Abstract
PII-like signal transmitter proteins, found in Bacteria, Archaea, and plants, are known to mediate control of carbon and nitrogen assimilation. They indirectly regulate the activity of key metabolic enzymes and transcription factors by protein-protein interactions with signal transduction proteins. Many Proteobacteria harbor two paralogous PII-like proteins, GlnB and GlnK, whereas a novel third PII paralogue (GlnY) was recently identified in Azoarcus sp. strain BH72, a diazotrophic endophyte of grasses. In the present study, evidence was obtained that the PII-like proteins have distinct roles in mediating nitrogen and oxygen control of nif gene transcription and nitrogenase activity. Full repression of nif gene transcription in the presence of a combined nitrogen source or high oxygen concentrations was observed in wild-type and glnB and glnK knockout mutants, revealing that GlnB and GlnK can complement each other in mediating the repression. In contrast, in a glnBK double mutant strain in the presence of only GlnY, nif gene transcription was still detectable, albeit at a lower level, on nitrate or 20% oxygen. As another level of control, nitrogenase activity was regulated by at least three types of mechanisms in strain BH72: covalent modification of dinitrogenase reductase (NifH), probably by ADP-ribosylation, and two other, unknown means. Functional inactivation upon ammonium addition (switch-off) required the putative high-affinity ammonium transporter AmtB and GlnK, but not GlnB or GlnY. Functional inactivation in response to anaerobiosis did not depend on AmtB, GlnK, or GlnB. In contrast, covalent modification of NifH required both GlnB and GlnK and AmtB as response to ammonium addition, whereas it required either GlnB or GlnK and not AmtB when cells were shifted to anaerobiosis. In a glnBK double mutant expressing only GlnY, NifH modification was completely abolished, further revealing functional differences between the three PII paralogues.
Bacteria encountering rapid changes in environmental conditions require complex regulatory networks to integrate the signals and to adapt the cellular machinery. The availability of nitrogen sources is an important parameter, resulting in rapid changes in enzyme activities and transcriptional activity. PII-like proteins are central signal transmitter proteins in this regulatory network and occur in many Proteobacteria as two paralogous gene copies, glnB and glnK (44). A third copy (glnY) has recently been identified in the beta subgroup proteobacterium Azoarcus sp. strain BH72 (39). Depending on the cellular nitrogen status of the cell, a bifunctional uridylyl-transferase/hydrolase covalently modifies or demodifies the PII protein. Under conditions of nitrogen deficiency, the PII-like proteins in enteric bacteria occur mainly in the uridylylated form (for a review, see reference 2).
Depending on its state of modification, the PII protein acts as a molecular switch by protein-protein interactions. One target in enteric bacteria is the adenylyltransferase, which regulates by covalent modification the activity of the key enzyme of ammonium assimilation, glutamine synthetase (30). Additionally, unmodified PII (GlnB) inhibits autophsphorylation and activates the phosphatase activity of the target NtrB (29), which is part of a two-component regulatory system, resulting in a decrease in phosphorylated transcriptional regulator NtrC, thus preventing the transcription of Ntr-dependent operons (for a review, see reference 44).
The diversity of cellular responses to nitrogen raises the question whether additional proteins might act as receptors for PII-like proteins. Most bacteria fixing N2 react to a supply of ammonium by repression of transcription of nitrogenase structural genes, nifHDK (10, 41), and more rapidly by inactivation of nitrogenase activity (47, 53). NifA is the specific transcriptional activator of σ54-dependent nif promoters, whose activity in enteric bacteria such as Klebsiella pneumoniae is regulated in response to combined nitrogen and oxygen by NifL (10).
The PII-like protein GlnK is involved in the signal transduction cascade by relieving the NifL-dependent inactivation of NifA when combined nitrogen is limiting (20, 27). In several nitrogen-fixing bacteria, nitrogenase activity is also regulated at the posttranslational level. The so-called nitrogenase switch-off by ammonium depends on two different mechanisms. In some diazotrophs, such as Rhodospirillum rubrum (47), Rhodobacter capsulatus (31), and Azospirillum brasilense (17), the iron protein of nitrogenase (NifH) is subject to posttranslational modification, a reversible mono-ADP-ribosylation at a specific arginine residue. The draT gene product, an ADP-ribosyltransferase, covalently modifies nitrogenase, while DraG removes the ADP-ribosyl residue and thus reactivates nitrogenase (37).
In Azoarcus sp. strain BH72, a gene encoding a DraT homologue (30% amino acid identity to DraT of A. brasilense) has been found upstream of nifH (Junker and Reinhold-Hurek, unpublished). The DraT/G system has recently been implicated as a target for PII-protein-mediated regulation of nitrogenase activity in R. rubrum (62). Additionally, a physiological switch-off mechanism exists in some bacteria which does not involve a covalent modification of nitrogenase (46). The mechanism is still unknown; however, it has been suggested that the electron flow to nitrogenase may be involved (46). This is consistent with our study on the role of the ferredoxin FdxN, an electron donor to nitrogenase, which was shown to be essential for fast nitrogenase inhibition upon ammonium addition (14).
In many Bacteria and Archaea, the PII paralogue glnK occurs in an operon with an amtB gene. amtB genes encode integral membrane proteins which were identified as high-affinity ammonium transporters in Saccharamyces cerevisiae (38) and Arabidopsis thaliana (45). An involvement in ammonium transport is also assumed for bacteria (9, 43, 54). It has been proposed that the conservation of physical linkage of glnK and amtB reflects a functional relationship and a physical interaction of these proteins (57).
An additional level of complexity is added to this regulatory system by the occurrence of two structurally and functionally similar PII paralogues. In Escherichia coli, in contrast to glnB, glnK is expressed only under conditions of nitrogen limitation (3, 58). In the diazotroph Klebsiella pneumoniae, only GlnK and not GlnB regulates the activity of NifA or NifL (20, 27). However, findings in enteric bacteria cannot be generalized, and differential functions may vary depending on the bacterial species. In the alpha-proteobacterial diazotrophs Azospirillum brasilense and Azorhizobium caulinodans, the paralogue essential for free-living nitrogen fixation is GlnB (9), or both paralogues can complement each other (43), respectively.
The diazotroph Azoarcus sp. strain BH72 is an endophyte of grasses which can also infect rice (24, 51). It is a strictly respiratory bacterium which fixes nitrogen under microaerobic conditions, reaching steady states in a chemostat at 0.5 to 25 μM dissolved O2 (23). Reporter gene studies have shown a regulation at the transcriptional level in response to O2 and ammonium; nifH::gus expression was not detectable at or above 4% O2 in the headspace or above 0.5 mM ammonium (12). Strain BH72 harbors three paralogous PII-like proteins, all of which can be uridylylated and are thus likely to have a function in nitrogen sensing (39). Like glnK, the novel third paralogue, glnY, is physically linked with an amount gene (amtY). Unlike in E. coli, both paralogues GlnB and GlnK are abundant in Azoarcus spp. under conditions of nitrogen excess.
Interestingly, neither GlnK nor GlnB is essential for nitrogen fixation when GlnY is still present (39). GlnY can only be detected in a glnK glnB double mutant strain; however, expression levels are low on ammonium as the nitrogen source (39). This novel paralogue was found to be unusual, as it only occurred in the uridylylated state in vivo as shown by mass spectrometric analysis (39). In the present study PII-like proteins were shown to have distinct roles for the physiological switch-off and posttranslational covalent modification of dinitrogenase reductase (NifH) upon addition of ammonium or anaerobiosis, underlining that they are paralogues and not homologues. Moreover, the AmtB protein was essential for ammonium-induced switch-off in Azoarcus sp. strain BH72, probably serving as an ammonium sensor transmitting the signal to membrane-associated GlnK.
(Preliminary accounts on the involvement of PII-like proteins in regulation of nitrogenase activity or their membrane association were presented on the 12th International Congress on Nitrogen Fixation, Brazil, 1999 [50], or at the 4th European Nitrogen Fixation Conference, Spain, 2000.)
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains, cloning vectors, and recombinant plasmids used
| Strain, vector, or plasmid | Relevant genotype or properties | Source or reference |
|---|---|---|
| E. coli DH5α | F′ recA1 endA1 hsdR17(rK− mK+) supE44 (λ−thi-1 relA1 φ80d lacZΔM15 Δ(lacZYA-argF′)U169 | 19 |
| Azoarcus sp. | ||
| BH72 | Wild type | 49 |
| BHGLKK | Kmr, BH72 glnK::Kmr | 39 |
| BHGLBS | Sm/Spr, BH72 glnB::Sm/Spr | 39 |
| BHGLKKBS | Kmr, Sm/Spr, BH72 glnK::Kmr, glnB::Sm/Spr | 39 |
| BHABK | Kmr, BH72 amtB::Kmr | This study |
| Cloning vectors | ||
| pBluescript SK | Apr, ColE1 origin | Stratagene |
| pBK-CMV | Kmr, Neor, ColE1 origin, fl(−) origin, simian virus 40 origin | Stratagene |
| pUC4K | Kmr, Neor, ColE1 origin | Pharmacia |
| pLAFR3 | Tetr, low-copy cosmid vector | 56 |
| Recombinant plasmids | ||
| pDZD17 | Kmr, glnk-amtB locus on a 5.8-kb chromosomal SauIIIAI fragment of strain BH72 in pBK-CMV | 39 |
| pDZD17.1 | Apr, 3.6-kb HindIII/SpeI fragment of pDZD17 cloned in pBluescript SK | This study |
| pDM3 | Apr, Kmr, 1.3-kb Kmr cassette of pUC4K cloned in SrfI restriction site of pDZD17.1 (600 bp downstream of amtB start codon) | This study |
| pEGN3.1 | Apr, nifH::uidA transcriptional fusion on pUC19 | 12 |
| pNHGus | Tetr, 3.3-kb HindIII/EcoRI fragment harboring nifH::uidA fusion of pEGN3.1 cloned in pLAFR3 | This study |
| pKKOM | Tetr, 1.5-kb SrfI/KpnI fragment of pDZD17 cloned in pLAFR3 | This study |
Media and growth conditions.
Growth of Azoarcus sp. strain BH72 on complex media for electroporation and mutant selection was performed as described earlier (11, 39). E. coli was grown in LB medium following standard protocols (4). To obtain N2-fixing cells of Azoarcus sp., bacteria were precultured on SM + N medium (48), washed twice in N-free SM medium (48), adjusted to an optical density at 578 nm (OD578) of 0.05 in the same medium, and incubated under microaerobic conditions in rubber stopper-sealed 1-liter Erlenmeyer flasks sparged with N2 and adjusted to 1.5% O2 and 1% acetylene in the headspace, with rotary shaking at 100 rpm.
For nitrogenase derepression analyses, precultivation was performed in the presence of either 10 mM NH4Cl or KNO3 as the sole nitrogen source, and cells were transferred to microaerobic conditions (0.8% O2) as described above at an initial OD578 of 0.05 in 30 ml of medium and grown for 10 h. For nitrogenase switch off (repression) analyses, N2-fixing cells were incubated as described above on N-free medium followed by addition of 0.2 and 2 mM NH4Cl or KNO3. Alternatively, cells were transferred to an anaerobic Erlenmeyer flask containing 1% acetylene using a 50-ml syringe.
Membrane isolation.
Cells were grown under conditions of N2 fixation or on VM-ethanol (complex) medium (52) as described above and harvested at 4°C by centrifugation for 10 min at 5,000 × g. The cell pellet was resuspended in 50 mM sodium phosphate buffer (pH 7.0) and sonicated five times (30-min pulses each, 40-W output) using a Branson Sonifier 250. Cellular debris was removed by centrifugation (20 min, 20,000 × g), and the remaining supernatant was used for membrane sedimentation by ultracentrifugation (2 h, 200,000 × g). The remaining supernatant was referred to as the soluble cytoplasmic fraction, whereas the isolated membrane fractions were washed twice with 50 mM sodium phosphate buffer (pH 7.0) followed by two salt washes using the same buffer supplemented with 600 mM NaCl to remove nonspecifically or loosely bound proteins. The insoluble pellet after ultracentrifugation was referred to as the membrane fraction.
Determination of nitrogenase and β-glucuronidase activity.
Nitrogenase activity of batch cultures was determined by using the acetylene reduction method (13). Activity of β-glucuronidase was measured quantitatively using the method described earlier (28) as modified (12) and expressed in Miller units, defined as E420 × 1,000/t (minutes) × OD600.
DNA and RNA analyses.
Isolation of chromosomal DNA was carried out as described previously (22). Other DNA techniques followed standard protocols (4). Genomic clones were characterized by restriction mapping and Southern blot analyses with digoxigenin-labeled gene probes (39). DNA was sequenced from both strands as described previously (26). Homology searches were carried out using the Blast program (1). Prediction of membrane protein topology of AmtB and AmtY was performed using the program TopPred2. The amtB and amtY sequences of Azoarcus sp. strain BH72 have been assigned GenBank accession numbers AF430400 and AF430401.
Construction of plasmids for marker exchange mutagenesis and a nifH::gusA reporter gene fusion.
Knockout mutants BHGLKK (glnK), BHGLBS (glnB), and BHGLKKBS (glnBK) had been obtained by insertion of resistance cartridges (39). The amtB gene was inactivated by cloning the kanamycin resistance cartridge (1.3 kb) of pUC4K into the Srf I restriction site (635 bp downstream of amtB start codon) of pDZD17.1, resulting in plasmid pDM3. Plasmid pDZD17.1 was generated by insertion of a 3.6-kb HindIII/SpeI fragment of pDZD17 (39) into pBluescriptSK. The orientation of the kanamycin cassette resulting in a nonpolar mutation was checked by restriction digestion (EcoRI/HindIII).
Marker exchange mutant strain BHABK (amtB) was obtained by transformation of Azoarcus sp. strain BH72 by electroporation with the suicide plasmid pDM3. Southern blot analysis using a glnK gene probe (39) verified the correct chromosomal integration of the resistance cartridge since the hybridizing 5.1-kb EcoRI fragment harboring the glnK-amtB operon shifted to 6.3 kb in size. Plasmid pNHGus harboring a transcriptional nifH::gusA fusion was generated by cloning a 3.3-kb HindIII/EcoRI fragment of pEGN3.1 (12) into the low-copy vector pLAFR3 (56). pHNGus was transformed into Azoarcus sp. strain BH72 wild-type, BHGLBS, BHGLKK, and BHGLKKBS by the method of triparental mating using E. coli(pRK2013) as the helper strain (11).
SDS-PAGE and Western blotting.
For the analysis of NifH modification, 1-ml aliquots of cell suspensions were removed from the culture, and proteins were precipitated immediately on ice with 100 μl of trichloroacetic acid (TCA) solution (1 g of TCA per ml) as described (61). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (24, 35). For separation of the two forms of NifH protein, a 10% (wt/vol) acrylamide low cross-linker gel with a ratio of acrylamide to N,N′-methylenebis(bisacrylamide) of 172:1 was used (32).
Electroelution of proteins onto nitrocellulose membranes was performed as described previously (24) for 45 min at 8 V with a semidry electroblotter (Bio-Rad, Munich, Germany). The NifH protein of Azoarcus sp. strain BH72 was detected using antiserum against NifH of Rhodospirillum rubrum, kindly provided by R. Ludden (Madison, Wis.) as outlined previously (24). GlnB, GlnK, and GlnY were immunodetected using antisera raised against purified fusion proteins of E. coli maltose-binding protein (MalE) and GlnB, GlnK, and GlnY, respectively (39). Proteins were visualized using ECL Western blotting detection reagents (Amersham Pharmacia Biotech). Protein concentrations were determined by the Bio-Rad protein assay based on the method of Bradford (6).
Biochemical analyses.
For analyses of NifH modification, protein extracts (sonicated cell extracts after centrifugation of cell debris; see membrane isolation) were incubated with RNase A (30 min at 30°C in 50 mM sodium phosphate buffer, pH 7.2), calf intestine alkaline phosphatase (30 min at 37°C in 50 mM Tris-HCl, pH 8.5, 0.1 mM EDTA), or snake venom phosphodiesterase I (30 min at 25°C in 100 mM Tris-HCl, pH 9.0, 100 mM NaCl, 15 mM MgCl) using approximately 1 U of enzyme and 20 μg of protein extract per assay. Protein extracts were also treated with 1 M hydroxylamine (pH 7.0), 10 mM HgCl2, 1 M HCl, or 1 M NaOH at 30°C for 6 h. Reaction mixtures were analyzed by SDS-PAGE, Western blotting, and immunodetection using NifH antiserum. Determination of ammonia concentrations in culture supernatants was performed using an enzymatic ammonium assay (Roche Molecular Biochemicals, no. 1112732) with a detection limit of 1 μM.
RESULTS
Effect of PII-like proteins on nitrogen-regulated nif gene expression.
Azoarcus sp. strain BH72 mutants in which the paralogues glnB or glnK had been inactivated showed equal growth rates on N2 compared to the wild type, indicating that nitrogenase gene expression under derepressing conditions was not abolished in a glnB or glnK knockout mutant (39). In order to test the influence of the PII paralogues on nifH gene transcription in response to combined nitrogen, nitrogenase gene expression was quantified using a nifH::gusA fusion on a stably replicated plasmid (pNHGus). Cells were cultivated aerobically on 10 mM of a combined nitrogen source (nitrate or ammonium), transferred to microaerobic conditions into medium containing 10 mM of the respective nitrogen source, and harvested for quantification of β-glucuronidase activity after 10 h of incubation. In wild-type cells, β-glucuronidase activity was strongly repressed by KNO3 or NH4Cl in comparison to N-free medium (Table 2). Mutants in which glnB or glnK was inactivated and which expressed GlnK or GlnB, respectively (39), reacted like the wild-type strain (Table 2). No acetylene reduction activity was detected in the presence of nitrate or ammonium, and in Western blot analysis using specific antibodies, the iron protein of nitrogenase was not detected in cell extracts (not shown), corroborating a strong repression of nifH.
TABLE 2.
Nitrogenase activity and nifH::gusA expression of Azoarcus sp. strain BH72 wild-type, BHGLBS (glnB), BHGLKK (glnK), and BHGLKKBS (glnBK) strains grown microaerobically in SM medium supplemented with 10 mM NH4Cl or KNO3a
| Genotype | N source | Nitrogenase activity (nmol of C2H4/mg of protein) | nifH::gusA expression (Miller units) in 1%/20% O2 |
|---|---|---|---|
| Wild type | NH4Cl | n.d. | 80 ± 10/<50 |
| KNO3 | n.d. | 120 ± 10/<50 | |
| N2 | 10,430 ± 530 | 7,200 ± 200/n.dm. | |
| glnB | NH4Cl | n.d. | 100 ± 10/<50 |
| KNO3 | n.d. | 150 ± 20/<50 | |
| N2 | 9,240 ± 150 | 6,320 ± 410/n.dm. | |
| glnK | NH4Cl | n.d. | 90 ± 10/<50 |
| KNO3 | n.d. | 140 ± 10/<50 | |
| N2 | 9,460 ± 460 | 6,910 ± 390/n.dm. | |
| glnBK | NH4Cl | 70 ± 10 | 710 ± 30/<50 |
| KNO3 | 4,010 ± 360 | 2,860 ± 60/700 ± 60 | |
| N2 | 7,320 ± 410 | 7,100 ± 450/n.dm. |
Data were obtained from three parallel measurements from two independent experiments and are given with standard deviations. n.d., not detected; n.dm., not determined (cells did not grow under atmospheric O2 concentrations (20%) with N2 as the sole nitrogen source).
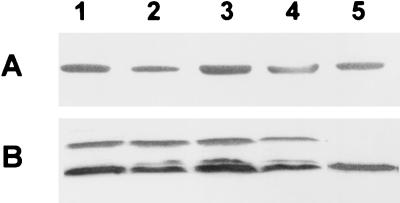
In contrast, in a glnBK double mutant which expresses GlnY (39), nifH::gusA expression was not completely repressed by nitrate and was still present at lower levels (10-fold repression) in the presence of ammonium (Table 2). Accordingly, acetylene reduction activity was high in this double mutant grown in the presence of nitrate and still detectable although at much lower levels during growth on ammonium (Table 2), when GlnY expression is very low (Fig. 1) (39). These results indicated that GlnB and GlnK were involved in efficient signal transduction leading to full nif gene repression in single mutants, while in the double mutant the presence of GlnY alone did not allow full repression.
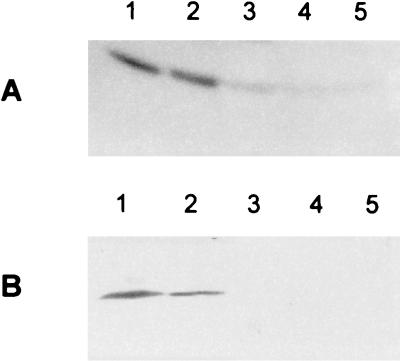
FIG. 1.
Western blot analysis of GlnY (A) and NifH (B) synthesis of a glnBK double mutant strain (BHGLKKBS) during growth on ammonium or nitrate. Equal amounts (20 μg) of protein extracts from cells grown microaerobically on 10 mM nitrate (lane 1), aerobically on 10 mM nitrate (lane 2), microaerobically on 10 mM ammonium (preculture on nitrate) (lane 3), aerobically on 10 mM ammonium (preculture on ammonium) (lane 4), or microaerobically on 10 mM ammonium (preculture on ammonium) (lane 5) in SM medium were separated by SDS-PAGE, followed by immunodetection of GlnY and NifH.
Additionally, Western blot analyses were carried out with antibodies specific to GlnY (39) or NifH (Fig. 1). As expected, in cells grown microaerobically on nitrate, both, GlnY and the iron protein of nitrogenase occurred in significant amounts (lane 1), in contrast to cells grown on ammonium (lanes 4 and 5). Surprisingly, cells grown aerobically on nitrate also showed nitrogenase expression (lane 2). Accordingly, nifH::gusA reporter gene activity was significantly higher in this strain compared to the wild type (Table 2), suggesting circumvention of the O2− control of nif gene expression.
GlnK and AmtB are involved in nitrogenase inhibition (switch-off) by ammonium.
Previously we have shown that upon addition of ammonium to nitrogen-fixing cells, nitrogenase activity is rapidly inhibited (14). We observed that upon ammonium addition to an N2-fixing culture in a biofermenter with mass-flow-controlled oxygen supply (39), oxygen consumption was dramatically decreased after addition of ammonium (not shown). To assess whether PII-like proteins are involved in the signaling pathway leading to nitrogenase inhibition, knockout mutants were tested in switch-off experiments using 2 mM NH4Cl (Fig. 2B). The glnB mutant expressing only GlnK showed a wild-type level inhibition of nitrogenase activity, while the glnK mutant expressing GlnB and the glnBK double knockout mutant expressing GlnY were not rapidly inhibited (Fig. 2A). N2 fixation of the latter mutant was almost not affected within 2 h. In the glnK mutant, acetylene reduction decreased slowly within 2 h, which can be explained by a decreased synthesis of nitrogenase due to strong repression of nifHDK transcription by ammonium, which is still observed in this mutant (see Table 2).
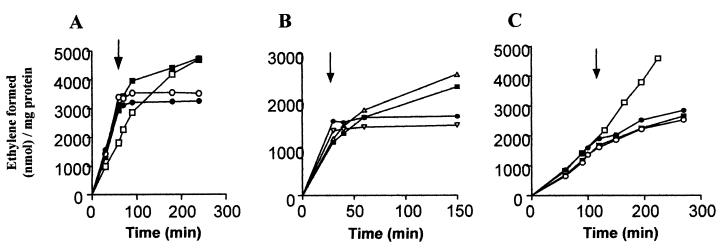
FIG. 2.
Effects of ammonia and nitrate addition on nitrogenase activity of N2-fixing cultures of wild-type Azoarcus sp. strain BH72 and glnB, glnK, glnBK, and amtB knockout mutants. Data are based on at least two independent experiments for all conditions tested. Arrows indicate time of ammonia or nitrate addition. (A and B) Influence of ammonium addition (2 mM NH4Cl) on acetylene reduction (nitrogenase activity) of wild-type Azoarcus sp. strain BH72 (•), glnB (○), glnK (▪), glnBK (□), and amtB (▵) mutant strains, and glnK complementation strain BHGLKK/pKKOM (▿). (C) Effect of nitrate addition (2 mM KNO3) on acetylene reduction (nitrogenase activity) of wild-type Azoarcus sp. strain BH72 (•) and glnB (○), glnK (▪) and glnBK (□) mutant strains.
The glnK gene is organized in an operon with amtB, encoding a putative membrane protein with high homology to ammonium transporters (50% identity to E. coli amtB) (39); typically, these proteins have 11 to 12 transmembrane helices; 12 membrane helices are also predicted for AmtB of strain BH72 (using programTopPred2). To rule out a polar effect of the glnK mutation on amtB function, the glnK gene including the upstream promoter region was complemented in trans on a plasmid (pKKOM). The resulting strain, BHGLKK(pKKOM), was functionally complemented, since nitrogenase activity was rapidly inhibited as in the wild-type Azoarcus sp. strain (Fig. 2A and B). Additionally, an insertion mutation inactivating the amtB gene (strain BHABK) resulted in loss of the rapid ammonium switch-off, similar to the glnK mutation (Fig. 2B). Therefore, both proteins, AmtB and GlnK, were essential for the signaling pathway leading to physiological nitrogenase switch-off by ammonium in Azoarcus sp. strain BH72.
The rapid nitrogenase switch-off of N2-fixing cells was not observed upon addition of the alternative nitrogen source nitrate (Fig. 2C) or glutamine (not shown), revealing that this response is specific to ammonium. Addition of nitrate led to a slow decrease in acetylene reduction activity in the wild type as well as in single glnB or glnK mutants (not shown), probably resulting from repression of nifHDK transcription by nitrate (see Table 2). In the glnBK double mutant, which expresses GlnY and is not subject to nitrogenase gene repression (see above), acetylene reduction activity remained at a high level (Fig. 2C).
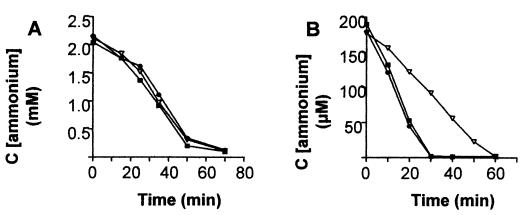
Since amtB encodes a putative ammonium transporter, the glnK knockout mutation may affect ammonium transport. Therefore, uptake of ammonium from the medium was monitored in the wild-type and in the mutant strains BHABK (amtB) and BHGLKK (glnK) after addition of 2 mM and 0.2 mM ammonium under the same conditions applied for switch-off experiments (Fig. 3). Wild-type and mutant strains assimilated ammonium from the medium at approximately the same rate at an initial concentration of 2 mM (70 to 75 nmol/min/mg of protein), and thus overall NH4+ transport was not affected by the amtB mutation under these conditions. However, ammonium uptake at low concentrations of ammonium (0.2 mM) was slightly higher in the wild-type strain and the glnK strain (10 nmol/min/mg of protein) compared to the amtB mutant (5 nmol/min/mg protein), which might indicate a function of AmtB for efficient NH4+ uptake during exposure to low ammonium concentrations.
FIG. 3.
Ammonium assimilation of Azoarcus sp. wild-type strain BH72, BHGLKK (glnK), and BHABK (amtB). Decrease of ammonium concentrations in supernatants after an initial addition of 2 mM NH4Cl (A) or 0.2 mM NH4Cl (B) to N2-fixing cultures of Azoarcus sp. wild-type strain BH72 (•) and glnK (▪) and amtB (▿) mutant strains.
Since GlnK and AmtB were shown to be essential for ammonium-induced switch-off and GlnK did not affect NH4+ transport via AmtB or other uptake systems under the conditions investigated, it is likely that AmtB fulfills a sensory function in detection of changes in the external ammonium concentration. These results suggest a dual role of AmtB as an ammonium transporter and ammonium sensor.
Effect of PII-like proteins and AmtB on posttranslational modification of dinitrogenase reductase.
To analyze the effect of mutations on the putative covalent modification of dinitrogenase reductase in Azoarcus sp. strain BH72, protein extracts of cells were subjected to Western blot analysis using antibodies against NifH (Fig. 4). Previously it was shown that the NifH protein of Azoarcus sp. strain BH72 was covalently modified in response to oxygen deficiency: a NifH protein of lower electrophoretic mobility accumulated (difference of approximately 1.5 kDa) which had identical amino acid sequences in the N terminus with the unmodified NifH protein usually observed (25).
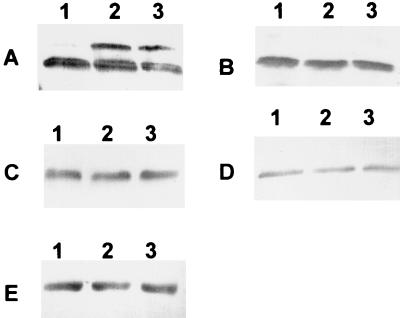
FIG. 4.
Western blot analysis of NifH modification after ammonium addition to nitrogen-fixing cultures of Azoarcus sp. wild-type strain BH72 (A) and BHGLBS (glnB) (B), BHGLKK (glnK) (C), BHGLKKBS (glnBK) (D), and BHABK (amtB) (E). Samples were taken before (lane 1) or 5 min (lane 2) or 15 min (lane 3) after addition of 2 mM NH4Cl. Experiments were performed at least two times.
After addition of ammonium, a modified NifH protein of higher apparent molecular weight accumulated (Fig. 4, lanes 2 and 3). To determine the type of NifH modification, protein extracts from an ammonium switch-off experiment were treated with either phosphodiesterase I or neutral hydroxylamine (NH2OH), which both led to the disappearance of the higher molecular weight protein in contrast to untreated controls in Western blots (not shown) and thus to a complete removal of the modifying group. Incubation with RNase A, alkaline phosphatase, 10 mM HgCl2, or 1 M HCl had no effect on NifH modification, whereas incubation with 1 M NaOH abolished the antigenicity of both the modified and the unmodified form of NifH (data not shown).
Phosphodiesterase I is known to cleave substrates harboring a phosphodiester bond and is therefore in general able to cleave protein-bound adenyl-, uridylyl-, or ADP-ribosyl residues (8, 36, 55). NH2OH has been shown to specifically hydrolyze ADP-ribose from arginine, whereas HgCl2 cleaves only cysteine-bound ADP-ribose (7). Therefore, it is very likely, by analogy to other reports, e.g., in R. rubrum and Rhodobacter capsulatus (31, 47), that the modifying group of NifH under conditions of ammonium switch-off represented an ADP-ribosyl moiety covalently linked to an arginine residue.
Inactivation of single genes glnB, glnK, or amtB abolished the alteration of the electrophoretic mobility and thus the modification (Fig. 4), indicating that the presence of both PII-like proteins as well as AmtB was necessary for this function.
We also analyzed the effect of oxygen limitation on NifH modification. All mutant strains investigated reacted with a fast and complete inhibition of nitrogenase activity upon transfer to anaerobic conditions (not shown). Since Azoarcus sp. strain BH72 has a strictly respiratory type of metabolism, nitrogenase inhibition was most likely due to energy limitation. Western blot analyses showed that nitrogenase modification upon oxygen limitation was not affected in single glnB, glnK, or amtB mutants, but was abolished in the glnBK double mutant (Fig 5). These results suggested that the signal transduction pathways leading to modification of NifH were different for the two stimuli tested, ammonium excess and oxygen limitation.
FIG. 5.
Western blot analysis of NifH modification upon anaerobiosis. N2-fixing cultures of the Azoarcus sp. were transferred from microaerobic conditions (0.8% oxygen) (A) to anaerobic conditions. Samples were taken after 5 min of anaerobic incubation (B). Lanes: wild-type strain BH72 (lane 1), glnB (lane 2), glnK (lane 3), amtB (lane 4), and glnBK (lane 5) mutant strains.
Apparently the presence of both PII paralogues, GlnB and GlnK, and AmtB was required for the response to ammonium, while for the response to anaerobiosis, AmtB was not required at all, and the presence of either GlnK or GlnB was necessary. Since nitrogenase inhibition still occurred in the double mutant, which expresses only GlnY and does not show a NifH modification, the modification is apparently not a prerequisite for inhibition.
GlnK and GlnY occur membrane associated, in contrast to GlnB.
As GlnK and the putative membrane protein AmtB were both found to be part of the signal transduction cascade for physiological nitrogenase switch-off upon ammonium addition, we speculated that GlnK, albeit a cytoplasmic protein, might occur in association with membranes due to interactions with integral membrane proteins. Therefore, membrane and cytoplasmic fractions were extracted from Azoarcus sp. strain BH72 cells, and the membranes were treated with salt (600 mM NaCl) to remove loosely attached proteins. Both fractions were compared in Western blot analysis using specific antibodies.
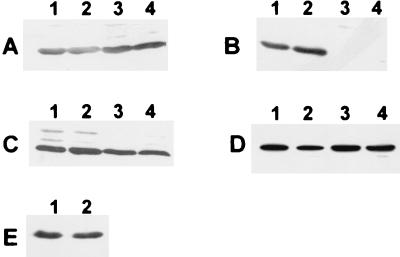
GlnB was detected exclusively in the cytoplasmic fractions (Fig. 6B, lanes 1 and 2), whereas GlnK and GlnY also occurred membrane associated: antibodies reacting with GlnK and GlnY localized a protein in the membrane as well as cytoplasmic fractions, which was GlnK in wild-type cells (Fig. 6A), as GlnY cannot be detected in wild-type Azoarcus sp. strain BH72 (39). In the glnBK double mutant, the reacting protein was GlnY (Fig. 6C). Membrane fractions of cells grown on SM medium with 10 mM NH4Cl (Fig. 6A, lanes 1 and 3), on complex medium (not shown), at conditions of N2 fixation (lanes 2 and 4), or at N2-fixing conditions after ammonium switch-off (Fig. 6E, lanes 1 and 2) contained GlnK, indicating that the nitrogen status of the cell did not strongly affect the binding of these proteins. Moreover, GlnK was still detectable in association with membranes in the amtB mutant strain BHABK (Fig. 6D).
FIG. 6.
Determination of cellular localization of GlnB, GlnK, and GlnY by Western blot analysis. Cytoplasmic (lanes 1 and 2) and membrane fractions (lanes 3 and 4) (20 μg of protein per lane) of Azoarcus sp. strain BH72 wild-type (A and B), strain BHGLKKBS (glnBK) (C), and strain BHABK (amtB) (D) grown on SM medium with 10 mM NH4Cl (lanes 1 and 3) or grown under conditions of nitrogen fixation on N-free SM medium (lanes 2 and 4) were analyzed using specific antisera against GlnK (A, D, and E), GlnB (B), and GlnY (C). (E) Membrane fractions of N2-fixing wild-type cells before (lane 1) and 15 min after (lane 2) ammonium switch-off.
DISCUSSION
Here we report evidence that the three paralogous PII-like proteins of Azoarcus sp. strain BH72 are involved differently in regulation of nif gene expression, nitrogenase activity and posttranslational modification of dinitrogenase reductase (NifH).
In several diazotrophic bacteria, one PII paralogue is essential for nitrogen fixation, however, the identity may differ. In Azotobacter vinelandii, which harbors only one gene encoding a PII-like protein (GlnK), a glnK null mutant could not be obtained up to now (40). In Azospirillum brasilense (9) and Herbaspirillum seropedicae (5), a null mutation of glnB results in a Nif− phenotype. In Azorhizobium caulinodans only a glnK glnB mutant is Nif−, since GlnK and GlnB can complement each other for free-living N2 fixation; however, symbiotic N2 fixation requires GlnB (43). In contrast, neither GlnB nor GlnK was essential for nitrogen fixation in Azoarcus sp. strain BH72; in the glnBK double mutant, GlnY is expressed, which might be sufficient to allow N2 fixation (39).
In accordance with these previous results, transcriptional activity of the nifH promoter was not affected by null mutations of glnK, glnB, or a glnBK double mutation under N-limiting conditions. However, the presence of GlnB or GlnK was necessary to confer full repression of nitrogenase gene transcription in response to combined nitrogen sources. In members of the gamma Proteobacteria such as Klebsiella pneumoniae (21, 42), in addition to the GlnB-dependent NtrBC pathway regulating nifA expression, NifL encoded in the nifLA operon inhibits NifA activity in the presence of high oxygen or ammonium concentrations. In K. pneumoniae, only GlnK and not GlnB is able to relieve this inhibition when ammonium is limiting (20, 27). In Azoarcus sp. strain BH72, nifLA homologues were also detected, their transcription being only moderately repressed in the presence of ammonium (Egener, Martin, and Reinhold-Hurek, submitted). NifLA might thus be an additional regulatory target for GlnK or GlnB.
The third PII paralogue in Azoarcus spp., GlnY, is unusual in several aspects. In a glnBK double mutant expressing only GlnY, the nifH gene transcription was only weakly repressed in the presence of nitrate. This may be due to the uridylylation state of GlnY. While both GlnK and GlnB are differentially uridylylated in response to changing ammonium concentrations, the third paralogue, GlnY, is not deuridylylated upon addition of ammonium or in the presence of nitrate but can only be detected in the uridylylated form in vivo (39). Therefore, GlnY is apparently not able to serve as a signal transmitter for changing nitrogen conditions.
Since GlnY is only detectable in a glnBK double mutant background, it cannot be excluded that deuridylylation might occur in the presence of another paralogue; however, such a dependence on a second paralogue would be very unusual. In single glnK or glnB mutants of A. brasilense (9) or E. coli (58), both proteins were modified or demodified in response to nitrogen in vivo. The structural reason for this unusual feature of GlnY still needs to be elucidated.
Transcription of nifH or nitrogenase protein synthesis was only observed under conditions which allowed high levels of GlnY expression, e.g., on nitrate (see Fig. 1 and Table 2). Surprisingly, in the glnBK mutant strain, the repression of nitrogenase synthesis by high oxygen concentrations was also relieved (see Fig. 1, lane 1, and Table 2). This indicates that either directly or indirectly by regulating expression of other genes involved, PII-like proteins may also play a role in the signal transduction pathway controlling nif gene expression in response to oxygen, presumably by interaction with the NifLA complex, which was also detected in strain BH72 (Egener, Martin, and Reinhold-Hurek, submitted).
In addition to control of nif gene expression, novel roles of PII-like proteins were identified with respect to the control of nitrogenase activity. In Azoarcus sp. strain BH72 a fast and reversible physiological switch-off mechanism was observed, similar to those in Rhodobacter capsulatus and Azospirillum brasilense in terms of speed and extent of nitrogenase inhibition (14). Previously we have shown that the ferredoxin FdxN may be involved in this process (14). As additional components involved in ammonium-induced nitrogenase switch-off, we identified AmtB and GlnK, whereas GlnB was not involved. Since at switch-off conditions (2 mM NH4Cl) the amtB and glnK mutants showed similar rates of NH4+ uptake as the wild type, the NH4+ transport via AmtB or other uptake systems was apparently not affected. Therefore, it is likely that AmtB fulfills a sensory function in detection of changes in the external ammonium concentration, rather than playing a mere role in the import of ammonium into the cell.
It is tempting to speculate that the GlnK and AmtB proteins interact physically. It is remarkable that the hydrophilic PII-like proteins showed association to membrane fractions at all, moreover, this was differential in Azoarcus. Only GlnK and GlnY, which are encoded in an operon with amtB homologues, occurred in association with membranes, but not GlnB. Previously it has been proposed that AmtB and GlnK might physically and functionally interact, based on conservation of an operon-like gene order of glnK and amtB in many different prokaryotes (57).
The cellular localization of the hydrophilic proteins at membranes in addition to the cytoplasmic fraction indeed suggests the existence of specific interactions with membrane proteins. Since we still observed membrane association of GlnK in amtB mutant strain BHABK, in which the gene is interrupted between putative transmembrane helices 4 and 5, GlnK might interact either with the N-terminal part or with as yet unidentified membrane proteins. Additionally, our observation implies that heterotrimers of GlnB and GlnK subunits, which have been proposed to fine tune nitrogen sensing in E. coli (59), are apparently not formed in vivo in Azoarcus spp., at least not at the cytoplasmic membrane.
In Azoarcus sp. wild-type strain BH72, inhibition of nitrogenase activity coincided with a covalent modification of NifH. Reversible modification of NifH is known to be catalyzed by the DraT/G system in several N2-fixing bacteria, which is controlled by ammonium and darkness (in phototrophs) or oxygen limitation and anaerobiosis (in A. brasilense). Chemical analyses of the nature and covalent linkage of the modifying group and the presence of a draT homologue upstream of the nifHDK operon (Junker and Reinhold-Hurek, unpublished) indicated the presence of NifH-ADP-ribosylation also in Azoarcus sp. strain BH72.
We found that PII-like proteins are also involved in the signal transduction of this process. Interestingly, ammonium-induced NifH modification required the presence of both GlnB and GlnK. In contrast, the rapid nitrogenase inactivation, which we refer to as the physiological switch-off, required only GlnK. Thus, the differential roles of PII-like proteins in Azoarcus spp. allow a clear distinction of these two mechanisms of nitrogenase inhibition: the glnB mutant showed rapid inactivation but lacked NifH modification. Also in R. capsulatus and A. brasilense, two mechanisms for nitrogenase inhibition were described, the physiological switch-off presumably accounting for the majority of nitrogenase inhibition, which is not the primary result of NifH modification (15, 16, 46, 60).
It has been proposed that this mechanism involves reduced electron flux to nitrogenase, resulting in fast inhibition of nitrogenase activity (46). The decrease in oxygen consumption which we observed after addition of ammonium to an N2-fixing culture of strain BH72 indicates a reduced flow of electrons towards O2 concomitant with decreased N2 fixation, which might result in changes in the redox status of cellular electron carriers. A regulatory role of the electron flux in nitrogenase inactivation is also suggested, since the ferredoxin FdxN, an electron donor to NifH, is essential for ammonium inactivation of nitrogenase in strain BH72 (14).
In accordance with these results, Halbleib et al. (18) showed that electron transport via NifF or NifJ and the redox state of NifH influences responses to NH4+ via ADP-ribosylation of NifH. Therefore, the sensory signals, e.g., the redox state of certain electron carriers, might be integrated, resulting in fast nitrogenase switch-off upon addition of ammonium or oxygen or energy limitation. However, cascades leading to nitrogenase modification in response to ammonium or anaerobiosis differ, since anaerobiosis-induced NifH modification did only require the presence of GlnB or GlnK, whereas ammonium-induced modification required the presence of both GlnB and GlnK as well as AmtB in strain BH72.
Involvement of PII-like proteins in regulation of nitrogenase activity was also observed in the methanogenic archaeon Methanococcus maripaludis (33). Klipp and coworkers also proposed a model for regulation of nitrogenase activity by PII-like proteins in R. capsulatus (34). Taken together, these results suggest that PII-protein-mediated regulation of nitrogenase activity might be widespread among diazotrophs.
In conclusion, our studies demonstrated that PII-like proteins in Azoarcus sp. strain BH72 are paralogues with distinct, characteristic functional properties, although they can complement some functions of each other. The most distantly related protein appears to be the third novel paralogue, GlnY, which cannot participate in certain signal transduction pathways in the absence of the other two proteins. Its specific functions and structure-function relationships still need to be elucidated. Moreover, our studies demonstrate the intriguing phenotypic diversity of PII-like proteins: the role of a particular paralogue in fine-tuning of nitrogen metabolism may vary considerably depending on the organisms studied.
Using mutant analyses, GlnK especially was shown to be involved in multiple signal transduction cascades. This involvement might be indirect, via transcriptional control of novel constituents of the cascade, or direct, by protein-protein interactions with novel receptors, for example, with AmtB (in physiological nitrogenase switch-off) or DraT/G and/or nitrogenase (in covalent modification of the iron protein of nitrogenase). In any case, the picture is emerging that PII-like proteins show a remarkable diversity of functions despite their small size.
Acknowledgments
We thank P. W. Ludden (University of Wisconsin-Madison) for antibodies against NifH, J. Plessl (University of Bremen) for initial experiments on membrane fractionation, and T. Hurek (Max Planck Institute for Marine Microbiology, Bremen) for valuable discussions.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) to B.R.-H. (Re 756/5-2 and 5-3).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arcondéguy, T., R. Jack, and M. Merrick. 2001. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65:80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, M. R., and A. J. Ninfa. 1998. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 29:431-447. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Benelli, E. M., M. Souza, S. Funayama, L. U. Rigo, and F. O. Pedrosa. 1997. Evidence for two possible glnB-type genes in Herbaspirillum seropedicae. J. Bacteriol. 179:4623-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cervantes-Laurean, D., D. E. Minter, E. L. Jacobson, and M. K. Jacobson. 1993. Protein glycation by ADP-ribose: studies of model conjugates. Biochemistry 32:1528-1543. [DOI] [PubMed] [Google Scholar]
- 8.Colonna-Romano, S., E. J. Patriarca, M. Amar, P. Bernard, G. Manco, A. Lamberti, M. Iaccarino, and R. Defez. 1993. Uridylylation of the PII protein in Rhizobium leguminosarum. FEBS Lett. 330:95-98. [DOI] [PubMed] [Google Scholar]
- 9.De Zamaroczy, M. 1998. Structural homologues PII and PZ of Azospirillum brasilense provide intracellular signalling for selective regulation of various nitrogen-dependent functions. Mol. Microbiol. 29:449-463. [DOI] [PubMed] [Google Scholar]
- 10.Dixon, R. 1998. The oxygen-responsive NIFL-NIFA complex: a novel two-component regulatory system controlling nitrogenase synthesis in gamma-Proteobacteria. Arch. Microbiol. 169:71-380. [DOI] [PubMed] [Google Scholar]
- 11.Dörr, J., T. Hurek, and B. Reinhold-Hurek. 1998. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol. Microbiol. 30:7-17. [DOI] [PubMed] [Google Scholar]
- 12.Egener, T., T. Hurek, and B. Reinhold-Hurek. 1999. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant-Microbe Interact. 12:813-819. [DOI] [PubMed] [Google Scholar]
- 13.Egener, T., T. Hurek, and B. Reinhold-Hurek. 1998. Use of green fluorescent protein to detect expression of nif genes of Azoarcus sp. BH72, a grass-associated diazotroph, on rice roots. Mol. Plant-Microbe Interact. 11:71-75. [DOI] [PubMed] [Google Scholar]
- 14.Egener, T., D. E. Martin, A. Sarkar, and B. Reinhold-Hurek. 2001. Role of a ferrodoxin gene cotranscribed with the nifHDK operon in N2 fixation and nitrogenase “switch off” of Azoarcus sp. strain BH72. J. Bacteriol. 183:3752-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedorov, A. S., O. U. Troshina, T. V. Laurinavichene, V. M. Glazer, M. M. Babykin, V. V. Zinchenko, A. F. Yakunin, and A. A. Tsygankov. 1998. Regulatory effect of ammonium on the nitrogenase activity of Rhodobacter sphaeroides and Rhodobacter capsulatus is not mediated by ADP-ribosylation of the Fe-protein of nitrogenase. Microbiology 67:610-615. [Google Scholar]
- 16.Förster, B., K. Maner, F. Fassbinder, and J. Oelze. 1999. Reversible inactivation of nitrogenase in Rhodobacter capsulatus strain W107I deleted in the draTG gene region. FEMS Microbiol. Lett. 170:167-171. [Google Scholar]
- 17.Fu, H., A. Hartmann, R. G. Lowery, W. P. Fitzmaurice, G. P. Roberts, and R. H. Burris. 1989. Posttranslational regulatory system for nitrogenase activity in Azospirillum spp. J. Bacteriol. 171:4679-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halbleib, C. M., Y. Zhang, G. P. Roberts, and P. W. Ludden. 2000. Effects of perturbations of the nitrogenase electron transfer chain on reversible ADP-ribosylation of nitrogenase Fe protein in Klebsiella pneumoniae strains bearing the Rhodospirillum rubrum dra operon. J. Bacteriol. 182:3681-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.He, L., E. Soupene, A. Ninfa, and S. Kustu. 1998. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J. Bacteriol. 180:6661-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, S., C. Kennedy, E. Kavanagh, R. B. Goldberg, and R. Hanau. 1981. Nitrogen fixation gene (nifL) involved in oxygen regulation of nitrogenase synthesis in K. pneumoniae. Nature 290:424-426. [DOI] [PubMed] [Google Scholar]
- 22.Hurek, T., S. Burggraf, C. R. Woese, and B. Reinhold-Hurek. 1993. 16S rRNA-targeted polymerase chain reaction and oligonucleotide hybridization to screen for Azoarcus spp., grass-associated diazotrophs. Appl. Environ. Microbiol. 59:3816-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurek, T., B. Reinhold, I. Fendrik, and E. G. Niemann. 1987. Root-zone-specific oxygen tolerance of Azospirillum spp. and diazotrophic rods closely associated with Kallar grass. Appl. Environ. Microbiol. 53:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurek, T., B. Reinhold-Hurek, M. Van Montagu, and E. Kellenberger. 1994. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J. Bacteriol. 176:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurek, T., M. Van Montagu, E. Kellenberger, and B. Reinhold-Hurek. 1995. Induction of complex intracytoplasmic membranes related to nitrogen fixation in Azoarcus sp. BH72. Mol. Microbiol. 18:225-236. [DOI] [PubMed] [Google Scholar]
- 26.Hurek, T., B. Wagner, and B. Reinhold-Hurek. 1997. Identification of N2-fixing plant- and fungus-associated Azoarcus species by PCR-based genomic fingerprints. Appl. Environ. Microbiol. 63:4331-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack, R., M. De Zamaroczy, and M. Merrick. 1999. The signal transduction protein GlnK is required for NifL-dependent nitrogen control of nif gene expression in Klebsiella pneumoniae. J. Bacteriol. 181:156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, P., and A. J. Ninfa. 1999. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol. 181:1906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of ntr gene transcription in Escherichia coli. Biochemistry 37:12795-12801. [DOI] [PubMed] [Google Scholar]
- 31.Jouanneau, Y., C. Roby, C. M. Meyer, and P. M. Vignais. 1989. ADP-ribosylation of dinitrogenase reductase in Rhodobacter capsulatus. Biochemistry 28:6524-6530. [Google Scholar]
- 32.Kanemoto, R. H., and P. W. Ludden. 1984. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J. Bacteriol. 158:713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessler, P. S., and J. A. Leigh. 1999. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152:1343-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klipp, W., T. Drepper, S. Groβ, and B. Masepohl. 2000. Genetics of nitrogen fixation in Rhodobacter capsulatus: ammonium and molybdenum control of both nitrogenase systems, p. 141-142. In F. O. Pedrosa, M. Hungria, M. G. Yates, and W. E. Newton (ed.), Nitrogen fixation: from molecule to crop productivity. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Y., and M. L. Kahn. 1995. ADP-ribosylation of Rhizobium melilot glutamine synthetase III in vivo. J. Biol. Chem. 70:624-1628. [DOI] [PubMed] [Google Scholar]
- 37.Ludden, P. W., and G. P. Roberts. 1989. Regulation of nitrogenase activity by reversible ADP-ribosylation. Curr. Top. Cell Regul. 30:23-55. [DOI] [PubMed] [Google Scholar]
- 38.Marini, A.-M., S. Vissers, A. Urrestarazu, and B. Andre. 1994. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 13:3456-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin, D., T. Hurek, and B. Reinhold-Hurek. 2000. Occurrence of three PII-like signal transmitter proteins in the diazotroph Azoarcus sp. BH72. Mol. Microbiol. 38:276-288. [DOI] [PubMed] [Google Scholar]
- 40.Meletzus, D., P. Rudnick, N. Doetsch, A. Green, and C. Kennedy. 1998. Characterization of the glnK-amtB operon of Azotobacter vinelandii. J. Bacteriol. 180:3260-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merrick, M., and R. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merrick, M., S. Hill, H. Hennecke, M. Hahn, R. Dixon, and C. Kennedy. 1982. Repressor properties of the nifL gene product in Klebsiella pneumoniae. Mol. Gen. Genet. 185:75-81. [Google Scholar]
- 43.Michel-Reydellet, N., N. Desnoues, C. Elmerich, and P. A. Kaminski. 1997. Characterization of Azorhizobium caulinodans glnB and glnA genes: involvement of the PII protein in symbiotic nitrogen fixation. J. Bacteriol. 179:3580-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ninfa, A. J., and M. R. Atkinson. 2000. PII signal transduction proteins. Trends Microbiol. 8:172-189. [DOI] [PubMed] [Google Scholar]
- 45.Ninnemann, O., J.-C. Jauniaux, and W. B. Frommer. 1994. Identification of a high affinity NH4+ transporter from plants. EMBO J. 13:3464-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierrard, J., P. W. Ludden, and G. P. Roberts. 1993. Posttranslational regulation of nitrogenase in Rhodobacter capsulatus: existence of two independent regulatory effects of ammonium. J. Bacteriol. 175:1358-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pope, M. R., S. A. Murell, and P. W. Ludden. 1985. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenine diphosphoribosylation of a specific arginine residue. Proc. Natl. Acad. Sci. USA 82:3173-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinhold, B., T. Hurek, and I. Fendrik. 1985. Strain-specific chemotaxis of Azospirillum spp. J. Bacteriol. 162:190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinhold, B., T. Hurek, E.-G. Niemann, and I. Fendrik. 1986. Close association of Azospirillum and diazotrophic rods with different root zones of kallar grass. Appl. Environ. Microbiol. 52:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinhold-Hurek, B., J. Dörr, T. Egener, D. Martin, and T. Hurek. 2000. Interactions of diazotrophic Azoarcus spp. with rice, p. 405-408. In F. Pedrosa, M. Hungria, M. G. Yates, and W. E. Newton (ed.), Nitrogen fixation: from molecules to crop productivity. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 51.Reinhold-Hurek, B., and T. Hurek. 1998. Life in grasses: diazotrophic endophytes. Trends Microbiol. 6:139-144. [DOI] [PubMed]
- 52.Reinhold-Hurek, B., T. Hurek, M. Claeyssens, and M. M. Van. 1993. Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J. Bacteriol. 175:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts, G. P., and P. W. Ludden. 1992. Nitrogen fixation by photosynthetic bacteria, p. 135-165. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 54.Soupene, E., L. He, D. Yan, and S. Kustu. 1998. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc. Natl. Acad. Sci. USA 95:7030-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stadtman, E. R. 1990. Discovery of glutamine synthetase cascade. Methods Enzymol. 182:793-809. [DOI] [PubMed] [Google Scholar]
- 56.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas, G., G. Coutts, and M. Merrick. 2000. The glnKamtB operon. Trends Genet. 16:11-14. [DOI] [PubMed] [Google Scholar]
- 58.Van Heeswijk, W. C., S. Hoving, D. Molenaar, B. Stegemann, D. Kahn, and H. V. Westerhoff. 1996. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol. Microbiol. 21:133-146. [DOI] [PubMed] [Google Scholar]
- 59.van Heeswijk, W. C., D. Wen, P. Clancy, R. Jaggi, D. L. Ollis, H. V. Westerhoff, and S. G. Vasudevan. 2000. The Escherichia coli signal transducers PII (GlnB and GlnK) form heterotrimers in vivo: fine tuning the nitrogen signal cascade. Proc. Natl. Acad. Sci. USA 97:3942-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yakunin, A. F., and P. C. Hallenbeck. 1998. Short-term regulation of nitrogenase activity by NH4+ in Rhodobacter capsulatus: multiple in vivo nitrogenase responses to NH4+ addition. J. Bacteriol. 180:6392-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y., R. H. Burris, P. W. Ludden, and G. P. Roberts. 1993. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J. Bacteriol. 175:6781-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y., E. L. Pohlmann, C. M. Halbleib, P. W. Ludden, and G. P. Roberts. 2001. Effect of PII and its homolog GlnK on reversible ADP-ribosylation of dinitrogenase reductase by heterologous expression of the Rhodospirillum rubrum dinitrogenase reductase ADP-ribosyltransferase-dinitrogenase reductase-activating glycohydrolase regulatory system in Klebsiella pneumoniae. J. Bacteriol. 183:1610-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]