Abstract
The pdhABCD operon of Bacillus subtilis encodes the pyruvate decarboxylase (E1α and E1β), dihydrolipoamide acetyltransferase (E2), and dihydrolipoamide dehydrogenase (E3) subunits of the pyruvate dehydrogenase multienzyme complex (PDH). There are two promoters: one for the entire operon and an internal one in front of the pdhC gene. The latter may serve to ensure adequate quantities of the E2 and E3 subunits, which are needed in greater amounts than E1α and E1β. Disruptions of the pdhB, pdhC, and pdhD genes were isolated, but attempts to construct a pdhA mutant were unsuccessful, suggesting that E1α is essential. The three mutants lacked PDH activity, were unable to grow on glucose and grew poorly in an enriched medium. The pdhB and pdhC mutants sporulated to only 5% of the wild-type level, whereas the pdhD mutant strain sporulated to 55% of the wild-type level. This difference indicated that the sporulation defect of the pdhB and pdhC mutant strains was due to a function(s) of these subunits independent of enzymatic activity. Growth, but not low sporulation, was enhanced by the addition of acetate, glutamate, succinate, and divalent cations. Results from the expression of various spo-lacZ fusions revealed that the pdhB mutant was defective in the late stages of engulfment or membrane fusion (stage II), whereas the pdhC mutant was blocked after the completion of engulfment (stage III). This analysis was confirmed by fluorescent membrane staining. The E1β and E2 subunits which are present in the soluble fraction of sporulating cells appear to function independently of enzymatic activity as checkpoints for stage II-III of sporulation.
When the medium is depleted of one or more certain essential nutrients, Bacillus subtilis cells cease normal division and initiate a highly organized developmental program culminating in the production of a dormant, resistant spore. The ordered sequence of sporulation events is strictly governed by regulatory networks which coordinate nutritional, transcriptional, and morphological signals (11, 35, 43, 46). One aspect of the physiological control of these networks is the metabolic and signaling role of the glycolytic pathway and the tricarboxylic acid (TCA) cycle. The function of the glycolytic pathway is probably not directly related to sporulation, since most of the pathway enzymes are not needed when cells start to sporulate (13).
In contrast, there is a link between TCA cycle function and sporulation (15). TCA cycle enzymes, unlike glycolytic enzymes, are maximally induced just before the onset of sporulation, and the absence of these activities results in a sporulation deficiency (16, 21, 23-25). The sporulation frequency of mutants defective in the TCA cycle could be elevated greatly by the addition of certain divalent cations or metabolites, by cultural pH alteration, or in one case by the introduction of a counterpart Escherichia coli gene (25). In such mutants, therefore, the abnormal accumulation of certain metabolites rather than other possible functions of these altered enzymes accounts for the sporulation defect.
The pyruvate dehydrogenase (PDH) complex, a member of the 2-oxo acid dehydrogenase family, catalyzes the irreversible oxidative decarboxylation of pyruvate to acetyl-coenzyme A, linking glycolysis with the TCA cycle (5, 15). The complex is composed of multiple copies of three different enzymes: pyruvate decarboxylase (E1), dihydrolipoamide dehydrogenase (E2), and lipoamide dehydrogenase (E3). E1 and E2 are unique to the complex, whereas E3 is shared with other 2-oxo acid dehydrogenase complexes (19, 33). The PDH complex of gram-negative bacteria is composed of a core of 24 E2 subunits arranged with octahedral symmetry, with 12 E1 and 6 E3 subunits associated (14). In contrast, PDH complexes of gram-positive bacteria consist of a core of 60 E2 subunits with icosahedral symmetry and 30 E1 and 6 E3 subunits associated. E1 of gram-negative bacteria is a homodimer, while this component from gram-positive bacteria is a heterotetramer (E1α2E1β2). In gram-positive bacteria, the PDH complex is encoded by the linked genes pdhA (E1α), pdhB (E1β), pdhC (E2), and pdhD (E3) (Fig. 1) (33).
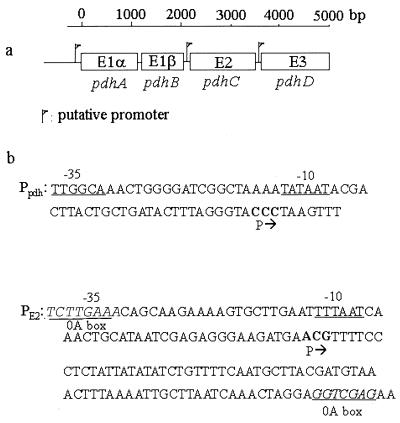
FIG. 1.
(a) Organization of the B. subtilis PDH operon. Three putative promoters were suggested on the basis of Northern blots (16), but only promoters in front of the pdhA and pdhC genes were found by reverse transcriptase mapping. (b) DNA sequences of Ppdh and PE2. −35 and −10 boxes are underlined. Transcription start points are marked P→. Potential spo0A boxes are underlined and italicized. The consensus spo0A box sequence is 5′-TGTCGAA-3′.
We initiated a study of the role of subunits of the PDH complex in sporulation on the basis of evidence that the PDH E2 subunit of Bacillus thuringiensis had a regulatory function in the expression of δ-endotoxin genes by binding to specific upstream sequences (52). While earlier attempts at insertional inactivation of the pdhABCD operon were unsuccessful in both B. subtilis and Staphylococcus aureus (1, 16), B. subtilis mutants defective in PDH had been isolated after chemical mutagenesis (13). Two such strains with mutations in the pdhAB genes required acetate for growth in a minimal medium and sporulated at a greatly reduced level. It is not known whether the mutations were in pdhA or pdhB. A pdhD mutant, deficient in both PDH and 2-oxoglutarate dehydrogenase (ODH) activities, has also been isolated but not further characterized (21), although disruption of the odgB gene, which encodes a subunit of the ODH complex, results in an early block in sporulation (A. Danchin, personal communication).
Given the potential importance of PDH in both growth and sporulation, we isolated mutants with disruptions of the pdhB, pdhC, and pdhD genes. All of the mutants lost PDH activity, but the reduction of sporulation in the pdhB and pdhC mutants was 10 times greater than that in the pdhD mutant. The basis for the difference and the stage of sporulation blocked in the former mutants was examined. The results indicate that the E1β and E2 subunits function in regulating sporulation independent of their participation as part of the PDH enzymatic complex.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. All strains were grown in Luria-Bertani (LB) medium or on LB agar plates at 37°C (32). Nutrient sporulation medium (NSM) and SM resuspension medium for sporulation of B. subtilis have been described elsewhere (13, 45). Spizizen's minimal medium (2) was used for growth on glucose and on certain metabolites. Growth was monitored in a Klett-Summerson colorimeter (660-nm filter) or by measuring absorbance at 600 nm in a Perkin-Elmer spectrophotometer (junior model 35). For both E. coli and B. subtilis, media were supplemented with antibiotics when selection was required: ampicillin, 50 μg/ml; neomycin, 20 μg/ml; and chloramphenicol, 7 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF) U169 endA1 recA1 hsdR1(rK− mK+) deoR thi-1 phoA supE44 λ−gyrA96 relA1 | Laboratory stock |
| B. subtilis | ||
| JH642 | trpC2 pheA1 | Laboratory stock |
| JCB50 | SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoVG′-lacZ) cat trpC2 pheA1 | 7 |
| JCB64 | Spβc2 Δ2::Tn917::pSK10Δ6::Φ(cotA′-lacZ) cat trpC2 pheA1 | 7 |
| KP66 | spoIIIG-lacZ spoIIIGΔ1 | 48 |
| SJB83 | SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoIIG′-lacZ) cat trpC2 pheA1 | 25 |
| SJB227 | Φ(sspB′-lacZ) cat trpC2 pheA1 | 7 |
| SJB253 | SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoIIA′-lacZ) cat trpC2 pheA1 | 25 |
| UOT-1101 | hisH metB Φ(spo0A′-lacZ) cat | 53 |
| AA001 | trpC2 pheA1 spoIIIG-lacZ spoIIIGΔ1 | KP66 DNA→JH642 |
| 501-75I | trpC2 pheA ???::neo | pUC18EAII-502→JH642 |
| 501-75I1 | trpC2 pheA ΔpdhA::neo pdhA | pUC18EAII-502→JH642 |
| 501-76 | trpC2 pheA1 ΔpdhB::neo | pDH87EB-502→JH642 |
| 501-760A | trpC2 pheA1 ΔpdhB::neo Φ(spo0A′-lacZ) cat | pDH87EB-502→UOT-1101 |
| 501-76IIA | trpC2 pheA1 ΔpdhB::neo SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoIIA′-lacZ) cat | pDH87EB-502→SJB253 |
| 501-76IIG | trpC2 pheA1 ΔpdhB::neo SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoIIG′-lacZ) cat | pDH87EB-502→SJB83 |
| 501-76IIIG | trpC2 pheA1 ΔpdhB::neo spoIIIG-lacZ spoIIIGΔ1 | pDH87EB-502→AA001 |
| 501-76VG | trpC2 pheA1 ΔpdhB::neo SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoVG′-lacZ) cat | pDH87EB-502→JCB50 |
| 501-76SSP | trpC2 pheA1 ΔpdhB::neo Φ(sspB′-lacZ) cat | pDH87EB-502→SJB227 |
| 50176COT | trpC2 pheAl ΔpdhB::neo SPβc2 Δ2::Tn917::pSK10Δ6::Φ(cotA′-lacZ) cat | pDH87EB-502→JCB64 |
| 501-77 | trpC2 pheA1 ΔpdhC::neo | pUC9EC-501→JH642 |
| 501-770A | trpC2 pheA1 ΔpdhC::neo Φ(spo0A′-lacZ) cat | pUC9EC-501→UOT-1101 |
| 501-77IIA | trpC2 pheA1 ΔpdhC::neo SPβc2Δ2::Tn917::pSK10Δ6::Φ(spoIIA′-lacZ) cat | pUC9EC-501→SJB253 |
| 501-77IIG | trpC2 pheA1 ΔpdhC::neo SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoIIG′-lacZ) cat | pUC9EC-501→SJB83 |
| 501-77IIIG | trpC2 pheA1 ΔpdhC::neo spoIIIG-lacZ spoIIIGΔ1 | pUC9EC-501→AA001 |
| 501-77VG | trpC2 pheA1 ΔpdhC::neo SPβc2 Δ2::Tn917::pSK10Δ6::Φ(spoVG′-lacZ) cat | pUC9EC-501→JCB50 |
| 501-77SSP | trpC2 pheA1 ΔpdhC::neo Φ(sspB′-lacZ) cat | pUC9EC-501→SJB227 |
| 501-77COP | trpC2 pheA1 ΔpdhC::neo SPβc2 Δ2::Tn917::pSK10Δ6::Φ(cotA′-lacZ) cat | pUC9EC-501→JCB64 |
| 501-78 | trpC2 pheA1 ΔpdhD::neo | pUC9ED-501→JH642 |
| Plasmids | ||
| pUC18 | E. coli cloning vector; Apr | Laboratory stock |
| pDH87 | B. subtilis integrational vector; Apr Camr | 18 |
| pUC18EA | B. subtilis HindIII-EcoRI fragment of pdhA in pUC18; Apr | This study |
| pUC18EA2 | B. subtilis pdhA in pUC18; Apr | This study |
| pDH87EB | B. subtilis HindIII-HindIII fragment of pdhAB in pDH87; Apr Camr | This study |
| pUC9EC | B. subtilis pKTH1877 fragment (containing pdhC) in pUC9; Apr | 16 |
| pUC9ED | B. subtilis pKTH1878 fragment (containing pdhD) in pUC9; Apr | 16 |
| pBEST501 | Neomycin gene donor vector; Apr Neor | 22 |
| pBEST502 | Neomycin gene donor vector; Apr Neor | 22 |
| pUC18EA-502 | Neomycin cassette at PstI site of pdhA of pUC18EA; Apr Neor | This study |
| pDH87EB-502 | Neomycin cassette at EcoRI site of pdhB of pDH87EB; Apr Neor | This study |
| pUC9EC-501 | Neomycin cassette at HpaI site of pdhC of pUC9EC; Apr Neor | This study |
| pUC9ED-501 | Neomycin cassette at SnaBI site of pdhC of pUC9ED; Apr Neor | This study |
| pMUTIN4 | Vector for systematic gene inactivation | 50 |
| pMUTIN4A | pMUTIN4 containing 800 bp of pdhA | This study |
DNA manipulations and transformations.
Methods of endonuclease digestion and ligation were as described by Sambrook et al. (38). Plasmid DNA was isolated from E. coli by alkaline lysis or with a QIAprep spin miniprep kit (Qiagen). Genomic DNA was prepared from E. coli and B. subtilis strains (3) for PCR and transformation. Restriction enzymes were obtained from New England Biolabs, Inc., and Life Technologies, Inc. (Gibco BRL). T4 DNA ligase was obtained from Boehringer Mannheim, Inc., and shrimp alkaline phosphatase was obtained from Promega, Inc. Tth DNA polymerase from Epicentre Technologies, Inc., was used for PCRs. The oligonucleotides were purchased from IDT, Inc. PCRs were carried out for 30 cycles under the reaction conditions specified with the minicycler (MJ Research, Inc.). Agarose gels were prepared and PCR products were electrophoresed in the buffers described by Sambrook et al. (38). E. coli strains were transformed by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories) (8). Competent B. subtilis cells were prepared and transformed by the techniques of Anagnostopoulos and Spizizen (2) Dubnau and Davidoff-Abelson (9).
Construction of mutants with insertions in pdhA, pdhB, pdhC, or pdhD.
The pdhAB genes were prepared by PCR with primers: 5′GTAAACTAATGCATGCTAAGCGGTG and 5′CTTCCCTCTAGATTATGCAGTTTG. Plasmid pUC18EAI was constructed by inserting a HindIII-EcoRI digest of the PCR product, which contains approximately 850 bp of pdhA (250 bp from the initiation codon) and 140 bp of pdhB, into a HindIII/EcoRI digest of pUC18 (51). A neomycin cassette, excised from pBEST502 (22) by digestion with PstI, was then inserted into the PstI site within pdhAB, resulting in plasmid pUC18EAI-502. Plasmid pUC18EAII was constructed by inserting a 1.3-kb fragment of the intact pdhA gene prepared by PCR with the primers (containing BglII sites) 5′-CCTACATAAAAGATCTTGGCAAACTGG and 5′-GCGTAACGCATCCGAGATCTCTTG into the BamHI site of pUC18. Plasmid pUC18EAII-502 is pUC18EAII with a neomycin cassette inserted at the PstI site in the pdhA gene.
Plasmid pDH87EB was constructed by inserting a HindIII fragment of the pdhAB genes, containing 850 bp of pdhA (250 bp from the initiation codon) and 550 bp of pdhB, into the HindIII site of pDH87 (20), resulting in pDH87EB. The neomycin cassette from pBEST501 (22) was isolated after digestion with EcoRI and introduced into pDH87EB, resulting in pDH87EB-501.
The cloned pdhC and pdhD genes in pUC9 were obtained from I. Palva (16) and are designated pUC9EC and pUC9ED, respectively. Plasmids pUC9EC-501 and pUC9ED-501 were constructed by inserting a neomycin cassette from pBEST501 digested with SmaI into the HpaI and SnaBI sites in pdhC and pdhD, respectively. Each plasmid was linearized with ScaI and transformed into B. subtilis JH642, selecting for neomycin resistance. Transformants carrying the pdhB, pdhC, and pdhD gene disruptions were obtained and designated strains 501-76, 501-77, and 501-78, respectively. Transformation with linearized pUC18EAII-502 (SalI) resulted in two types of colonies, designated strains 501-75I and 501-75II.
Since the gene replacement method did not work for constructing a pdhA mutant, an alternative plasmid, pMUTIN4, was used to attempt to disrupt the pdhA gene and place the wild-type gene under the control of the lac promoter (50). An EcoRI-BamHI fragment of 750 bp from the amino end of pUC18EAII was subcloned into pMUTIN4. The resulting plasmid, pMUTIN4A, was transformed into B. subtilis strain JH642, but no transformants were obtained even in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside).
Introduction of the pdhB and pdhC mutations into lacZ fusion strains.
Strain AA001 was constructed by introducing chromosomal DNA from strain KP66 into JH642, screening for chloramphenicol resistance and low sporulation. Chromosomal DNA from strains 501-76 and 501-77 was used for transformation of strains UOT-1101, SJB253, SJB83, AA001, JCB50, SJB227, and JCB64 (Table 1), screening for resistance to both neomycin and chloramphenicol. The resulting strains were designated 501-760A, 501-76IIA, 501-76IIG, 501-76IIIG, 501-76VG, 501-76SSP, 501-76COT, and 501-770A, 501-77IIA, 501-77IIG, 501-77IIIG, 501-77VG, 501-77SSP, and 501-77COT.
Assay of spore formation.
Heat resistance was measured 10, 15, 20, and 25 h after the end of exponential growth in NSM, and the maximum sporulation frequency was recorded. Samples were heated at 80°C for 20 min, and serial dilutions were plated on LB medium containing the appropriate antibiotic.
Enzyme assay.
Cells for PDH assays were grown at 37°C in 25 ml of LB containing the appropriate antibiotics until the beginning of the stationary phase, harvested, and washed twice with a 0.04 M potassium phosphate buffer (pH 7.5). The resulting pellets were frozen rapidly and stored at −80°C. Cell extracts were prepared by resuspending the thawed pellets in 2 ml of the same buffer prior to sonication with a microtip in a Branson model 200 Sonifier (2 min total, with 40-s pulses at 20-s intervals). Cell debris was removed by centrifugation (10 min at 12,000 × g and 4°C), and the supernatants were used for PDH assays at 25°C. The decrease in absorbance was measured at 366 nm by recording the reduction of 3-acetyl-NAD (14) in a Spectronic Genesys 5 spectrophotometer. Protein concentrations were determined by the bicinchoninic acid protein assay reagent (Pierce Chemical Co.). Specific activities (changes of absorbance per minute per milligram of protein) were determined in the range of proportionality between initial reaction velocity and protein concentration.
Cells for β-galactosidase assays were grown at 37°C in 30 ml of NSM containing the appropriate antibiotics until approximately 2 h before the beginning of the stationary phase. Subsequently, 1-ml samples were taken every 2 h for a total of 20 h, and sporulation was monitored by using a phase-contrast microscope. The samples were centrifuged, and the resulting pellets were frozen rapidly and stored at −80°C. Cell extracts were prepared by resuspending the thawed pellets in 200 μl of Z buffer (32) containing 1% (vol/vol) toluene immediately prior to sonication with a microtip in a Branson model 200 Sonifier (1 min total, with 20-s pulses at 20-s intervals). β-Galactosidase activity was assayed at 25°C and expressed as units per absorbance of the culture at 600 nm as described elsewhere (4).
Polyacrylamide gel electrophoresis and immunoblotting.
Cells were grown at 37°C in 3 ml of LB medium containing the appropriate antibiotic until the beginning of the stationary phase, harvested, and washed with 1 ml of distilled water. The resulting pellets were frozen rapidly and stored at −80°C. Cell extracts were prepared by resuspending the thawed pellets in 80 μl of 6 M urea-1% sodium dodecyl sulfate-5 mM dithiothreitol-2 mM phenylmethylsulfonyl fluoride (pH 9.6) prior to sonication (2 min total, with 40-s pulses at 20-s intervals). Cell debris was removed by centrifugation (10 min at 12,000 × g and 4°C), and the supernatant was used for electrophoresis in sodium dodecyl sulfate-10% polyacrylamide gels (38). Samples in Laemmli sample buffer were boiled for 2 min prior to loading onto the gel (28). Procedures for Western transfer and the detection of bound antibody by alkaline phosphatase staining were as described elsewhere (27, 49). The S. aureus PDH antibody and the B. subtilis E2 subunit antibody were from I. Palva (16) and W. Firshein (44), respectively.
Cell fractionation.
B. subtilis JH642 was grown in 300 ml of NSM in a 2-liter flask at 37°C in a New Brunswick shaker-incubator. Eighty milliliters of cells was harvested during exponential growth and at the end of exponential growth and centrifuged at 12,000 × g in a Sorvall SS34 rotor. The pellets were resuspended in 10 ml of 0.04 M KH2PO4-K2HPO4, pH 7.0, and centrifuged as above. The washing was repeated, and the pellets were finally suspended in 1 ml of buffer for passage through a French press at 9,000 lb/in2 (twice). MgCl2 was added to 1 mM, and RNase-free DNase (Boehringer Mannheim, Inc.) was added to 5 U. The extracts were centrifuged at 4,000 × g to remove intact cells. The cell extracts (overlaid with mineral oil) were then centrifuged in a Beckman SW50.1 rotor at 35,000 rpm for 1 h. The supernatants were carefully removed, and these are designated the soluble fraction. The pellets were suspended in phosphate buffer plus 0.2% sodium deoxycholate to the same volume as the soluble fraction and are designated the insoluble fraction. Equal volumes of each fraction (5 to 10 μl) were electrophoresed in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. Immunoblotting and treatment with the E2 antibody were as described above.
Membrane fluorescent staining.
Sporulation was induced by nutrient exhaustion, and samples were harvested and stained as previously described (36, 40).
RESULTS
Transcription of the pdh operon.
On the basis of Northern hybridizations, it was reported that there may be three promoters, one for the entire operon and two that are internal (16) (Fig. 1). The sequence indicated possible σA-type −10 and −35 regions upstream of the pdhA, pdhC, and pdhD genes. The potential promoter for the pdhC gene was unusual, since the spacing was 21 bp rather than the canonical 17 bp. The presence of promoters upstream of the pdhA and pdhC genes was confirmed by reverse transcriptase mapping, but none was found upstream of the pdhD gene (A. Aronson, unpublished results). The former promoter is probably for the entire operon, and the latter is probably for the pdhC and pdhD genes. This organization may reflect the stoichiometry of the PDH subunits in the complex, since 60 E2 subunits are required to form the PDH core (versus 30 tetramers of E1α2E1β2 and 6 dimers of E3). The E3 subunit also functions with other dehydrogenases (5, 19). PDH functions during growth and in the early stages of sporulation for the catabolism of pyruvate, which is excreted during exponential growth (13). Subsequently, the PDH complex dissociates (with some degradation), and there is a relatively greater concentration of the E2 protein in the crude soluble fraction of cell extracts (Fig. 2). This pattern of change agrees with that reported previously (10).
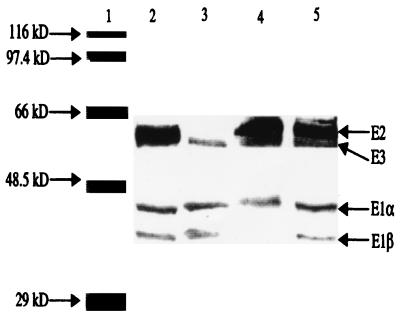
FIG. 2.
Immunoblot of the E2 antigen obtained from the soluble (lanes 1 and 3) and insoluble (lanes 2 and 4) fractions of B. subtilis JH642 grown and sporulated in NSM. Cells were harvested at the end of exponential growth (lanes 1 and 2) and during exponential growth (lanes 3 and 4). Lane 5 contains purified His6-E2 from B. thuringiensis (arrow). Cells were treated as described in Materials and Methods. The ratio for lanes 1 and 2 is 0.54, and that for lanes 3 and 4 is 0.11.
Disruptions of the pdhA, pdhB, pdhC, and pdhD genes.
pdhB, pdhC, and pdhD mutations were constructed by insertion of a neomycin cassette as described in Materials and Methods. These insertions were confirmed by PCR (H. Gao, unpublished results). The PCR products of the pdhB and pdhC genes from the wild type were about 1.3 kb smaller than those from the mutants, indicating that the neomycin cassette of 1.3 kb had been inserted. A similar result was obtained with the pdhD gene. In the case of the pdhA gene, neither insertion of the neomycin cassette nor use of pMUTIN was successful. There was no evidence from PCR of a disrupted pdhA gene in most neomycin-resistant isolates, so either resistance was spontaneous or the cassette was inserted into an unknown location. In about 5% of the neomycin-resistant isolates, there was a duplication such that a copy of an intact pdhA gene and a copy with an insertion were present. Similar duplications have been found in attempts to inactivate “essential” genes with pMUTIN (E. Dervyn, personal communication), indicating that the pdhA gene may be essential for B. subtilis.
No PDH activity was detected in strains 501-76, 501-77, and 501-78 (Table 2). Mixtures of cell extracts from strains 501-76 and 501-77 and from 501-77 and 501-78 restored PDH activity to a low level, suggesting that the mutations were not polar. The absence of only the corresponding subunit of PDH in each mutant was established in immunoblots (Fig. 3), confirming that the mutations were not polar.
TABLE 2.
PDH activity of the wild type and pdh mutants
| Strain | Relevant genotype | PDH activitya |
|---|---|---|
| JH642 | pdh+ | 0.111 ± 0.002 |
| 501-76 | ΔpdhB | 0.000 ± 0.002 |
| 501-77 | ΔpdhC | 0.000 ± 0.002 |
| 501-78 | ΔpdhD | 0.000 ± 0.002 |
| 501-76 + 501-77b | ΔpdhB + ΔpdhC | 0.013 ± 0.002 |
| 501-77 + 501-78b | ΔpdhC + ΔpdhD | 0.009 ± 0.002 |
Change in absorbance at 366 nm per minute per milligram of protein. Values are means ± standard errors.
Mixture of equal amounts (protein concentration) of cell extract from each mutant.
FIG. 3.
Immunoblot with anti-PDH antibody. Lane 1, protein markers; lane 2, 501-78 (ΔpdhD); lane 3, 501-77 (ΔpdhC); lane 4, 501-76 (ΔpdhB); lane 5, JH642 (pdh+).
Growth and sporulation of the pdh mutants.
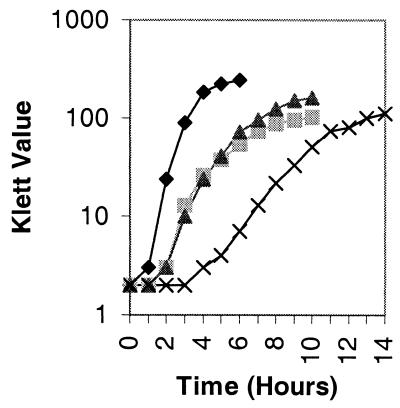
Growth rates (Fig. 4) and sporulation frequencies (Table 3) were measured as described in Materials and Methods. Strains 501-76, 501-77, and 501-78 were unable to grow in a glucose minimal medium, indicating that there are no additional genes encoding subunits which could function for PDH activity. The mutants grew at a lower rate than the wild type and reached stationary phase at a lower cell density. Moreover, the wild type and strain 501-78 lost 20 to 25% of their viable cells in stationary phase, whereas strains 501-76 and 501-77 lost approximately 60% (Table 3). Surprisingly, the maximum cell density of strain 501-78 was higher than that of strain 501-76 and 501-77, although it had a much lower growth rate. In strains 501-76 and 501-77, only 3 to 5% of cells formed heat-resistant spores, whereas sporulation was approximately 50% of the wild-type level for strain 501-78. The same result was obtained by resuspending cells to equivalent optical densities (600 nm) in SM resuspension medium (45), indicating that the neither the medium nor the cell density played a significant role in sporulation. The difference in sporulation frequencies among strains 501-76, 501-77, and 501-78 implied that PDH enzyme activity per se was not essential for sporulation in B. subtilis.
FIG. 4.
Growth in NSM of strains JH642 (pdh+; ⧫), 501-76 (ΔpdhB;  ), 501-77 (ΔpdhC; ▴), and 501-78 (ΔpdhD; ×). For each strain, the starting cell concentration was adjusted to 2 Klett units from an overnight culture.
), 501-77 (ΔpdhC; ▴), and 501-78 (ΔpdhD; ×). For each strain, the starting cell concentration was adjusted to 2 Klett units from an overnight culture.
TABLE 3.
Sporulation of the wild type and pdh mutants in NSM
| Strain | Relevant genotype | Maximum titer (108)a | Titer (108) at t20b
|
Sporulation efficiencyc | Relative sporulation frequencyd | |
|---|---|---|---|---|---|---|
| Cell | Spore | |||||
| JH642 | pdh+ | 6.1 | 4.9 | 4.1 | 0.67 | 1.00 |
| 501-76 | ΔpdhB | 2.1 | 0.8 | 0.05 | 0.03 | 0.05 |
| 501-77 | ΔpdhC | 1.9 | 0.7 | 0.05 | 0.03 | 0.05 |
| 501-78 | ΔpdhD | 3.8 | 2.9 | 1.4 | 0.38 | 0.54 |
Concentration of viable cells at the highest cell density.
The time of onset of stationary phase is designated t0. The sporulation frequency of each strain at t20 was almost the same as the maximum frequency (<5%).
Ratio of spore titer to maximum titer.
Ratio of the sporulation frequency of each strain to that of the wild type.
Effects of divalent cations, pH alterations, and supplementation of metabolites.
Sporulation of most mutants defective in TCA cycle enzymes can be restored by changes of the physiological conditions. We therefore added excess MnCl2, MgSO4, Ca(NO3)2, and FeSO4, separately or together, to the pdhB or pdhC mutants in NSM. The concentration of each divalent cation was raised from the values in NSM [MnCl2, 0.01 M; MgSO4, 0.05 M; Ca(NO3)2, 0.05 M; FeSO4, 0.001 mM] to a 50-fold-higher level, an increase which was effective for rescuing the sporulation of certain TCA cycle mutant strains (7, 12, 15, 21, 23-25). The sporulation frequency was determined by direct counts in the phase-contrast microscope with a Petroff-Hausser chamber. No significant changes in sporulation frequency of the mutants were observed under any of these conditions. Adjustments of the pH of cell cultures (pH 6.0, 6.5, 7.0, 7.5, and 8.0) by addition of Tris-HCl buffers to a final concentration of 0.01 M to NSM when cells entered the stationary phase also did not change the sporulation frequency. When the mutants were grown in NSM supplemented with 0.2% acetate, 0.2% glutamate, 0.2% succinate, and 0.2% citrate, they grew to a higher density (close to half that of the wild-type), but the sporulation frequency did not change.
Expression of sporulation genes in the pdhB and pdhC mutants.
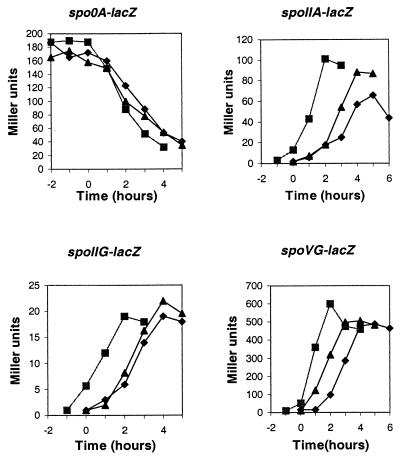
In order to determine the stage in sporulation at which a pdhB or pdhC mutant was blocked, the pdhB and pdhC mutant genes were introduced into a series of strains containing lacZ fusions to promoters transcribed at different stages (Fig. 5). The expression of β-galactosidase under the control of the spo0A promoters (6, 7) showed that vegetative expression of the gene was normal and was shut off at the onset of stationary phase in both strains 501-76 and 501-77. Transcription of the spoIIA, spoIIG, and spoVG genes was activated by σA (spoIIG)- or σH (spoIIA and spoVG)-associated RNA polymerase within 60 min after the sporulation process started. In both mutants, production of β-galactosidase from these early sporulation genes was the same as in the wild type.
FIG. 5.
Effects of pdhB and pdhC mutations on expression of spo0A-lacZ, spoIIA-lacZ, spoIIG-lacZ, and spoVG-lacZ fusions. ▪, JH642 (pdh+); ▴, 501-76 (ΔpdhB); ⧫, 501-77 (ΔpdhC). Strains grown in NSM were taken at the indicated times and assayed for β-galactosidase activity. As indicated for each panel, the strains carried a transcriptional fusion of a spo promoter region to the E. coli lacZ gene. Time zero is the onset of stationary phase.
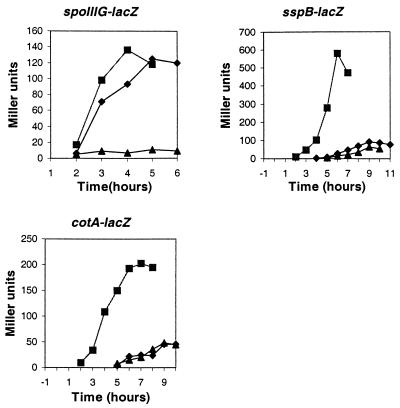
The spoIIIG gene is transcribed by a σF-associated RNA polymerase about the time engulfment is completed (11, 34). Expression of β-galactosidase from this gene in strain 501-76 was barely detectable, whereas it reached the same level in strain 501-77 as in the wild type (Fig. 6). Transcription of the sspB gene typically begins approximately 2 to 3 h after initiation of sporulation in the forespore and depends on σG (47). The σK-directed transcription of the cotA gene is normally induced about 4 h after initiation of sporulation in the mother cell and depends on stage III and most stage V sporulation genes (41). Accumulation of β-galactosidase from sspB-lacZ or cotA-lacZ fusions was greatly reduced in strains 501-76 and 501-77. These results indicated that sporulation of the pdhB mutant was blocked before the completion of engulfment (stage II), whereas sporulation of the pdhC mutant was blocked after engulfment was completed (stage III).
FIG. 6.
Effects of pdhB and pdhC mutations on the expression of spoIIIG-lacZ, sspB-lacZ, and cotA-lacZ fusions. ▪, JH642 (pdh+); ▴, 501-76 (ΔpdhB); ⧫, 501-77 (ΔpdhC).
Assay for membrane fusion during sporulation.
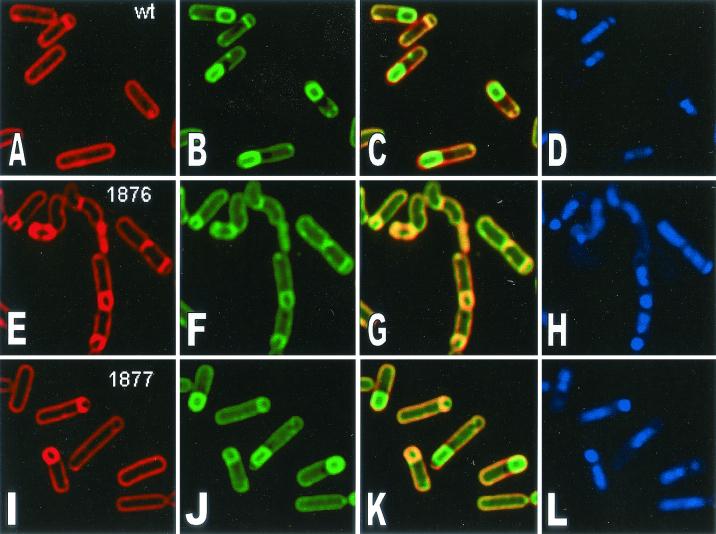
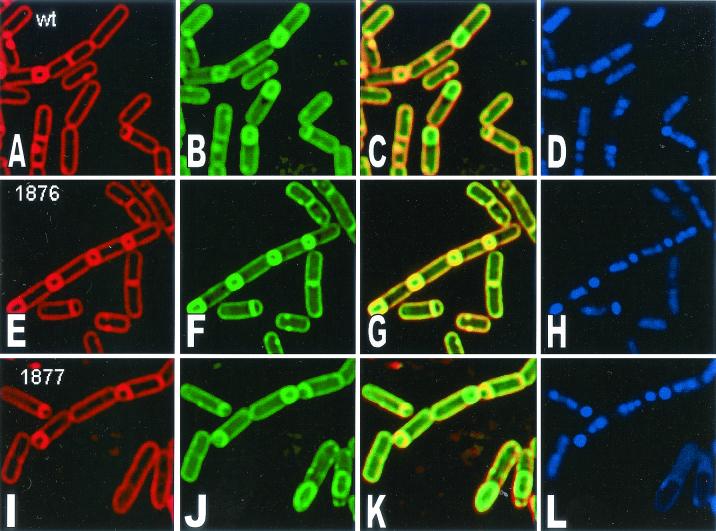
In order to determine more precisely the block in sporulation, a membrane fusion assay that detects the completion of engulfment was employed (40). The fusion assay relies on the fact that the fluorescent membrane dye FM 4-64 is unable to cross lipid bilayers and therefore fails to stain the forespore membranes following the completion of engulfment. A second membrane-permeative stain, MTG (Mitotracker Green FM; Molecular Probes) was used to visualize forespores in fully engulfed and fused sporangia. The chromosomes in both the forespore and the mother cell were stained with DAPI (4′,6′-diamidino-2-phenylindole). In the wild type, most cells formed sporulation septa, and some (30%) finished engulfment and membrane fusion 2 h after the initiation of sporulation (Fig. 7A to D; Table 4). By 4 h after sporulation, 50% had completed engulfment (Fig. 8). However, strain 501-76 failed to complete membrane fusion at either 2 or 4 h after sporulation (Fig. 7 and 8; Table 4), as indicated by staining of the forespore in panels E and F (Fig. 7 and 8). Furthermore, by 4 h, some cells were bent and appeared to be lysing (Fig. 8; Table 4). Strain 501-77 was able to complete engulfment and membrane fusion (Fig. 8, compare panels E and F and panels I and J), but in some of the fused sporangia, the mother cell appeared to be lysing, as judged by the appearance of membrane vesicles and the somewhat collapsed appearance of the mother cell membrane (Fig. 8). Chromosome translocation into the forespore was unaltered in both mutants. These results suggest that E1β but not E2 is essential for the final stages of engulfment.
FIG. 7.
Time course of sporulation in the wild-type strain and pdh mutants. Cells (2 h after the initiation of sporulation) were stained with FM 4-64 (A, E, and I), MTG (B, F, and J), FM 4-64 and MTG (C, G, and K) (membrane staining), and DAPI (D, H, and L) (DNA staining). wt, JH642 (pdh+) (A to D); 1876, 501-76 (ΔpdhB) (E to H); 1877, 501-77 (ΔpdhC) (I to L).
TABLE 4.
Scoring of membrane morphology in the wild type, 501-76, and 501-77
| Membrane morphology | No. (%) of sporangia showing morphologya
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| JH642
|
501-76
|
501-77
|
|||||||
| t2 | t3 | t4 | t2 | t3 | t4 | t2 | t3 | t4 | |
| Flat | 7 (3) | 3 (2) | 4 (2) | 15 (5) | 3 (1.5) | 16 (6) | 1 (<1) | 0 | 0 |
| Curve | 8 (3) | 6 (4) | 11 (7) | 26 (8) | 10 (5) | 8 (3) | 4 (2.5) | 8 (4) | 2 (1) |
| Engulfed | 30 (12) | 17 (11) | 34 (20) | 22 (7) | 19 (10) | 36 (14) | 25 (16) | 36 (20) | 24 (15) |
| Fused | 37 (15) | 36 (24) | 50 (30) | 2 (<1) | 2 (1) | 2 (<1) | 16 (10) | 47 (26) | 30 (19) |
| Lysed | 0 | 6 (4) | 10 (6) | 3 (1) | 10 (5) | 25 (10) | 4 (5) | 20 (11) | 19 (12) |
| Total | 248 | 150 | 166 | 325 | 201 | 251 | 161 | 181 | 160 |
t2, t3, and t4, 2, 3, and 4 h after the onset of sporulation.
FIG. 8.
Time course of sporulation in the wild-type strain and pdh mutants. Cells (4 h after the initiation of sporulation) were stained with FM 4-64 (A, E, and I), MTG (B, F, and J), FM 4-64 and MTG (C, G, and K) (membrane staining), and DAPI (D, H, and L) (DNA staining). wt, JH642 (pdh+) (A to D); 1876, 501-76 (ΔpdhB) (E to H); 1877, 501-77 (ΔpdhC) (I to L).
DISCUSSION
The B. subtilis pdh operon has two promoters, one for the whole operon and an internal one upstream of the pdhC gene. Both promoters are probably recognized by vegetative σA RNA polymerase. The pdhC promoter has a spacing of 21 bp between its −10 and −35 elements rather than the consensus 17 bp. Several other genes in B. subtilis have this pattern. In the case of the spoIIG operon, the −10 and −35 regions are separated by 22 bp (39) and transcription requires binding of the Spo0A protein. Presumably, the binding helps to twist the DNA and bring the −10 and −35 elements to the same face of the DNA helix. Since Spo0A is available in its active form during sporulation (42, 43), this promoter may function only when both σA and Spo0A are present. There are two possible “Spo0A boxes” upstream of the pdhC gene, with one at the −35 region of the promoter (Fig. 1), but they each differ from the consensus by two base pairs. This internal promoter could ensure the synthesis of adequate E2 and E3 for enzymatic function and may also provide additional E2 during sporulation.
The inability to obtain deletions of the pdhA gene by insertion of a neomycin cassette or with the integrational plasmid pMUTIN4 was unexpected. The basis for the neomycin-resistant strains with only the wild-type copy of the pdhA gene is not known. Resistance may be due to a spontaneous mutation or insertion of the cassette at other sites. In about 5% of the resistant isolates, there were two copies of the pdhA gene: one was intact, and the other had an insertion. Similar duplications were found with pMUTIN4 for presumptive essential B. subtilis genes (Dervyn, personal communication). The gene duplication and the inability to obtain gene replacement imply that the pdhA gene may be essential for B. subtilis.
No deletions of the genes encoding the E1 subunit have been reported. However, a partial deletion resulting in an amino-terminus-truncated E1 from Azotobacter vinelandii was obtained (17). The truncated protein could not bind to the E2 core (E1β function in B. subtilis) but retained its catalytic properties (E1α function in B. subtilis) for the oxidative decarboxylation of pyruvate (29, 31). Perhaps such a catalytic function is essential in order to reduce the internal concentration of pyruvate. It is also conceivable that E1α may have a second, nonenzymatic role required for growth.
The pdhB, pdhC, and pdhD genes were insertionally inactivated. Nonpolar mutations were obtained, and all of the mutants grew in an enriched medium but not in a glucose minimal medium. This confirmed that the pdh operon provides the only copy of genes for PDH activity in B. subtilis (33). In contrast to previous reports (1, 16), the pdhC gene was not found to be essential for growth. Strain 501-78 had the lowest growth rate, and the growth rates of strains 501-76 and 501-77 were similar. Surprisingly, strain 501-78 reached a higher cell density than the other two mutants. In addition to the differences in growth and cell density, strain 501-78 sporulation was 10 times higher than that of strains 501-76 and 501-77. The sporulation defect in the latter mutants could not be overcome by the addition of acetate, glutamate, succinate, or divalent cations or by changing the pH of the medium. However, both the growth rate and cell density were greatly improved by the addition of such metabolites. These results confirm that the sporulation and the enzymatic defects in these mutants are separable.
The ability of a pdhD mutant to sporulate could be attributed to the loss of both PDH and ODH activity and thus restoration of a “metabolic imbalance” resulting from inactivation of just PDH activity. Disruption of the odgB gene, however, results in an early block in sporulation (Danchin, personal communication). In addition the odgB pdhC double mutant did not sporulate (Aronson, unpublished); i.e., there was no apparent return to a metabolic balance.
Insertional inactivation of either the pdhB or pdhC gene resulted in inhibition of sporulation at stage II-III. The block in the pdhB mutant was earlier, based on the lack of expression of the spoIIIG-lacZ fusion and the incomplete closure of the forespore membrane (Fig. 6 to 8). In contrast, expression of the spoIIIG-lacZ fusion in the pdhC mutant strain was as in the wild type, and a substantial fraction of the cells completed closure of the forespore membrane. Since σG is under both transcriptional and posttranslational control (26, 30), it is possible that the pdhB mutant is defective at the transcriptional stage, whereas the pdhC mutant is altered at the posttranslational stage.
There is a relative increase in the amount of E2 in the crude soluble cell fraction at the end of exponential growth (Fig. 2) which is sustained until at least stage II of sporulation. There is also a similar increase in soluble E1β (Aronson, unpublished). At about stage II, pyruvate, which had been excreted during growth on glucose, was exhausted, so there was no further major catalytic function for PDH. PDH activity decreases markedly at this time, so these subunits, perhaps as a complex, may have arisen from the dissociation of PDH. In this capacity, they could serve as regulators (perhaps checkpoints) for the continuation of sporulation beyond stage II-III. Each could function at a slightly different time and stage of sporulation or together to regulate critical stage II-III genes. In the absence of E1β, the expression of a subset of the regulated genes involved in the completion of the forespore could be blocked, whereas loss of E2 would permit expression of these genes with repression of those functioning somewhat later.
There is precedence for regulatory functions for PDH subunits. In A. vinelandii, the E1 subunit has a helix-turn-helix motif and binds specifically to the fpr promoter region (37). The E2 subunit from B. subtilis binds to a region of the DNA close to the origin of replication (44). In B. thuringiensis, the E2 subunit was identified as a protein binding specifically to a region about 200 bp upstream from the start of translation (52). Only in the case of the A. vinelandii E1 protein was a regulatory function clearly established. In B. subtilis, suppressors of the sporulation defect in the pdhC mutant strain have been isolated. In one case, suppression resulted from the insertion of Tn10 in the rpoE gene encoding the δ subunit of RNA polymerase (H. Gao and A. Aronson, unpublished results). This result implies a functional interaction of E2 (and perhaps E1β) with RNA polymerase and thus regulation at the level of transcription.
Acknowledgments
We thank A. L. Sonenshein for making lacZ fusion strains available.
This work was supported by research grant MCB-9600584 from the National Science Foundation to A.I.A. H.G. was supported in part by a fellowship from the Purdue Research Foundation.
REFERENCES
- 1.Adler, L.-Å., and S. Arvidson. 1988. Cloning and expression in Escherichia coli of genes encoding a multiprotein complex involved in secretion of proteins from Staphylococcus aureus. J. Bacteriol. 170:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson, A. I. 1994. Flexibility in the protoxin composition of Bacillus thuringiensis. FEMS Microbiol. Lett. 117:21-28. [DOI] [PubMed] [Google Scholar]
- 4.Belitsky, B. R., P. G. Janssen, and A. L. Sonenshein. 1995. Sites required for GltC-dependent regulation of Bacillus subtilis glutamate synthase expression. J. Bacteriol. 177:5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, A., and A. de Kok. 1997. 2-Oxo acid dehydrogenase multienzyme complexes. The central role of the lipoyl domain. Biol. Chem. 378:617-634. [PubMed] [Google Scholar]
- 6.Chibazakura, T., F. Kawamura, and H. Takahashi. 1991. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J. Bacteriol. 173:2625-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig, J. E., M. J. Ford, D. C. Blaydon, and A. L. Sonenshein. 1997. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J. Bacteriol. 180:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 10.Eident-Wilkinson, B., L. Mele, J. Laffan, and W. Firshein. 1992. Temporal expression of a membrane-associated protein putatively involved in repression of initiation of DNA replication in Bacillus subtilis. J. Bacteriol. 174:477-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortnagel, P. 1993. Glycolysis, p. 171-180. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 13.Freese, E., and P. Fortnagel. 1969. Growth and sporulation of Bacillus subtilis mutants blocked in the pyruvate dehydrogenase complex. J. Bacteriol. 99:745-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guest, J. R., and I. T. Creaghan. 1973. Gene-protein relationships of the α-keto acid dehydrogenase complexes of Escherichia coli K12: isolation and characterization of lipoamide dehydrogenase mutants. J. Gen. Microbiol. 75:197-210. [DOI] [PubMed] [Google Scholar]
- 15.Hederstedt, L. 1993. The Krebs citric acid cycle, p. 181-197. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 16.Hemilä, H., A. Palva, L. Paulin, S. Arvidsion, and I. Palva. 1990. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J. Bacteriol. 172:5052-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengeveld, A. F., S. E. Schoustra, A. H. Westphal, and A. de Kok. 1999. Pyruvate dehydrogenase from Azotobacter vinelandii: properties of the N-terminally truncated enzyme. Eur. J. Biochem. 265:1098-1107. [DOI] [PubMed] [Google Scholar]
- 18.Henner, D. J. 1990. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 185:223-228. [DOI] [PubMed] [Google Scholar]
- 19.Hoch, J. A., and H. J. Coukoulis. 1978. Genetics of the α-ketoglutarate dehydrogenase complex of Bacillus subtilis. J. Bacteriol. 133:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodgson, J. A., P. N. Lowe, and R. N. Perham. 1983. Wild-type and mutant forms of the pyruvate dehydrogenase multienzyme complex from Bacillus subtilis. Biochem. J. 211:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ireton, K., S. Jin, A. D. Grossman, and A. L. Sonenshein. 1995. Krebs cycle function is required for activation of the Spo0A transcription factor in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92:2845-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, S., and A. L. Sonenshein. 1994. Identification of two distinct Bacillus subtilis citrate synthase genes. J. Bacteriol. 176:4669-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, S., M. de Jesús-Berríos, and A. L. Sonenshein. 1996. A Bacillus subtilis malate dehydrogenase gene. J. Bacteriol. 178:560-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, S., P. A. Levin, K. Matsuno, A. D. Grossman, and A. L. Sonenshein. 1997. Deletion of the Bacillus subtilis isocitrate dehydrogenase gene causes a block at stage I of sporulation. J. Bacteriol. 179:4725-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karmazyn-Campelli, C., C. Bonamy, B. Savelli, and P. Stragier. 1989. Tandem genes encoding σ-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 3:150-157. [DOI] [PubMed] [Google Scholar]
- 27.Knecht, D. A., and R. L. Diamond. 1984. Visualization of antigenic proteins on Western blots. Anal. Biochem. 136:180-184. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lessard, I. A. D., and R. N. Perham. 1995. Interaction of component enzymes with the peripheral subunit-binding domain of the pyruvate dehydrogenase complex of Bacillus stearothermophilus and assembly of a functional E1 component (α2β2) in vitro. J. Biol. Chem. 269:10378-10383. [PubMed] [Google Scholar]
- 30.Mason, J. M., R. H. Hackett, and P. Setlow. 1988. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J. Bacteriol. 170:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattevi, A., A. D. Kok, and R. N. Perham. 1992. The pyruvate dehydrogenase multienzyme complex. Curr. Opin. Struct. Biol. 2:877-887. [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Neveling, U., S. Bringer-Meyer, and H. Sahm. 1998. Gene and subunit organization of bacterial pyruvate dehydrogenase complexes. Biochem. Biophys. Acta 1385:367-372. [DOI] [PubMed] [Google Scholar]
- 34.Partridge, S. R., and J. Errington. 1993. Importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945-955. [DOI] [PubMed] [Google Scholar]
- 35.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-de Mello, A. Perez, Y.-L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regnström, K., S. Sauge-Merle, K. Chen, and B. K. Burgess. 1999. In Azotobacter vinelandii, the E1 subunit of the pyruvate dehydrogenase complex binds fpr promoter region DNA and ferredoxin I. Proc. Natl. Acad. Sci. USA 96:12389-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Satola, S., P. A. Kirchman, and C. P. Moran, Jr. 1991. Spo0A binds to a promoter used by σA RNA polymerase during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 88:4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slack, F. J., J. P. Mueller, M. A. Strauch, C. Mathiopoulos, and A. L. Sonenshein. 1991. Transcriptional regulation of a Bacillus subtilis dipeptide transport operon. Mol. Microbiol. 5:1915-1925. [DOI] [PubMed] [Google Scholar]
- 42.Smith, I. 1989. Initiation of sporulation, p. 185-210. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of prokaryotic development. American Society for Microbiology, Washington, D.C.
- 43.Sonenshein, A. L. 1989. Metabolic regulation of sporulation and other stationary-phase phenomena, p. 109-130. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of prokaryotic development. American Society for Microbiology, Washington, D.C.
- 44.Stein, A., and W. Firshein. 2000. Probable identification of a membrane-associated repressor of Bacillus subtilis DNA replication as the E2 subunit of the pyruvate dehydrogenase complex. J. Bacteriol. 182:2119-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 47.Sun, D., P. Stragier, and P. Setlow. 1989. Identification of a new sigma-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 3:141-149. [DOI] [PubMed] [Google Scholar]
- 48.Sun, Y., M. D. Sharp, and K. Pogliano. 2000. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J. Bacteriol. 182:2919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 51.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 52.Walter, T., and A. I. Aronson. 1999. Specific binding of the E2 subunit of pyruvate dehydrogenase to the upstream region of Bacillus thuringiensis protoxin genes. J. Biol. Chem. 274:7901-7906. [DOI] [PubMed] [Google Scholar]
- 53.Yamashita, S., H. Yoshikawa, F. Kawamura, H. Takahashi, T. Yamamoto, Y. Kobayashi, and H. Saito. 1986. The effect of spo0 mutations on the expression of spo0A- and spo0F-lacZ fusions. Mol. Gen. Genet. 205:28-33. [DOI] [PubMed] [Google Scholar]