proteobacteria have different gene arrangements. The genes coding for the enzymes in alkane metabolism in P. oleovorans are clustered in an operon (alk) (43), and in Acinetobacter sp. strain ADP1, the genes for alkane metabolism are spread through its chromosome (28, 29). The genetic arrangement of the genes for alkane metabolism in P. butanovora has not yet been determined. However, in P. butanovora, the genes for alkane metabolism may be arranged in different operons as in Acinetobacter sp. strain ADP1, since each enzyme activity in the butane oxidation pathway is induced independently by the substrate being oxidized (33).
This study suggests that the NAD+-independent PQQ alcohol dehydrogenase BOH (a quinoprotein) is linked to butane metabolism in conjunction with the previously characterized BDH (a quinohemoprotein [45]). The inferred amino acid sequence of the gene coding for BOH (boh) showed a polypeptide similar to periplasmic PQQ-containing ADHs in other bacteria. The expression of boh was compared to the expression of bdh (encoding BDH). Inactivation of each gene coding for BOH or BDH decreased the rate of growth on butane, and inactivation of both genes eliminated the growth of P. butanovora on butane and on 1-butanol.
MATERIALS AND METHODS
Cell culture and assay conditions.
Cells of P. butanovora (ATCC 43655) were grown in sealed serum bottles as previously described (2, 19, 40) but with the omission of yeast extract and CO2. A headspace of at least 50% of the total volume was used in the bottles to assure an adequate supply of O2 to the cells. For growth with butane, the gas was added as overpressure (10% [vol/vol] of the headspace). Butane gas (99%) was purchased from Airgas, Inc. (Randor, Pa.). For growth in sodium lactate, the substrate was added to the sterile basal media at concentrations of 5 to 10 mM. The bottles were incubated with shaking at 30°C for 1 to 3 days until turbidities (optical densities) at 600 nm of about 0.5 were observed. All chemicals were of analytical grade.
Activity assays.
Butane monooxygenase activity was assayed by monitoring the accumulation of ethylene oxide from ethylene, an alternative substrate for the monooxygenase (33). Ethylene oxidation was determined in capped serum vials (10 ml) with 20% (vol/vol) ethylene, 5 mM sodium butyrate, and 1 ml of cell suspension (0.5 mg of protein). By using a relatively high concentration of ethylene, the inactivation of the monooxygenase by ethylene oxide was prevented (19). The accumulation of ethylene oxide was determined by injecting 100 μl of the headspace into a gas chromatograph as described below.
The in vivo assay for 1-butanol consumption was carried out by using serum vials (10 ml) with a 1 mM concentration of the substrate and 1 ml of cell suspension (0.5 to 5 mg of protein). The vials were capped with gray-butyl rubber stoppers and aluminum crimp seals. The capped vials were mixed by shaking in a reciprocating water bath at 30°C. Substrate consumption during the incubation was determined by injecting 5 μl of the liquid phase into a gas chromatograph as described below. Control cells not exposed to butane or 1-butanol were grown in lactate and did not have butane, ethylene, or 1-butanol consumption activities.
The in vitro oxidations of 1-butanol and other alcohols and aldehydes were measured as phenazine methosulfate (PMS)-mediated, dichlorophenolindophenol (DCPIP) reduction. DCPIP reduction was monitored spectrophotometrically at 600 nm in a reaction mixture consisting of 25 mM MOPS (morpholinepropanesulfonic acid) at pH 7.0, 0.7 mM PMS, 0.1 mM DCPIP, 4 mM NH4Cl, 0.8 mM KCN, 1 mM alcohol or aldehyde substrate, and 0.1 to 0.5 mg of protein in a total volume of 3 ml. BOH required a 1-h incubation with 5 mM PQQ for activation.
Analytical techniques.
The concentrations of the substrates were determined by gas chromatography (GC-8A; Shimadzu Corporation, Tokyo, Japan) with the appropriate pure compounds as standards. The gas chromatograph was equipped with a flame ionization detector and a 60-cm-long by 0.1-cm-inside-diameter stainless steel column packed with Porapak Q (Waters, Milford, Mass.). The oven temperature was 90°C for ethylene oxide and 160°C for 1-butanol analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described previously (12). The protein content in the cell suspensions was determined by using the bicinchoninic acid protein assay reagent as described by the manufacturer (Pierce, Rockford, Ill.). The samples were added directly to the protein assay reagent with 0.01% Triton X-100 (Sigma, St. Louis, Mo.) added to help cell lysis. Alternatively, protein determination was performed by the protein-dye binding assay as described previously (5). Bovine serum albumin was used as a protein standard.
Plasmids, bacterial strains, DNA manipulations, and library screening.
Table 1 summarizes the plasmids and strains used in this work. DNA isolation, cloning, agarose gel electrophoresis, and Southern hybridization were performed by standard protocols (32). Total RNA was isolated by the direct addition of acid-phenol, 100 mM sodium acetate, and 1% SDS to a 500-μl cell suspension (2 to 5 mg of cell protein). The cell suspension was mixed thoroughly and centrifuged 5 min at 16,000 × g. The RNA was then recovered by ethanol precipitation and dissolved in 50 μl of diethyl pyrocarbonate-treated water. The isolated total RNA was stored at −70°C until used. DNA probes were labeled by random priming with a kit (Prime-a-gene; Promega Co., Madison, Wis.) and [α32P]dCTP (3,000 Ci/mmol; DuPont NEN products, Wilmington, Del.) following the directions of the manufacturers. Northern hybridization was carried out as described previously (34). The hybridization signals were visualized and analyzed by using phosphorimaging and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). The genomic library of P. butanovora was constructed in LambdaGEM-11 (Promega) by using Escherichia coli LE392 as a host and screened as described previously (32). The PCR was carried out using Taq DNA polymerase (Promega) and standard protocols (21). The PCR-amplified fragments were cloned into pGEM-T Easy (Promega) following the directions of the manufacturer and with E. coli strain JM101 as host.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli JM101 | F′ traD36 proA+proB+lacIqlacZΔM15/supE thi Δ(lac-proAB) | 47 |
| E. coli LE392 | hsdR514(rK− mK+) supE44 supF58 lacY1 galK2 galT22 metB1 trpR55 | 25 |
| P. butanovora ATCC 43655 | Type strain | American Type Culture Collection |
| P. butanovora boh::tet | Mutant strain with the gene for BOH inactivated; tetracycline resistant | This work |
| P. butanovora bdh::kan | Mutant strain with the gene for BDH inactivated; kanamycin resistant | This work |
| P. butanovora boh::tet-bdh::kan | Mutant strain with the genes for BOH and BDH inactivated; tetracycline and kanamycin resistant | This work |
| Plasmids and λ clones | ||
| λPbu1 | λ clone (14 kb) with the gene for BOH | This work |
| pPbu1 | 7-kb clone containing boh in pBluescript II SK(+) | This work |
| pPbu11 | pPbu1 with the tet cassette inserted into the EcoRI site in boh | |
| pAV1 | Partial bdh clone (0.7 kb) into pGEM-T Easy | This work |
| pAV2 | Subclone of λPbu5 containing bdh with the kan cassette inserted | This work |
| λPbu5 | λ clone (20 kb) with the gene for BDH | This work |
| Cloning plasmids | ||
| LambdaGEM-11 | λ vector used to construct the genomic library of P. butanovora | Promega |
| pBluescript II SK(+) | 2,961-bp phagemid derived from pUC19; multiple-cloning cassette; Ampr | GenBank no. X52328 |
| pGEM-T Easy | PCR product cloning vector; multiple-cloning cassette that includes flanking EcoRI sites; Ampr | Promega |
DNA sequencing and oligonucleotide syntheses were performed at the Central Services Laboratory of the Center for Gene Research and Biotechnology in Oregon State University. Sequence analysis was performed using software from the Wisconsin Package Version 10.0 (Genetics Computer Group [GCG], Madison, Wis).
Peptide purification, N terminus determination, and enzyme enrichment.
For the purification of the peptide of the putative aldehyde dehydrogenase, cell extracts from butane-grown P. butanovora were prepared by passing a cell suspension through a French pressure cell disrupter at 5,000 lbs/cm2. The cell extract was then subjected to ultracentrifugation (45,000 × g), and the supernatant was fractionated with a 1.6- by 20-cm Q-Sepharose FF column (Amersham Pharmacia, Piscataway, N.J.) using a gradient from 0 to 1.0 M NaCl at 1.5 ml/min. The enriched peptide (55 kDa) was separated by SDS-PAGE and then electroblotted onto polyvinylidene difluoride membrane (Millipore Corp., Bedford, Mass.). The N-terminal amino acid sequence of the peptide for the aldehyde dehydrogenase was determined by the Biotechnology Laboratory of the Institute of Molecular Biology at the University of Oregon. The partial BDH amino acid sequences were previously reported (45). The partial purification of BOH was performed using the bdh::kan mutant strain of P. butanovora, which lacks BDH. The soluble cell extract of the bdh::kan mutant strain, which contained 1-butanol and PQQ-dependent DCPIP reductase activity, was purified through Q-Sepharose FF column using an NaCl linear gradient (0 to 1.0 M) in 25 mM MOPS (pH 7.0). The active BOH-containing fraction (eluted at 0.19 to 0.3 M) was concentrated with a centrifugal filter membrane (Centricon YM-30, Amicon; Millipore Corp.) which removed molecules smaller than 30 kDa. The partially purified BOH was used to determine the substrate specificity of the enzyme.
DNA constructs and generation of the mutant strains.
For the inactivation of boh, the tetracycline gene from pALTER-1 (Promega) with the restriction site-modified tetracycline resistance gene (tet) was isolated by PCR and cloned into pGEM-T Easy. The tet gene was then subcloned into the EcoRI restriction site 622 nucleotides (nt) downstream of the ATG start codon of boh in plasmid pPbu11 (Table 1). For the inactivation of bdh, the EZ::TN <KAN> kit from Epicentre (Madison, Wis.) was used to insert a transposon conferring kanamycin resistance (kan) into the coding region of bdh following the directions of the manufacturer. The insertion of the kan gene was localized by nucleotide sequence determination at 715 nt downstream of the ATG start codon of bdh in plasmid pAV2. The antibiotic resistance alcohol dehydrogenase constructs were then introduced into P. butanovora cells by electroporation. P. butanovora cells for electroporation were harvested in early stationary phase and washed three times in sterile distilled H2O and chilled in ice. Cell electroporation was performed with an ElectroPorator (Invitrogen, Carlsbad, Calif.) in 1-mm-gap cuvettes (Invitrogen). Electroporation conditions were 1,300 V, 71 μF, and 200 Ω. In a prechilled cuvette, 120 μl of cells (10 μg/ml) were premixed with the plasmid construct (∼0.5 μg in 1 μl) and pulsed. The cells were transferred to basal media with lactate and allowed to grow under nonselective conditions for 3 h at 30°C while shaking. Cells were then challenged with tetracycline (7 μg/ml) or kanamycin (20 μg/ml) and plated in lactate-antibiotic plates for selection. To obtain the mutant with both genes inactivated, the mutant with boh inactivated was subjected to electroporation with the bdh-kan construct pAV2. The resultant double mutant, boh::tet-bdh::kan, was selected in a basal-lactate medium supplemented with tetracycline and kanamycin.
Nucleotide sequence accession numbers.
The DNA sequences for the genes of the two 1-butanol dehydrogenases have been deposited in the GenBank database. The nucleotide sequence of boh has the GenBank (NCBI) accession number AF326086. The nucleotide sequence of bdh has the GenBank (NCBI) accession number AF355798.
RESULTS
Isolation of DNA fragments coding for 1-butanol dehydrogenase BOH and for 1-butanol dehydrogenase BDH.
The isolation of a DNA fragment containing the gene for BOH was achieved indirectly through the isolation of a gene cluster containing an aldehyde dehydrogenase. Analysis of polypeptide patterns of cell extracts by SDS-PAGE showed a prominent 55-kDa polypeptide expressed in butane-grown cells but not in lactate-grown cells. This 55-kDa polypeptide was purified by column chromatography, and its N-terminal amino acid sequence (MIYAMPGQSGAAV) was determined. Database searches showed that the amino acid sequence was similar to the N terminus of the aldehyde dehydrogenases from Pseudomonas aeruginosa (strain PAO1) (69% identity; accession no. AE004625 [38]) and Alteromonas sp. KE10 (61% identity; accession no. AB009654 [22]). The degenerate oligonucleotide ATG-ATH-TAY-GCN-ATG-CCN-GGN-GAR-T was synthesized and used to screen the library of P. butanovora. A 9-kb genomic λ clone (λPbu1) was isolated. A 7.1-kb BamHI DNA fragment from λPbu1 was cloned to form pPbu1. Preliminary sequence determination of the DNA fragment in pPbu1 showed a gene cluster coding for the aldehyde dehydrogenase (from which the oligonucleotide sequence was deduced), a transcription regulator, and an alcohol dehydrogenase (BOH in this study). Because our interest was in characterizing the alcohol dehydrogenase activity in P. butanovora, we focused on the determination of the nucleotide sequence of the gene coding for BOH. The deduced amino acid sequence indicated a quinoprotein but was distinct from the sequence of BDH (see below and reference 45).
To isolate a DNA fragment of the gene for BDH, degenerate PCR primers were synthesized after the two known internal amino acid sequences of BDH (MSYAPQTGLAYFPAQNIPL and KGGGIPNLGYSTAETIAHLDQFVFK [45]). The degenerate PCR primers (forward primer, 5′ATG AGC TAC GCC CCA CAG ACC GGC CTG GCC TAC TTY CCN GCN CAR AAY ATH TTY YT 3′, and reverse primer, 5′ TT CAA GAC CAA CTG GTC CAG ATG CGC GAT GGT CTC CGC GGT GCT RTA NCC NAR RTT NGG DAT NCC 3′) were designed using the consensus-degenerate hybrid oligonucleotide primer approach (CODEHOP [30]). A 0.7-kb DNA fragment of the gene for BDH was amplified and cloned into pGEM-T Easy to form pAV1. The nucleotide sequence of the DNA fragment in pAV1 was determined and showed similarity to other PQQ-containing alcohol dehydrogenases. This DNA fragment was used to screen the genomic library of P. butanovora for the gene of BDH. The genomic clone λPbu5, with a 20-kb insert, hybridized to the probe and was used to obtain the complete sequence of the gene coding for BDH. The nucleotide sequence of bdh was determined directly from the λPbu5 genomic clone. These results were the first indication of the presence of two quinoprotein ADHs in P. butanovora.
Analysis of the nucleotide sequences for BOH and BDH.
A summary of the sequence comparisons of boh and bdh to other bacterial ADHs is shown in Table 2. The open reading frame (ORF) of boh is composed of 1,872 nt. A clear start codon for boh was found with a Shine-Dalgarno-like ribosome binding site sequence (GGAG) five bases upstream. A nucleotide sequence that started 96 bases upstream of the start codon was indicative of a σ54-dependent promoter (CTG GCA CGC TCT TTG CCA) in boh as well. The nucleotide sequence of this putative σ54-dependent promoter in P. butanovora has 83% identity with the consensus sequence for σ54-dependent promoters (24). The deduced amino acid sequence of boh indicates a type I NAD+-independent ADH such as the type I quinoprotein ethanol dehydrogenase from P. aeruginosa (accession no. 10120672 [8]), a homodimer with subunits of relative molecular mass of 60 kDa. The calculated molecular mass of the complete polypeptide encoded by boh is 67,553 Da. The BOH polypeptide without the putative 29-amino-acid leader sequence has a calculated molecular mass of 64,666 Da, and a pI of 6.15 is indicated. The nucleotide sequence and the requirement of PQQ in the activity assay of BOH suggest that PQQ is the prosthetic group of BOH (see below).
TABLE 2.
Sequence comparison of BOH, BDH, and other PQQ-containing ADHs
| PQQ-containing ADH | Organism | No. of amino acids | % Identity (% similarity) to BOH | % Identity (% similarity) to BDH | Accession no. |
|---|---|---|---|---|---|
| BOH | P. butanovora | 623 | 35 (47) | AF326086 | |
| BDH | P. butanovora | 691 | 35 (47) | AF355798 | |
| Methanol dehydrogenase | Methylophilus methylotrophus W3A1 | 573 | 33 (46) | 30 (43) | 1942860 |
| Methanol dehydrogenase | Methylobacterium extorquens | 626 | 34 (46) | 29 (42) | AAA25380 |
| EDH | P. aeruginosa | 623 | 70 (80) | 35 (49) | CAA08896 |
| EDH | Comamonas testosteroni | 708 | 36 (50) | 60 (70) | CAA57464 |
| Tetrahydrofurfuryl ADH | R. eutropha | 698 | 37 (51) | 60 (72) | AAF86335 |
The ORF of bdh is composed of 2,078 nt having a theoretical molecular mass of 70.9 kDa, compared to an experimental mass determination of 66 kDa (45). The experimental mass determination is in close agreement with a putative leader sequence of 37 residues in BDH (4.1 kDa). The calculated pI is 6.66. A Shine-Dalgarno-like ribosome binding site sequence (GGAG) is localized six bases upstream of the start codon. The amino acid sequence deduced from bdh is similar to other quinohemoproteins (Table 2) (37).
Upstream of the ORF coding for BDH, an ORF of 929 nt with no clear Shine-Dalgarno-like ribosome binding site sequence is present. The deduced amino acid sequence has 43% similarity to that of orf1 in the ntn gene cluster of Pseudomonas sp. strain TW3 (20) and 40% similarity to ChnX encoded by a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 (7), both of which are ORFs of unknown function. The nucleotide sequence CTG GCA TGG CTT CTG CA is located 163 nt from the putative start codon of this unknown ORF in P. butanovora. This sequence has 82% identity to the consensus sequence of a σ54-dependent promoter (CTG GCA CGG CCT TTG CA [24]).
BDH and BOH have seven Trp residues in positions equivalent to those implied in the formation of the β-propeller fold of PQQ-containing alcohol dehydrogenases (26, 46). In BOH the Trp at positions 345 (9 residues from the consensus position) and 438 (16 residues from the consensus position) offer alternatives for the Trp residues in the W7 and W8 β-propeller folds of the methanol dehydrogenase from Methylophilus W3A1 (46). The contiguous Cys residues implied in the interaction with PQQ and Ca2+ (46) were located at positions 134 and 135 of BOH and positions 130 and 131 of BDH. Comparison of the deduced amino acid sequence of BOH and the deduced amino acid sequence of BDH shows 35% identity and 47% similarity (Table 2).
Time course of boh and bdh expression upon exposure to butane.
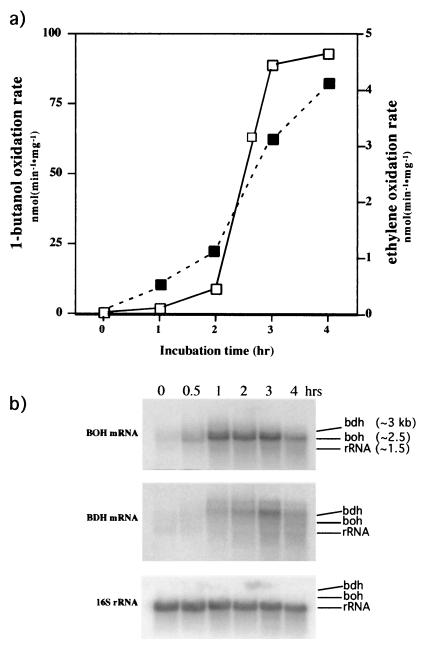
To corroborate and test the involvement of BOH and BDH in butane metabolism, lactate-grown cells were washed and incubated in basal medium plus butane. In a time course experiment, we monitored the development of ethylene oxidation activity (butane monooxygenase activity) and 1-butanol consumption. A typical induction of the butane monooxygenase and 1-butanol oxidation activities by butane was observed (33) (Fig. 1a). During the induction by butane, cell samples were withdrawn at intervals and their total RNA was extracted and blotted onto nylon membranes for analysis by Northern hybridization. The probe for bdh was derived from the plasmid pAV1. The probe for boh was generated by PCR using primers that flanked the ORF for boh and pPbu1 as a template. The probe for the 16S rRNA was obtained by PCR using eubacterial universal primers (15) and genomic DNA as a template. The 16S rRNA probe was used to show that equivalent mRNA amounts were loaded into the gel for each treatment. The same membrane blot was hybridized to the three probes separately after stripping the membrane (Fig. 1b). Cells exposed to butane first showed induction of boh in about 30 min followed by the induction of bdh, which was first detected at 60 min (Fig. 1b). At 240 min of incubation, a decrease of the total amount of mRNA for both ADHs was observed. This decrease of mRNA levels occurred as the maximum 1-butanol oxidation activity was reached (Fig. 1a). These results suggest that BOH and BDH are involved in butane metabolism. Furthermore, under the conditions of this study, BOH and BDH were differentially expressed upon exposure to butane.
FIG. 1.
Induction of BOH and BDH total activity and mRNAs by butane. (a) Development of ethylene oxidation (dashed line, solid squares) and 1-butanol oxidation (solid line, open squares) activities. Lactate-grown cells were washed and then incubated in basal medium with butane for the indicated times. (b) BOH and BDH mRNA levels during the induction of butane oxidation. The same blot was used for the three hybridizations after probe stripping. The three arrows represent the relative position of each mRNA with respect to the other two. The numbers in the first frame are the estimated sizes of the mRNAs and of the 16S rRNA.
We also measured 1-butanol consumption and BOH and BDH mRNA levels during cell growth on butane. The specific activity for 1-butanol consumption increased as the cell mass of P. butanovora increased, reaching a maximum of 68 nmol min−1 mg of protein−1 at early stationary phase (optical density at 600 nm, ∼0.55) at 35 h (not shown). Specific activity of 1-butanol consumption in butane-grown cells typically reaches 50 to 100 nmol min−1 mg of protein−1 at optical densities of 0.5 to 0.7 (33). The mRNA levels of BDH and BOH were determined by using Northern hybridizations. Both genes were expressed, and their mRNA levels increased until the cells reached stationary phase. We were not able to detect clear differences between the patterns of boh and bdh expression during growth on butane (not shown).
BOH and BDH mRNA induction by various alcohols.
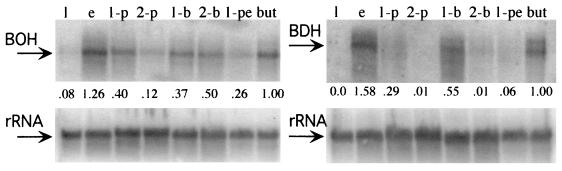
The DNA probes derived from the gene sequences also permitted us to determine the levels of induction of the mRNAs of BOH and BDH in response to various alcohols and butane. Lactate-grown cells were washed and then exposed to basal medium containing 2 mM ethanol, 1-propanol, 2-propanol, 1-butanol, 2-butanol, or 1-pentanol. After 2 h of incubation in a given alcohol, the cells were harvested and their total RNA was extracted. The same membrane blot was hybridized with probes for the mRNAs of bdh and boh and the 16S rRNA separately after probe stripping (Fig. 2). In these experiments the cells showing induction of BOH or BDH mRNAs also showed 1-butanol consumption (10 to 30 nmol min−1 mg of protein−1). Although the relative levels of the BOH and BDH mRNAs varied among replicate incubations, the trends were consistent. Primary C2 and C4 alcohols were the most effective inducers of boh and bdh. Some differences in the levels of BOH and BDH mRNAs produced were also observed when cells were exposed to C2 to C5 alcohols (Fig. 2). For example, 2-butanol is as effective as 1-butanol in inducing boh expression, but 2-butanol is much less effective than 1-butanol in inducing bdh. Compared to BDH mRNA, BOH mRNA was induced by a wider range of alcohols.
FIG. 2.
Induction of the mRNA for BOH and BDH upon incubation with different alcohols and butane. The same RNA preparation was probed for the presence of the BOH and BDH mRNAs. For comparison among treatments, the blots were stripped and hybridized to a probe for the 16S rRNA. Lactate-grown cells were washed and then incubated for 2 h in medium containing the indicated substrate and then tested for the presence of BOH and BDH mRNAs by Northern hybridization. Cells were incubated with lactate (1), ethanol (e), 1-propanol (1-p), 2-propanol (2-p), 1-butanol (1-b), 2-butanol (2-b), 1-pentanol (1-pe), and butane (but). The numbers above the 16S rRNA blot were calculated from two batches of cells and are the ratios of the BOH or BDH mRNA signal to the rRNA signal normalized to the ratios obtained with cells exposed to butane.
Gene inactivation of BOH and BDH.
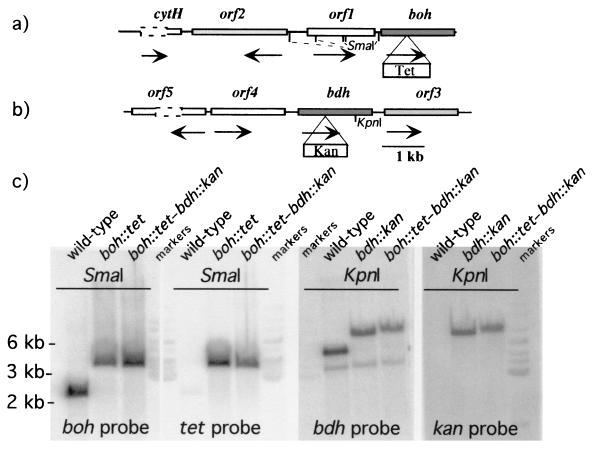
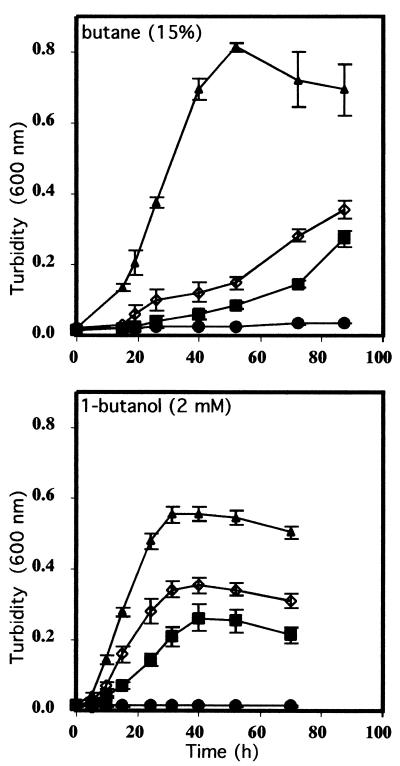
To address the involvement of BOH and BDH separately in the metabolism of butane, the genes for BOH and BDH were inactivated by insertion mutagenesis. The tetracycline resistance cassette inserted into boh (pPbu11) and the kanamycin resistance cassette inserted into bdh (pAV2) were introduced by electroporation into P. butanovora, producing the mutant strains boh::tet and bdh::kan, respectively. The mutant strain with both boh and bdh inactivated was also produced (boh::tet-bdh::kan). The mutations were confirmed with probes for boh and bdh or for the antibiotic markers in Southern hybridizations (Fig. 3), where the sizes of the restriction fragments containing each gene were increased by the sizes of the antibiotic markers. Growth on butane was delayed when boh or bdh was inactivated. After a lag period of about 12 h both mutant cells began to grow and eventually reached optical densities similar to those observed in the wild-type cells. The lack of BOH slowed cell growth more than the lack of BDH did (Fig. 4). Cells with boh or bdh inactivated growing on 1-butanol reached final optical densities that were only half of that observed with the wild type (Fig. 4). When both genes were inactivated, growth on butane and 1-butanol was eliminated (Fig. 4). Citrate-grown cells of the boh::tet-bdh::kan strain accumulated 1-butanol (102.5 ± 10.6 nmol [mean ± standard deviation]) when incubated for 4.5 h with 20% butane, indicating induction of the monooxygenase.
FIG. 3.
Maps of the loci of boh (a) and bdh (b) and phosphorimage of the Southern blot of DNA (c) from the wild-type and mutant strains of P. butanovora. The maps show the locations of the adjacent genes and the sites of insertion of the antibiotic-conferring cassettes. The arrows under the genes show the direction of transcription. The nucleotide sequences of the genes adjacent to the alcohol dehydrogenase-encoding genes are incomplete but show similarity to a regulatory element (orf1) and to genes coding for aldehyde dehydrogenase (orf2), to another aldehyde dehydrogenase (orf3), to an orf of unknown function (orf4), and to a regulatory element (orf5). The dashed lines represent undetermined sequences. In the Southern hybridization two restriction digests were used for clarity to show the different loci of boh and bdh and the increase in size as a result of the cassette insertion. The strains, restriction enzymes, and probes used are indicated.
FIG. 4.
Growth of the wild-type, boh::tet mutant, bdh::kan mutant, and boh::tet-bdh::kan mutant strains of P. butanovora. The growth substrates were butane and 1-butanol as indicated. Symbols: ▴, wild-type; ▪, boh::tet; ◊, bdh::kan; and •, boh::tet-bdh::kan P. butanovora strains.
Biochemical characterization of BOH.
The bdh::kan mutant strain facilitated the characterization of BOH in cell extracts. Quinoproteins lacking heme c (i.e., methanol dehydrogenase and ethanol dehydrogenase [EDH] from P. aeruginosa and P. putida and soluble glucose dehydrogenase from E. coli) do not react with ferricyanide (9, 13, 17); thus, ferricyanide reductase activity in the bdh::kan mutant strain should be absent. As expected, cell extract of the mutant strain lacking BDH showed <2 nmol min−1 mg of protein−1 of ferricyanide reductase activity towards 1-butanol, a fraction of that typically observed with the wild-type P. butanovora (102 ± 19.6 nmol min−1 mg of protein−1). BOH was partially purified (3.4-fold increase in specific activity) from the soluble cell extract of butane-grown bdh::kan mutant strain. Following activation with PQQ, this BOH preparation was used to examine the substrate range of BOH (Table 3). As expected, 1-butanol was a good substrate. However, 2-butanol supported 15% higher rates of activity. In contrast, BDH does not oxidize 2-butanol (45). BOH also exhibited slightly higher activity with 2-propanol and 2-pentanol than with 1-propanol and 1-pentanol (Table 3). The partially purified preparation of BOH also oxidized butyraldehyde, the product of 1-butanol oxidation (Table 3). This PMS-mediated butyraldehyde oxidation activity was not likely due to another enzyme in the partially purified BOH preparation because cells lacking both BOH and BDH (boh::tet-bdh::kan mutant strain) did not show this activity, even when incubated with 1-butanol or butyraldehyde for 6 h to induce activity.
TABLE 3.
Specific activitya of BOH towards primary alcohols, secondary alcohols, and aldehydes
| Substrate | Mean ± SD of sp act (nmol min−1 mg of protein−1) | Relative activity (%) |
|---|---|---|
| Methanol | 2 ± 0.5 | 0.7 |
| Ethanol | 82 ± 12 | 30 |
| 1-Propanol | 61 ± 7 | 22 |
| 1-Butanol | 239 ± 17 | 87 |
| 1-Pentanol | 92 ± 10 | 34 |
| 1-Octanol | 59 ± 10 | 22 |
| 2-Propanol | 84 ± 9 | 30 |
| 2-Butanol | 274 ± 9 | 100.0 |
| 2-Pentanol | 167 ± 6 | 61 |
| 2-Octanol | 42 ± 1 | 15 |
| Propionaldehyde | 91 ± 5 | 33 |
| Butyraldehyde | 68 ± 22 | 25 |
PMS-mediated, DCPIP reduction activity of the partially purified BOH (see text).
DISCUSSION
In this study, we show that P. butanovora has two distinct genes (boh and bdh) encoding two PQQ-containing alcohol dehydrogenases (a quinoprotein and a quinohemoprotein) and that both alcohol dehydrogenases participate in butane metabolism. The nucleotide sequences of the genes and Southern hybridizations argue for different genetic loci for boh and bdh. Northern analyses and gene inactivation experiments link each gene to the metabolism of butane. The involvement of boh and bdh in butane metabolism comes from several lines of evidence. First, neither of the genes is expressed in lactate-grown cells (which are devoid of butane metabolism). Second, both genes are expressed in butane-grown cells. Third, when lactate- or citrate-grown cells are washed and subsequently exposed to butane, both genes are expressed as cells develop butane and 1-butanol oxidation activities. Fourth, gene inactivation experiments, where either gene was inactivated, showed that P. butanovora grew more slowly on butane than did the wild-type strain. When both boh and bdh were inactivated, growth on butane and 1-butanol was eliminated. The involvement of BDH in the butane oxidation pathway in P. butanovora was previously established at the biochemical and physiological levels as well (45).
The involvement of diverse alcohol dehydrogenases in alcohol or alkane metabolism is not without precedent (42, 44). For example, Pseudomonas putida produces three distinct PQQ-containing ADHs (a quinoprotein and two quinohemoproteins), each with different substrate ranges, and each induced primarily by a different alcohol (42). Rhodococcus rhodochrous PNKb1 produces two distinct NAD+-dependent ADHs, one for primary and another for secondary alcohols, and each is required for the metabolism of propane (4). However, the situation with P. butanovora seems to be different because genes for two distinct PQQ-containing alcohol dehydrogenases were expressed in response to 1-butanol which was generated in the oxidation of butane.
To gain additional insight into the role of each of these ADHs in P. butanovora, we investigated the expression of each gene in response to different alcohols (C2 to C5) and butane. The highest levels of mRNA for both BOH and BDH were found in cells exposed to ethanol, 1-butanol, and butane. Differences in the expression patterns were also apparent. For example, compared to bdh, boh was induced by a broader range of alcohols, and higher levels of mRNA for BOH were induced in the presence of 1-propanol and 2-butanol than for BDH (Fig. 3). The induction patterns reflected the substrate ranges of each enzyme. The induction of boh expression by 2-butanol and the higher specific activity of BOH for 2-butanol suggest a possible role for BOH. Perhaps BOH oxidizes both 1- and 2-butanol, both of which are potential products of butane oxidation. While 1-butanol was shown to be the predominant product, production of a low level of the subterminal oxidation product, 2-butanol, was not demonstrated (2). The deduced amino acid sequence of BOH is 80% similar to that of the quinoprotein EDH from P. aeruginosa, which also exhibits a wide substrate range. In addition to ethanol, EDH oxidizes both primary (1-propanol and 1-butanol) and secondary (2-propanol and 2-butanol) alcohols efficiently (31). BOH oxidizes 1-butanol and 2-butanol efficiently (Table 3). In contrast, BDH, which has only a 30% similarity with EDH, efficiently oxidizes 1-propanol and 1-butanol, but not 2-propanol or 2-butanol (45). The deduced amino acid sequence of BDH shows 72% similarity to quinohemoprotein tetrahydrofurfuryl ADH of Ralstonia eutropha. Tetrahydrofurfuryl ADH and BDH have high activity towards 1-butanol and 2-pentanol, but little or no activity with 2-butanol (45, 48).
The lack of growth of the boh::tet-bdh::kan mutant strain on either butane or 1-butanol suggests that there are only two primary alcohol dehydrogenases involved in the butane metabolism of P. butanovora. Furthermore, citrate-grown boh::tet-bdh::kan cells when incubated in butane tended to accumulate 1-butanol, reinforcing the notion of only two primary alcohol dehydrogenases. Perhaps the presence in P. butanovora of these two distinct ADHs that are induced in the butane oxidation pathway reflects the need of the cells to treat 1-butanol as both a source of energy and a toxic compound. One ADH may respond to low levels of 1-butanol as a metabolite, and the second ADH may respond to higher levels of 1-butanol as a toxin. Cells of the boh::tet-bdh::kan mutant strain did die when incubated in 1-butanol for extended periods. Another possible reason for the two distinct ADHs induced by the same substrate is that of specialization for different bioenergetic roles. BOH (the quinoprotein) is expected to require a cytochrome as the immediate acceptor which can then transfer electrons to the respiratory chain (16). BDH, with a heme as a prosthetic group, could transfer electrons to azurin as is the case with the quinoprotein of P. putida (41).
This work demonstrates the involvement of two similar alcohol dehydrogenases for the oxidation of 1-butanol in the butane metabolism of P. butanovora. In addition we show that BOH and BDH have different substrate specificities and that their mRNAs are expressed in response to similar stimuli but to different extents. The explanation for the existence of this dual pathway in P. butanovora awaits further experimentation into the bioenergetics and toxicity of 1-butanol.
Acknowledgments
This research was supported by National Institutes of Health grant no. GM56128 to D.J.A and L.A.S.S. and the Oregon Agricultural Experiment Station.
REFERENCES
- 1.Anzai, Y., H. Kim, J. Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. E vol. Microbiol. 50:1563-1589. [DOI] [PubMed] [Google Scholar]
- 2.Arp, D. J. 1999. Butane metabolism by butane-grown Pseudomonas butanovora. Microbiology 145:1173-1180. [DOI] [PubMed] [Google Scholar]
- 3.Ashraf, W., A. Mihdhir, and J. C. Murrell. 1994. Bacterial oxidation of propane. FEMS Microbiol. Lett. 122:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Ashraf, W., and J. C. Murrell. 1992. Genetic, biochemical and immunological evidence for the involvement of two alcohol dehydrogenases in the metabolism of propane by Rhodococcus rhodochrous PNKb1. Arch. Microbiol. 157:488-492. [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:2482-2488. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarty, A. M., G. Chou, and I. C. Gunsalus. 1973. Genetic regulation of octane dissimilation plasmids in Pseudomonas. Proc. Natl. Acad. Sci. USA 70:1137-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, Q., S. M. Thomas, K. Kostichka, J. R. Valentine, and V. Nagarajan. 2000. Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 by in vitro transposition. J. Bacteriol. 182:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl, A., F. von Wintzingerode, and H. Gorisch. 1998. Quinoprotein ethanol dehydrogenase of Pseudomonas aeruginosa is a homodimer—sequence of the gene and deduced structural properties of the enzyme. Eur. J. Biochem. 257:409-419. [DOI] [PubMed] [Google Scholar]
- 9.Docker, P., J. Frank, and J. A. Duine. 1986. Purification of quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus L.M.D. 79.41. Biochem. J. 23:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggink, G., H. Engel, W. G. Meijer, J. Otten, J. Kingma, and B. Witholt. 1988. Alkane utilization in Pseudomonas oleovorans. Structure and function of the regulatory locus alkR. J. Biol. Chem. 263:13400-13405. [PubMed] [Google Scholar]
- 11.Eggink, G., P. H. van Lelyveld, A. Arberg, N. Arfman, C. Witteveen, and B. Witholt. 1987. Structure of the Pseudomonas putida alkBAC operon. Identification of transcription and translation products. J. Biol. Chem. 262:6400-6406. [PubMed] [Google Scholar]
- 12.Gallagher, S. R. 1999. One dimensional SDS gel electrophoresis of proteins, p. 10.2A.1-10.2A.34. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology, vol. 2. John Wiley & Sons, New York, N.Y. [Google Scholar]
- 13.Geerlof, A., J. J. Rakels, A. J. Straathof, J. J. Heijnen, J. A. Jongejan, and J. A. Duine. 1994. Description of the kinetic mechanism and the enantioselectivity of quinohaemoprotein ethanol dehydrogenase from Comamonas testosteroni in the oxidation of alcohols and aldehydes. Eur. J. Biochem. 226:537-546. [DOI] [PubMed] [Google Scholar]
- 14.Geissdörfer, W., R. G. Kok, A. Ratajczak, K. J. Hellingwerf, and W. Hillen. 1999. The genes rubA and rubB for alkane degradation in Acinetobacter sp. strain ADP1 are in an operon with estB, encoding an esterase, and oxyR. J. Bacteriol. 181:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. Wileys, New York, N.Y.
- 16.Goodwin, P. M., and C. Anthony. 1998. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv. Microb. Physiol. 40:1-80. [DOI] [PubMed] [Google Scholar]
- 17.Groen, B., J. Frank, Jr., and J. A. Duine. 1984. Quinoprotein alcohol dehydrogenase from ethanol-grown Pseudomonas aeruginosa. Biochem. J. 223:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamamura, N., C. Page, T. Long, L. Semprini, and D. J. Arp. 1997. Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB 5 and methane-grown Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 63:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamamura, N., R. T. Storfa, L. Semprini, and D. J. Arp. 1999. Diversity in butane monooxygenases among butane-grown bacteria. Appl. Environ. Microbiol. 65:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James, K. D., M. A. Hughes, and P. A. Williams. 2000. Cloning and expression of ntnD, encoding a novel NAD(P)(+)-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J. Bacteriol. 182:3136-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer, M. F., and D. M. Coen. 1999. Enzymatic amplification of DNA by PCR: standard procedures and optimization, p. 15.1.1-15.1.15. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology, vol. 3. John Wiley & Sons, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 22.Maeda, T., I. Yoshinaga, T. Shiba, M. Murakami, A. Wada, and Y. Ishida. 2000. Cloning and sequencing of the gene encoding an aldehyde dehydrogenase that is induced by growing Alteromonas sp. strain KE10 in a low concentration of organic nutrients. Appl. Environ. Microbiol. 66:1883-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeng, J. H., Y. Sakai, Y. Tani, and N. Kato. 1996. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J. Bacteriol. 178:3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morett, E., and L. Segovia. 1993. The σ54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, N. E., W. J. Brammar, and K. Murray. 1977. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol. Gen. Genet. 150:53-61. [DOI] [PubMed] [Google Scholar]
- 26.Oubrie, A., H. J. Rozeboom, K. H. Kalk, A. J. Olsthoorn, J. A. Duine, and B. W. Dijkstra. 1999. Structure and mechanism of soluble quinoprotein glucose dehydrogenase. EMBO J. 18:5187-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry, J. J. 1980. Propane utilization by microorganisms. Adv. Appl. Microbiol. 26:89-115. [Google Scholar]
- 28.Ratajczak, A., W. Geissdorfer, and W. Hillen. 1998. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl. Environ. Microbiol. 64:1175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratajczak, A., W. Geissdörfer, and W. Hillen. 1998. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J. Bacteriol. 180:5822-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rupp, M., and H. Gorisch. 1988. Purification, crystallisation and characterization of quinoprotein ethanol dehydrogenase from Pseudomonas aeruginosa. Biol. Chem. Hoppe-Seyler. 369:431-439. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sayavedra-Soto, L. A., C. M. Byrd, and D. J. Arp. 2001. Induction of butane consumption in Pseudomonas butanovora. Arch. Microbiol. 176:114-120. [DOI] [PubMed] [Google Scholar]
- 34.Sayavedra-Soto, L. A., N. G. Hommes, J. J. Alzerreca, D. J. Arp, J. M. Norton, and M. G. Klotz. 1998. Transcription of the amoC, amoA, and amoB genes in Nitrosomonas europaea and Nitrosospira sp. NpAV. FEMS Microbiol. Lett. 167:81-88. [DOI] [PubMed] [Google Scholar]
- 35.Singer, M. E., and W. R. Finnerty. 1985. Alcohol dehydrogenases in Acinetobacter sp. strain HO1-N: role in hexadecane and hexadecanol metabolism. J. Bacteriol. 164:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens, G. M., and H. Dalton. 1986. The role of the terminal and subterminal oxidation pathways in propane metabolism by bacteria. J. Gen. Microbiol. 132:2453-2462. [Google Scholar]
- 37.Stoorvogel, J., D. E. Kraayveld, C. A. Van Sluis, J. A. Jongejan, S. De Vries, and J. A. Duine. 1996. Characterization of the gene encoding quinohaemoprotein ethanol dehydrogenase of Comamonas testosteroni. Eur. J. Biochem. 235:690-698. [DOI] [PubMed] [Google Scholar]
- 38.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, J. 1980. Production of intracellular and extracellular protein from n-butane by Pseudomonas butanovora sp. nov. Adv. Appl. Microbiol. 26:117-127. [Google Scholar]
- 40.Takahashi, J., Y. Ichikawa, H. Sagae, I. Komura, H. Kanou, and K. Yamada. 1980. Isolation and identification of n-butane-assimilating bacterium. Agric. Biol. Chem. 44:1835-1840. [Google Scholar]
- 41.Toyama, H., N. Aoki, K. Matsushita, and O. Adachi. 2001. Azurin involved in alcohol oxidation system in Pseudomonas putida HK5: expression analysis and gene cloning. Biosci. Biotechnol. Biochem. 65:1617-1626. [DOI] [PubMed] [Google Scholar]
- 42.Toyama, H., A. Fujii, K. Matsushita, E. Shinagawa, M. Ameyama, and O. Adachi. 1995. Three distinct quinoprotein alcohol dehydrogenases are expressed when Pseudomonas putida is grown on different alcohols. J. Bacteriol. 177:2442-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Beilen, J. B., M. G. Wubbolts, and B. Witholt. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161-174. [DOI] [PubMed] [Google Scholar]
- 44.Van der Linden, A. C., and R. Huybregtse. 1969. Occurrence of inducible and NAD(P)-independent primary alcohol dehydrogenases in an alkane-oxidizing Pseudomonas. Antonie Leeuwenhoek 35:344-360. [DOI] [PubMed] [Google Scholar]
- 45.Vangnai, A. S., and D. J. Arp. 2001. An inducible 1-butanol dehydrogenase, a quinohemoprotein, is involved in the oxidation of butane by Pseudomonas butanovora. Microbiology 147:745-756. [DOI] [PubMed] [Google Scholar]
- 46.Xia, Z., W. Dai, Y. Zhang, S. A. White, G. D. Boyd, and F. S. Mathews. 1996. Determination of the gene sequence and the three-dimensional structure at 2.4 angstroms resolution of methanol dehydrogenase from Methylophilus W3A1. J. Mol. Biol. 259:480-501. [DOI] [PubMed] [Google Scholar]
- 47.Yanish-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 48.Zarnt, G., T. Schrader, and J. R. Andreesen. 1997. Degradation of tetrahydrofurfuryl alcohol by Ralstonia eutropha is initiated by an inducible pyrroloquinoline quinone-dependent alcohol dehydrogenase. Appl. Environ. Microbiol. 63:4891-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]