Abstract
The interaction of Streptococcus pyogenes (group A streptococcus [GAS]) with its human host requires several surface proteins. In this study, we isolated mutations in a gene required for the surface localization of protein F by transposon mutagenesis of the M6 strain JRS4. This gene (srtA) encodes a protein homologous to Staphylococcus aureus sortase, which covalently links proteins containing an LPXTG motif to the cell wall. The GAS srtA mutant was defective in anchoring the LPXTG-containing proteins M6, protein F, ScpA, and GRAB to the cell surface. This phenotype was complemented when a wild-type srtA gene was provided in trans. The surface localization of T6, however, was unaffected by the srtA mutation. The M1 genome sequence contains a second open reading frame with a motif characteristic of sortase proteins. Inactivation of this gene (designated srtB) in strain JRS4 affected the surface localization of T6 but not M6, protein F, ScpA, or GRAB. This phenotype was complemented by srtB in trans. An srtA probe hybridized with DNA from all GAS strains tested (M types 1, 3, 4, 5, 6, 18, 22, and 50 and nontypeable strain 64/14) and from streptococcal groups C and G, while srtB hybridized with DNA from only a few GAS strains. We conclude that srtA and srtB encode sortase enzymes required for anchoring different subsets of proteins to the cell wall. It seems likely that the multiple sortase homologs in the genomes of other gram-positive bacteria have a similar substrate-specific role.
Streptococcus pyogenes (group A streptococcus [GAS]) is a gram-positive pathogen capable of causing a wide variety of diseases (13). The majority of these are mild, suppurative infections, including pharyngitis and pyoderma, which are sometimes followed by serious sequelae such as rheumatic fever, acute glomerulonephritis, and reactive arthritis. More recently, this organism has become notorious for its ability to cause severe invasive diseases with high mortality rates. These include streptococcal toxic shock syndrome, septicemia, and the “flesh-eating” disease necrotizing fasciitis. The pathogenesis of GAS infections is attributed to the ability of these organisms to produce a wide array of virulence factors, including secreted toxins and superantigens, which are responsible for many of the symptoms characteristic of GAS disease and cell-associated molecules that are used to adhere to and interact with the host.
Surface proteins of gram-positive bacteria are attached to the cell surface by one of several mechanisms (11). Some surface proteins are noncovalently attached to teichoic and lipoteichoic acids, anchored directly to the cytoplasmic membrane, or covalently attached to the cell wall cross-bridge. In GAS, many surface proteins appear to be covalently anchored to the cell wall, although others, such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and α-enolase, are anchored by an as yet unidentified mechanism. Covalent attachment of proteins to the cell wall cross-bridge is catalyzed by the enzyme sortase, whose mechanism of action was recently elucidated using Staphylococcus aureus protein A as a model system (39, 40, 65, 66).
Protein A and other sortase-anchored proteins have a conserved LPXTG motif near their C terminus, followed by a hydrophobic stretch of amino acids and a small, positively charged tail (17). During secretion via the Sec system, the hydrophobic region and charged tail retain the protein in the cytoplasmic membrane and prevent its release into the medium. This allows sortase to cleave the protein between the threonine and glycine residues of the LPXTG motif, and the carboxyl group of threonine is then linked to the free amino group of the pentaglycine cross-bridge via a two-step transpeptidation reaction (43, 67). The universal nature of this process is reflected by the ability of S. aureus to efficiently anchor protein A fused to the cell wall sorting signals of any of several other proteins from gram-positive bacteria (58).
The sequence of sortase from S. aureus has been used to identify homologs in the genomes of several other gram-positive bacteria (27, 47). These proteins possess a motif (TLXTC) that has been shown to be at the active site in the S. aureus enzyme. Substitution of the cysteine residue with an alanine abolishes the ability of this enzyme to anchor protein A to the cell wall (65). This explains the observation that sulfhydryl-modifying reagents can inhibit the sortase reaction (67).
Potential sortase substrates in GAS include its major virulence factor, M protein (23), which is responsible for resistance to phagocytosis (33) and binding to fibrinogen (55, 70); the fibronectin-binding proteins protein F (18, 61) and Fba (64); a protein G-related α2-macroglobulin-binding protein (GRAB) (54); C5a peptidase (ScpA) (7), an enzyme responsible for cleaving the complement-driven chemotaxin C5a (8); two recently described collagen-like proteins, Scl1 and Scl2, that may function as adhesins (34, 35, 52, 53, 69); and the trypsin-resistant protein T6, which is the target of the T serotyping scheme (57).
In this study, we used a transposon insertion approach to isolate mutants and screened these with a monoclonal antibody for mutations affecting the ability of GAS strain JRS4 to localize proteins to the cell surface. We assayed the surface localization of M6, protein F, ScpA, GRAB, and T6 to characterize these mutants.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strains DH5α and JM109 were used as hosts for molecular cloning experiments. GAS strain JRS4 is a spontaneous streptomycin-resistant derivative of D471 (59). JRS145 (6) and SAM1 (18) are isogenic emm and prtF derivatives of JRS4, respectively, and were used as negative controls for M protein and protein F assays, respectively. Plasmid pJRS973 (M. J. Federle and J. R. Scott, unpublished data) was used as a source of the transposon TnSpc (37). This plasmid contains TnSpc in a vector that is unable to replicate in GAS. DNA fragments used for complementation analysis were cloned into pNZ276 under the Lactococcus lactis lacA promoter (15, 51).
Culture conditions.
E. coli strains were grown in Luria-Bertani (LB) medium, and GAS strains were grown in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY) at 37°C. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml for E. coli; spectinomycin, 50 μg/ml; streptomycin, 1,000 μg/ml for GAS; chloramphenicol, 20 μg/ml for E. coli and 5 μg/ml for GAS.
DNA manipulation.
Chromosomal DNA was extracted from GAS using the MasterPure DNA extraction kit (Epicentre). Plasmid DNA was isolated from E. coli using anion exchange columns (Qiagen) or Qiaprep spin columns (Qiagen). Probes for Southern hybridization were generated by PCR using primers SRTA-F6 (5′-CAAACCTATCCGAAATACATTAATTGCTCG-3′) and SRTA-R6 (5′-CTGTTTTTAGTTCTCCTTTGACAATAATACG-3′) (srtA); SRTB-F3 (5′-GGTGTGGCAAAAGGCTAAGG-3′) and SRTB-R3 (5′-GCACACACTACTTCTGCCC-3′) (srtB); and SPEC-S1 (5′-ATGTTTGGATCAGGAGTTGAGA-3′) and SPEC-A1 (5′-GTGTTTCCACCATTTTTTCAAT-3′) (aad9). These probes were labeled using the Decaprime labeling kit (Ambion).
Transposon mutagenesis and screening.
A transposon insertion library was constructed by electroporating pJRS973 into JRS4 and selecting for spectinomycin resistance. This library was screened with a monoclonal immunoglobulin M (IgM) antibody (2-2F4) raised against a GAS surface protein, GAPDH, produced in E. coli (14). We found that this antibody causes cultures of JRS4 to aggregate and fall out of solution. This phenomenon was found to be specific for protein F on JRS4, since the wild-type strain and an emm derivative (JRS145) precipitate normally, whereas an isogenic prtF mutant strain (SAM1 [18]) failed to immunoprecipitate (data not shown). Therefore, we used this antibody to enrich and screen for mutants defective in the surface localization of protein F.
Colonies from each TnSpc library were scraped into phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 5.4 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]) to give a concentration of approximately 100 colonies/ml. The cells were harvested by centrifugation (6,500 × g, 15 min), resuspended in the same volume of PBS containing 2% (wt/vol) bovine serum albumin (BSA; Sigma) and 10% (vol/vol) normal goat serum (Sigma) and incubated at 4°C for 1 h with end-over-end mixing to block nonspecific protein- and immunoglobulin-binding sites. The cells were again recovered by centrifugation and resuspended in PBS, and the 2-2F4 monoclonal antibody was added to a final concentration of 1:100 to immunoprecipitate cells with protein F on their surface. The cells were incubated at 4°C with end-over-end mixing for 1 h, and immunoprecipitated cells were removed by low-speed centrifugation (100 × g, 2 min). The remaining cells were subsequently recovered by centrifugation at a higher speed (6,500 × g, 15 min). This process was repeated three to four times until the majority of cells expressing protein F had been removed. The nonprecipitated cells were then plated onto THY agar with spectinomycin. Overnight cultures of individual spectinomycin-resistant colonies were screened to confirm the protein F-negative phenotype by a scaled-down version of the same method in 96-well plates. The number of TnSpc insertions in each strain was determined by Southern hybridization using the aad9 gene (encoding spectinomycin resistance) from pJRS973 as a probe.
TnSpc insertion sites were mapped by arbitrary PCR essentially as described previously (46). The first round of PCR was performed with primers ARB2-2 (5′-GCCGACCGCTGGACTGTACGNNNNNNNNNNGTAGC-3′) and OUT3 (5′-GCGTGCCTACACGTGTCG-3′). The second round of PCR utilized 5 μl of the first-round product as the template DNA and primers ARB2-1 (5′-GCCGACCGCTGGACTGTACG-3′) and OUT1 (5′-GTCCTCCTGGGTATGTTTTT-3′). Second-round PCR products were gel purified and sequenced using primer SEQ1 (5′-GTACCGTAAAAGGACTGTTATATGGCC-3′) at the Microchemical Facility, Emory University. The position of TnSpc insertion into the JRS4 chromosome was determined by searching the GAS M1 genome sequence at http://www.genome.ou.edu/strep_blast.html.
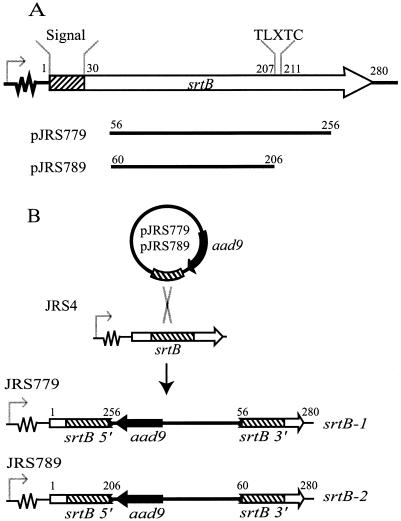
Construction of pJRS779 and pJRS789 for insertional inactivation of srtB.
The srtB-1 and srtB-2 alleles were constructed by single-crossover mutagenesis using plasmids pJRS779 and pJRS789, respectively (Fig. 1). For construction of pJRS779, a PCR fragment containing an internal region of srtB was amplified using the high-fidelity enzyme Herculase (Stratagene) and primers SRTB-F3 and SRTB-R3. The srtB PCR fragment was blunt end ligated into EcoRV-digested pZErO-2.1 (Invitrogen) to create pJRS778. The aad9 gene from pAH256 (21) was isolated as an EcoRI-HindIII fragment and cloned into EcoRI- and HindIII-digested pJRS778, producing pJRS779. For construction of pJRS789, an internal fragment of srtB was amplified by PCR using primers SRTB-F5 (5′-GCAAAAGCTTAAGGCGTATAACGCTAGGC-3′) and SRTB-R5 (5′-CGCAGAATTCTAATCTTTCCCCATGACTCC-3′). The resulting fragment was digested with HindIII and EcoRI and cloned into pUCSpec (26) digested with the same enzymes.
FIG. 1.
Construction of srtB-1 and srtB-2 alleles. (A) Internal fragments of srtB were amplified by PCR and cloned into pZErO-2.1 (pJRS779) and pUCSpec (pJRS789). Numbers above the srtB open reading frame and the fragments cloned into pJRS779 and pJRS789 refer to the locations of encoded amino acid residues of the JRS4 srtB gene. (B) Integration of pJRS779 and pJRS789 into the chromosome of JRS4 results in one copy of srtB (srtB 5′) encoding a protein truncated after amino acid 256 (pJRS779) or 206 (pJRS789) and a second copy (srtB 3′) lacking a promoter and the first 55 (pJRS779) or 59 (pJRS789) amino acids. The signal sequence of SrtB is indicated by a striped box. Open and solid arrows represent srtB and aad9, respectively. The srtB promoter is indicated with a bent arrow preceding a region of unknown length. The internal region of srtB cloned into pJRS779 and pJRS789 is indicated by a striped box.
To inactivate srtB, pJRS779 and pJRS789 were introduced into JRS4 by electroporation and plated on THY agar, with selection for spectinomycin resistance. From each transformation, one spectinomycin-resistant colony resulting from insertion of pJRS779 or pJRS789 into the chromosome of JRS4 and inactivation of srtB was isolated and named JRS779 and JRS789, respectively. Inactivation of srtB in JRS779 (srtB-1) and JRS789 (srtB-2) was confirmed by PCR across the insertion site with primers SRTB-F4 (5′-CGCTGCAGGTAGTCAGTGGAATGCTATCGTGATCG-3′), M13EXT-F (5′-TTTCACACAGGAAACAGCTATGACCATG-3′), SRTB-R4 (5′-CGATGCATGTAACCGAAGGAGCTCTTCTGTACG-3′), and M13EXT-R (5′-GTCACGACGTTGTAAAACGACGGCCAGT-3′). These insertions were also confirmed by Southern hybridization using a PCR product generated with primers SRTB-F3 and SRTB-R3 as a probe.
Construction of plasmids for complementation of srtA and srtB.
The srtA and srtB genes were amplified by PCR using Herculase and primers SRTA-F2 (5′-CCTTGGTTCAGCCTGCAGACAGTAGTATCGC-3′) and SRTA-R3 (5′-GTAGGATCCGATAAATTTTCTCTATGGTCC-3′) (srtA) and SRTB-F7 (5′-GCTTAATGCCTGCAGAAGTCGAAATCAATCGAGACTG-3′) and SRTB-R4 (srtB). The srtA PCR product was digested with PstI and cloned into pNZ276 (51) that had been digested with PstI and MscI, creating pJRS757. The srtB PCR product was digested with PstI and EcoRV and cloned into pNZ276 that had been digested with PstI and MscI to create pJRS797.
Cell fractionation.
Culture supernatant and cell wall protein fractions were prepared from JRS4 and derivatives as described previously (4). Protein extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 12% gradient gels (Invitrogen) and stained for total protein (SimplyBlue stain; Invitrogen) or transferred to a nitrocellulose membrane (Hybond C; Amersham) for immunoblot analysis.
Western immunoblot and protein-binding studies.
Whole-cell dot blots and detection of proteins with antisera and digoxigenin (DIG)-labeled proteins were performed as described previously (4). The following concentrations of each reagent were used: M6, 1:2,000 dilution of monoclonal antibody 10B6 (32); T6, 1:250 dilution of anti-T6 polyclonal antibody (57); ScpA, 1:1,000 dilution of anti-ScpA antibody (30); protein F (18), 1 μg of DIG-labeled fibronectin per ml; and GRAB (54), 1 μg of DIG-labeled α2-macroglobulin per ml. For detection of ScpA on whole-cell dot blots, cells were incubated at room temperature for 2 h in 10 mM Tris-HCl-160 mM NaCl-2% sodium dodecyl sulfate (pH 8.0) prior to spotting onto nitrocellulose membrane to remove non-cell-wall-anchored protein.
RESULTS
Isolation of mutations in srtA by transposon mutagenesis.
To isolate mutants defective in the surface expression of protein F, we used the transposon TnSpc, a derivative of Tn4001 that inserts into the genome of gram-positive organisms, including GAS, with a high degree of randomness (36, 37). The transposon was introduced into JRS4 on plasmid pJRS973 in two independent transformation experiments, resulting in approximately 6 to 8,000 independent spectinomycin-resistant colonies. The colonies from each experiment were pooled, enriched by five cycles of immunoprecipitation with monoclonal antibody 2-2F4 (14) and screened for protein F mutants (see Materials and Methods). A total of 30 mutants from the first transformation and 18 mutants from the second transformation were selected for further analysis.
Clumping of GAS cells in liquid culture is usually associated with the presence of M protein on the bacterial cell surface. All 30 mutants from the first transformation failed to clump as rapidly as the wild type following overnight growth in liquid medium. Southern blot analysis revealed two different classes of mutants in this library, one with a single insertion of TnSpc and one with at least two insertions. A combination of arbitrary PCR and direct sequencing of PCR products revealed that all mutants with a single insertion had the transposon inserted in a gene of unknown function immediately downstream of gyrA, which corresponds to open reading frame Spy1154 in the recently annotated GAS M1 genome (16). Due to the nature of the mutagenesis strategy, it is likely that these mutants are all siblings. One of these mutants, selected for further study, was named JRS758. The mutants with two or more insertions appeared to have one transposon in a region encoding a macrolide efflux pump (either Spy0549 or downstream of Spy0543 in the M1 genome) and were not characterized further. All 18 mutants from the second transformation clumped normally and had a single mutation in prtF, encoding protein F.
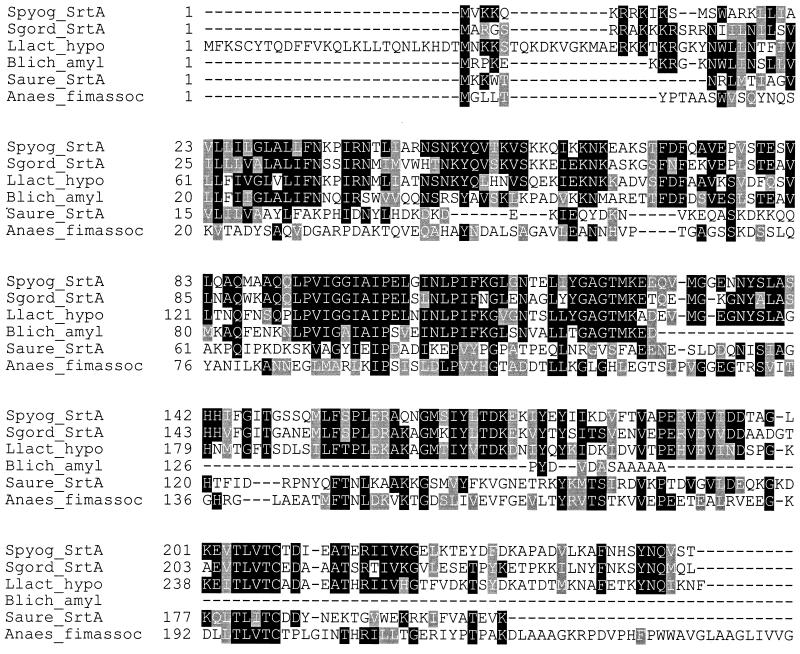
The TnSpc insertion in Spy1154 was located between bases 84 and 85 of the predicted open reading frame. Comparison of the predicted 249 amino acids encoded by this gene against the DDBJ/EMBL/GenBank databases identified a 127-amino-acid region in the N terminus of the predicted protein with a high degree of homology to α-amylase from Bacillus licheniformis (49.61% identity, 72.44% similarity) and homology throughout the entire protein with sortase from Streptococcus gordonii (57.94% identity, 75.4% similarity), a hypothetical protein from L. lactis (48.96% identity, 66.67% similarity), a putative fimbria-associated protein from Actinomyces naeslundii (11.73% identity, 28% similarity), and sortase (SrtA) from S. aureus (19.6% identity, 41.6% similarity) (Fig. 2). Like the S. aureus and S. gordonii srtA genes, Spy1154 encodes a potential 39-amino-acid signal peptide and a region containing a TLXTC motif (amino acids 204 to 208), which is conserved among sortase homologs from gram-positive bacteria. Based on its homology to the srtA genes from S. aureus and S. gordonii, we named Spy1154 srtA.
FIG. 2.
Multiple sequence alignment of SrtA with homologous proteins identified by Blast search. The alignment was performed using ClustalW. The GenBank accession numbers for the sequences are as follows: S. pyogenes (Spyog) SrtA, AAK34025; S. gordonii (Sgord) SrtA, AAG41778; L. lactis (Llact) hypO, AAK05211; B. licheniformis (Blich) Amyl, AAA73122; S. aureus (Saure) SrtA, AAD48437; and A. naeslundii (Anaes) fimbria-associated protein (fimassoc), AAC13546 (truncated after amino acid 251).
Mutation of srtA affects the cell wall anchoring of a subset of GAS surface proteins.
Since JRS758 was not immunoprecipitated by monoclonal antibody 2-2F4 and overnight cultures of this strain in liquid medium failed to clump, it seemed likely that the TnSpc insertion in srtA affected the surface localization of proteins containing an LPXTG motif. A search of the M1 genome sequence identified 12 proteins containing this motif sufficiently near the C terminus to be involved in cell wall anchoring (data not shown) (28). Some of these have been characterized previously, and suitable assays were available for their detection.
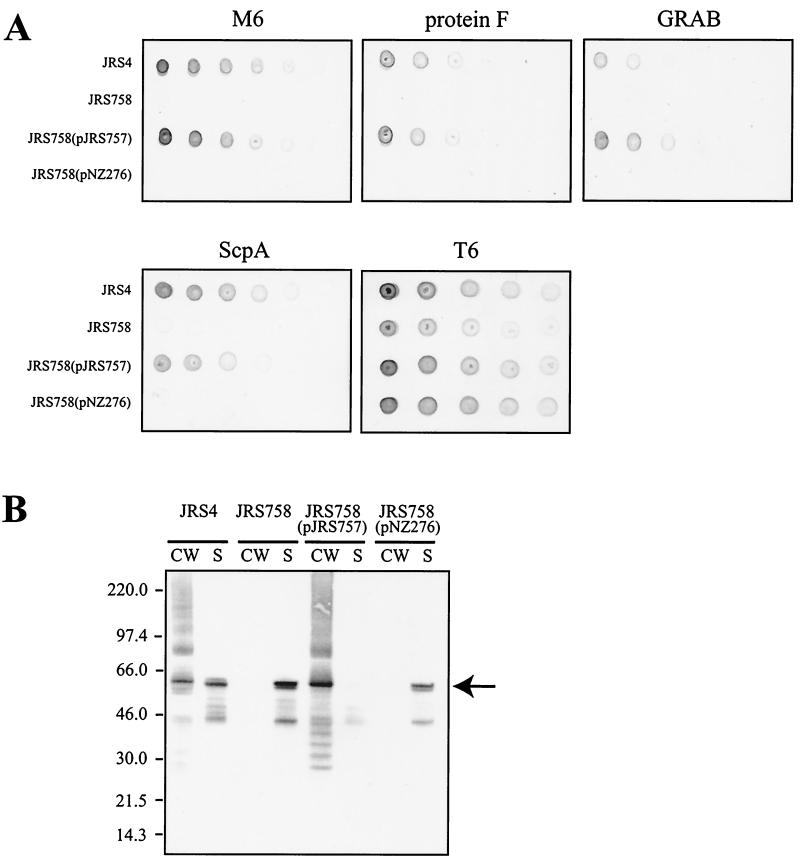
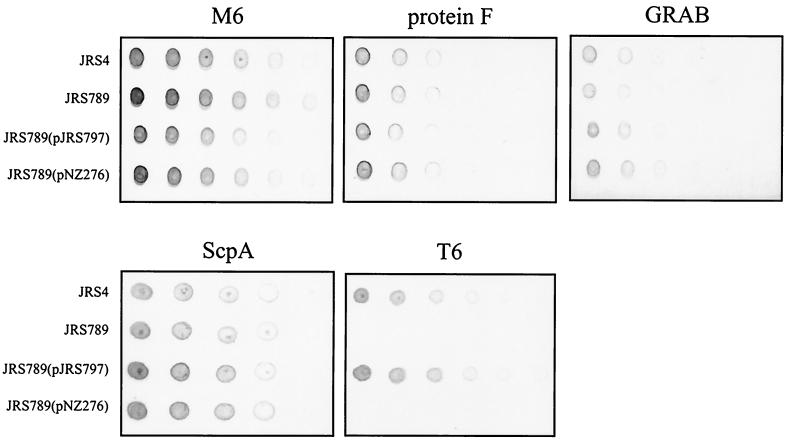
To determine the role of SrtA in anchoring proteins to the cell surface of JRS4, dilutions of overnight cultures of JRS4 and derivatives were analyzed for the presence of M6, protein F, ScpA, GRAB, and T6 by whole-cell dot blots using specific reagents that recognize each protein. Whole cells of JRS4 contain M6, protein F, ScpA, GRAB, and T6 on their surface, as shown by the dot blot analysis (Fig. 3A). In the srtA mutant strain JRS758, no M6, protein F, ScpA, or GRAB was detectable on the cell surface, whereas the presence of T6 was unaffected in this strain.
FIG. 3.
Effect of the srtA mutation on surface anchoring of LPXTG proteins by GAS. (A) Dot blot analysis for surface localization of M6, protein F, ScpA, GRAB, and T6 proteins in JRS4 and srtA derivatives. Each strain was grown overnight in THY broth, and cell density was standardized to an optical density at 600 nm of 2.0. Individual spots represent 10-μl aliquots of twofold serial dilutions of each strain. (B) Western immunoblot analysis of JRS4 and srtA derivatives with monoclonal antibody 10B6. CW, cell wall fraction; S, culture supernatant fraction. Proteins were separated by SDS-PAGE on 4 to 12% gradient gels, transferred to nitrocellulose, and detected with 10B6 as described in Materials and Methods. The sizes of molecular mass standards are indicated to the left (in kilodaltons). The arrow to the right represents the position of mature M6 protein.
To determine whether the phenotypes observed in JRS758 are caused by the TnSpc insertion and not an unknown point mutation elsewhere in the genome, complementation analysis was used. A copy of the entire srtA gene was cloned into the expression vector pNZ276 (51) to give plasmid pJRS757. This plasmid was transformed into JRS758 to complement the srtA mutation (see Materials and Methods). Complementation of the srtA mutant with pJRS757 but not the vector pNZ276 alone relieved the defect in surface protein anchoring (Fig. 3A).
Expression from the lacA promoter in pNZ276 is activated in the presence of lactose due to the inactivation of the LacR repressor encoded by pNZ276 and repressed to low levels in the presence of glucose by catabolite repression (15, 68). We obtained the highest levels of complementation when cells were grown in the presence of glucose (11 mM in THY broth), presumably because pNZ276 derivatives are present in multiple copies, so high levels of transcription are not necessary to complement the chromosomal srtA mutation. Overexpression of srtA appeared to be toxic to JRS758, since overnight cultures of this strain grown in the presence of lactose and absence of glucose did not reach the same cell density as JRS4 or JRS758 containing pNZ276.
To determine the location of surface proteins in the srtA mutant strain, cell wall and culture supernatant fractions were analyzed for the presence of M protein by Western immunoblotting (Fig. 3B). The wild-type strain, JRS4, had M protein present in both the cell wall and culture supernatant fractions. However, in the srtA mutant strain JRS758, there was no observable M protein in the cell wall fraction, although this protein was still secreted into the culture supernatant. When srtA was provided in trans on pJRS757, the M protein produced by JRS758 was attached to the cell wall. This was not observed for JRS758 containing pNZ276 alone. The multiple bands of M protein from cell wall extracts detected with monoclonal antibody 10B6 are indicative of cross-linkage to fragments of peptidoglycan and are characteristic of cell wall preparations obtained by digestion with muralytic enzymes (44). The results in Fig. 3 demonstrate that SrtA is required for the anchoring of M6, protein F, and GRAB but not T6 to the cell wall of JRS4.
Second sortase gene, srtB, revealed by a database search.
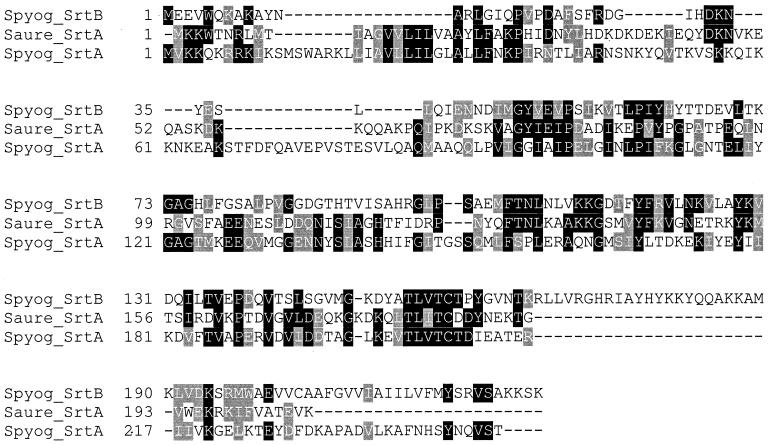
When the amino acid sequence of the entire S. aureus SrtA protein was used in a Blast search of the SF370 (M1) sequence at http://dna1.chem.ou.edu/strep.html, the srtA gene (Spy1154) was not found, but a different gene was identified (Spy0135). Spy0135 is predicted to encode a 227-amino-acid protein with a high degree of homology (31.16% identity, 47.1% similarity) across a 133-amino-acid region (amino acids 32 to 165) to amino acids 56 to 191 of the S. aureus SrtA protein (Fig. 4). However, the global alignment of these two proteins showed much lower homology (18.43% identity, 32.94% similarity). This appears to be due, at least in part, to an approximately 33- amino-acid region homologous to the N terminus of S. aureus SrtA being present at the C terminus of Spy0135. Consequently, Spy0135 is not predicted to have a signal sequence, but has a hydrophobic region from amino acids 199 to 219 that may anchor this protein in the cytoplasmic membrane. There is a region of net positive charge (amino acids 216 to 227) downstream of this hydrophobic region and a small region with a net negative charge from amino acids 201 to 204. Spy0135 also encodes a TLXTC motif (amino acids 154 to 158). Based on the homology of Spy0135 with S. aureus srtA and the presence of a TLXTC motif, this gene was designated srtB.
FIG. 4.
Multiple sequence alignment of SrtB using ClustalW with homologous proteins identified by Blast search. The GenBank accession numbers for the sequences are as follows: S. aureus (Saure) SrtA, AAD48437; S. pyogenes (Spyog) SrtA, AAK34025; and S. pyogenes SrtB, AAK33243.
The region upstream of srtB in JRS4 is different from the corresponding region in SF370. Analysis of the sequence of this region from JRS4 (3) revealed that SrtB from SF370 is truncated at the N terminus. SrtB from JRS4 contains an additional 53 N-terminal amino acids that are predicted to encode a signal sequence that would be cleaved after amino acid 30 to produce the mature form of this protein (Fig. 1A).
Mutation of srtB affects the cell wall anchoring of T6 but not SrtA-dependent proteins.
Our initial experiments to inactivate the srtB gene used plasmid pJRS779 to create the srtB-1 allele (Fig. 1A). This plasmid contains an internal fragment of srtB, encoding amino acids 56 to 256 of the protein encoded by the JRS4 srtB sequence, that was used to disrupt the chromosomal copy of srtB by insertional inactivation (see Materials and Methods), generating strain JRS779. The insertion in JRS779, which would truncate SrtB at amino acid 256 (Fig. 1B), did not affect the ability of this strain to anchor M6, protein F, GRAB, or T6 to the cell wall (data not shown). The protein encoded by this allele should still contain the TLXTC motif, thought to be in the active site of this protein. Therefore, we remade the srtB mutation using plasmid pJRS789, which would produce a copy of srtB truncated after amino acid 206 (Fig. 1B). This protein does not contain the active-site TLXTC motif (amino acids 207 to 211).
The ability of JRS789 and various derivatives to effectively anchor proteins to the cell wall was determined by dot blot analysis as described above for srtA (Fig. 5). In the srtB mutant JRS789, the amount of T6 on the cell surface was reduced, while the presence of M6, protein F, ScpA, and GRAB remained unaffected by this mutation. The defect in surface protein expression was relieved by providing a wild-type copy of srtB in trans on plasmid pJRS797, but not by the vector pNZ276 alone. As described above for complementation of the srtA mutation, sufficient srtB expression to complement the mutation in JRS789 was achieved when the cells harboring pJRS797 were grown in the presence of glucose.
FIG. 5.
Dot blot analysis for the surface localization of M6, protein F, ScpA, GRAB, and T6 proteins in JRS4 and srtB derivatives. Dot blots were prepared as described for Fig. 3.
srtA and srtB are distributed differently throughout the Streptococcus genus.
SrtA and SrtB appear to be important for anchoring several proteins to the cell wall of GAS. The importance of srtA may be reflected by the conservation of this gene in the genomes of strains of both the M1 and M5 serotypes. However, a search of the M5 genome database (http://www.sanger.ac.uk/Projects/S_pyogenes/) did not identify a homolog of srtB, although the gene immediately downstream of srtB in M1 (Spy0136) was present in the M5 genome. This suggests that there might be some variation in the presence of srt genes among strains of different M types.
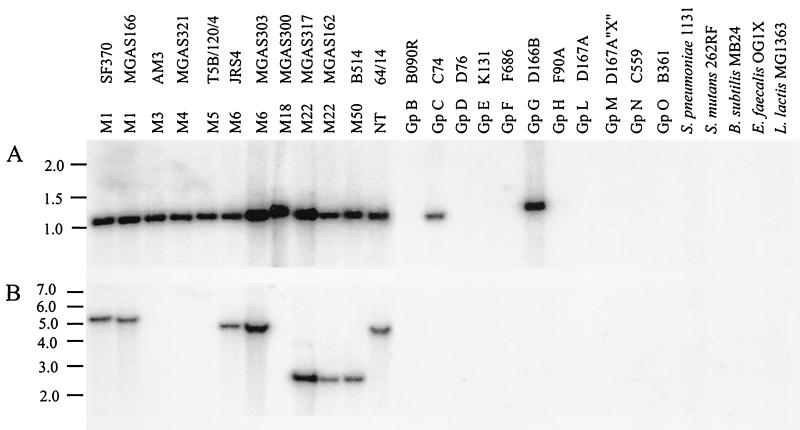
To investigate the distribution of srtA and srtB further, chromosomal DNA from a variety of GAS strains, strains representing other species of Streptococcus, and strains of other related gram-positive bacteria was analyzed by Southern hybridization for the presence of srtA and srtB (Fig. 6). An internal fragment of srtA (corresponding to amino acids 34 to 225) hybridized with a ∼1.1-kb HindIII fragment from all GAS strains tested, demonstrating that srtA is well conserved in this species. In addition, the srtA probe also hybridized with fragments from representatives of group C and group G streptococci but not from group B, D, E, F, H, L, M, N, or O, S. pneumoniae, S. mutans, or any of the other gram-positive bacteria examined. In contrast, the srtB probe (corresponding to amino acids 3 to 203) hybridized only with DNA from GAS strains of M types 1, 6, 22, and 50 and then nontypeable strain 64/14. These results confirmed the absence of an srtB homolog in the M5 genome.
FIG. 6.
Southern hybridization analysis for the presence of srtA (A) and srtB (B) genes in strains of GAS and other gram-positive bacteria. DNA was digested with HindIII and separated by 0.8% agarose gel electrophoresis. Following transfer to a nylon membrane, srtA- and srtB-homologous sequences were detected by hybridization with an internal fragment labeled with [α-32P]dCTP. Gp, group; NT, nontypeable.
DISCUSSION
Surface proteins are required by gram-positive bacteria for adherence to and colonization of host surfaces and for evasion of the host's immune response. Many of these proteins possess conserved C-terminal sequences that are likely to be covalently linked to the cell wall cross-bridge via a transpeptidation reaction catalyzed by the enzyme sortase (39). Consequently, sortase represents an important target for the development of new antimicrobial compounds against gram-positive bacteria. In this work, we have identified and mutated each of two sortase gene homologs of GAS, srtA and srtB, and investigated their role in anchoring surface proteins to the cell wall. We found that SrtA is required for the localization of M protein, protein F, ScpA, and GRAB to the GAS cell surface, while SrtB is required for anchoring T6 to the GAS surface.
Sortase was originally identified and characterized in S. aureus, and since then a functional sortase has also been found in the oral pathogen S. gordonii (5). Sortase enzymes (SrtA) from both of these bacteria contain a conserved sequence (TLXTC) near their C termini that appears to be located at the active site (27, 39, 65). This motif is also conserved in sortase homologs identified in genome sequences of several other gram-positive organisms (27, 47), including the enzymes described here from GAS. We found that the GAS mutant carrying the srtB-1 allele, in which SrtB was truncated after the TLXTC motif, was unaffected in its ability to anchor T6, while the srtB-2 allele (truncated before the TLXTC sequence) caused a deficiency in T6 anchoring. This is consistent with the TLXTC motif being needed for the function of SrtB.
Like SrtA from S. aureus and S. gordonii, SrtA and SrtB from JRS4 are predicted to possess an N-terminal signal peptide. In S. aureus, this signal peptide is not cleaved and is predicted to anchor SrtA to the cytoplasmic membrane (38). It is likely that the GAS SrtA and SrtB proteins are also anchored to the cytoplasmic membrane by this mechanism. The GAS SrtA protein differs from that of S. aureus (but is similar to that of S. gordonii) in that it possesses a region following the signal peptide with high homology to the N-terminal domain of AmyL, a thermostable α-amylase from Bacillus licheniformis (25, 50). Although the function of the N-terminal domain of AmyL has not been determined, α-amylases have a high affinity for and catalyze the hydrolysis of glucose α-1,4 linkages (63). Therefore, if the AmyL-homologous domain of SrtA from S. gordonii and GAS has affinity for carbohydrates containing glucose α-1,4 linkages on the streptococcal surface, these SrtA proteins may be attached by this domain rather than by the N-terminal signal peptide, as described above.
While there were no major differences between T6- and SrtA-dependent proteins in the hydrophobic region or in the distance between the LPXTG motif and the charged tail, SrtA-dependent GAS surface proteins differ from the SrtB-dependent T6 protein in the amino acid following the LPXTG motif. The SrtA-dependent proteins M6, protein F, ScpA, and GRAB have an acidic amino acid residue (aspartate or glutamate) immediately following the LPXTG motif (Fig. 7), while T6 has serine at this position. In the GAS genome, all but one of the 12 proteins with a potential LPXTG cell wall-anchoring domain have an acidic amino acid following the LPXTG sequence (28). The exception is Spy0843, which, like T6, is predicted to encode LPXTGS. Since the predicted product of this open reading frame has not yet been identified, we were unable to determine whether it is covalently attached to the GAS cell wall by SrtA or SrtB. While the amino acid immediately following the LPXTG motif may be essential for recognition by SrtA, the glycine residue within this sequence does not appear to be required for SrtA recognition. Unlike other SrtA-dependent proteins, ScpA has an asparagine residue in place of a glycine in its LPXTG motif. When the signals determining the target specificity of individual sortase enzymes are identified, it seems likely that specific amino acid residues in the region downstream of the LPXTG motif will be involved.
FIG. 7.
Alignment of the C-terminal amino acid sequences of GAS proteins containing an LPXTG motif. Alignments were performed using ClustalW. Spy designations are annotations from the genome sequence of the M1 GAS strain SF370 (16). The sequence of the corresponding region of the M6 protein analyzed in this study is identical to the M1 sequence (23). References for the sequences are as follows: SPy130, -416, -747, -843, -872, -1494, and -1972 (57); Scl2 (35, 52, 69); GRAB (54); Scl1 (34, 53); Fba (64); ScpA (7); M1 (20). Protein F (61) and T6 (57) sequences are from the M6 strain JRS4 and are not present in the M1 genome.
The importance of SrtA for anchoring proteins to the streptococcal cell wall is reflected by the presence of srtA in all 12 GAS strains representing different M types and in representatives of group C and G streptococci. Conservation of srtA at the DNA sequence level is indicated by hybridization of their DNA with a srtA gene probe under the high-stringency conditions employed in this study. Minor sequence divergence from GAS srtA would have prevented identification in this screen, as demonstrated by the lack of hybridization to DNA from S. gordonii (group H) under the conditions used despite the presence of a srtA homolog in the genome of this organism. In contrast, srtB was present in only 5 of the 12 GAS strains examined, suggesting that srtB is important for strains of only a limited number of M types. Perhaps these strains encode a surface protein(s), such as the T6 protein examined in this study, which is anchored by SrtB and is not present in strains lacking the srtB gene.
The conservation of srtA in group A, C, and G streptococci may reflect the similarity of the cell wall structure in these closely related human pathogens. The cell wall cross-bridges of these organisms are virtually identical (56), and it seems likely that the peptidoglycan structure is also similar. The muralytic lysin enzyme from phage A25 is active on the cell walls of group A, C, and G streptococci (22), and group A and C streptococci have been shown to possess the receptor for phage A25 (9). Similarly, group A and C streptococci are sensitive to the C1 phage lysin (45). The proteins cross-linked to the cell wall of group A, C, and G streptococci are also likely to show significant similarities. For example, group C and G streptococci express proteins genetically and functionally homologous to the GAS M protein (31, 60, 62), and like the GAS M protein, these proteins contain the sequence LPXTGE near their C termini (10, 42).
Although SrtA appears to be required for attachment of M6 to the cell wall of GAS, we found a significant amount of this protein released into the culture medium by JRS4, as did Piard et al. (49). It is possible that the amount of M protein produced is too great to be successfully anchored by the amount of SrtA present in the cell. In support of this, we found that the amount of M protein in the culture supernatant was reduced by providing srtA on the multicopy plasmid pNZ276, suggesting that overproduction of SrtA might compensate for inefficient anchoring. It is also possible that the sequence of the cell wall-anchoring domain of the M6 protein is not optimal. In S. aureus, anchoring of the N-terminal portion of protein A fused to the M protein sorting domain was increased from approximately 20 to 85% by increasing the spacing between the LPXTG motif and the positively charged amino acid tail from 30 to 32 amino acids (58). If the M protein anchor domain is not optimal, it may provide a mechanism by which GAS can partition this protein between the cell wall and the surrounding medium. Released M protein may provide a selective advantage to the bacterium by diverting the immune response away from the invading organism.
The results presented in this study show that SrtA is necessary for the surface localization of several important virulence factors of GAS. These include the M protein (1, 2, 24, 33, 48), GRAB (54), protein F (19), and ScpA (29, 30). Many of the GAS surface proteins that we did not test also possess an LPXTG motif followed by an acidic amino acid, which makes it likely that they require SrtA as well. These include serum opacity factor (12), streptococcal protective antigen (41), and the collagen-like protein Scl1 (34), all of which have been shown in one or more model systems to have possible roles in GAS pathogenesis. The role of SrtB in virulence is less clear. The only substrate for SrtB thus far identified, the T6 protein, has no known role in pathogenesis, although T6 is a surface protein and appears to be regulated by the CovR regulon (T. C. Barnett and J. R. Scott, unpublished observations).
Given that the genome sequences of many gram-positive bacteria, including pathogenic organisms such as S. aureus, Streptococcus pneumoniae, and Bacillus anthracis, have been shown to possess multiple sortase homologs, it seems likely that, as in S. pyogenes, these are required for anchoring different subsets of proteins to the cell wall via their LPXTG motif and that these enzymes do not serve redundant functions. It is possible that SrtA and SrtB are differentially regulated in vivo, which would allow the bacterium to modulate the display of different proteins on the cell surface. SrtA, and possibly SrtB, represents an important target for the design of antimicrobial compounds. Inhibitors of sortases or vaccines directed against these proteins may prevent colonization by GAS, and it is possible that inhibitors of SrtA and SrtB might prevent established GAS infections from progressing to more severe and invasive disease.
Acknowledgments
We thank Michael Boyle, Vince Fischetti, Pat Cleary, and Richard Facklam for supplying antibodies, Michael Caparon for supplying strains, and Debra Bessen for providing sequence data prior to publication.
This work was supported by grant R37-AI20723 from the National Institutes of Health.
REFERENCES
- 1.Ashbaugh, C. D., T. J. Moser, M. H. Shearer, G. L. White, R. C. Kennedy, and M. R. Wessels. 2000. Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cell. Microbiol. 2:283-292. [DOI] [PubMed] [Google Scholar]
- 2.Ashbaugh, C. D., H. B. Warren, V. J. Carey, and M. R. Wessels. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Investig. 102:550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding for extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, I., P. Germon, K. McDade, and J. R. Scott. 2001. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect. Immun. 69:7029-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolken, T. C., C. A. Franke, K. F. Jones, G. O. Zeller, C. H. Jones, E. K. Dutton, and D. E. Hruby. 2001. Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion. Infect. Immun. 69:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. C., and P. P. Cleary. 1990. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J. Biol. Chem. 265:3161-3167. [PubMed] [Google Scholar]
- 8.Cleary, P. P., U. Prahbu, J. B. Dale, D. E. Wexler, and J. Handley. 1992. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect. Immun. 60:5219-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary, P. P., L. W. Wannamaker, M. Fisher, and N. Laible. 1977. Studies of the receptor for phage A25 in group A streptococci: the role of peptidoglycan in reversible adsorption. J. Exp. Med. 145:578-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, C. M., A. Kimura, and A. L. Bisno. 1992. Group G streptococcal M protein exhibits structural features analogous to those of class I M protein of group A streptococci. Infect. Immun. 60:3689-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cossart, P., and R. Jonquieres. 2000. Sortase, a universal target for therapeutic agents against gram-positive bacteria? Proc. Natl. Acad. Sci. USA 97:5013-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtney, H. S., D. L. Hasty, Y. Li, H. C. Chiang, J. L. Thacker, and J. B. Dale. 1999. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol. Microbiol. 32:89-98. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Costa, S. S., H. Wang, D. W. Metzger, and M. D. Boyle. 1997. Group A streptococcal isolate 64/14 expresses surface plasmin-binding structures in addition to Plr. Res. Microbiol. 148:559-572. [DOI] [PubMed] [Google Scholar]
- 15.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 18.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanski, E., P. A. Horwitz, and M. G. Caparon. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbaugh, M. P., A. Podbielski, S. Hugl, and P. P. Cleary. 1993. Nucleotide substitutions and small-scale insertion produce size and antigenic variation in group A streptococcal M1 protein. Mol. Microbiol. 8:981-991. [DOI] [PubMed] [Google Scholar]
- 21.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, J. E., and L. W. Wannamaker. 1981. Identification of a lysin associated with a bacteriophage (A25) virulent for group A streptococci. J. Bacteriol. 145:696-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J. Biol. Chem. 261:1677-1686. [PubMed] [Google Scholar]
- 24.Hollingshead, S. K., J. W. Simecka, and S. M. Michalek. 1993. Role of M protein in pharyngeal colonization by group A streptococci in rats. Infect. Immun. 61:2277-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hols, P., A. Baulard, D. Garmyn, B. Delplace, S. Hogan, and J. Delcour. 1992. Isolation and characterization of genetic expression and secretion signals from Enterococcus faecalis through the use of broad-host-range alpha-amylase probe vectors. Gene 118:21-30. [DOI] [PubMed] [Google Scholar]
- 26.Husmann, L. K., J. R. Scott, G. Lindahl, and L. Stenberg. 1995. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect. Immun. 63:345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilangovan, U., H. Ton-That, J. Iwahara, O. Schneewind, and R. T. Clubb. 2001. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6056-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janulczyk, R., and M. Rasmussen. 2001. Improved pattern for genome-based screening identifies novel cell wall-attached proteins in gram-positive bacteria. Infect. Immun. 69:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji, Y., B. Carlson, A. Kondagunta, and P. P. Cleary. 1997. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 65:2080-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji, Y., L. McLandsborough, A. Kondagunta, and P. P. Cleary. 1996. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect. Immun. 64:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, K. F., and V. A. Fischetti. 1987. Biological and immunochemical identity of M protein on group G streptococci with M protein on group A streptococci. Infect. Immun. 55:502-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, K. F., S. A. Khan, B. W. Erickson, S. K. Hollingshead, J. R. Scott, and V. A. Fischetti. 1986. Immunochemical localization and amino acid sequences of crossreactive epitopes within the group A streptococcal M6 protein. J. Exp. Med. 164:1226-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lancefield, R. C. 1962. Current knowledge of the type specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 34.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, R. M. Ireland, S. D. Reid, G. G. Adams, and J. M. Musser. 2000. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 68:6542-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, B. J. Shelvin, E. A. Graviss, and J. M. Musser. 2001. Identification and characterization of a second extracellular collagen-like protein made by group A Streptococcus: control of production at the level of translation. Infect. Immun. 69:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lunsford, R. D., and A. G. Roble. 1997. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence factor-dependent DNA transformation in Streptococcus gordonii. J. Bacteriol. 179:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 97:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 40.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 41.McLellan, D. G., E. Y. Chiang, H. S. Courtney, D. L. Hasty, S. C. Wei, M. C. Hu, M. A. Walls, J. J. Bloom, and J. B. Dale. 2001. Spa contributes to the virulence of type 18 group A streptococci. Infect. Immun. 69:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meehan, M., P. Nowlan, and P. Owen. 1998. Affinity purification and characterization of a fibrinogen-binding protein complex which protects mice against lethal challenge with Streptococcus equi subsp. equi. Microbiology 144:993-1003. [DOI] [PubMed] [Google Scholar]
- 43.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 44.Navarre, W. W., H. Ton-That, K. F. Faull, and O. Schneewind. 1998. Anchor structure of staphylococcal surface proteins. II. COOH-terminal structure of muramidase and amidase-solubilized surface protein. J. Biol. Chem. 273:29135-29142. [DOI] [PubMed] [Google Scholar]
- 45.Nelson, D., L. Loomis, and V. A. Fischetti. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 98:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 47.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9:97-101. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1992. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenes restores its ability to resist phagocytosis. Res. Microbiol. 143:549-558. [DOI] [PubMed] [Google Scholar]
- 49.Piard, J. C., I. Hautefort, V. A. Fischetti, S. D. Ehrlich, M. Fons, and A. Gruss. 1997. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J. Bacteriol. 179:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piggott, R. P., A. Rossiter, S. A. Ortlepp, J. T. Pembroke, and J. F. Ollington. 1984. Cloning in Bacillus subtilis of an extremely thermostable alpha amylase: comparison with other cloned heat-stable alpha amylases. Biochem. Biophys. Res. Commun. 122:175-183. [DOI] [PubMed] [Google Scholar]
- 51.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rasmussen, M., and L. Bjorck. 2001. Unique regulation of SclB—a novel collagen-like surface protein of Streptococcus pyogenes. Mol. Microbiol. 40:1427-1438. [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen, M., A. Eden, and L. Bjorck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasmussen, M., H. P. Muller, and L. Bjorck. 1999. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial surface by binding alpha2-macroglobulin. J. Biol. Chem. 274:15336-15344. [DOI] [PubMed] [Google Scholar]
- 55.Ringdahl, U., H. G. Svensson, H. Kotarsky, M. Gustafsson, M. Weineisen, and U. Sjobring. 2000. A role for the fibrinogen-binding regions of streptococcal M proteins in phagocytosis resistance. Mol. Microbiol. 37:1318-1326. [DOI] [PubMed] [Google Scholar]
- 56.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneewind, O., K. F. Jones, and V. A. Fischetti. 1990. Sequence and structural characteristics of the trypsin-resistant T6 surface protein of group A streptococci. J. Bacteriol. 172:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scott, J. R., W. M. Pulliam, S. K. Hollingshead, and V. A. Fischetti. 1985. Relationship of M protein genes in group A streptococci. Proc. Natl. Acad. Sci. USA 82:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sela, S., A. Aviv, A. Tovi, I. Burstein, M. G. Caparon, and E. Hanski. 1993. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10:1049-1055. [DOI] [PubMed] [Google Scholar]
- 62.Srivastava, S. K., D. A. Barnum, and J. F. Prescott. 1985. Production and biological properties of M-protein of Streptococcus equi. Res. Vet. Sci. 38:184-188. [PubMed] [Google Scholar]
- 63.Stredansky, M., L. Kremnicky, E. Sturdik, and A. Feckova. 1993. Simultaneous production and purification of Bacillus subtilis alpha-amylase. Appl. Biochem. Biotechnol. 38:269-276. [DOI] [PubMed] [Google Scholar]
- 64.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed] [Google Scholar]
- 65.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ton-That, H., S. K. Mazmanian, K. F. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase-catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2Gly3 substrates. J. Biol. Chem. 275:9876-9881. [DOI] [PubMed] [Google Scholar]
- 67.Ton-That, H., and O. Schneewind. 1999. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J. Biol. Chem. 274:24316-24320. [DOI] [PubMed] [Google Scholar]
- 68.van Rooijen, R. J., M. J. Gasson, and W. M. de Vos. 1992. Characterization of the Lactococcus lactis lactose operon promoter: contribution of flanking sequences and LacR repressor to promoter activity. J. Bacteriol. 174:2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whatmore, A. M. 2001. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology 147:419-429. [DOI] [PubMed] [Google Scholar]
- 70.Whitnack, E., and E. H. Beachey. 1982. Antiopsonic activity of fibrinogen bound to M protein on the surface of group A streptococci. J. Clin. Investig. 69:1042-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]