Abstract
The Ffh protein of Escherichia coli is a 48-kDa polypeptide that is homologous to the SRP54 subunit of the eukaryotic signal recognition particle (SRP). Efforts to understand the function of Ffh in bacteria have depended largely on the use of E. coli strains that allow depletion of the wild-type gene product. As an alternative approach to studying Ffh, a temperature-sensitive ffh mutant was isolated. The ffh-10(Ts) mutation results in two amino acid changes in conserved regions of the Ffh protein, and characterization of the mutant revealed that the cells rapidly lose viability at the nonpermissive temperature of 42°C as well as show reduced growth at the permissive temperature of 30°C. While the ffh mutant is defective in insertion of inner membrane proteins, the export of proteins with cleavable signal sequences is not impaired. The mutant also shows elevated expression of heat shock proteins and accumulates insoluble proteins, especially at 42°C. It was further observed that the temperature sensitivity of the ffh mutant was suppressed by overproduction of 4.5S RNA, the RNA component of the bacterial SRP, by stabilizing the thermolabile protein. Collectively, these results are consistent with a model in which Ffh is required only for localization of proteins integral to the cytoplasmic membrane and suggest new genetic approaches to the study of how the structure of the SRP contributes to its function.
The signal recognition particle (SRP) is a ribonucleoprotein complex that was first described for eukaryotic cells as being important for targeting secretory proteins and integral membrane proteins to the translocation apparatus (67, 68; for a recent review see reference 27). Following the molecular characterization of a 54-kDa protein component of the SRP from mammals (SRP54), it was discovered that Escherichia coli encodes a protein with striking homology to this SRP subunit (7, 52). The gene encoding this protein was identified as an open reading frame with no known function (13) and was termed ffh for “fifty-four homologue.” Amino acids with a high degree of conservation were located throughout the protein and included an amino-terminal domain of unknown function, a GTP binding domain (G domain), and a methionine-rich carboxy-terminal M domain. These features led to a model of Ffh function in which the M domain binds to signal sequences of exported proteins by contacts between the relatively flexible side chains of methionine and the hydrophobic amino acids of a typical export signal (7).
Other E. coli gene products were also discovered that displayed significant homologies with components of the eukaryotic SRP. Specifically, the product of ftsY, a poorly characterized gene thought to be important for cell division (19), has homology with the α subunit of the SRP receptor (SRα) from higher cells. FtsY also possesses a G domain that is highly homologous to that of Ffh (7). Furthermore, the 7S RNA component of the mammalian SRP has sequence and structural similarities with a 4.5S RNA species from E. coli (50). As a result of these discoveries, it was proposed that E. coli also possesses an SRP that functions in bacterial protein localization. These conclusions were made solely from arguments based on homology comparisons, however, as none of the SRP homologues had ever been identified in genetic screens designed to reveal components of the protein localization machinery (2, 4).
A series of subsequent studies provided evidence of additional similarities between the E. coli SRP and its counterpart in eukaryotic cells. For example, it was shown previously that Ffh and 4.5S RNA form a ribonucleoprotein complex in vivo (38, 49, 51) and that Ffh could functionally replace the SRP54 protein in specific in vitro assays (8). Interaction between Ffh and hydrophobic signal peptides was also shown elsewhere by in vitro cross-linking (5, 30, 64, 65).
The first genetic approach to exploring the possibility that E. coli employs an SRP for protein targeting was made with reverse genetics. An E. coli mutant was constructed in which the sole functioning copy of ffh in the cells was placed under the control of the arabinose (araBAD) promoter and operator. Growth of this strain without inducer revealed that ffh is an essential E. coli gene (47). However, experiments to determine the role of Ffh in protein export yielded equivocal results. Although the export of several proteins became inefficient upon depletion of Ffh, the defects were not nearly as severe as when genes encoding members of the sec-dependent protein export pathway were mutationally disrupted (32, 45, 55, 58). These results suggested that Ffh does not participate equally in the export of all E. coli proteins. Characterization of an E. coli mutant in which FtsY, the SRP receptor homologue, was depleted, as well as of a strain that expressed a dominant-negative allele of ffs, encoding 4.5S RNA, yielded similar results (39, 49). Despite the limited availability of ffh mutants, the role of the SRP in E. coli was shown indeed to be limited to a subset of exported proteins, specifically proteins that are targeted to the inner membrane (16, 41). A screen with a plasmid library that searched for clones with an increased requirement for Ffh further confirmed these conclusions and also suggested that not all inner membrane proteins require Ffh for membrane targeting (63).
These important insights into Ffh function notwithstanding, it must also be acknowledged that there exist inherent weaknesses in using depletion strains to study an essential cellular process such as protein localization. Extensive depletion of a gene product can require the lapse of several generations before a phenotype is observed (36). During the prolonged growth period necessary to sufficiently deplete a gene product, secondary effects can occur that may obscure the true function of the gene product. Consistent with this notion is the observation that depletion of Ffh leads to a defect in cell division (47); however, given the experimental evidence presented to date, Ffh does not play a direct role in this process. Another potential indirect effect that could lead to misinterpretation of the role of the SRP is that SecY, an essential E. coli protein required for efficient protein export, is itself a membrane-bound protein whose localization utilizes the SRP pathway (30, 57). Caution is also warranted when interpreting the results of depletion of gene products that normally function in relatively low abundance, such as Ffh (6). In this case, exhaustive depletion is required since low, residual levels of the gene product could still support physiological processes. Indeed, these concerns prompted the revisiting of a genetic screen that eventually yielded weak mutations in SRP components (61, 62).
To more clearly understand the function of the SRP in bacterial protein localization, we have sought an alternative approach to conditional expression of Ffh. We have isolated a temperature-sensitive ffh mutant that is rapidly inactivated upon shift to the nonpermissive temperature. Growth of this mutant at the nonpermissive temperature leads to rapid inactivation of Ffh function and, therefore, provides a means to study the consequences of Ffh inactivation that does not require prolonged growth to deplete the wild-type gene product. This mutant has been used to confirm the role of SRP in membrane protein localization and to better elucidate how the 4.5S RNA interacts with Ffh in assembly of the SRP.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The genotypes and sources of the E. coli K-12-derived strains and plasmids used in this study are shown in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| WAM100 | MC4100; ara+ | 47 |
| WAM113 | Source of ffh-1::kan | 47 |
| SKP1101 | WAM100 ffh-1::kan [pSKPP10] | This study |
| SKP1102 | WAM100 ffh-1::kan [pSKPP11] | This study |
| SKP1103 | WAM100 ffh-1::kan [pSKPP12] | This study |
| SKP1104 | WAM100 ffh-1::kan [pSKPP13] | This study |
| CAG629 | F−lacZ(Am) phoA(Am) lon supC(Ts) trp(Am) rpsL rpoH(Am)165 zhg::Tn10 mal(Am) | 1; New England Biolabs |
| SKP1105 | SKP1101 [pHP42] | This study |
| SKP1106 | SKP1102 [pHP42] | This study |
| SKP1107 | SKP1101 [pBAP576] | This study |
| SKP1108 | SKP1102 [pBAP576] | This study |
| S1693 | HfrH lacIq relA1 spoT1 lacY::Tn10 mini-tet ffs::kan-591 [λimm-434 nin-5 XhoI::φ(Ptac-ffs)] | 10 |
| Plasmids | ||
| pSKPP10 | ffh-10(Ts) cloned into the RSF1030-derived cloning vector pLCC29; Cmr | This study |
| pSKPP11 | ffh+ cloned into the RSF1030-derived cloning vector pLCC29; Cmr | This study |
| pSKPP12 | ffh-10(Ts) cloned into the RSF1030-derived cloning vector pLCA29; Apr | This study |
| pSKPP13 | ffh+ cloned into the RSF1030-derived cloning vector pLCA29; Apr | This study |
| pHP42 | ftsQ-PBST in pBAD18; Apr | 62 |
| pHP44 | Source of acrB576-PBST | 62 |
| pBADTopo | Cloning vector for arabinose-regulated gene expression; araC+ Apr | Invitrogen |
| pBAP576 | acrB576-PBST cloned into pBADTopo; Apr | This study |
| pT7T3PhoA123 | Expression of alkaline phosphatase and β-lactamase; Apr | 31 |
| pINGE-Lep | Expression of Lep; araC+ Apr | 15 |
| pSB832 | ffs+ pBR327 derivative; Apr | S. Brown |
| pSB832ΔM | ffsΔMluI (ffs mutant) pBR327 derivative | This study |
| pSB432 | ffs+; pACYC184 derivative; Cmr | 11 |
| pSB579 | ffs+; pACYC184 derivative; Cmr | 11 |
| pSB586 | ffs mutant; pACYC184 derivative; Cmr | 11 |
| pSB832ΔA | ffsΔApaI (ffs+); pSB832 derivative | This study |
| pSB832-59B | ffs-A59G (ffs+); pSB832 derivative | This study |
| pSB832-61H | ffs-G61U (ffs mutant); pSB832 derivative | This study |
| pSB832-62D | ffs-C62A (ffs mutant); pSB832 derivative | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance.
Reagents.
Specific antibodies either were obtained from commercial sources or were generous gifts. Antibodies to alkaline phosphatase, β-lactamase, and chloramphenicol transferase were obtained from 5 Prime→3 Prime Inc. (Boulder, Colo.); maltose binding protein (MBP) was obtained from Research Diagnostics (Flanders, N.J.); and DnaK and GroEL were obtained from Stress Gen (Victoria, British Columbia, Canada).
Growth media were obtained from Difco (Detroit, Mich.), and all antibiotics and amino acid and vitamin supplements were obtained from Sigma Chemical Co. (St. Louis, Mo.). Restriction enzymes and other reagents for cloning were obtained from various vendors, including New England Biolabs (Beverly, Mass.); MBI Fermentas, Inc. (Hanover, Md.); and Gibco-BRL (Gaithersburg, Md.).
Cell growth and general techniques.
Cells were cultured in either Luria broth (LB) or supplemented minimal E medium (66). When required, antibiotics were added to the culture media at the following concentrations: 100 μg of ampicillin/ml, 20 μg of chloramphenicol/ml, and 30 μg of kanamycin/ml.
For growth characterization experiments, cells were diluted from cultures grown overnight at 30°C. Cultures were grown with aeration at 30°C until an optical density at 600 nm (OD600) of 0.05 to 0.10 was reached, upon which half of the cell volume was shifted to 42°C. Samples were removed at 1-h intervals from both the 30 and 42°C cultures for measurement of optical density and counts of viable cells. For viable cell counts, samples were diluted in LB medium, plated in duplicate on LB agar, and incubated at 30°C overnight. Colonies were counted after 48 h of incubation, and the values from three independent experiments were averaged.
Individual cells were visualized by observing wet mounts with phase-contrast microscopy with an Olympus BH-2 microscope. Digital images were captured with a charge-coupled device camera.
Transformations were performed as described by Inoue et al. (22). Generalized transductions using bacteriophage P1 vir were done as described by Miller (42). Plasmid DNA was prepared by a modification of the technique of Carter and Milton (14).
DNA sequence analysis of the ffh-10(Ts) allele was done by first cloning a 1.6-kb EcoRI-SalI fragment from a pBR322-derived vector on which the mutation was isolated into appropriately digested pUC19 (72). DNA sequence analysis was performed by dideoxy dye-termination reactions at the Nucleic Acid Synthesis and Sequencing Facility at Iowa State University.
Plasmid constructions.
pSKPP10 and pSKPP11 were constructed by cloning a 1.6-kb EcoRI-to-SalI fragment, encoding the thermolabile and the wild-type Ffh proteins, respectively, into a medium-copy-number cloning vector, pLCC29 (G. J. Phillips, unpublished data). This vector is a derivative of pDHC29 and replicates by using the origin of replication from RSF1030 that is compatible with most other cloning vectors, including other ColE1-like plasmids (46). In a similar manner, pSKPP12 [ffh-10(Ts)] and pSKPP13 (ffh+) were constructed by cloning the appropriate EcoRI-to-SalI fragment into an RSF1030-derived plasmid that imparts Apr (Phillips, unpublished).
pBAP576 was constructed by PCR amplification of the acrB576-PSBT gene fusion described by Tian and Beckwith (62) with the primers 5′-CCTAATTTCTTTATCGATCGCCCG-3′ and 5′-ACCCTGACCGCCCTGCAC-3′. The 1.9-kb product was cloned into pBADTopo (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. PCR primers were synthesized at the Iowa State University DNA Synthesis and Sequencing facility.
Strain constructions.
The E. coli K-12 strain WAM100, an ara+ derivative of MC4100 (47), was used for construction of the temperature-sensitive ffh mutant SKP1101 and an isogenic ffh+ control strain, SKP1102. These strains were constructed by first introducing either pSKPP10 (SKP1101) or pSKPP11 (SKP1102) to WAM100 by selection for Cmr at 30°C. The chromosomal copy of ffh was replaced with ffh1::kan from WAM113 (47) by P1 vir-mediated transduction. The Kanr transductants were tested for growth at both 30 and 42°C to confirm the temperature-sensitive phenotype of SKP1101 and the ability of SKP1102 to grow at both temperatures.
Pulse-chase experiments and immunoprecipitation.
SKP1101 and SKP1102 were grown overnight at 30°C in minimal E medium containing 1 mg of vitamin B1/ml; a mixture of 18 amino acids, excluding methionine and cysteine; appropriate antibiotics; and 0.4% glucose. Maltose, at a concentration of 0.4% in combination with 0.4% glycerol, was used for induction of MBP and LamB. For detection of β-lactamase and alkaline phosphatase, SKP1101 and SKP1102 were transformed with pT7T3PhoA123.
Overnight cultures were resuspended in the same medium to an OD600 of 0.02 and cultured with aeration at 30°C until the cells reached early logarithmic growth. One half of the culture was then transferred to a sterile flask, and growth was continued at 42°C. At various times (typically 1 or 2 h) after division of the culture, 1-ml aliquots were removed from each culture and pulse-labeled for 20 s with 10 mCi of Tran35S-label (Amersham Pharmacia Biotech, Piscataway, N.J.; specific activity, 1,000 Ci/mmol)/ml. An equal volume of prewarmed chase solution (minimal E medium supplemented with 0.8% methionine and cysteine) was added and chased for various times at the same temperature as that for the growth of cultures. Immunoprecipitations were performed as described previously (18). Autoradiograms were visualized with the GS-363 Molecular Imager System (Bio-Rad Laboratories, Hercules, Calif.). Half-life (T1/2) determinations of Ffh were made from the densitometric scans of the gel images with chloramphenicol transacetylase as an internal standard to normalize the amount of protein detected in each lane.
Cellular fractionation.
The B-PER bacterial protein extraction reagent (Pierce Chemical Co., Rockford, Ill.) was used for the extraction of insoluble proteins. Cultures were grown to late logarithmic phase, and the cells were concentrated in a 1.5-ml volume to an OD600 of 1.5 in a microcentrifuge. For lysis, cells were resuspended in 150 μl of B-PER reagent by vigorous vortexing until the cell suspension was homogenous, followed by vortexing for an additional minute. The separation of insoluble and soluble proteins was done by centrifugation at 13,000 rpm in a microcentrifuge for 5 min with the pellet resuspended in an additional 150 μl of B-PER II reagent. Lysozyme was added to the resuspended pellet at a final concentration of 400 mg/ml, and the mixture was vortexed for 1 min. An additional 1 ml of a 1:20 dilution of the B-PER II reagent was added to the suspension and vortexed for 1 min. Insoluble material was collected by centrifugation at 13,000 rpm for 10 min. The pellet was then resuspended in an additional 1 ml of the diluted (1:20) reagent for washing. After two more washings with the diluted reagent, the pellet was resuspended in 300 μl of sodium dodecyl sulfate (SDS) sample buffer prior to electrophoresis. Gels were scanned using a Bio-Rad GS-700 Imaging Densitometer.
Immunoblot analysis.
Protein fractions were separated by electrophoresis through an SDS-12% polyacrylamide gel and visualized by staining with Coomassie blue dye. Proteins were transferred onto a nitrocellulose membrane (Osmonics Inc., Westborough, Mass.) with a Semidry Electroblotter (Owl Scientific, Portsmouth, N.H.). Filters were blocked in 5% dried nonfat milk in Tris-buffered saline with 0.2% Tween 20 at room temperature for 1 h and washed with Tris-buffered saline. Appropriate antibody at an optimum dilution was added and incubated at room temperature for 1 h. Detection of bound antibody was done with the Opti-4CN detection kit (Bio-Rad Laboratories).
Detection of biotinylated proteins.
Detection of biotinylated fusion proteins was done essentially as described previously (62). Cells were grown overnight at 30°C in LB supplemented with 50 mM biotin and 100 mg of ampicillin/ml. To induce FtsQ-PBST from pHP42 and AcrB-PBST from pBAP576, cells were subcultured in the same medium supplemented with 0.02% arabinose and grown to early log phase prior to shifting a portion of the culture to 42°C for an additional 1 to 2 h. The cells were concentrated to an OD600 of 1.5 in a 1-ml volume by centrifugation at 12,000 rpm in a microcentrifuge. The pellets were resuspended in 100 μl of SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, boiled for 3 min, and then recentrifuged. After electrophoresis, the proteins were transferred to a nitrocellulose membrane, and the membranes were blocked as described above. To detect biotinylated fusion proteins, streptavidin-horseradish peroxidase conjugate (Amersham Pharmacia Biotech) was added to the membrane and incubated at room temperature for 1 h. To detect expressed fusion protein, the membrane was probed with appropriate antibody by using the ECL Western blotting detection system (Amersham Pharmacia Biotech) per the manufacturer's instructions.
Northern hybridization.
Total cellular RNA was isolated from E. coli transformants expressing either wild-type or mutant 4.5S RNA species with RNAWiz RNA isolation reagent (Ambion, Austin, Tex.). The RNA was resolved on an 8% polyacrylamide gel and transferred to a nitrocellulose membrane with a Semidry Electroblotter. A 0.6-kb HindIII-to-BamHI DNA fragment from pSB832 was gel purified, labeled with [32P]dCTP by random priming (Hexalabel DNA labeling kit; MBI Fermentas), and used as a hybridization probe. Hybridization and detection of the blots were done essentially as described previously (53).
PCR-based mutagenesis.
Site-directed changes to ffs were made by PCR mutagenesis with the primers 5′-GTCCGGAAGGGAGCAGCCAAGGCA-3′ (ffs-A59G), 5′-GTCCGGAAGGAATCAGCCAAGGCA-3′ (ffs-G61U), and 5′-GTCCGGAAGGAAGAAGCCAAGGCA-3′ (ffs-C62A). The underlined base represents the mutational alteration of each mutant allele. These primers were used in combination with the antisense primer 5′-CTGGGTTGAAGGCTCTCAAGGGC-3′ to amplify a 0.19-kb fragment that was subsequently digested with BspEI and BamHI and cloned into pSB832 digested with the same enzymes. The mutational alterations were confirmed by DNA sequencing. Random mutagenesis of ffs was done by PCR amplification with the sense primer 5′-TATCATCGATAAGCTTGTCGCTGACGC-3′ in combination with the antisense primer described above under conditions that decreased the fidelity of Taq DNA polymerase (Diversity PCR Random Mutagenesis kit; Clontech, Inc., Palo Alto, Calif.). The PCR products were gel purified, digested with HindIII and BamHI, and cloned into pSB832.
Protease accessibility assays.
SKP1101 and SKP1102 were transformed with pINGE-Lep (Table 1) and grown as described above prior to the expression of Lep being induced with arabinose. After 2 h at the nonpermissive temperature, cells were pulse-labeled and the protease accessibility of the large periplasmic domain of Lep was measured as previously described (56).
RESULTS
Characterization of the ffh-10(Ts) allele.
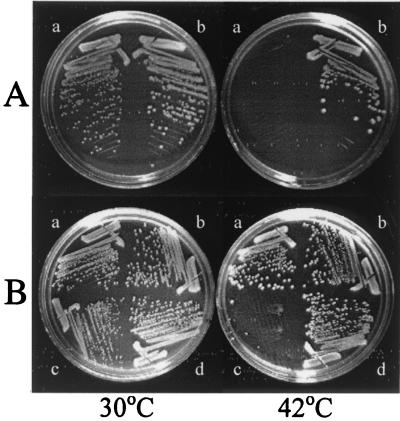
A temperature-sensitive ffh mutant whose viability was strictly dependent on growth temperature was isolated by use of a plasmid-shuffling scheme (S.-K. Park, B. Graham, and G. J. Phillips, unpublished data). To ensure that the temperature-sensitive phenotype was due to a mutational alteration in the essential ffh gene, a 1.6-kb EcoRI-to-SalI restriction fragment was cloned from the pBR322-derived plasmid used to isolate the mutant into the medium-copy-number cloning vector pLCC29 (see Materials and Methods), creating pSKPP10. A control plasmid, pSKPP11, was also assembled to carry ffh+. These plasmids were used to construct SKP1101, in which the sole source of Ffh was encoded by the plasmid-borne ffh-10(Ts) allele, and SKP1102, an isogenic ffh+ control strain, respectively (Table 1). Figure 1A shows the distinct temperature-sensitive phenotype of SKP1101 in comparison with SKP1102.
FIG. 1.
(A) Temperature-sensitive growth of the ffh-10(Ts) mutant. SKP1101 [ffh-10(Ts)] (a) and the isogenic control strain SKP1102 (ffh+) (b) were streaked onto LB plates and cultured overnight at 30 and 42°C, as indicated. (B) Suppression of the temperature sensitivity of the ffh-10(Ts) mutant by 4.5S RNA overproduction. SKP1101 (a and c) and SKP1102 (b and d) were transformed with pSB832 (ffs+) (a and b) or pSB832ΔM (ffh mutant) (c and d) and incubated at either 30 or 42°C.
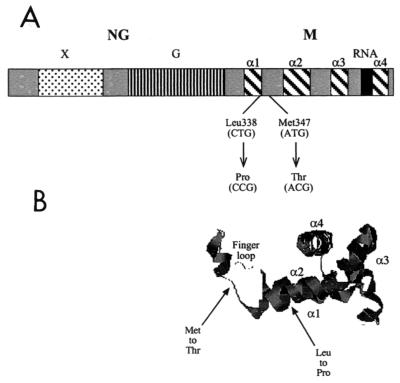
To identify the base pair changes in ffh responsible for the temperature-sensitive phenotype, the 1.6-kb EcoRI-to-SalI fragment was sequenced. As summarized in Fig. 2, two changes to ffh clustered in the region encoding the carboxy-terminal M domain of the protein were found. These mutations resulted in a change from Leu338 to Pro and from Met351 to Thr. While the Leu-to-Pro change introduced a helix-breaking amino acid into a region that has been predicted elsewhere to be in an α-helical conformation (3, 28), the substitution of Thr occurs at a conserved Met residue (20). PCR-based site-directed mutagenesis was used to construct ffh mutants altered at each position individually and revealed that both amino acid substitutions were necessary for the strong temperature-sensitive phenotype (data not shown).
FIG. 2.
Domain organization and structure of Ffh. (A) Functional domains of Ffh are as labeled and include the NG domain including the X domain of unknown function and the G domain with GTPase activity. The carboxy-terminal M domain includes four α-helical structures (α1 to α4) important for contacting the signal sequence and binding 4.5S RNA. Also shown are the locations of the two amino acid changes and their corresponding base pair alterations, resulting in the temperature-sensitive phenotype of the ffh-10(Ts) mutant. (B) Predicted location of the two amino acid changes superimposed on the three-dimensional view of the M domain of Ffh from Thermus aquaticus. The figure was generated using the program RasMol with coordinates from 2FFH from the Protein Data Base.
Growth characteristics of the temperature-sensitive ffh mutant.
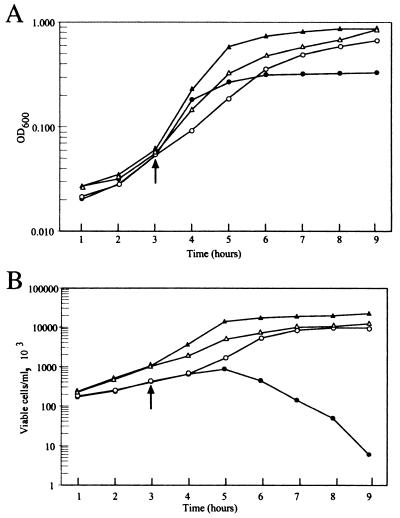
To further characterize the temperature-sensitive mutant, growth of SKP1101 was monitored spectrophotometrically. As shown in Fig. 3A, the control strain showed typical growth kinetics at both 30°C (open triangles) and 42°C (closed triangles), while the mutant continued growth only at the lower temperature (compare open and closed circles in Fig. 3A). At 42°C, the mutant cells increased in density for only 1 h postshift and had completely ceased growth by 2 h. Given the initial doubling time of SKP1101 at 42°C of 30 min, Ffh relinquishes function within two generations after shift to 42°C. Similar results were seen when cells were shifted at either higher or lower optical densities. It was also noted that the mutant grew more slowly than the control even at 30°C, suggesting that the mutant protein was not fully functional even at the permissive temperature. Additional experiments described below confirm this prediction.
FIG. 3.
Growth characteristics of SKP1101 and SKP1102. Both SKP1101 and the control SKP1102 were cultured as described in Materials and Methods to measure the changes in OD600 (A) and viable cell counts (B) at the permissive and nonpermissive growth temperatures. (A) OD600 of SKP1101 (circles) and SKP1102 (triangles) at 30°C (open symbols) or 42°C (closed symbols). (B) Viable cell counts of SKP1101 and SKP1102 with the same symbols as in panel A. The arrow indicates the point of shift of the cultures from 30 to 42°C.
A similar pattern was observed by measuring the number of viable cells recovered following growth at the nonpermissive temperature. When SKP1101 was grown at 30°C (open circles in Fig. 3B), the number of viable cells increased at a rate somewhat lower than that of the wild-type strain (open triangles). However, cells rapidly lost their ability to form colonies at 30°C after growth at 42°C for as little as 2 h (closed circles).
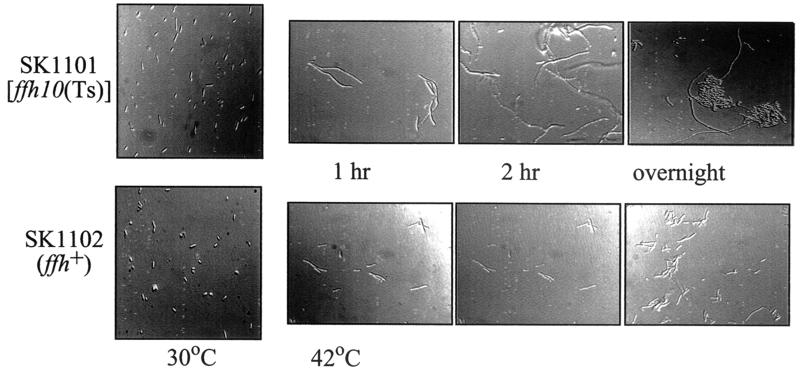
Previous studies using an Ffh depletion system revealed that the cells acquired an elongated morphology within a few hours of growth without inducer of ffh expression (47). We also examined SKP1101 microscopically at different growth temperatures (Fig. 4). In comparison with the control strain, the temperature-sensitive mutant revealed marked cell elongation within 1 h of shift to 42°C. Inspection of the cells at 30°C also revealed that, even at this permissive temperature, the ffh mutant exhibited an elongated cell shape. Further growth at the nonpermissive temperature resulted in formation of inclusion bodies, suggesting that insoluble proteins were accumulating in the cytoplasm. This observation was explored further by the use of the cell fractionation methods described below.
FIG. 4.
Morphological changes of E. coli strains cultured at different times following growth at the nonpermissive temperature. Wet mounts of cells were prepared from cultures of SKP1101 or SKP1102 grown at either 30 or 42°C for the times indicated and observed by microscopy.
Attempts to introduce ffh-10(Ts) into the chromosome in single copy (48) to further restrict the temperature-sensitive phenotype were also made. However, these efforts failed repeatedly, indicating that elevated levels of the thermolabile Ffh protein were necessary to support growth at the permissive temperature.
Signal sequence processing of cell envelope proteins in the ffh mutant.
Previous experiments using an Ffh depletion system yielded equivocal results when used to determine the role of Ffh in E. coli protein export (47). Similar results were obtained when the synthesis of other components of the SRP pathway, including FtsY (39, 57) or 4.5S RNA (49, 51), was disrupted. A separate in vivo approach showed no evidence of a protein export defect upon depletion of 4.5S RNA (25). However, biochemical approaches showed that Ffh interacts with signal sequences from a number of exported proteins (64, 65, 73), suggesting a biological significance to the interaction between SRP and exported protein. The efficiency of cross-linking, however, was dependent upon the hydrophobicity of the signal peptide (5, 30, 64), further suggesting additional selectivity in substrate recognition. To more precisely determine the role of the SRP in generalized protein export in E. coli, we took advantage of the rapid inactivation of Ffh in SKP1101 to monitor the efficiency of signal sequence cleavage of selected periplasmic and outer membrane proteins.
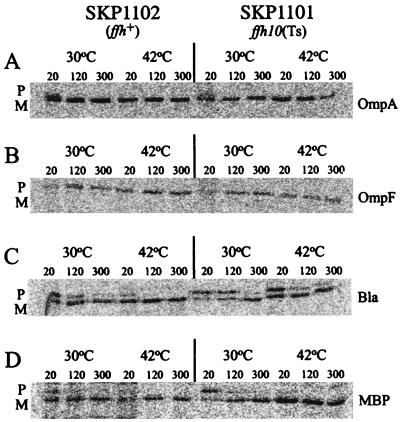
Shown in Fig. 5 are the results of a number of assays used to measure the kinetics of signal sequence processing of the outer membrane proteins OmpA (Fig. 5A) and OmpF (Fig. 5B) and the periplasmic proteins β-lactamase (Fig. 5C) and MBP (Fig. 5D). Signal sequence processing was measured after growth at the nonpermissive temperature for 2 h, a time when cell growth was no longer increasing and viability was declining (Fig. 3). In all cases, the efficiency of signal sequence processing in SKP1101 was comparable to that found in the wild-type control. Low-level accumulation of the precursor form of MBP was detected at early chase times (20 s and 2 min), but virtually all preproteins of MBP were chased out within 5 min, indicating that MBP preprotein does not require Ffh for its translocation. In addition to the proteins shown in Fig. 5, no differences in the rate of signal sequence processing of alkaline phosphatase, OmpC, or LamB were observed between SKP1101 and SKP1102 (data not shown).
FIG. 5.
In vivo processing of presecretory proteins. The efficiency of signal sequence processing in SKP1101 or SKP1102 of selected periplasmic and outer membrane proteins was measured after growth at 42°C for 2 h by pulse-labeling the cells, followed by a chase at the indicated times (in minutes). Bla, β-lactamase.
Processing of pre-β-lactamase has been reported previously to be inhibited at some level in strains where the SRP pathway components are disrupted (39, 47, 49, 51). Likewise, the temperature-sensitive ffh mutant showed evidence of delayed processing at the 20- and 120-s chase times (Fig. 5C). However, this result is most likely an indirect consequence of perturbation of cell physiology in the mutant strain at 42°C, since export of β-lactamase is sensitive to alterations in the levels of heat shock proteins (33, 34, 69).
Insertion of inner membrane proteins.
Studies have suggested that the E. coli SRP functions specifically in the localization of proteins to the cytoplasmic membrane (16, 41, 63). To directly test the importance of Ffh in targeting specific proteins to the inner membrane, we utilized a simple assay developed by Jander et al. based on the efficiency with which biotinylation of polypeptides can occur in vivo (24). Normally, protein localization is so efficient that a membrane-bound polypeptide fused at its carboxy-terminal end to a biotin-accepting domain is transported out of the cytoplasm faster than it can be modified by biotin ligase (BirA). A delay in membrane localization, however, allows the polypeptide to reside in the cytoplasm long enough to be biotinylated. The detection of a biotinylated protein, therefore, is a sensitive indicator of the efficiency of membrane protein targeting. This assay has been effectively used to characterize membrane protein insertion defects of nonconditional ffh and ffs (encoding 4.5S RNA) alleles (61, 62).
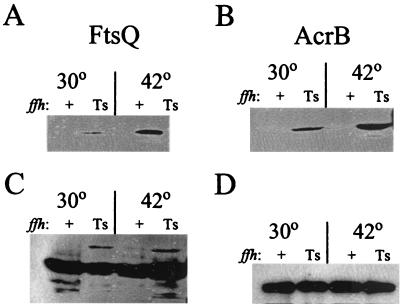
FtsQ, an inner membrane protein involved in cell division, is oriented in the cytoplasmic membrane with a short cytoplasmic amino terminus, a single transmembrane segment, and a relatively large carboxy-terminal end exposed to the periplasmic space. The plasmid pHP42 encodes ftsQ fused to the biotin-accepting domain from Propionibacterium shermania transcarboxylase at its carboxy terminus (FtsQ-PBST) (62). pHP42 was transformed into both SKP1101 and SKP1102, and synthesis of FtsQ-PBST was induced by arabinose as described in Materials and Methods. Following induction, total cellular proteins from the temperature-sensitive mutant and the wild-type control strain were resolved by SDS-PAGE and transferred onto a nitrocellulose membrane for detection of biotinylated FtsQ. As shown in Fig. 6A, no biotinylated protein was detected in the control strain, although Western immunoblotting revealed that the protein was synthesized (Fig. 6C). In contrast, biotinylated FtsQ was detected in SKP1101 even at 30°C, again consistent with the observation that the Ffh protein encoded by ffh-10(Ts) is defective at the permissive temperature. Significantly more biotinylated protein was detected in the mutant at 42 than at 30°C.
FIG. 6.
Efficiency of localization of integral membrane proteins. Localization of FtsQ-PBST (A and C) and AcrB-PBST (B and D) fusion proteins was monitored using a biotinylation assay (62). SKP1105 [ffh-10(Ts)] and SKP1106 (ffh+) were constructed to express FtsQ-PBST, and SKP1107 [ffh-10(Ts)] and SKP1108 (ffh+) were constructed for expression of AcrB-PBST (Table 1). Synthesis of the fusion proteins was induced with arabinose. Cells were grown at the indicated temperatures. (A) Biotinylated FtsQ-PBST. (B) Biotinylated AcrB-PBST. (C) Immunoblot detection of FtsQ-PBST. (D) Immunoblot detection of AcrB-PBST.
AcrB is a polytopic membrane protein with 12 transmembrane segments that is essential for multidrug resistance (40). A fusion protein (AcrB-PBST) has been constructed that fuses PBST after the 576th amino acid residue in a region of the protein localized to the periplasmic space (62). To determine the effect of Ffh inactivation on membrane targeting of this membrane protein, the plasmid pHP44, encoding the constitutively expressed AcrB-PBST fusion protein, was introduced into SKP1101 and SKP1102. While healthy transformants were obtained in the control strain, transformants into the temperature-sensitive mutant grew very poorly. Attempts to grow these transformants to detect AcrB-PBST protein expression were unsuccessful even at the permissive temperature. We predicted that high-level expression of the polytopic membrane protein was toxic to the temperature-sensitive mutant. Consequently, we placed expression of AcrB-PBST under the control of the arabinose operator and promoter, permitting regulated expression of the toxic fusion protein. As shown in Fig. 6B and consistent with the results obtained with FtsQ, AcrB was found to be inefficiently localized in the ffh mutant at both the permissive and the nonpermissive temperatures, with a significantly higher amount of biotinylation detected at 42 than at 30°C.
To quantitatively measure the efficiency of membrane insertion, we also tested the efficiency of leader peptidase (Lep) localization by proteinase K mapping studies. Lep has previously been shown to be dependent upon the SRP for efficient membrane targeting (16, 17). SKP1101 and SKP1102 were transformed with pINGE-Lep (15), encoding leader peptidase (Lep) under arabinose control. Following induction of Lep synthesis at both 30 and 42°C, control and mutant cells were pulse-labeled, and the accessibility of the large periplasmic domain of Lep to protease digestion was determined (56).
As expected, essentially 100% of Lep was localized in the control strain at both growth temperatures, as indicated by the protease sensitivity of the polypeptide. However, even at 30°C, a significant portion (22%) of Lep was inaccessible to protease digestion in the ffh-10(Ts) mutant. At 42°C the portion of protease-resistant polypeptide increased, as two-thirds (67%) of the carboxy-terminal domain did not properly insert into the inner membrane. Interestingly, however, one-third (33%) of Lep did become protease accessible at the nonpermissive temperature, indicating that some portion of the protein properly inserted into the membrane in the absence of functional Ffh.
Detection of insoluble proteins and heat shock proteins.
As described above, prolonged incubation of the ffh mutant at the nonpermissive temperature resulted in inclusion body formation. The formation of insoluble protein complexes in the ffh mutant is a predicted consequence of the accumulation of aggregated hydrophobic proteins resulting from failed protein localization. Indeed, heat shock protein expression has been observed elsewhere for other mutants defective in SRP component expression (6, 9). To determine the extent of insoluble and aggregated proteins accumulating in the mutant, we used a reagent (B-PER) designed for purification of insoluble inclusion bodies typically associated with the overexpression of recombinant proteins (60).
Cells were fractionated by the B-PER reagent, as described in Materials and Methods, and analyzed by SDS-PAGE. Significant increases in the total amount of insoluble proteins were observed for the ffh mutant at both the permissive and the nonpermissive growth temperatures (data not shown). The B-PER reagent was used to analyze the protein profiles for an rpoH mutant that does not synthesize heat shock proteins in response to elevated growth temperatures (1) as an independent means to induce protein misfolding and aggregation (12). Significant increases in the total amount of insoluble proteins were observed for the rpoH mutant, indicating that the B-PER reagent is sensitive to changes in the amounts of nonrecombinant, insoluble proteins in E. coli (data not shown).
We further predicted that such an increase in insoluble proteins should trigger a strong heat shock response, as observed elsewhere for other conditions in which Ffh or 4.5S RNA was depleted (6, 9). To test this, we used Western blotting to compare the levels of two heat shock proteins, DnaK and GroEL, in the whole-cell fraction of the strains at the different growth temperatures. In comparison with the wild-type control, the levels of DnaK and GroEL were elevated in SKP1101 at all growth temperatures, with the most significant increases being noted at 42°C (data not shown).
The ffh-10(Ts) gene product is unstable.
To better understand the nature of the defect of the ffh-10(Ts) gene product, the stability of Ffh was measured by a pulse-chase assay. Calculations of the T1/2 of both the mutant and wild-type proteins are summarized in Table 2. At the nonpermissive temperature, 42°C, the thermolabile protein had a T1/2 of 8 min. Even at 30°C, the mutant protein showed heightened instability in comparison with that of the wild-type protein at the same temperature (T1/2 of 10 min versus 40 min for the wild-type protein). This latter value is comparable to previous determinations of the stability of wild-type Ffh expressed from a recombinant plasmid (26). Even the wild-type Ffh protein, however, showed instability at 42°C, as its T1/2 was reduced to 15 min.
TABLE 2.
Ffh protein stability
| Strain | ffh allele | In vivo T1/2 (min)a at:
|
|
|---|---|---|---|
| 30°C | 42°C | ||
| SKP1101 | ffh-10(Ts) | 10 | 8 |
| SKP1102 | ffh+ | 40 | 15 |
| SKP1101(pSB832) | ffh-10(Ts) | >30b | >30b |
| SKP1101(pSB832ΔM) | ffh-10(Ts) | 12 | 8 |
| SKP1102(pSB832) | ffh+ | >30b | >30b |
| SKP1102(pSB832ΔM) | ffh+ | 38 | 18 |
Strains were grown at either 30 or 42°C prior to pulse-labeling. Labeling was terminated by addition of chase solution for 5, 15, and 30 min as described in Materials and Methods.
No significant reduction in Ffh protein was detected even after the 30-min chase time.
Suppression of a temperature-sensitive ffh mutant by 4.5S RNA overproduction.
Since the SRP is a ribonucleoprotein complex comprised of Ffh and 4.5S RNA, we investigated the consequences of overproducing the RNA component of the SRP for the phenotype of SKP1101. As shown in Fig. 1B, when SKP1101 was transformed with pSB832, a pBR327-derived plasmid expressing 4.5S RNA, the temperature sensitivity of the mutant was completely suppressed. The cellular morphology of the SKP1101/pSB832 transformants was also indistinguishable from that of the wild-type cells shown in Fig. 4. Although it has not been tested, we also predict that the protein localization defect of SKP1101 is reversed as well. In contrast, a nonfunctional allele of ffs (ffsΔMluI), created by deleting an MluI fragment from pSB832, had no effect on the temperature sensitivity of SKP1101 (Fig. 1B). To confirm that overproduction of 4.5S RNA was necessary to suppress the ffh-10(Ts) mutant, ffs+ carried by derivatives of pACYC184, a plasmid whose copy number is lower than that of pSB832, was transformed into SKP1103, an isogenic derivative of SKP1101 in which ffh-10(Ts) is carried by a plasmid that imparts ampicillin resistance. In contrast to the results with pSB832, ffs expression from the plasmids pSB432 and pSB579 suppressed the temperature sensitivity of SKP1101 no better than did that from pSB586, which has the intact ffs gene deleted (11) (Table 3).
TABLE 3.
Suppression and complementation patterns with ffs alleles expressed from recombinant plasmids
| ffs allelea | Plasmid | Suppression of ffh-10(Ts)b | Complementation of ffs::kan-591c |
|---|---|---|---|
| ffs+ | pSB832 | + | + |
| ffsΔMluI | pSB832ΔM | − | − |
| ffs+ | pSB432 | − | + |
| ffs+ | pSB579 | − | + |
| ffsΔMluI | pSB586 | − | − |
| ffsΔApaI | pSB832ΔA | + | + |
| ffs-A59G | pSB832-59B | + | + |
| ffs-G61U | pSB832-61H | − | + |
| ffs-C62A | pSB832-62D | − | − |
The ffs alleles are as described in the text.
Plus signs indicate that the transformants of SKP1101 formed colonies on LB agar at 42°C, comparable to that shown in Fig. 1. The isogenic, Apr derivative SKP1103 was used as the host strain for the Cmr plasmids pSB432, pSB579, and pSB586.
Plus signs indicate that the transformants of S1693 formed colonies on LB agar plates in the absence of IPTG at 37°C.
To further assess the expression of ffs from recombinant plasmids, we also tested their ability to complement the knockout mutation ffs::kan-591. Plasmids were transformed into strain S1693 (10), a mutant whose sole functional copy of ffs is under the control of the tac promoter. Since ffs is an essential gene (11), colonies formed without IPTG (isopropyl-β-d-thiogalactopyranoside) only when the plasmids expressed functional 4.5S RNA. As summarized in Table 3, pSB832, pSB432, and pSB579 complemented ffh-10(Ts), while pSB586 and pSB832ΔM did not. Since only pSB832 suppressed the ffh-10(Ts) mutant, significant overproduction must be necessary to permit growth of the temperature-sensitive mutant, while complementation of ffs::kan-591 can occur with lower levels of 4.5S RNA.
One explanation of how the ffh-10(Ts) mutant is suppressed is that the thermolabile Ffh protein is stabilized by the increased levels of its interactive partner. To test this, we again measured the stability of Ffh in SKP1101, as well as in the isogenic ffh+ control strain SKP1102, in the presence of different levels of 4.5S RNA. It was determined that, when 4.5S RNA was overproduced, the stability of the thermolabile protein was even greater than that of the wild-type Ffh protein expressed without 4.5S RNA (Table 2), as no appreciable turnover of Ffh was detected after a 30-min chase. These results are consistent with previous studies that showed that in vivo wild-type Ffh is itself unstable when not associated with 4.5S RNA (26, 46) and with data from experiments that revealed that interaction with 4.5S RNA dramatically reduces the susceptibility of Ffh to protease cleavage in vitro (73). The ffh-10(Ts) mutation appears to heighten the instability that is inherent in wild-type Ffh in the absence of 4.5S RNA. The protein becomes stabilized, however, as more of the protein is driven into a ribonucleoprotein complex in the presence of elevated levels of 4.5S RNA.
Isolation of ffs mutants.
It became apparent from these results that a genetic screen could be developed to isolate ffs mutants that have lost the ability to suppress the temperature sensitivity of the ffh-10(Ts) mutant. These ffs mutants would likely be defective in interaction with Ffh and should, therefore, be useful in addressing questions about the structure and function of the SRP. Initially we used mutagenic PCR conditions to amplify ffs and clone the PCR products into pSB832. SKP1101 transformants were first selected at 30°C and then replica plated to 42°C to identify clones that failed to yield viable colonies. Several temperature-sensitive mutants were isolated in this screen.
Given the essential nature of both SRP components in E. coli (11, 47), we predicted that mutations in ffs that prevented interaction of 4.5S RNA with Ffh would also fail to complement an ffs knockout mutation. Surprisingly, all mutants tested supported growth of S1192 without inducer. DNA sequence analysis of two of these mutants revealed that both had alterations outside the 4.5S RNA coding region and in the putative promoter of ffs (21). Since suppression of the temperature-sensitive phenotype of SKP1101 requires sufficient overproduction of 4.5S RNA, we hypothesized that the promoter mutations resulted in the reduction of 4.5S RNA levels below that required for suppression but that levels were still sufficient to support viability. Indeed, Northern hybridization experiments revealed that the levels of 4.5S RNA were significantly reduced in the promoter mutants in comparison with the pSB832 control (data not shown). Since these ffs promoter mutations did not directly alter 4.5S RNA, they were not characterized further. These results did, however, further confirm that sufficient overproduction of 4.5S RNA is necessary for ffh-10(Ts) suppression.
To characterize mutations localized within the 4.5S RNA coding region for their ability to suppress ffh-10(Ts), we used PCR-based site-directed mutagenesis to generate specific changes to ffs. These changes resulted in alterations to positions 61 (G61U) and 62 (C62A) of the mature 4.5S RNA molecule, both of which have been shown elsewhere to be important for Ffh interaction (71). Two other variants were also constructed that altered bases not believed to directly participate in Ffh binding (71). These included an alteration to position 59 (A59G) and a four-base deletion at the extreme 3′ end of 4.5S RNA, creating ffsΔApaI. As summarized in Table 3, the single-base changes to positions 61 and 62 eliminated suppression of ffh-10(Ts), consistent with the importance of the bases at these positions in interacting with Ffh. Changes to the other bases, on the other hand, did not affect the ability of the plasmids to suppress temperature sensitivity. The plasmids bearing the different ffs alleles were also transformed into S1693 to determine if they could complement the ffs knockout mutation in this strain. As anticipated, ffs+, along with ffs-A59G and ffsΔApaI, complemented S1693, while ffsΔMluI did not complement it. Interestingly, although ffs-C62A failed to complement ffs::kan-591, ffs-G61U did support growth of S1693. Northern hybridizations revealed that all of the ffs alleles (ffs+, ffs-C62A, ffs-G61U, ffs-A59G, and ffsΔApaI) expressed equivalent amounts of 4.5S RNA (data not shown).
These results are, therefore, consistent with previous studies that showed the essentialness of cytosine at position 62 in contacting Ffh (71). Although guanine at position 61 has also been implicated in Ffh interaction, 4.5S RNA with other bases at this position can apparently still function in some capacity if the RNA is sufficiently overproduced. Although we have not tested the idea, we predict that overproduction of the ffs-G61T gene product is necessary for the observed complementation of S1192.
Correlation of Ffh stability and ffs allele.
To further characterize the ffs mutants, an assay to indirectly measure Ffh-4.5S RNA interaction was used to determine if any of the 4.5S RNA molecules, when expressed from pBR327, could stabilize the Ffh protein in vivo. As described above, previous results indicated that the Ffh protein is naturally unstable when not part of a ribonucleoprotein complex (26, 46); therefore, elevated steady-state levels of Ffh should be detected only if expressed with functional 4.5S RNA.
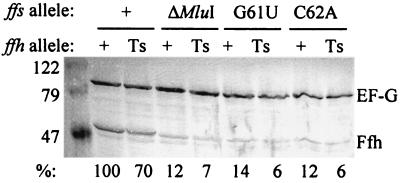
We further predicted that only those ffs alleles that suppressed the ffh-10(Ts) mutant would also stabilize the Ffh protein in vivo. To test these assumptions, SKP1101 and SKP1102 were transformed with pBR327-derived plasmids encoding wild-type 4.5S RNA and different ffs mutants. As shown in Fig. 7, only when ffs+ was expressed from pSB832 were the products of either ffh+ or ffh-10(Ts) readily detectable by immunoblot analysis. As indicated at the bottom of Fig. 7, when the level of detectable wild-type Ffh coexpressed with wild-type 4.5S RNA was assigned a value of 100%, the levels of both the wild-type and the thermolabile Ffh proteins were reduced to nearly 10% of this amount when expressed with the different ffs mutant alleles. Elongation factor G was used as an internal standard for these comparisons. Although the thermolabile Ffh protein was reduced to 70% of the level of the wild-type Ffh protein, it too was stabilized when expressed with the wild-type 4.5S RNA species. These results confirm our prediction that ffs mutants that do not efficiently interact with Ffh are incapable of suppressing ffh-10(Ts) and also cannot stabilize Ffh expressed in vivo. This experiment also allowed us to compare the steady-state levels of mutant and wild-type Ffh proteins. Consistent with the kinetic data, the steady-state amounts of these proteins do differ, reflecting the inherent instability of the ffh-10(Ts) product.
FIG. 7.
Stabilization of both wild-type and thermolabile Ffh proteins requires synthesis of functional 4.5S RNA. SKP1101 (Ts) and SKP1102 (+) were transformed with plasmids expressing different ffs alleles, as shown. Cell lysates were resolved by electrophoresis and immunoblotted. The locations of elongation factor G and Ffh, along with molecular weight standards (in thousands), are indicated. The percent values shown at the bottom indicate the relative amounts of Ffh protein detected by densitometric scanning in comparison with Ffh detected in the control stain (SKP1102 transformed with pSB832, second lane from the left).
DISCUSSION
The SRP of E. coli was discovered by homology comparisons and not by selection for mutants displaying a specific phenotype. One consequence of this approach is that the repertoire of mutants is often limited for experimental analysis. Until recently the only reported alleles of ffh, ftsY, and ffs were complete gene knockouts. Through the careful application of a genetic screen for defective localization of an inner membrane protein, new ffs, ftsY, and ffh mutants have now been isolated (61, 62). Prior to these studies, however, most of the insights into the function of the SRP in general, and Ffh specifically, were gleaned by use of strains that allow regulated expression of the wild-type protein (47). Although careful use of these depletion systems has been informative about the function of Ffh (6, 16, 41, 44, 63), there could potentially be problems in interpreting results due to secondary effects caused by depletion of this essential gene product. It was with this caveat in mind that an alternative approach was sought for depleting E. coli of functional Ffh to further elucidate its cellular function.
Our strategy was to isolate a temperature-sensitive ffh mutant by performing localized chemical mutagenesis on a recombinant plasmid and then to screen for transformants that could complement an ffh knockout mutation only at 30°C. In order to identify a temperature-sensitive, recessive mutation in ffh, however, a complementing copy of the gene had to be provided prior to introduction of the mutagenized plasmid DNA pool. To facilitate this, we developed a plasmid-shuffling system to allow displacement of one plasmid by another (Park et al., unpublished data). The results of this screen yielded a temperature-sensitive ffh mutant that lost viability as a consequence of Ffh inactivation within two generations after shift to the nonpermissive temperature (Fig. 3). Consistent with previous observations, the ffh mutant cells also displayed filamentation at the nonpermissive temperature (Fig. 4), likely resulting from improper localization of membrane-bound cell division proteins.
Our initial studies of the plasmid-borne ffh-10(Ts) allele included efforts to introduce the mutant gene into the chromosome to further reduce the gene dosage of the gene product. However, despite repeated attempts, we were unable to introduce the ffh-10(Ts) mutation to the chromosome at the attachment site for bacteriophage λ (48) as the cell's sole functional copy of ffh. Consequently, overproduction of the thermolabile Ffh protein from a medium-copy-number plasmid was required to complement the ffh knockout mutation. This result, coupled with other characteristics of SKP1101, such as slow growth (Fig. 3), an elongated cell morphology (Fig. 4), and heat shock protein induction, indicated that the mutant protein is defective even at the permissive temperature of 30°C.
It has been proposed elsewhere that the SRP plays a limited role in generalized protein export, since no SRP mutants were ever isolated from genetic screens and selections that successfully revealed other components of the protein export pathway (2). Furthermore, the disruption of the SRP pathway components, including Ffh (47, 63), 4.5S RNA (25, 49, 51), and FtsY (39, 57), either by regulated synthesis of the wild-type protein or by use of dominant-lethal mutations, resulted in only minor defects in signal sequence processing of several exported proteins. In addition, a genetic screen to identify the substrates for Ffh in vivo also failed to uncover any proteins with cleavable signal sequences (63). However, in vitro cross-linking experiments (64, 65), and other studies designed to study interactions between Ffh and its substrates, showed that Ffh can interact with a variety of signal sequences, perhaps suggesting a broader function of Ffh in protein localization (17, 29).
Characterization of SKP1101 confirmed that the role of the SRP in E. coli protein localization is highly specific for cytoplasmic membrane proteins, with no evidence that Ffh functions in the export of proteins synthesized with cleavable signal sequences. Three integral membrane proteins were tested, and all showed reduced efficiency of membrane localization (Fig. 6), while several envelope proteins targeted to the periplasmic space and outer membrane were efficiently exported (Fig. 5). Localization of membrane proteins in the ffh mutant was also inefficient at 30°C (Fig. 6), confirming that the product of ffh-10(Ts) is defective at all temperatures.
In constructing derivatives of SKP1101 to determine protein localization efficiency, we observed that transformants constitutively expressing an AcrB-PBST hybrid protein grew extremely poorly. Expression of this polytopic membrane protein could be accomplished only by placing it under the control of the tightly regulated ara promoter. This result is reminiscent of the Slo phenotype reported by Ulbrandt et al. (63), in which increased expression of membrane proteins from recombinant plasmids increased the cell's requirement for Ffh. These studies led to the conclusion that Ffh is required only for localization of a subset of membrane proteins (44, 63) with the recognition of targeting signals apparently being made on the basis of the hydrophobicity of the transmembrane domain (17, 35). The observation that the ffh-10(Ts) mutant is defective in protein localization even at the permissive temperature of 30°C should prove useful in identifying and characterizing E. coli proteins with differential requirements for the SRP in membrane targeting.
Surprisingly, insertion of Lep, as measured by a quantitative proteinase accessibility assay, revealed that insertion of the large carboxy-terminal periplasmic domain of this membrane protein continued even after prolonged growth at the nonpermissive temperature. Likewise, with the use of other systems for disruption of the SRP pathway, inner membrane protein insertion has been observed to occur at some low level even after significant depletion of SRP (16, 17, 44, 63). However, with these systems it is difficult to confirm that essentially all of the functional Ffh has been depleted from the cells. With the temperature-sensitive mutant, however, it is unlikely that Lep insertion is the result of residual Ffh activity at the nonpermissive temperature, since it has been reported previously that E. coli requires only very low concentrations of Ffh to survive (6), and yet at the time at which the localization measurements were made the cells had ceased growth and were inviable (Fig. 3). Perhaps some of the Lep fraction can be targeted by an SRP-independent pathway (54), or other Sec proteins can facilitate translocation of Lep by maintaining it in an export-competent conformation. It may also be that some proportion of the Lep population fortuitously arrives at the membrane as a ribosome-associated inner membrane protein (6).
A secondary consequence of disrupting the synthesis of SRP components is the induction of the heat shock response (6, 9, 49). Induction of a subset of heat shock proteins, specifically proteases, is actually required for viability of Ffh-depleted cells, likely due to the toxic effects of accumulation of extremely hydrophobic proteins in the cytoplasm (6). Characterization of the ffh-10(Ts) mutant revealed that, even at the permissive growth temperature of 30°C, moderate induction of the heat shock proteins DnaK and GroEL was observed, with significant induction observed following growth at 42°C (data not shown). Using a detergent-based reagent to fractionate cellular proteins, we observed a significant increase in the number of insoluble proteins that accumulated in SKP1101, particularly at the nonpermissive temperature (data not shown). These results are likely explicable in light of other observations that showed that the failure to accurately localize proteins destined for noncytoplasmic locations triggers heat shock protein induction (23, 70). Experiments are in progress to better characterize these insoluble proteins.
The rapidity with which the temperature-sensitive mutant ceased growth indicates that the protein undergoes a conformational change that inactivates previously synthesized molecules of Ffh. The conformational change of Ffh renders the protein unstable and susceptible to proteolysis, as evidenced by a T1/2 of only 8 min at 42°C. Although calculation of the T1/2 of Ffh is complicated by the fact that overproduced protein that is not in a complex with 4.5S RNA is itself unstable (26), these results are consistent with our observation that the altered Ffh protein rapidly loses function upon shift to a nonpermissive temperature.
Since Ffh is found in a ribonucleoprotein complex in vivo, we investigated the consequences of increased expression of the RNA for the phenotype of SKP1101 and found that the temperature sensitivity of this strain was completely reversed upon sufficient overproduction of 4.5S RNA (Fig. 1B). The suppression is the result of stabilization of the thermolabile product of ffh-10(Ts), as turnover of mutant protein was negligible during a 30-min chase (Table 2). These results are consistent with previous studies that showed that Ffh not associated with 4.5S RNA in vivo is unstable, with a significantly shortened T1/2 (26, 46). Apparently, the mutant protein is incapable of efficiently binding 4.5S RNA at the nonpermissive temperature and, as a consequence, becomes extremely labile. We further exploited this observation to genetically characterize Ffh-4.5S RNA interaction in vivo by isolating ffs mutants that lost the ability to interact with Ffh. Overproduction of 4.5S RNA altered at positions known to be important for contacting Ffh, including G61 and C62 (71), failed to support growth of SKP1101 at 42°C and was also incapable of stabilizing steady-state levels of wild-type or mutant Ffh protein (Fig. 7). RNA mutants altered at positions not implicated in Ffh interaction did not, however, eliminate suppression (Table 3).
These results provide additional genetic evidence for formation of a ribonucleoprotein complex between Ffh and 4.5S RNA. Although previous studies have provided insights into how this interaction occurs in vitro (37, 43, 59, 71), including determination of the three-dimensional structure of the bacterial SRP (3), the observation that overproduction of 4.5S RNA can suppress the temperature-sensitive phenotype of an ffh-10(Ts) mutant provides a new genetic approach to isolating and characterizing ffs mutants that should prove useful for understanding how the structure of the SRP contributes to function in vivo.
In conclusion, the temperature-sensitive ffh mutant described in this study has provided an independent approach to testing the function of the SRP in E. coli. In agreement with previous insights primarily obtained by depletion of the wild-type protein, Ffh participates specifically in the localization of proteins to the inner membrane. The rapid inactivation of functional Ffh at the nonpermissive temperature, however, lessens the likelihood that these conclusions are based on the consequences of prolonged depletion of this essential gene product. Since the mutant also displays a moderate defect in Ffh function at 30°C and yet remains viable, it will facilitate construction of double mutants to test the potential requirements of the SRP pathway for other components of the Sec protein export machinery. Further characterization of this mutant should also prove useful in addressing how the structure of the SRP contributes to its molecular function.
Acknowledgments
We thank the following individuals for generous gifts of antisera, plasmids, or bacterial strains: Arnold Driessen, Jon Beckwith, Stanley Brown, Harris Bernstein, Rajeev Misra, Hiroshi Nikaido, Tom Silhavy, P. C. Tai, and Hong Ping Tien.
This work was supported by grants from the National Institutes of Health (GM50836-01A2, G.J.P.) and the National Science Foundation (R.E.D.).
REFERENCES
- 1.Baker, T. A., A. D. Grossman, and C. A. Gross. 1984. A gene regulating the heat shock response in Escherichia coli also affects proteolysis. Proc. Natl. Acad. Sci. USA 81:6779-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassford, P., J. Beckwith, K. Ito, C. Kumamoto, S. Mizushima, D. Oliver, L. Randall, T. Silhavy, P. C. Tai, and B. Wickner. 1991. The primary pathway of protein export in E. coli. Cell 65:367-368. [DOI] [PubMed] [Google Scholar]
- 3.Batey, R. T., R. P. Rambo, L. Lucast, B. Rha, and J. A. Doudna. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287:1232-1239. [DOI] [PubMed] [Google Scholar]
- 4.Beckwith, J. 1991. Sequence gazing? Science 251:1161-1162. [DOI] [PubMed] [Google Scholar]
- 5.Behrmann, M., H.-G. Koch, T. Hengelage, B. Wieseler, H. K. Hoffschulte, and M. Muller. 1998. Requirements of the translocation of elongation-arrested, ribosome-associated OmpA across the plasma membrane of Escherichia coli. J. Biol. Chem. 273:13898-13904. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, H. D., and J. B. Hyman. 2001. Physiological basis for conservation of the signal recognition particle targeting pathway in Escherichia coli. J. Bacteriol. 183:2187-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, H. D., M. A. Poritz, K. Strub, P. J. Hoben, S. Brenner, and P. Walter. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 340:482-486. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein, H. D., D. Zopf, D. M. Freymann, and P. Walter. 1993. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc. Natl. Acad. Sci. USA 90:5229-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourgaize, D. B., T. A. Phillips, R. A. VanBogelen, P. G. Jones, F. C. Neidhardt, and M. J. Fournier. 1990. Loss of 4.5S RNA induces the heat shock response and lambda prophage in Escherichia coli. J. Bacteriol. 172:1151-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, S. 1989. Time of action of 4.5 S RNA in Escherichia coli translation. J. Mol. Biol. 209:79-90. [DOI] [PubMed] [Google Scholar]
- 11.Brown, S., and M. J. Fournier. 1984. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J. Mol. Biol. 178:533-550. [DOI] [PubMed] [Google Scholar]
- 12.Bukau, B., F. X. Schmid, and J. Buchner. 1999. Chaperones and folding catalysts. Regulation, cellular function and mechanism. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 13.Bystrom, A. S., K. J. Hjalmarsson, P. M. Wikstrom, and G. R. Bjork. 1983. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 2:899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter, M. J., and I. D. Milton. 1993. An inexpensive and simple method for DNA purifications on silica particles. Nucleic Acids Res. 21:1044.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalbey, R. E., and W. Wickner. 1985. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J. Biol. Chem. 260:15925-15931. [PubMed] [Google Scholar]
- 16.de Gier, J.-W., P. Mansournia, Q. A. Valent, G. J. Phillips, J. Luirink, and G. von Heijne. 1996. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399:307-309. [DOI] [PubMed] [Google Scholar]
- 17.de Gier, J.-W., P. A. Scotti, A. Saaf, Q. A. Valent, A. Kuhn, J. Luirink, and G. von Heijne. 1998. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:14646-14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feilmeier, B. J., G. Iseminger, D. Schroeder, H. Webber, and G. J. Phillips. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill, D. R., G. F. Hatfull, and G. P. Salmond. 1986. A new cell division operon in Escherichia coli. Mol. Gen. Genet. 205:134-145. [DOI] [PubMed] [Google Scholar]
- 20.Gorodkin, J., B. Knudsen, C. Zweib, and T. Samuelsson. 2001. SRPDB (signal recognition particle database). Nucleic Acids Res. 29:169-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu, L. M., J. Zagorski, and M. J. Fournier. 1984. Cloning and sequence analysis of the Escherichia coli 4.5 S RNA gene. J. Mol. Biol. 178:509-531. [DOI] [PubMed] [Google Scholar]
- 22.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 23.Ito, K., Y. Akiyama, T. Yura, and K. Shiba. 1986. Diverse effects of the MalE-LacZ hybrid protein on Escherichia coli cell physiology. J. Bacteriol. 167:201-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jander, G., J. E. Cronan, Jr., and J. Beckwith. 1996. Biotinylation in vivo as a sensitive indicator of protein secretion and membrane protein insertion. J. Bacteriol. 178:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, C. G., S. Brown, and S. Pedersen. 1994. Effect of 4.5S RNA depletion on Escherichia coli protein synthesis and secretion. J. Bacteriol. 176:2502-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen, C. G., and S. Pedersen. 1994. Concentrations of 4.5S RNA and Ffh protein in Escherichia coli: the stability of Ffh protein is dependent on the concentration of 4.5S RNA. J. Bacteriol. 176:7148-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keenan, R. J., D. M. Freymann, R. M. Stroud, and P. Walter. 2001. The signal recognition particle. Annu. Rev. Biochem. 70:755-775. [DOI] [PubMed] [Google Scholar]
- 28.Keenan, R. J., D. M. Freymann, P. Walter, and R. M. Stroud. 1998. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94:181-191. [DOI] [PubMed] [Google Scholar]
- 29.Kim, J., S. Rusch, J. Luirink, and D. A. Kendall. 2001. Is Ffh required for export of secretory proteins? FEBS Lett. 505:245-248. [DOI] [PubMed] [Google Scholar]
- 30.Koch, H. G., T. Hengelage, C. Neumann-Haefelin, J. MacFarlane, H. K. Hoffschulte, K. L. Schimz, B. Mechler, and M. Muller. 1999. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10:2163-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohl, J., F. Ruker, G. Himmler, D. Mattanovich, and H. Katinger. 1990. Engineered gene for Escherichia coli alkaline phosphatase for the construction of translational fusions. Nucleic Acids Res. 18:1069.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumamoto, C. A., and J. Beckwith. 1983. Mutations in a new gene, secB, cause defective protein localization in Escherichia coli. J. Bacteriol. 154:253-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusukawa, N., T. Yura, C. Ueguchi, Y. Akiyama, and K. Ito. 1989. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 8:3517-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laminet, A. A., T. Ziegelhoffer, C. Georgopoulos, and A. Pluckthun. 1990. The Escherichia coli heat shock proteins GroEL and GroES modulate the folding of the β-lactamase precursor. EMBO J. 9:2315-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, H. C., and H. D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 98:3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerner, C. G., and M. Inouye. 1991. Pleiotropic changes resulting from depletion of Era, an essential GTP-binding protein in Escherichia coli. Mol. Microbiol. 5:951-957. [DOI] [PubMed] [Google Scholar]
- 37.Liao, X., D. Selinger, S. Althoff, A. Chiang, D. Hamilton, M. Ma, and J. A. Wise. 1992. Random mutagenesis of Schizosaccharomyces pombe SRP RNA: lethal and conditional lesions cluster in presumptive protein binding sites. Nucleic Acids Res. 20:1607-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luirink, J., S. High, H. Wood, A. Giner, D. Tollervey, and B. Dobberstein. 1992. Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature 359:741-743. [DOI] [PubMed] [Google Scholar]
- 39.Luirink, J., C. M. ten Hagen-Jongman, C. C. van der Weijden, B. Oudega, S. High, B. Dobberstein, and R. Kusters. 1994. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 13:2289-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 41.Macfarlane, J., and M. Muller. 1995. The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur. J. Biochem. 233:766-771. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Nakamura, K., H. Miyamoto, S. Suzuma, T. Sakamoto, G. Kawai, and K. Yamane. 2001. Minimal functional structure of Escherichia coli 4.5 S RNA required for binding to elongation factor G. J. Biol. Chem. 276:22844-22849. [DOI] [PubMed] [Google Scholar]
- 44.Newitt, J. A., N. D. Ulbrandt, and H. D. Bernstein. 1999. The structure of multiple polypeptide domains determines the signal recognition particle targeting requirement of Escherichia coli inner membrane proteins. J. Bacteriol. 181:4561-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliver, D. B., and J. Beckwith. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25:765-772. [DOI] [PubMed] [Google Scholar]
- 46.Phillips, G. J., S. K. Park, and D. Huber. 2000. High copy number plasmids compatible with commonly used cloning vectors. BioTechniques 28:400-406. [DOI] [PubMed] [Google Scholar]
- 47.Phillips, G. J., and T. J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744-746. [DOI] [PubMed] [Google Scholar]
- 48.Platt, R., C. Drescher, S. K. Park, and G. J. Phillips. 2000. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid 43:12-23. [DOI] [PubMed] [Google Scholar]
- 49.Poritz, M. A., H. D. Bernstein, K. Strub, D. Zopf, H. Wilhelm, and P. Walter. 1990. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science 250:1111-1117. [DOI] [PubMed] [Google Scholar]
- 50.Poritz, M. A., K. Strub, and P. Walter. 1988. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell 55:4-6. [DOI] [PubMed] [Google Scholar]
- 51.Ribes, V., K. Romisch, A. Giner, B. Dobberstein, and D. Tollervey. 1990. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell 63:591-600. [DOI] [PubMed] [Google Scholar]
- 52.Romisch, K., J. Webb, J. Herz, S. Prehn, R. Frank, M. Vingron, and B. Dobberstein. 1989. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature 340:478-482. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 54.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 55.Schatz, P. J., P. D. Riggs, A. Jacq, M. J. Fath, and J. Beckwith. 1989. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev. 3:1035-1044. [DOI] [PubMed] [Google Scholar]
- 56.Schuenemann, T. A., V. M. Delgado-Nixon, and R. E. Dalbey. 1999. Direct evidence that the proton motive force inhibits membrane translocation of positively charged residues within membrane proteins. J. Biol. Chem. 274:6855-6864. [DOI] [PubMed] [Google Scholar]
- 57.Seluanov, A., and E. Bibi. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053-2055. [DOI] [PubMed] [Google Scholar]
- 58.Shiba, K., K. Ito, T. Yura, and D. P. Ceretti. 1984. A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of a new temperature-sensitive secY mutation. EMBO J. 3:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuma, S., K. Hayashi, K. Nakamura, and K. Yamane. 1999. Analysis of Escherichia coli 4.5S RNA binding affinity to Ffh and EF-G. FEMS Microbiol. Lett. 180:271-277. [DOI] [PubMed] [Google Scholar]
- 60.Thomas, J. G., and F. Baneyx. 1996. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing heat-shock proteins. J. Biol. Chem. 271:11141-11147. [DOI] [PubMed] [Google Scholar]
- 61.Tian, H., and J. Beckwith. 2002. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 184:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian, H., D. Boyd, and J. Beckwith. 2000. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. USA 97:4730-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187-196. [DOI] [PubMed] [Google Scholar]
- 64.Valent, Q. A., J.-W. de Gier, G. von Heijne, D. A. Kendall, C. M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1997. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol. Microbiol. 25:53-64. [DOI] [PubMed] [Google Scholar]
- 65.Valent, Q. A., P. A. Scotti, S. High, J. W. de Gier, G. von Heijne, G. Lentzen, W. Wintermeyer, B. Oudega, and J. Luirink. 1998. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17:2504-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogel, H. J., and D. M. Bonner. 1955. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 67.Walter, P., and G. Blobel. 1980. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 77:7112-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walter, P., and G. Blobel. 1982. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299:691-698. [DOI] [PubMed] [Google Scholar]
- 69.Wild, J., P. Rossmeissl, W. A. Walter, and C. A. Gross. 1996. Involvement of the DnaK-DnaJ-GrpE chaperone team in protein secretion in Escherichia coli. J. Bacteriol. 178:3608-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wild, J., W. A. Walter, C. A. Gross, and E. Altman. 1993. Accumulation of secretory protein precursors in Escherichia coli induces the heat shock response. J. Bacteriol. 175:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wood, H., J. Luirink, and D. Tollervey. 1992. Evolutionary conserved nucleotides within the E. coli 4.5S RNA are required for association with P48 in vitro and for optimal function in vivo. Nucleic Acids Res. 20:5919-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 73.Zheng, N., and L. M. Gierasch. 1997. Domain interactions in E. coli SRP: stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol. Cell 1:79-87. [DOI] [PubMed] [Google Scholar]