Abstract
Expression of the two Bacillus subtilis genes encoding l-asparaginase is controlled by independent regulatory factors. The ansZ gene (formerly yccC) was shown by mutational analysis to encode a functional l-asparaginase, the expression of which is activated during nitrogen-limited growth by the TnrA transcription factor. Gel mobility shift and DNase I footprinting experiments indicate that TnrA regulates ansZ expression by binding to a DNA site located upstream of the ansZ promoter. The expression of the ansA gene, which encodes the second l-asparaginase, was found to be induced by asparagine. The ansA repressor, AnsR, was shown to negatively regulate its own expression.
l-Asparaginase catalyzes the conversion of l-asparagine to l-aspartate and ammonium. In Bacillus subtilis, two different studies indicate that the synthesis of asparaginase is regulated in response to the nutritional status of the cell. In one study, elevated levels of asparaginase were shown to be present in cells growing either in rich medium or in glucose minimal medium containing aspartate or asparagine as the sole nitrogen source (25). In addition, the ansA gene was shown to encode an l-asparaginase. The ansA gene is located in an operon with ansB, which encodes l-aspartase (25). Expression of the ansAB operon is repressed by AnsR, and the activity of AnsR has been proposed to be regulated by either asparagine or aspartate (26).
Asparaginase synthesis was also shown to be elevated in the absence of asparagine or aspartate when cells are grown with a limiting nitrogen source: e.g., glucose minimal medium containing glutamate, proline, or urea as the sole nitrogen source (1). High-level expression of many B. subtilis genes during nitrogen-limited growth is due to regulation by GlnR or TnrA (10). TnrA and GlnR are members of the MerR family of transcription factors. Since the sequences of their DNA-binding domains are highly similar, both proteins bind to DNA sites that have a common consensus sequence (TGT-A-------T-ACA) (10, 34). GlnR represses transcription when cells are grown in the presence of excess nitrogen, while TnrA functions as an activator or repressor of gene expression when nitrogen is limited (3, 23, 30, 31, 35). Although asparaginase expression is activated during nitrogen-limited growth, no TnrA or GlnR binding sites are present in the promoter region of the ansAB operon. This observation raised the possibility that either TnrA or GlnR indirectly regulates ansAB expression or that B. subtilis contains a second asparaginase, the synthesis of which is controlled by TnrA or GlnR. In this report, we demonstrate that ansZ (formerly yccC) encodes a functional l-asparaginase, the expression of which is activated during nitrogen-limited growth by TnrA. In addition, we demonstrate that AnsR regulates both ansAB and its own expression in response to asparagine availability.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists B. subtilis strains used in this study. Derivatives of strains 168, SF168G, and SF62 containing the lacZ transcriptional fusions were constructed with plasmid DNA as previously described (32). Transformation with selection for spectinomycin resistance was used to transfer the ansB::spc and ansZ::spc mutations. The codY mutation was transferred by transformation with selection for the genetically linked spectinomycin resistance gene. Transformants containing the codY mutation were identified by poor growth on Difco sporulation plates. Transformation with selection for erythromycin and tetracycline resistance, respectively, was used to transfer the ansR::erm and ansA::tet mutations.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotypea | Source, reference, or derivativeb |
|---|---|---|
| 168 | trpC2 | This laboratory |
| SF62 | tnrA62::Tn917 trpC2 | 31 |
| PS37 | unkU::spc ΔcodY trpC2 | P. Serror |
| SF168G | ΔglnR57 trpC2 | 30 |
| PS1833 | ansR::erm trpC2 | 26 |
| SF168NR | ansR::erm trpC2 | 168 × PS1833 DNA |
| SF168NZ | ansZ::spc trpC2 | 168 × pANS48 DNA |
| SF168NB | ansB::spc trpC2 | 168 × pANS30 DNA |
| SF168NA | ansA::tet | 168 × pANS29 DNA |
Genotype symbols are those of Biaudet et al. (4), with ansZ (formerly yccC) added.
Strains were derived by transforming the first strain listed with DNA from the second strain or plasmid listed.
Cell growth, media, and enzyme assays.
The methods used for bacterial cultivation in the MOPS (morpholinepropanesulfonic acid) minimal medium of Neidhardt et al. (20) have been described previously (2). Glucose was added to a final concentration of 0.5%. All nitrogen sources were added at 0.2% to minimal growth medium, except where noted.
Extracts for enzyme assays were prepared from cells grown to mid-log growth phase (70 to 90 Klett units). β-Galactosidase was assayed in crude extracts as previously described (2). β-Galactosidase activity was always corrected for endogenous β-galactosidase activity present in B. subtilis cells containing the promoterless lacZ gene from pSFL7 integrated at the amyE site. Cells harvested for l-asparaginase assays were washed twice with buffer I (50 mM KPO4 [pH 8.4], 10 mM EDTA [pH 8.0]) containing 150 mM NaCl and stored at −80 until assayed. Cell extracts were prepared by resuspending the thawed cells in 1 ml of buffer I, incubating the cells with 0.2 mg of lysozyme per ml for 20 min at 4°C, and sonication. After lysis, the cell debris was removed by centrifugation, and the supernatant was used for enzyme assays. l-Asparaginase activity was measured as the l-asparagine-dependent production of ammonium. The cell extracts were added to the reaction mixture (25 mM KPO4 [pH 8.4], 10 mM EDTA [pH 8.0], 20 mM l-asparaginase) and incubated for 20 min at 30°C, and then the ammonium concentrations were determined by using a modified Berthelot reaction (29). One unit of l-asparaginase activity produced 1 μmol of NH4 per min.
Plasmids and lacZ fusions.
Three different subclones of the ansA promoter were constructed. Plasmids pANS4, pANS6, and pANS15 contain, respectively, BspHI-HincII, BspHI-XmnI, and BspHI-MslI DNA fragments from pPS1367 (25) cloned between the NcoI and StuI sites of pLEW424 (32). To construct ansA-lacZ fusions, EcoRI-HindIII ansA promoter fragments from pANS6, pANS4, and pANS15 were cloned into pSFL7 to give pANS10, pANS12, and pANS19, respectively. Plasmid pANS5 contains the ansR promoter and was constructed by inserting a HincII-BspHI DNA fragment from pPS1367 between the Ecl136II and NcoI sites of pLEW424. The ansR-lacZ fusion plasmid pANS25 was constructed by cloning the EcoRI-HindIII ansR promoter fragment from pANS5 into pSFL7.
A 417-bp DNA fragment containing the ansZ promoter region was obtained by PCR amplification of B. subtilis chromosomal DNA with Pwo DNA polymerase by using primers ANSZ1 (5′-TCCATAACTCATAACATTCCCACC) and ANSZ2 (5′-AGCTATCGTGCCTCCTGTCG). Plasmid pANS41 contains a 200-bp MfeI-BsaWI fragment from the PCR-generated DNA cloned into pMTL23P (7). The ansZ promoter DNA in pANS41 was sequenced to confirm the fidelity of the PCR amplification. Plasmid pANS43 contains an XbaI-SphI DNA fragment of the ansZ promoter from pANS41 cloned into pJDC9 (8). An ansZ-lacZ fusion was constructed by cloning an EcoRI-HindIII DNA fragment from pANS43 into pSFL7 (30).
Inactivation of the chromosomal ansA, ansB, and ansZ genes.
A DNA fragment containing the ansA gene was obtained by PCR amplification of B. subtilis chromosomal DNA with primers ANSA2 (5′-TACGGAATTCATACCTTCTCGCAACCCATC) and ANSA3 (5′-CGTAAAGCTTAAGATCCGAAGACTGACCTAGC). Plasmid pANS28 was constructed by cloning an SnaBI-HindIII DNA fragment from the PCR product into pJDC9. Located within the ansA coding sequence of pANS28 is a unique PstI site. Plasmid pANS29 contains a PstI-PstI tetracycline resistance gene cassette from pBEST309 (16) inserted into the PstI site of pANS28. Plasmid pANS30 contains a disruption of the ansB gene and was constructed by inserting a spectinomycin resistance gene cassette into the NsiI site of pPS1304 (25). The chromosomal ansA and ansB genes were disrupted by transforming B. subtilis cells with DNA from plasmids pANS29 and pANS30, respectively.
A DNA fragment containing the ansZ gene was obtained by PCR amplification of B. subtilis DNA with primers ANSZ3 (5′-CGTAGAATTCATATAAAGCAGGTGTTGTCGGCG) and ANSZ4 (5′-TTTTAAGATTCAGAGGTCATTATTGGTCC). Plasmid pANS47 contains an EcoRI-HindIII fragment from the PCR-generated DNA cloned into pJDC9. A unique EcoRV site lies within the ansZ coding sequence. Plasmid pANS48 was constructed by inserting a spectinomycin-resistant EcoRV-HincII DNA fragment from pDG1726 (13) into the EcoRV site of pANS47. The chromosomal ansZ gene was disrupted by transforming B. subtilis cells with pANS48 DNA.
In vitro analysis of DNA binding.
The binding of purified TnrA to the ansZ promoter region utilized a 208-bp XbaI-Acc65I DNA fragment from pANS41. The downstream end of this DNA fragment was labeled by a fill in reaction with Klenow DNA polymerase and [α-32P]dGTP. Gel mobility shift and DNase I footprinting experiments were performed as previously described (34). The binding of TnrA to the ansA promoter region was tested with a 232-bp BamHI-PstI DNA fragment from pANS4.
Primer extension experiments.
RNA was isolated from B. subtilis cells grown to mid-log growth phase (70 to 90 Klett units) by extraction with guanidine thiocyanate and CsCl centrifugation (11). Primer extensions were performed as previously described (11) with two oligonucleotide primers, ANSZ6 (5′-AAAAACAAACAATAGTGCGG) and ANSZ7 (5′-GTGCGGTAAAAAGTACGAGC), which are complementary to the 5′ end of the ansZ coding region.
RESULTS
ansZ encodes a functional l-asparaginase.
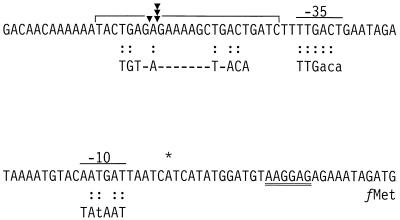
The ansZ gene (previously called yccC) was identified by the B. subtilis genome sequencing project and was proposed to encode an l-asparaginase (18). The deduced amino acid sequence of ansZ shows 59% identity to the l-asparaginase from Erwinia chrysanthemi (19) and 53% identity to l-asparaginase II from Escherichia coli (5). The N-terminal amino acid sequence of YccC has been predicted to function as a lipoprotein signal peptide (27). An inverted repeat sequence with structural similarity to factor-independent transcriptional terminators is located immediately downstream of the ansZ coding region, while the divergently transcribed lmrAB operon is located upstream of ansZ. Together, these observations suggest that ansZ is expressed as a monocistronic transcript. A DNA sequence with similarity to the TnrA binding site consensus sequence is centered 76 nucleotides upstream of the ansZ translational start codon (Fig. 1).
FIG. 1.
Nucleotide sequence of the ansZ promoter region. The consensus sequences for the −10 and −35 regions of σA-dependent promoters (14) and the TnrA binding site (33) are shown below the nucleotide sequence. The uppercase letters in the −10 and −35 consensus sequences are used to indicate bases that are highly conserved in B. subtilis σA-dependent promoters (14). The transcriptional start site is indicated by an asterisk. The putative ribosomal binding site is double underlined, and the translational start codon for ansZ has the label fMet located below it. The region protected from DNase I digestion by TnrA is indicated by the horizontal bracket above the DNA sequence. The DNase I-hypersensitive sites are shown as solid triangles.
To determine whether ansZ encodes a functional l-asparaginase the synthesis of which is nitrogen regulated, the ansZ gene was mutationally inactivated and asparaginase activity was determined for wild-type and ΔansZ cells grown in glucose minimal medium containing either excess (glutamate plus ammonium) or limiting (glutamate) nitrogen. In wild-type cells, asparaginase levels are 10-fold higher in extracts of glutamate-grown cells than in cells grown with glutamate plus ammonium as the nitrogen source (Table 2). These data agree with previously published results indicating that asparaginase expression is nitrogen regulated in B. subtilis (1). The levels of asparaginase present in extracts of glutamate-grown ΔansZ cells are 10-fold lower than that seen in extracts of wild-type cells (Table 2). In contrast, similar levels of l-asparaginase are present in extracts of wild-type and ΔansZ cells grown in medium containing glutamate plus ammonium as the nitrogen source (Table 2). These results indicate that ansZ encodes an l-asparaginase that is preferentially expressed in nitrogen-limited cells. As noted above, the deduced amino acid sequence of AsnZ contains a putative protein secretion sequence. Nonetheless, ansZ-encoded asparaginase activity was detected in soluble cell extracts. No experiments were performed to determine whether AnsZ is also secreted.
TABLE 2.
Asparaginase expression in wild-type or ansA and ansZ mutant strains
| Relevant genotypea | Asparaginase sp act (U/mg of protein) in cells grown onb:
|
|
|---|---|---|
| Excess nitrogen (glutamate + NH4Cl) | Limited nitrogen (glutamate) | |
| Wild type | 5.1 | 51.8 |
| ΔansZ | 3.4 | 5.5 |
| ΔansA ΔansZ | ≤1.6 | ≤1.6 |
All strains are derivatives of strain 168.
Data are the average of three or more determinations, which did not vary by more than 15%. Cultures were grown in MOPS glucose minimal medium containing the indicated nitrogen sources.
Because the B. subtilis ansA gene also encodes an l-asparaginase (25), the level of asparaginase activity was also examined in a ΔansA ΔansZ double mutant. Since no detectable asparaginase activity was present in extracts of the ΔansA ΔansZ double mutant (Table 2), the low levels of asparaginase present in extracts of ΔansZ cells must be due to ansA expression. Thus, ansA and ansZ encode the only asparaginases detectable in B. subtilis under these conditions.
Identification of the ansZ transcriptional start site.
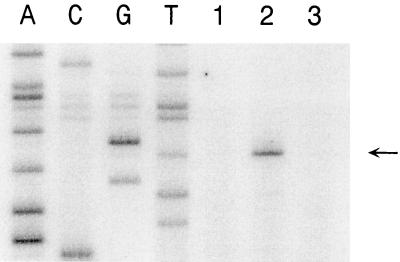
Primer extension analysis was used to identify the apparent transcriptional start site for the ansZ promoter. These experiments used RNA isolated from cells grown in glucose minimal medium containing either limiting (glutamate) or excess (glutamate plus ammonium) nitrogen sources and an oligonucleotide complementary to the ansZ coding region. The primer extension product obtained with RNA from glutamate-grown cells (Fig. 2) corresponds to an mRNA with a 5′ end located 27 bp upstream of the ansZ start codon (Fig. 1). In contrast, no primer extension product was obtained with RNA isolated from cells grown with excess nitrogen (glutamate plus ammonium) (Fig. 2). The transcriptional regulation observed in the primer extension experiments is in agreement with the results obtained with asparaginase assays (Table 2) and with transcriptional ansZ-lacZ fusion studies described below. Examination of the DNA sequence upstream of the transcriptional start site reveals that while the −35 promoter region is highly similar to the σA consensus sequence (14), the −10 region contains two mismatches with highly conserved nucleotides (Fig. 1). This observation suggests that the ansZ promoter is a nonoptimal σA-dependent promoter with a low level of intrinsic transcriptional activity.
FIG. 2.
Primer extension analysis of ansZ. B. subtilis RNA used in the primer extension was isolated from wild-type cells grown in glucose minimal medium containing either glutamate plus ammonium (lane 1) or glutamate (lane 2) as the nitrogen source. S. cerevisiae RNA was used in the primer extension shown in lane 3. The ANSZ7 oligonucleotide primer was used for dideoxynucleotide sequencing of pANS41 (lanes A, C, G, and T) and for primer extensions. The primer extension product is denoted by the arrow. Identical results were obtained with the ANS6 oligonucleotide primer (data not shown).
Identification of the ansZ TnrA binding site.
Gel mobility shift analysis was used to determine whether TnrA directly interacts with the ansZ promoter region. In these experiments, purified TnrA bound to ansZ promoter region DNA with an equilibrium dissociation binding constant (Kd) of 340 nM (data not shown). The affinity of TnrA for its ansZ binding site is significantly less than its affinity for the amtB (previously called nrgA) promoter, which has a Kd of 8 nM (34). The TnrA binding site in the ansZ promoter region has two mismatches with the TnrA consensus sequence (Fig. 1), which may account for the low in vitro affinity of TnrA for this site.
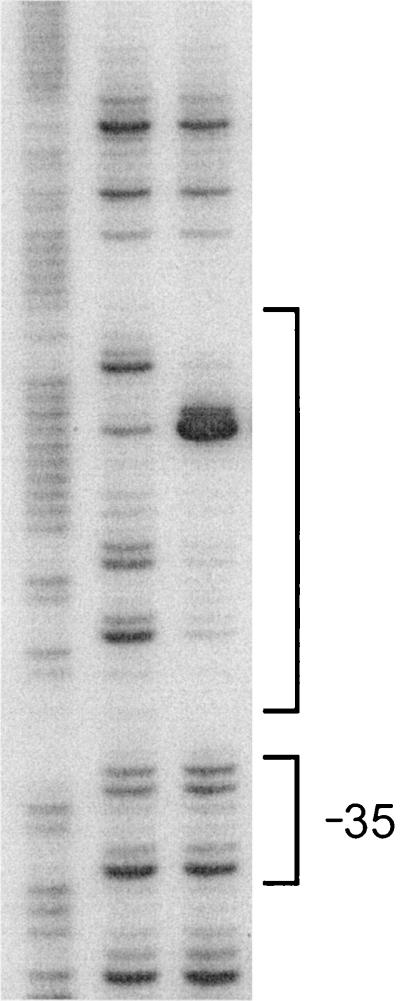
A DNase I protection experiment demonstrated that purified TnrA protects DNA upstream of the ansZ promoter region from DNase I digestion (Fig. 3). This protected region includes the TnrA consensus sequence and extends from position −37 to −70 on the nontemplate strand (Fig. 1). In addition, the binding of TnrA to the ansZ promoter DNA gives rise to DNase I-hypersensitive sites at positions −52 and −53 on the nontemplate strand. DNase I-hypersensitive sites were previously observed at the same relative position in the TnrA footprint of the amtB promoter (34). The TnrA-binding site in the ansZ promoter is centered 49 bp upstream of the transcriptional start site, which is similar to what has been observed for other TnrA-activated genes (33).
FIG. 3.
DNase I footprinting of the ansZ promoter with TnrA. Lane R, G+A molecular size standards. Lanes 1 and 2 were treated with DNase I in the absence (lane 1) or presence (lane 2) of 2.5 μM TnrA. The TnrA-protected region and the −35 promoter region are bracketed.
Nitrogen regulation of ansZ expression.
The expression of ansZ was examined in more detail with the (ansZ-lacZ)45 transcriptional fusion. This lacZ fusion contains an ansZ promoter fragment that extends from −121 to +83 with respect to the transcriptional start site. Since β-galactosidase levels were 3,800-fold higher in cells grown with limiting nitrogen (glutamate) than in extracts of cells grown with excess nitrogen (glutamate plus ammonium) (Table 3), expression of the ansZ-lacZ fusion is nitrogen regulated. The observation that expression of the (ansZ-lacZ)45 fusion is not elevated in a tnrA mutant grown with glutamate as the nitrogen source (Table 3) indicates that TnrA is required for the activation of ansZ expression during nitrogen-limited growth. GlnR does not regulate ansZ expression, because similar levels of ansZ expresion were seen in wild-type and glnR cells grown with either excess or limiting nitrogen (data not shown). The CodY repressor protein regulates the expression of many genes involved in nitrogen metabolism (10, 21). The highest levels of CodY-dependent repression occur in cells grown with amino acids. Two experimental observations indicate that ansZ expression is not subject to nutritional regulation by CodY. First, the addition of 0.2% Casamino Acids to cells growing in glucose minimal medium containing glutamate plus ammonium as the nitrogen source did not alter ansZ-lacZ expression (data not shown). Second, no difference in ansZ-lacZ expression was observed in wild-type and codY cells grown in glucose minimal medium containing glutamate plus ammonium plus 0.2% Casamino Acids as the nitrogen source (data not shown).
TABLE 3.
β-Galactosidase expression of lacZ fusions in wild-type and mutant strains
| lacZ fusiona | Relevant genotype | β-Galactosidase sp act (U/mg of protein) in cells grown onb:
|
||
|---|---|---|---|---|
| Excess nitrogen | Limiting nitrogen
|
|||
| Glutamate + NH4Cl | Aspartate | Glutamate | ||
| (ansZ-lacZ)45 | Wild type | 0.1 | 124 | 382 |
| tnrA | 0.1 | 0.1 | 0.1 | |
| ansR | 0.2 | NDc | 111 | |
| ansR ansB | 0.2 | ND | 269 | |
| ansB | 0.1 | ND | 303 | |
| (amtB-lacZ)416 | Wild type | 0.06 | 119 | 167 |
| (amtB-lacZ)435 | Wild type | 0.05 | 11.7 | 29.4 |
All strains are derivatives of strain 168 containing lacZ fusions integrated as a single copy at the amyE locus. The (amtB-lacZ)416 and (amtB-lacZ)435 lacZ fusions have been described previously as nrgA-lacZ fusions (33).
Data are the average of three or more determinations, which did not vary by more than 20%. Cultures were grown in MOPS glucose minimal medium containing the indicated nitrogen sources.
ND, not determined.
Although aspartate is a limiting nitrogen source, expression of the (ansZ-lacZ)45 fusion was threefold higher in glutamate-grown cultures than in aspartate-grown cultures (Table 3). Two observations indicate that glutamate-grown cultures are more nitrogen limited than aspartate-grown cultures. First, glutamate-grown cells grow more slowly than aspartate-grown cells (Table 4). Second, the level of glutamine synthetase, a key enzyme in nitrogen assimilation, is higher in glutamate-grown B. subtilis cultures than in aspartate-grown cultures (1). Thus, increased expression of the (ansZ-lacZ)45 fusion in glutamate-grown cells compared to aspartate-grown cells most likely results from glutamate being a poorer source of nitrogen than aspartate. To test this hypothesis, the expression of two other TnrA-dependent nitrogen-regulated lacZ fusions was examined. The wild-type amtB promoter in the (amtB-lacZ)416 fusion contains an optimal TnrA binding site (Kd of 8 nM) (34). The TnrA binding site in the (amtB-lacZ)435 fusion contains 2-bp changes that reduce the in vitro TnrA affinity (Kd of 650 nM) and in vivo expression (33, 34). The (amtB-lacZ)416 and 435 fusions have β-galactosidase levels that are 1.4- and 2.5-fold higher, respectively, in glutamate-grown cells than in aspartate-grown cells (Table 3). Thus, the increased level of ansZ expression in glutamate-grown cells compared to aspartate-grown cells most likely results from glutamate being a poorer source of nitrogen than aspartate rather than from some intrinsic property of the ansZ promoter.
TABLE 4.
Culture doubling time for wild-type and mutant strains
| Relevant genotype | Doubling time (min) of cells grown ona:
|
||
|---|---|---|---|
| Glutamate + NH4Cl | Aspartate | Glutamate | |
| Wild type | 65 | 90 | 160 |
| ansR | 62 | NDb | 85 |
| ansR ansB | 65 | ND | 275 |
| ansB | 60 | ND | 315 |
Cells were grown in MOPS minimal medium containing glucose as the carbon source and the indicated nitrogen sources. Doubling times are the averages of two to four independent determinations, which did not vary by more than 25%.
ND, not determined.
Regulation of ansA expression by AnsR.
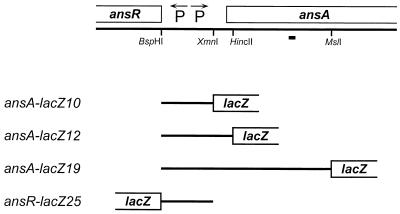
The AnsR-dependent regulation of ansA expression was examined with the (ansA-lacZ)12 transcriptional fusion (Fig. 4). The ansA promoter fragment in this lacZ fusion extends from −118 to +87 with respect to the transcriptional start site. As expected, the (ansA-lacZ)12 fusion is negatively regulated by AnsR, because high-level β-galactosidase expression from the (ansA-lacZ)12 fusion occurs in the ansR mutant during growth on all nitrogen sources (Table 5). The expression of ansA has been proposed to be induced by either asparagine or aspartate (25). Asparagine was found to induce ansA expression, because β-galactosidase levels in wild-type cells are 23- and 40-fold higher, respectively, in cells grown with either asparagine or glutamate plus ammonium plus asparagine as the nitrogen source than in cells grown with glutamate plus ammonium as the nitrogen source (Table 5). Since similar levels of ansA expression occurred in cells grown in medium containing glutamate plus ammonium, glutamate plus ammonium plus aspartate, or aspartate alone as the nitrogen source (Table 5), aspartate does not appear to induce ansA expression. Thus, our data indicate that AnsR regulates ansA expression in response to asparagine availability.
FIG. 4.
Physical structure of the ansR-ansA promoter region. The ansR and ansA promoters are indicated by the letter P with an arrow above it. The black box below the line indicates the putative cre site. The DNA fragments used to construct ansA- and ansR-lacZ fusions are diagrammed at the bottom.
TABLE 5.
β-Galactosidase expression from ansA-lacZ and ansR-lacZ fusions
| lacZ fusiona | Relevant genotype | β-Galactosidase sp act (U/mg of protein) in cells grown onb:
|
|||||
|---|---|---|---|---|---|---|---|
| Excess nitrogen
|
Limiting nitrogen
|
||||||
| Glutamate + NH4Cl | Glutamate + NH4Cl + Asparaginec | Glutamate + NH4Cl + Aspartatec | Asparagine | Aspartate | Glutamate | ||
| (ansA-lacZ)12 | Wild type | 13 | 301 | 16 | 523 | 16 | 10 |
| ansR | 574 | 639 | NDd | 634 | 589 | ND | |
| (ansA-lacZ)10 | Wild type | 1.0 | 76 | 1.6 | ND | ND | 0.9 |
| (ansR-lacZ)25 | Wild type | 4 | 28 | 5 | 49 | 3 | ND |
| ansR | 55 | 55 | ND | 50 | 49 | ND | |
All strains are derivatives of strain 168 containing the indicated lacZ fusions integrated as a single copy at the amyE locus (see Fig. 4).
Data are the average of three or more determinations, which did not vary by more than 15%. Cultures were grown in MOPS glucose minimal medium containing the indicated nitrogen sources.
Minimal medium containing glutamate plus ammonium as nitrogen sources was supplemented with either 0.02% aspartate or 0.02% asparagine.
ND, not determined.
It has been suggested that AnsR represses expression of ansA by binding to an inverted repeat sequence located between nucleotides +9 and +33 (25). To determine if this sequence is required for the AnsR-dependent regulation of ansA, the (ansA-lacZ)10 fusion was constructed (Fig. 4). This lacZ fusion contains an ansA promoter DNA fragment that extends from −118 to +23 and lacks the downstream half of the inverted repeat sequence that is present in the (ansA-lacZ)12 fusion. Since asparagine induces the expression of β-galactosidase from both the (ansA-lacZ)10 and (ansA-lacZ)12 fusions (Table 5), this sequence does not appear to be required for AnsR repression.
Role of carbon and nitrogen transcriptional regulatory factors in ansA expression.
Carbon catabolite repression of many B. subtilis genes is mediated at cis-acting sites called carbon repression elements (cre) (24). The cre sites for several B. subtilis genes are located considerable distances downstream of their transcriptional start sites (6, 12, 17, 32, 37). It has been previously noted that a putative cre site (TGATAACGATTACA) is located 185 bp downstream of the ansA transcriptional start site (15). To see whether this is a functional cre site, β-galactosidase expression from the (ansA-lacZ)19 fusion, which contains this cre site (Fig. 4), was examined in cells grown in minimal medium containing glutamine as the nitrogen source and either excess (glucose) or limited (citrate) carbon. Since carbon limitation did not elevate ansA expression in either uninduced or asparagine-induced cultures (data not shown), ansA expression does not appear to be subject to regulation by carbon catabolite repression.
The observation that similar levels of β-galactosidase expression from the (ansA-lacZ)12 fusion were seen in wild-type cells grown with the excess (glutamate plus ammonium) and limiting (glutamate) nitrogen indicates that TnrA and GlnR most likely do not regulate ansA expression (data not shown). Moreover, no significant binding of TnrA to the ansA promoter region was observed in gel mobility shift experiments (data not shown). There is also no evidence that CodY regulates ansA expression. The addition of 0.2% Casamino Acids to medium containing glutamate plus ammonium as the nitrogen source did not repress ansA expression. Similar levels of ansA expression were seen in wild-type and codY cells grown with glutamate plus ammonium plus 0.2% Casamino Acids as the nitrogen source (data not shown).
AnsR regulates its own expression.
The AnsR protein belongs to a family of transcriptional repressors which includes some phage repressors that are known to be autogenously regulated (28). The (ansR-lacZ)25 fusion (Fig. 4) was used to examine the regulation of ansR expression. β-Galactosidase expression from the (ansR-lacZ)25 fusion is induced by asparagine in wild-type cells and is derepressed in an ansR mutant (Table 5). These results indicate that AnsR regulates its own expression in response to asparagine availability.
Role of AnsR in ansZ expression.
The ability of AnsR to regulate ansZ expression in response to asparagine availability was examined. Two experimental observations argue that the expression of ansZ is not induced by asparagine. First, β-galactosidase expression from the (ansZ-lacZ)45 fusion is not altered by addition of 0.02% asparagine to glucose minimal medium containing glutamate plus ammonium as the nitrogen source (data not shown). Second, (ansZ-lacZ)45 cells grown with either glutamate plus ammonium or asparagine as the nitrogen sources contain the same low levels of β-galactosidase (Table 3) (data not shown). However, when expression of the (ansZ-lacZ)45 fusion was examined in an ansR mutant strain, a surprising result was obtained. While the ansR mutation had no effect on ansZ expression under conditions of nitrogen excess, the level of ansZ expression in glutamate-grown cells is 3.5-fold lower in the ansR mutant than in the wild-type strain (Table 3).
There are two possible explanations for the altered ansZ expression seen in the ansR mutant during nitrogen-limited growth. First, AnsR might function as a transcriptional activator of ansZ expression during nitrogen-limited growth. Alternatively, the ansR mutation could indirectly alter ansZ expression under these growth conditions. The latter hypothesis is supported by the observation that the ansR mutant, which has higher levels of asparaginase and aspartase than wild-type cells, grows more rapidly than the wild-type cells in glutamate-grown cultures (Table 4). In contrast, wild-type and ansR cultures grew with similar doubling times when glutamate plus ammonium was the nitrogen source (Table 4). Since the amino group of glutamate can be readily transferred to aspartate by transamination, the elevated levels of aspartase in the ansR mutant may be responsible for altered ansZ expression in glutamate-grown ansR cells. To test this hypothesis, ansZ expression was examined in ansR ansB and ansB mutant strains. During growth in medium containing glutamate as the sole nitrogen source, the expression of the (ansZ-lacZ)45 fusion is 2.5- to 3-fold higher, respectively in the ansR ansB and ansB mutants than in the ansR strain (Table 3). Moreover, the ansR ansB and ansB cultures grow more slowly than the wild-type cultures with glutamate as the sole nitrogen source (Table 4). All of this indicates that lower levels of ansZ expression in glutamate-grown ansR cells result from overexpression of aspartase and that aspartase plays a significant role in the utilization of glutamate as a nitrogen source in B. subtilis.
DISCUSSION
Two genes, ansA and ansZ, encode functional l-asparaginase enzymes in B. subtilis. The expression of these two genes is regulated in response to different nutritional signals. When cells are grown in the presence of inducing amounts of asparagine, AnsR repression of the ansAB operon is relieved and AnsA is expressed at high levels. Since the ansAB operon encodes both l-asparaginase and l-aspartase, high-level ansAB expression causes asparagine to be catabolized to fumarate and two molecules of ammonium. In contrast, expression of the monocistronic ansZ gene is activated during nitrogen-limited growth by the nitrogen regulatory factor TnrA. Under these growth conditions, asparagine would be degraded to ammonium and aspartate by l-asparaginase. Other microorganisms are known to contain multiple l-asparaginases. Both Escherichia coli and Klebsiella aerogenes contain two l-asparaginases, a high-affinity periplasmic enzyme and a low-affinity cytoplasmic enzyme (22). In E. coli, synthesis of the cytoplasmic asparaginase I is constitutive, while expression of the periplasmic asparaginase II is activated during anaerobiosis, where fumarate generated from asparagine degradation functions as an electron acceptor. In contrast, the K. aerogenes periplasmic asparaginase is subject to nitrogen regulation. Two l-asparaginases are also present in Saacharomyces cerevisiae (9). The cytoplasmic asparaginase I is synthesized constitutively, while expression of extracellular asparaginase II is elevated during nitrogen-limited growth (9). Interestingly, the amino acid sequence of the B. subtilis AnsA enzyme closely resembles the sequence of E. coli asparaginase I, while the sequence of AnsZ is more similar to that of E. coli asparaginase II. There is no evidence that expression of the B. subtilis asparaginase genes is elevated under anaerobic conditions (36).
During growth on minimal medium containing an amino acid as the sole nitrogen source, ammonium must be generated by amino acid catabolism, because glutamine synthesis requires ammonium. If degradation of an amino acid used as the sole nitrogen source produces ammonium inefficiently, the growth rate of the culture slows due to nitrogen limitation. For instance, the doubling time of B. subtilis cells grown in glucose minimal medium containing glutamate as the sole nitrogen source is considerably longer than that of cultures growing with both glutamate and ammonium as nitrogen sources (Table 4). The observation that the doubling time of a glutamate-grown ansB strain, which lacks aspartase, is significantly longer than that of a wild-type strain (Table 4) argues that aspartase plays a major role in ammonium generation in glutamate-grown cells. This pathway most likely involves the conversion of glutamate to aspartate by glutamate-oxaloacetate transaminase, followed by the degradation of aspartate to fumarate and ammonium by aspartase. Since ansB cells still are able to grow with glutamate as the sole nitrogen source, albeit at a slower rate than wild-type cells, other pathways for generating ammonium from glutamate must also be present in B. subtilis.
The role of aspartase in the generation of ammonium from glutamate provides an explanation for the reduced ansZ expression seen in the ansR mutant (Table 3). Since the ansR mutant contains higher levels of aspartase than wild-type cells, ammonium would be generated from glutamate at a faster rate in ansR cells than in wild-type cells. The increased flux of nitrogen from glutamate to ammonium in glutamate-grown ansR cells would partially relieve nitrogen limitation, resulting in faster growth rates (Table 4) and lower levels of ansZ expression (Tables 3). Since introduction of the ansB mutation into the ansR strain blocks production of ammonium from glutamate via aspartate, growth of the ansB ansR double culture becomes more nitrogen-limited than that of the ansR culture. As a result, ansZ is expressed at higher levels in glutamate-grown ansB ansR cells than in ansR cells.
While aspartate and glutamate are limiting nitrogen sources, TnrA-dependent promoters are not activated in aspartate-grown cells to the same level as glutamate-grown cells (Table 3). As noted above, this difference reflects the fact that aspartate-grown cells are not as limited for nitrogen as glutamate-grown cells. It is noteworthy that the relative levels of expression in aspartate-grown cells compared to that in glutamate-grown cells are not the same for all TnrA-activated promoters. The wild-type amtB promoter is expressed in aspartate-grown cells at 70% of the level seen in glutamate-grown cells. In contrast, the ansZ and amtB435 promoters are expressed at relatively lower levels (32 and 40%, respectively) in aspartate-grown cells than in glutamate-grown cells (Table 3). These differences in expression reflect the affinity of these promoters for TnrA. The wild-type amtB promoter has a much higher affinity for TnrA than the ansZ and amtB435 promoters. Thus, in aspartate-grown cells, promoters with high-affinity TnrA-binding sites are activated to a greater extent than promoters with low affinity for TnrA. These differences in the affinity of various promoters for TnrA would result in a graded response to nitrogen availability and raise the possibility that during a shift to nitrogen-limited conditions, some TnrA-activated promoters would be expressed before others.
Acknowledgments
We are grateful to Peter Setlow for strain PS1833 and plasmids pPS1304 and pPS1367. We thank Jaclyn Brandenburg and Erin Bitner for providing technical assistance.
This research was supported by Public Health Service research grant GM51127 from the National Institutes of Health.
REFERENCES
- 1.Atkinson, M. R., and S. H. Fisher. 1991. Identification of genes and gene products whose expression is activated during nitrogen-limited growth in Bacillus subtilis. J. Bacteriol. 173:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky, B. R., L. V. Wray, Jr., S. H. Fisher, D. E. Bohannon, and A. L. Sonenshein. 2000. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J. Bacteriol. 182:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biaudet, V., F. Samson, C. Anagnostopoulos, S. D. Erlich, and P. Bessières. 1996. Computerized map of Bacillus subtilis. Microbiology 142:2669-2729. [DOI] [PubMed] [Google Scholar]
- 5.Bonthron, D. T. 1990. l-Asparaginase II of Escherichia coli K-12: cloning, mapping and sequencing of the ansB gene. Gene 91:101-105. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, E. M., B. W. Beall, and C. P. Moran, Jr. 1996. A σE-dependent operon subject to catabolite repression during sporulation in Bacillus subtilis. J. Bacteriol. 178:4778-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155-164. [DOI] [PubMed] [Google Scholar]
- 9.Dunlop, P. C., G. M. Meyer, D. Ban, and R. J. Roon. 1978. Characterization of two forms of asparaginase in Saccharomyces cerevisiae. J. Biol. Chem. 253:1297-1304. [PubMed] [Google Scholar]
- 10.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, S. H., and L. V. Wray, Jr. 1989. Regulation of glutamine synthetase in Streptomyces coelicolor. J. Bacteriol. 171:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy, F. J., A. J. Turinsky, and T. M. Henkin. 1994. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J. Bacteriol. 176:4527-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 14.Helman, J. D. 1995. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hueck, C. J., W. Hillen, and M. H. Saier. 1994. Analysis of cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res. Microbiol. 145:503-518. [DOI] [PubMed] [Google Scholar]
- 16.Itaya, M. 1992. Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci. Biotech. Biochem. 56:685-686. [DOI] [PubMed] [Google Scholar]
- 17.Kraus, A., C. Hueck, D. Gärtner, and W. Hillen. 1994. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J. Bacteriol. 176:1738-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumano, M., A. Tamakoshi, and K. Yamane. 1997. A 32 kb nucleotide sequence from the region of the lincomycin-resistance gene (22°-25°) of the Bacillus subtilis chromosome and identification of the site of the lin-2 mutation. Microbiology 143:2775-2782. [DOI] [PubMed] [Google Scholar]
- 19.Minton, N. P., H. M. S. Bullman, M. D. Scawen, T. Atkinson, and H. J. Gilbert. 1986. Nucleotide sequence of the Erwinia chrysanthemi NCPPB 1066 l-asparaginase gene. Gene 46:25-35. [DOI] [PubMed] [Google Scholar]
- 20.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratnyke-Lecamwassam, M., P. Serror, K.-W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reitzer, L. J. 1996. Sources of nitrogen and their utilization. p. 380-390. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 23.Schreier, H. J., S. W. Brown, K. D. Hirschi, J. F. Nomellini, and A. L. Sonenshein. 1989. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J. Mol. Biol. 210:51-63. [DOI] [PubMed] [Google Scholar]
- 24.Stülke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Sun, D., and P. Setlow. 1991. Cloning, nucleotide sequence, and expression of the Bacillus subtilis ans operon, which codes for l-asparaginase and l-aspartase. J. Bacteriol. 173:3831-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun, D., and P. Setlow. 1993. Cloning and nucleotide sequence of the Bacillus subtilis ansR gene, which encodes a repressor of the ans operon coding for l-asparaginase and l-aspartase. J. Bacteriol. 175:2501-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldor, M. K., E. J. Rubin, G. D. N. Pearson, H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTXϕ are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]
- 29.Wong, B. L., and C. R. Shobe. 1974. Single-step purificationof urease by affinity chromatography. Can. J. Microbiol. 20:623-630. [DOI] [PubMed] [Google Scholar]
- 30.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wray, L. V., Jr., A. E. Ferson, K. Rohrer, and S. H. Fisher. 1996. TnrA, a transcriptional factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:8841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wray, L. V., Jr., F. K. Pettengill, and S. H. Fisher. 1994. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J. Bacteriol. 176:1894-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wray, L. V., Jr., J. M. Zalieckas, A. E. Ferson, and S. H. Fisher. 1998. Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabP promoter regions. J. Bacteriol. 180:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2000. Purification and in vitro activities of the Bacillus subtilis TnrA transcription factor. J. Mol. Biol. 300:29-40. [DOI] [PubMed] [Google Scholar]
- 35.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2001. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 107:427-435. [DOI] [PubMed] [Google Scholar]
- 36.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng, X., A. Galiner, and H. H. Saxild. 2000. Catabolite repression of dra-nupC-pdp operon expression in Bacillus subtilis. Microbiology 146:2901-2908. [DOI] [PubMed] [Google Scholar]