Abstract
A number of bacteriophages belonging to the Microviridae have been described infecting chlamydiae. Phylogenetic studies divide the Chlamydiaceae into two distinct genera, Chlamydia and Chlamydophila, containing three and six different species, respectively. In this work we investigated the biological properties and host range of the recently described bacteriophage Chp2 that was originally discovered in Chlamydophila abortus. The obligate intracellular development cycle of chlamydiae has precluded the development of quantitative approaches to assay bacteriophage infectivity. Thus, we prepared hybridomas secreting monoclonal antibodies (monoclonal antibodies 40 and 55) that were specific for Chp2. We demonstrated that Chp2 binds both C. abortus elementary bodies and reticulate bodies in an enzyme-linked immunosorbent assay. Monoclonal antibodies 40 and 55 also detected bacteriophage Chp2 antigens in chlamydia-infected eukaryotic cells. We used these monoclonal antibodies to monitor the ability of Chp2 to infect all nine species of chlamydiae. Chp2 does not infect members of the genus Chlamydia (C. trachomatis, C. suis, or C. muridarum). Chp2 can infect C. abortus, C. felis, and C. pecorum but is unable to infect other members of this genus, including C. caviae and C. pneumoniae, despite the fact that these chlamydial species support the replication of very closely related bacteriophages.
Chlamydiae are important pathogens of humans and animals. In humans they cause a range of diseases, including trachoma (the world's leading cause of preventable blindness) (25, 30), nonspecific urethritis and pneumonitis (24). Chlamydiae are obligate intracellular bacteria that have a developmental cycle alternating between an inert but infectious extracellular form, the elementary body (EB), and a metabolically active, replicating form, the reticulate body (RB), that resides within a specialized host cell compartment termed an inclusion.
Genome sequences are now available for a number of different chlamydiae. However, there are still significant barriers to investigating chlamydial genes and their regulation and consequently little molecular detail about biological function of gene products is known. The obligate intracellular developmental cycle means there is no host-free system for chlamydial propagation. Thus, only a limited number of phenotypic markers have been available to select a few chlamydial mutants. Recently the taxonomy of chlamydiae has been revised, and the family Chlamydiaceae is now recognized as comprising two separate genera, Chlamydia and Chlamydophila, containing three and six recognized chlamydial species, respectively (10).
The lack of a stable gene transfer system is the most significant barrier to using genetic approaches to understand gene structure/functional relationships in chlamydiae. Recently, a number of Microviridae bacteriophages have been reported (15, 18, 21, 29), which could be used to develop a genetic transfer system. These phages, along with one which infects Bdellovibrio bacteriovorus (4) and Spiroplasma melliferum (8, 23), appear to form a distinct subfamily of microviruses, related, albeit distantly, to coliphage φX174. Within the group infecting various species of chlamydiae, φCPAR39, φCPG1, and Chp2 are very closely related, sharing overall genome identities in excess of 90% (22). Since these phages were isolated from different hosts, C, pneumoniae (φCPAR39), C. caviae (φCPG1), and C. abortus (Chp2), it may be possible to correlate small differences in amino acid sequences with tropism determinants.
While Microviridae tropism is partly governed by extracellular factors affecting host cell recognition (5, 16, 20, 27, 33) intracellular factors also play a critical role. During φX174 DNA packaging, a complex containing two viral proteins, proteins A and C, and the host cell Rep protein must physically interact with the viral procapsid. This interaction is particularly sensitive to small structural variations in the viral protein A, coat, and host cell Rep proteins (9). In addition φX174 does not encode a true lysozyme. Lysis is dependent on the inhibition, by the viral E protein, of translocase I, of a host cell enzyme involved in peptidoglycan biosynthesis (2).
The primary aim of this study was to investigate factors affecting the tropism of the Microviridae infecting chlamydiae. The results of binding studies suggest that host cell recognition is governed only by protein-protein interactions. This represents a fundamental difference from the φX174-like phages, in which a sugar-binding step is also required. In addition, an intracellular tropism factor affecting lysis was also uncovered.
MATERIALS AND METHODS
Cells and chlamydiae.
BGMK cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS). Cells were infected with chlamydiae by centrifugation at 1,000 × g for 1 h in medium containing cycloheximide (1 μg/ml) and gentamicin (25 μg/ml). Infected monolayers were detached with phosphate-buffered saline (PBS) containing 0.125% trypsin-0.02% EDTA and pelleted in DMEM containing 10% FCS at 3,000 × g for 10 min. The infected cell pellet was suspended in PBS-H2O (1:10) and homogenized in a Dounce homogenizer to break open cells and release the EBs. Cell debris was sedimented at 250 × g for 5 min, and the supernatant containing partially purified chlamydiae was mixed with an equal volume of phosphate buffer containing 0.4 M sucrose, stored at −80°C, and used for Chp2 challenge studies.
Further purification was performed by overlaying impure EBs onto 18% Nycodenz (Nycomed, Oslo, Norway) in 5 mM Tris-HCl buffer (pH 7.2) containing 3 mM KCl, 0.3 mM CaNa2EDTA, and 0.13 M NaCl and centrifuging at 55,000 × g for 2.5 h in a Beckman SW28 rotor. A band containing EBs was collected and pelleted at 35,000 × g for 40 min. The pellet was resuspended in PBS and stored in aliquots at −80°C. RBs were prepared from C. abortus strain B577 by two cycles of density gradient centrifugation as previously described (3). Preparations of chlamydiae were verified by PCR using primers U23F and 23SIGR, followed by DNA sequence analysis and BLAST searching of the GenBank database as previously described (11).
Phage preparation and purification.
BGMK cells were grown as monolayers in 25-cm2 flasks in DMEM supplemented with 10% (vol/vol) fetal calf serum. Cells were infected with the C. abortus (strain MA) bearing the Chp2 bacteriophage by centrifugation at 1,000 × g for 1 h in medium containing cycloheximide (1 μg/ml) and gentamicin (25 μg/ml).
At 72 h postinfection the culture medium was replaced with a small volume of phosphate-buffered saline (PBS) and the flasks were frozen at −70°C. One hundred flasks of Chp2-infected chlamydiae were prepared, stored frozen, and then processed as a single batch. Flasks were frozen and thawed three times to lyse the chlamydial RBs and release the Chp2 particles. Any monolayer that had not detached after this procedure was scraped off. The suspension was centrifuged at 2,000 × g for 15 min to sediment cell debris. The supernatant was passed through a 0.45-μm filter followed by a 0.22-μm filter. The filtrate was centrifuged at 100,000 × g in a Beckman SW28 rotor for 3 h and the resultant pellet was washed with PBS and centrifuged at 80,000 × g for 40 min. The pellet was finally suspended in PBS, vortexed with glass beads, and stored at −70°C as a semipurified bacteriophage preparation.
Further purification was performed by cesium chloride equilibrium density gradient centrifugation. Bacteriophages were diluted in 10 mM Tris-150 mM sodium chloride (pH 7.5). Caesium chloride was added to the suspension to a final concentration of 0.48 g/cm3 and centrifuged at 150,000 × g for 24 h in a Beckman SW40 rotor. Optical density at 280 nm (OD280) values, weight measurements, and immunoblot analyses were performed using 1-ml fractions of the gradient.
A sample from each gradient fraction was dotted onto a nitrocellulose membrane and blocked in Tris-HCl (pH 7.5)-0.5 M NaCl-0.05% Tween (TTBS) containing 5% milk powder for 2 h at 37°C. The membrane was washed in TTBS and then incubated overnight at room temperature with Chp2-specific hybridoma culture medium diluted in TTBS containing 10% goat serum. Bound antibody was detected using an anti-mouse alkaline phosphatase conjugate diluted in TTBS containing 5% milk powder and the color was developed with BCIP (15 mg/ml in dimethyl formamide) and NBT (30 mg/ml in 70% dimethyl formamide) substrate in carbonate buffer (0.1 M NaHCO3, 1 mM MgCl2, pH 9.0).
Monoclonal antibody production and fluorescence antibody screening.
BALB/c mice were immunized intraperitoneally (i.p.) with the semipurified phage preparation in complete Freund's adjuvant followed by two i.p. boosts in incomplete Freund's adjuvant. Spleen cells were fused with BALB/c P3-NS-1 plasmacytoma cells in polyethylene glycol.
BGMK cells were grown either in 96-well trays or on 13-mm cover slips in 24-well trays. Cells were infected with the strain C. abortus MA (bearing Chp2) and a strain of C. abortus without bacteriophage infection. From 48 to 72 h postinfection, the culture medium was removed, and monolayers were washed twice in PBS and fixed in ice-cold methanol for 15 min. Medium from the resultant hybridomas was incubated with fixed cells for 40 min at 37°C and washed three times in PBS. Bound antibody was detected with an anti-mouse fluorescein-conjugated antibody (ISL, Paignton, United Kingdom) diluted in 0.0025% Evans Blue dye in PBS. Only those hybridomas that gave positive fluorescence with the Chp2-bearing strain of C. abortus were selected for cloning by limiting dilution and subsequent expansion for monoclonal antibody production.
Host range of Chp2.
A standard inoculum of Chp2 capable of infecting >99% of C. abortus B577 by inclusion staining was mixed with each of a range chlamydial species for 30 min at room temperature. The mixture was diluted in DMEM containing 10% FCS, cycloheximide (1 μg/ml), and gentamicin (25 μg/ml), and BGMK cells cultured on coverslips were infected by centrifugation at 1,000 × g for 1 h. For strain C. pneumoniae AR39 the above procedure was followed except HEP2 cells were used for infection. A set of cover slips were prepared in parallel with unchallenged chlamydiae. When inclusions were clearly visible varying from 48 to 72 h postinfection, monolayers were fixed in ice-cold methanol. The ability of chlamydiae to become infected with Chp2 was monitored by fluorescence microscopy using Chp2-specific monoclonal antibodies.
Neutralization assay.
Equal volumes of Chp2 were mixed with culture medium from bacteriophage-specific monoclonal antibodies 40 and 55 for 1 h at room temperature, and the Chp2-susceptible strain C. abortus (B577) was added to the neutralizing mixture and incubated for 30 min at room temperature. BGMK cells grown on cover slips were infected as above. Monolayers were fixed with ice-cold methanol, and Chp2 infection was detected by fluorescence using the monoclonal antibodies described above.
ELISA and Chp2 binding assays.
The semipurified Chp2 preparation was diluted to 10 μg/ml in 0.05 M carbonate-bicarbonate buffer (pH 9.6) and used to coat immunoasssay trays at room temperature overnight. Wells were washed four times in 0.9% sodium chloride containing 0.05% Tween (enzyme-linked immunosorbent assay [ELISA] wash) and then blocked in 5% milk powder in PBS at 37°C for 1 h, followed by four washes in ELISA wash. A dilution series of monoclonal antibodies 40 and 55 was made in 1% milk powder in PBS containing 0.05% Tween and incubated at 37°C for 3 h. Wells were washed four times, and bound antibody was detected with anti-mouse horseradish peroxidase (HRP) conjugate (ISL, Paignton, United Kingdom) and TMB substrate. The level of antibody was optimized for subsequent binding assays.
Purified EBs of B577 (C. abortus), VR628 (C. pecorum), VR1310 (C. pneumoniae), and L1 and UW1 (C. trachomatis) were diluted to 10 μg/ml in 0.05 M carbonate-bicarbonate buffer (pH 9.6) and used to coat immunoassay trays at room temperature overnight. These ELISA trays were washed and blocked as above. Semipurified Chp2, diluted in PBS, was added to individual wells for 1 h at 37°C. Wells were washed four times, and the monoclonal antibody was added, diluted in 1% milk powder in PBS containing 0.05% Tween, and incubated for 3 h at 37°C. Wells were washed four times, and antibody was detected as described above.
Enzyme and chemical treatments of EBs and RBs.
Preparations of purified EBs and RBs from C. abortus (B577) and C. trachomatis (L2) were adjusted to 1 mg/ml in PBS. Reactions were set up by taking 50 μl of EB or RB suspension, to which were added (i) proteinase K, 1 mg/ml; (ii) phospholipase C, 50 U/ml; (iii) sodium periodate, 2 mM; and (iv) PBS. Samples were incubated at 37°C for 1 h, except the proteinase K sample, which was incubated at 65°C for 30 min. Proteinase K enzyme activity was terminated by the addition of phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM. Treated samples were diluted to 10 μg/ml in carbonate buffer and used to coat immunoassay trays. Wells were washed and blocked as described above. Semipurified Chp2 was added to wells at a concentration of 10 μg/ml in PBS and incubated at 37°C for 1 h. Chp2 bound to treated chlamydiae was detected with monoclonal antibodies 40 and 55 and a standard ELISA procedure.
PCR amplification and cloning of VP1, -2, and -3 genes for in vitro expression.
Oligonucleotide primers were synthesized using β-cyanoethylphosphoramidite chemistry on an automated synthesizer (Expedite 8909; PerCeptive Biosystems). The primers used for amplification of the VP1 gene were Vp1F (5′-ATTTCTTCACTTGGATCCATGGTTAGGA10-3′ ) and VP1R (5′-ATTTCTTCACTTCAGCTGTTAGAAATGAT1687-3′). To amplify the VP2 and VP3 genes, the following pairs of primers were used: Vp2F (5′-CGTAAGGGATCCATGAATCCCGAACAACTTACG1824-3′), Vp2R (5′-GCAGTA AAGCTTCTACCTTCCTTTTCTTGA2347-3′), Vp3F (5′-GCATACGGATCCATGTTTAAGTCGGCATATTCC2385-3′), and Vp3R (5′-GAGCGTAAGCTTTCATTTTTGGGCTAATCCAGG2791-3′).
The forward primers contain a BamHI restriction site (italic), and the reverse primer contains either a PvuII or a HindIII restriction site to facilitate directional cloning of the amplification products into the expression vector pRSET A (Invitrogen, Amsterdam, The Netherlands). Additional nucleotides were added to the 5′ terminus of each oligonucleotide primer to allow efficient restriction enzyme cleavage of the PCR amplicon for subsequent cloning. Amplifications, using Bio-X-Act DNA polymerase (Bioline, London, United Kingdom) with purified Chp2 RF as the template, were performed in a Perkin Elmer 9600 thermal cycler for 30 cycles of denaturation at 94°C for 15 s; annealing at 56°C for 15 s; and extension at 72°C for 40 s. PCR amplicons were purified using the Wizard PCR Prep system (Promega, Southampton, United Kingdom). Following restriction endonuclease cleavage, PCR amplicons were cloned into pRSETA, and recombinant plasmids were transformed into Escherichia coli strain JM101 maintained on Luria broth (LB) containing ampicillin (50 μg/ml).
SDS-PAGE and analysis of monoclonal antibody specificity by radioimmunoprecipitation assay.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the discontinuous buffer system method of Laemmli (17a) with 10% acrylamide gels (acrylamide-bisacrylamide, 38.5:1, wt/wt). In vitro protein synthesis was performed by using a T7 RNA polymerase-coupled reticulocyte lysate system (TNT; Promega, Southampton, United Kingdom) in accordance with the manufacturer's instructions. The TNT rabbit reticulocyte system is an in vitro eukaryotic transcription-translation system. It requires a circular DNA or RNA template containing a T7 promoter. The recombinants we used were circular templates containing a T7 promoter that can be expressed in the TNT system, yielding a product which is free of background proteins which occur in the E. coli S30 system.
Translation products from the TNT system (6 μl) were incubated with 2 μl of undiluted antisera in 600 μl of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.15 M NaCl, 0.1% SDS, 0.5% Empigen BB [N-dodecyl-N,N-dimethylglycine], 0.1 mM phenylmethylsulfonyl fluoride). After incubation at 37°C for 1 h, goat anti-mouse or goat anti-rabbit immunoglobulin G attached to beaded agarose (Sigma) was added to adsorb immune complexes. The beads were washed three times in RIPA buffer and once in phosphate-buffered saline before derivatization in sample-dissociating buffer and separation by SDS-PAGE. Gels were stained and prepared for autoradiography by treatment with 1 M sodium salicylate-50% methanol for 30 min at room temperature and then dried under vacuum and exposed to Kodak XAR-5 film at −70°C.
Electron microscopy.
BGMK cells or HEP 2 cells were infected with C. abortus MA and C. pneumioniae AR 39, respectively, in a six-well tray. At 48 to 72 h postinfection monolayers were washed in PBS and fixed in 3% glutaraldehyde in 0.1% cacodylate buffer and processed for negatively stained thin-section grids (18).
RESULTS AND DISCUSSION
Monoclonal antibodies to Chp2.
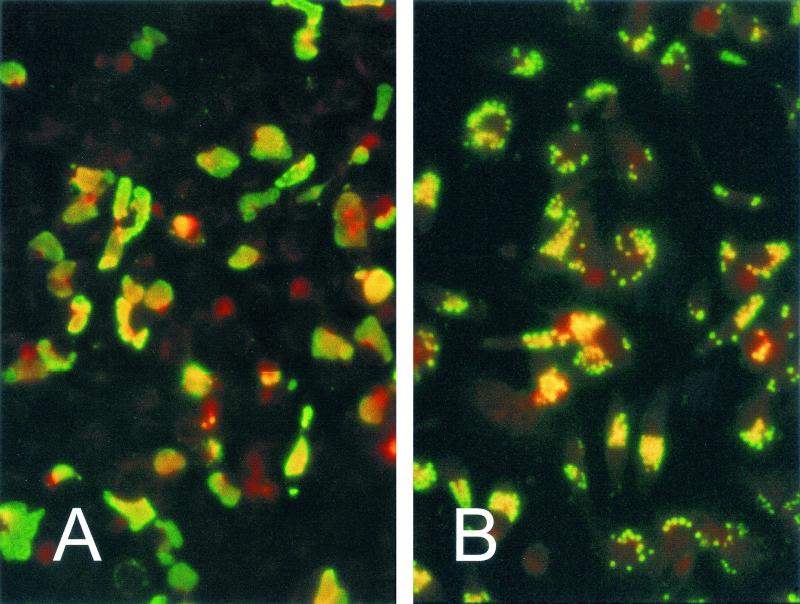
The obligate intracellular nature of the chlamydial developmental cycle has severely restricted studies on this microorganism. It is not possible to measure chlamydiaphage infectivity using traditional quantitative approaches or to determine the concentration of Chp2 preparations as specific titers because there is no plaque assay system. An alternative method to monitor infection by Chp2 is to use specific antisera. Our approach was to develop monoclonal antibodies to investigate the infection process using immunofluorescence (IF) techniques. A key factor in our plan was to produce monoclonal antibodies specific only for Chp2 that gave no fluorescent backgrounds with either chlamydiae or eukaryotic host cells. Our experimental design was to hyperimmunize mice with semipurified Chp2 and produce monoclonal antibodies by standard techniques. Hybridomas that were secreting antibodies specific for Chp2 were selected by IF screening of BGMK cells infected with the MA strain of C. abortus carrying Chp2 and the uninfected C. abortus strain A22. Two hybridomas were chosen that showed similar immunofluorescence (IF) patterns only with the Chp2-bearing C. abortus (Fig. 1A).
FIG. 1.
Immunofluorescence staining of Chp2-infected C. abortus (panel A) and CPAR39-infected C. pneumoniae (panel B). Monoclonal antibody 55 (which recognizes Chp2 VP1) was visualized with FITC-conjugated IgG. In C. pneumoniae, enlarged infected chlamydiae can be identified within inclusions; by contrast, in C. abortus, the whole inclusion is stained. Host cells are counterstained with Evans Blue, and both samples were taken at 72 h postinfection.
The properties of the monoclonal antibodies secreted by the two hybridomas were investigated further by IF, ELISA, RIPA, immunoblotting, and neutralization studies. While both monoclonal antibodies could detect Chp2 infection by immunofluorescence, monoclonal antibody 55 bound specifically to recombinant coat protein VP1 by RIPA (Fig. 2A), whereas monoclonal antibody 40 did not react with recombinant Chp2 structural proteins VP1, -2, or -3. Neither monoclonal antibody recognized Chp2 antigens in Western blotting experiments (data not shown) and thus, they recognize different conformation-dependent epitopes on the Chp2 surface. The monoclonal antibodies did not block Chp2 binding to susceptible host chlamydiae, indicating that these are not neutralizing antibodies.
FIG. 2.
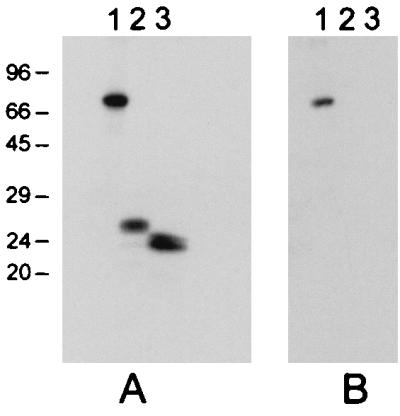
In vitro transcription-translation of Chp2 structural protein genes. The positions of the molecular weight markers are indicated to the left of panel A (in kilodaltons). Lane 1, VP1. Lane 2, VP2. Lane 3, VP3. In panel B, monoclonal antibody 55 immunoprecipitated the coat protein VP1, as indicated in lane 1.
Properties of Chp2.
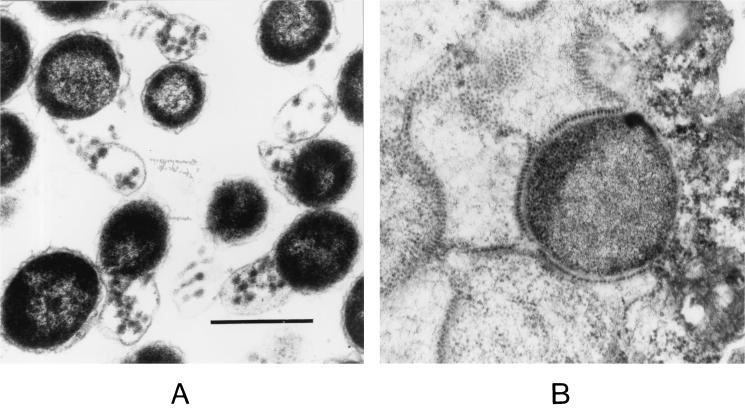
Initially C. pneumoniae AR39 was also tested with both these monoclonal antibodies because the complete genome of this isolate was recently sequenced and shown to carry a small Chp2-like replicon closely related to Chp2 (21). In contrast to C. abortus infected with Chp2, inclusions of C. pneumoniae carrying φCPAR39 did not stain with monoclonal antibody 40. However, IF staining with monoclonal antibody 55 showed a characteristic staining pattern (Fig. 1B). In C. pneumoniae carrying φCPAR39 individual large reticulate bodies could be seen within inclusions; this was very different from C. abortus (MA) infected by Chp2, where whole inclusions fluoresced (Fig. 1A). These results suggest that φCPAR39 may not be able to efficiently lyse host cells. In addition, the nature of Chp2- and φCPAR39-infected chlamydiae was investigated at higher magnification by thin section EM analysis. Chp2-infected C. abortus showed the characteristic lysis pattern of RBs previously described (Fig. 3A). This was in sharp contrast to C. pneumoniae, where the virions formed sheets and beaded patterns within membranes of RBs (Fig. 3B).
FIG. 3.
Thin-section transmission electron micrographs of Chp2-infected chlamydiae. (A) Chp2/C. abortus and (B) φCPAR39/C.pneumoniae. Both samples were taken at 72 h postinfection. The scale bar represents 0.5 μm.
To investigate further the physical properties of Chp2, it was purified by cesium chloride equilibrium density centrifugation. A band representing purified Chp2 could not be seen by direct light illumination of the gradient; therefore, the gradient was divided into 1 ml fractions and tested for the presence of Chp2 antigen by dot blot analysis with monoclonal antibodies 40 and 55. Maximum color reaction was observed in gradient fractions at a buoyant density of 1.3 g/cm3. These gradient fractions were combined and Chp2 particles were pelleted by centrifugation. The presence of bacteriophage Chp2 particles in the pellet was confirmed by direct negative-stain electron microscopy (data not shown).
Host range of Chp2.
A standard inoculum of Chp2 was prepared and optimized by dilution and used initially to infect the susceptible host C. abortus strain B577. The standard inoculum infected >99% of B577 inclusions as judged by IF. Chlamydial strain titers were determined, and inocula were prepared that allowed infection of all cells in the monolayer. Twenty-two different chlamydial strains representing all nine currently recognized chlamydial species in both genera from either our own chlamydial culture collection, the American Type Culture Collection (ATCC) or other sources were initially screened for the presence of Chp2 related bacteriophages by IF with both monoclonal antibodies 40 and 55 (Table 1). The inclusions were screened for the presence of Chp2 infection by IF at 48 to 72 h postinfection, a negative result was scored as no fluorescent inclusion staining. The results are summarized in Table 1, which shows that Chp2 is able to infect other C. abortus strains, C. felis, and C. pecorum.
TABLE 1.
Susceptibility of chlamydial strains to infection with bacteriophage Chp2
| Genus | Species | Source or reference | Strain | Susceptibility to Chp2 infectiona
|
|||
|---|---|---|---|---|---|---|---|
| Prechallenge
|
Postchallenge
|
||||||
| MAb 40 | MAb 55 | MAb 40 | MAb 55 | ||||
| Chlamydophila | C. abortus | VR-656 | B577 | − | − | + | + |
| 28 | A22 | − | − | + | + | ||
| 19 | S26/3 | − | − | + | + | ||
| Jones | S95/3 | − | − | + | + | ||
| 13 | BA1 | − | − | + | + | ||
| C. psittaci | VR-125 | 6BC | − | − | − | − | |
| 12 | CAL 10 | − | − | − | − | ||
| C. felis | 7 | FP | − | − | + | + | |
| C. caviae | VR813 | GPIC | − | − | − | − | |
| C. pecorum | VR-628 | E58 | − | − | + | + | |
| Griffiths | BE53 | − | − | + | + | ||
| C. pneumoniae | VR 1310 | CWL 029 | − | − | − | − | |
| 6 | IOL 207 | − | − | − | − | ||
| VR-2282 | TW 183 | − | − | − | − | ||
| ATCC 53592 | AR 39 | − | + | − | + | ||
| Chlamydia | C. trachomatis | 32 | TW-5 | − | − | − | − |
| 1 | UW-1 | − | − | − | − | ||
| 17 | NL-D | − | − | − | − | ||
| 31 | Nl 1 | − | − | − | − | ||
| 26 | L1/440/LN | − | − | − | − | ||
| C. suis | Sachse | DC 6 | − | − | − | − | |
| C. muridarum | VR123 | MoPn | − | − | − | − | |
MAb, monoclonal antibody.
The six remaining chlamydial species were refractory to Chp2 infection. Three C. trachomatis serovars (B-TW5, C-UW1 and E-NI 1) showed some residual, low-level fluorescence staining following the primary challenge with Chp2. However, this disappeared completely following subsequent passages of these chlamydiae. Failure of a bacteriophage to infect a potential host can be accounted for by several explanations: no attachment, attachment but no penetration, penetration of the host bacterium but no replication, or active production of virions.
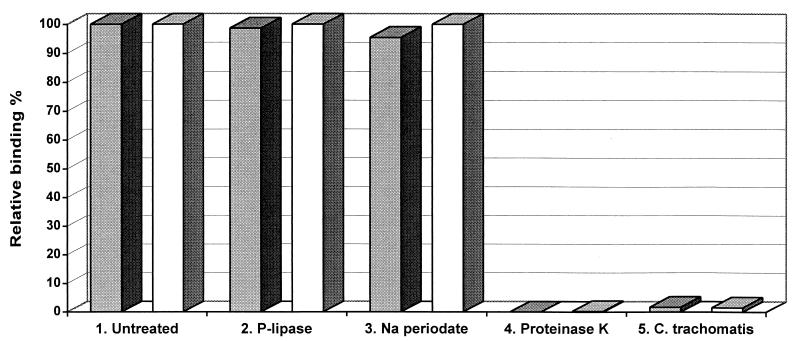
To investigate whether Chp2 could bind to non-susceptible chlamydiae a range of purified EBs and RBs were tested for binding in an ELISA format. The results showed similar strong binding to C. abortus (both RBs and EBs; Fig. 4), C. pecorum and C. felis (data not shown) and no significant binding to other non-susceptible chlamydiae. These results are consistent with the observation, from thin section EM, that the bacteriophage C. caviae binds EBs and RBs through a specific receptor molecule present on the outer membrane surface of susceptible host chlamydial strains (14).
FIG. 4.
Effect of cell surface modification on binding of Chp2 to purified EBs and RBs. Binding of Chp2 to chlamydial EBs (dark columns) and RBs (light columns) was measured by ELISA with monoclonal antibody 55. The untreated control (data set 1) is compared with various treatments (data sets 2, 3, and 4) indicated beneath the columns. Data set 5 shows binding of Chp2 to C. trachomatis EBs and RBs.
To characterize the nature of C. abortus cell surface molecules participating in the binding of Chp2, purified RBs and EBs were incubated with phospholipase C, sodium periodate, and proteinase K. The binding of Chp2 to EBs and RBs each treated separately with phospholipase C, sodium periodate and proteinase K together with appropriate controls was assessed by ELISA with monoclonal antibodies 40 and 55. The results (Fig. 4) showed that binding of Chp2 was unaffected by phospholipase C and sodium periodate but abolished following proteinase K treatment. These experiments demonstrated that the major determinant of tropism is external and the chlamydial Chp2 receptor is a protein.
Host cell recognition by φX174 is a two-step process. Initial, and reversible, attachment involves a coat protein sugar binding site located near the threefold axis of symmetry (16). However, the results of host range studies (5, 20, 27, 33) indicate the involvement of a second receptor, which most likely interacts with the fivefold related spike and DNA pilot proteins. The results of the cell binding experiments suggest that the Chp-like members of the Microviridae have evolved a different cell recognition mechanism, one that circumvents the initial sugar-binding site. Accordingly the residues which constitute the sugar-binding site in the φX174-like phages are not conserved in the coat proteins of the Chp2-like phages.
Chipman et al. (8) hypothesized that the mushroom-like protrusions found at the threefold axes of symmetry in the SpV4 cryoimage reconstruction may play a role in receptor recognition. This hypothesis is attractive due to the loss of the fivefold related spike proteins and threefold related sugar-binding sites found in the φX174-like phages. In addition the Chp2-like viral coat proteins exhibit the greatest divergence in this region. However, direct involvement of these protrusions has yet to be demonstrated. Further host range studies and/or the selection of bacteriophage mutants with expanded host range phenotypes will either prove or refute this hypothesis.
At present we know little about the molecular architecture and function of the chlamydial outer membrane; thus, the identification of the Chp2 receptor and viral anti-receptor remains an important goal. Moreover, there is no genetic transfer system for the chlamydiae and this has placed severe constraints on advancing our knowledge of the biological function of chlamydial gene products. The chlamydial microviruses are the only vectors for these obligate intracellular bacteria; thus, a detailed understanding of their basic biology is crucial for the development of these bacteriophages as potential chlamydial gene transfer vehicles.
Acknowledgments
We are grateful to G. Jones, P. Griffiths, and K. Sachse for the kind gift of Chlamydophila spp.
This work was supported by Wellcome Trust grant no. 063882.
REFERENCES
- 1.Alexander, E. R., S. P. Wang, and J. T. Grayston. 1967. Further classification of TRIC agent from ocular trachoma and other sources by the mouse toxicity test. Am. J. Ophthalmol. 63:1469-1478. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt, T. G., D. K. Struck, and R. Young. 2001. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J. Biol. Chem. 276:6093-6097. [DOI] [PubMed] [Google Scholar]
- 3.Birkelund, S., A. G. Lundenmose, and G. Christiansen. 1988. Chemical cross-linking of Chlamydia trachomatis. Infect. Immun. 56:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brentlinger, K. L., S. Hafenstein, C. R. Novak, B. A. Fane, R. Borgon, R. McKenna, and M. Agbandje-McKenna. 2002. Microviridae, a family divided: isolation, characterization, and genome sequence of φMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J. Bacteriol. 184:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, J. J., M. R. Badgett, H. A. Wichman, J. P. Huelssenbeck, D. M. Hillis, A. Gulati, C. Ho, and I. J. Molineux. 1997. Exceptional evolution in a virus. Genetics 147:1497-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, M. W. A., S. A. H. Almahdawi, I. G. Giles, J. D. Treharne, M. E. Ward, and I. N. Clarke. 1991. Nucleotide-sequence and taxonomic value of the major outer-membrane protein gene of Chlamydia pneumoniae IOL-207. J. Gen. Microbiol. 137:465-475. [DOI] [PubMed] [Google Scholar]
- 7.Cello, R. M. 1967. Ocular infections in animals with PLT (Bedsonia) group agents. Am. J. Opthalmol. 63:1270-1273. [DOI] [PubMed] [Google Scholar]
- 8.Chipman, P. R., M. Agbandje-McKenna, J. Renaudin, T. S. Baker, and R. McKenna. 1998. Structural analysis of the spiroplasma virus, SpV4: implications for evolutionary variation to obtain host diversity among the Microviridae. Structure 6:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekechukwu, M. C., D. J. Oberste, and B. A. Fane. 1995. Host and phiX174 mutations which affect the morphogenesis or stabilization of the 50S complex, a single stranded DNA synthesizing intermediate. Genetics 140:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, K. D. E., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 11.Everett, K. D. E., L. J. Hornung, and A. A. Andersen. 1999. Rapid detection of the Chlamydiaceae and other families in the order Chlamydiales: three PCR tests. J. Clin. Microbiol. 37:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis, T. J., and T. P. Magill. 1938. An unidentified virus producing acute meningitis and pneumonitis in experimental animals. J. Exp. Med. 68:147-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths, P. C., J. M. Plater, T. C. Martin, S. L. Hughes, K. J. Hughes, R. G. Hewinson, and M. Dawson. 1995. Epizootic bovine abortion in a dairy-herd-characterization of a Chlamydia psittaci isolate and antibody-response. Br. Vet. J. 151:683-693. [DOI] [PubMed] [Google Scholar]
- 14.Hsia, R. C., H. Ohayon, P. Gounon, A. Dautry-Varsat, and P. M. Bavoil. 2000. Phage infection of the obligate intracellular bacterium. Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect. 2:761-772. [DOI] [PubMed] [Google Scholar]
- 15.Hsia, R. C., L. M. Ting, and P. M. Bavoil. 2000. Microvirus of Chlamydia psittaci strain guinea pig inclusion conjunctivitis: isolation and molecular characterization. Microbiology 146:1651-1660. [DOI] [PubMed] [Google Scholar]
- 16.llag, L. L., R. McKenna, M. P. Yadav, J. N. BeMiller, N. L. Incardona, and M. G. Rossmann. 1994. Calcium induced structural changes in bacteriophage phiX174. J. Mol. Biol. 244:291-300. [DOI] [PubMed] [Google Scholar]
- 17.Johansson, M., K. Schon, M. Ward, and N. Lycke. 1997. Genital tract infection with Chlamydia trachomatis fails to induce protective immunity in gamma interferon receptor-deficient mice despite a strong local immunoglobulin A response. Infect. Immun. 65:1032-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Liu, B. L., J. S. Everson, B. Fane, P. Giannikopoulou, E. Vretou, P. R. Lambden, and I. N. Clarke. 2000. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J. Virol. 74:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClenaghan, M., A. J. Herring, and I. D. Aitken. 1984. Comparison of Chlamydia psittaci isolates by DNA restriction endonuclease analysis. Infect. Immun. 45:384-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newbold, J. E., and R. L. Sinsheimer. 1970. Process of infection with bacteriophage phiX174. J. Virol. 5:427-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read, T. D., C. M. Fraser, R. C. Hsia, and P. M. Bavoil. 2000. Comparative analysis of Chlamydia bacteriophages reveals variation localized to putative receptor binding domain. Microb. Comp. Genomics 5:223-231. [DOI] [PubMed] [Google Scholar]
- 23.Renaudin, J., M. Pascarel, and J. Bové. 1987. Spiroplasm virus 4: nucleotide sequence of the viral DNA, regulatory signals, and proposed genome organization. J. Bacteriol. 169:4950-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, and P. H. Makela. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii:983-986. [DOI] [PubMed]
- 25.Schachter, J., and C. R. Dawson. 1990. The epidemiology of trachoma predicts more blindness in the future. Scand. J. Infect. Dis. 69(Suppl.):55-62. [PubMed] [Google Scholar]
- 26.Schachter, J., and K. F. Meyer. 1969. Lymphogranuloma venereum. II. Characterization of some recently isolated strains. J. Bacteriol. 99:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinsheimer, R. L. 1968. Bacteriophage phi-X174 and related viruses. Prog. Nucleic Acid Res. Mol. Biol. 8:115-169. [PubMed] [Google Scholar]
- 28.Stamp, J. T., A. D. McEwen, J. A. A. Watt, and D. I. Nisbet. 1950. Enzootic abortion in ewes. I. Transmission of the disease. Vet. Rec. 62:251-256. [DOI] [PubMed] [Google Scholar]
- 29.Storey, C. C., M. Lusher, and S. J. Richmond. 1989. Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J. Gen. Virol. 70:3381-3390. [DOI] [PubMed] [Google Scholar]
- 30.Thylefors, B., A. D. Negrel, R. Pararajasegaram, and K. Y. Dadzie. 1995. Global data on blindness. Bull. W. H. O. 73:115-121. [PMC free article] [PubMed] [Google Scholar]
- 31.Tuffrey, M., P. Folder, J. Gale, and D. Taylor-Robinson. 1986. Salpingitis in mice induced by human strains of Chlamydia trachomatis. Br. J. Exp. Pathol. 67:605-616. [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, S. P., and J. T. Grayston. 1963. Classification of trachoma virus strains by protection of mice from toxic death. J. Immunol. 90:849-856. [PubMed] [Google Scholar]
- 33.Weisbeek, P. J., J. H. Van de Pol, and G. A. Van Arkel. 1973. Mapping of host range mutants of bacteriophage φX174. Virology 52:408-416. [DOI] [PubMed] [Google Scholar]