Abstract
TraG-like proteins are potential NTP hydrolases (NTPases) that are essential for DNA transfer in bacterial conjugation. They are thought to mediate interactions between the DNA-processing (Dtr) and the mating pair formation (Mpf) systems. TraG-like proteins also function as essential components of type IV secretion systems of several bacterial pathogens such as Helicobacter pylori. Here we present the biochemical characterization of three members of the family of TraG-like proteins, TraG (RP4), TraD (F), and HP0524 (H. pylori). These proteins were found to have a pronounced tendency to form oligomers and were shown to bind DNA without sequence specificity. Standard NTPase assays indicated that these TraG-like proteins do not possess postulated NTP-hydrolyzing activity. Surface plasmon resonance was used to demonstrate an interaction between TraG and relaxase TraI of RP4. Topology analysis of TraG revealed that TraG is a transmembrane protein with cytosolic N and C termini and a short periplasmic domain close to the N terminus. We predict that multimeric inner membrane protein TraG forms a pore. A model suggesting that the relaxosome binds to the TraG pore via TraG-DNA and TraG-TraI interactions is presented.
Bacterial conjugation is responsible for the spread of genetic traits among a broad range of bacterial species. It is the primary mechanism for dissemination of antibiotic resistance among human pathogens (60). Nearly all functions required to mediate bacterial conjugation are encoded by conjugative plasmids, which are usually further endowed with antibiotic resistance genes (64). In general, transfer (Tra) proteins are grouped into functional classes, defined as those involved in mating pair formation (Mpf) and DNA processing (Dtr). Secretion systems used by some pathogens, such as Agrobacterium tumefaciens, Bordetella pertussis, Helicobacter pylori, and Legionella pneumophila, for delivering effector molecules to eukaryotic cells are composed of protein components evolutionarily related to those of Mpf complexes (12). Such macromolecular transfer systems ancestrally related to the conjugative Mpf complexes are called type IV secretion systems, as originally proposed by Salmond (46). Each of these systems secretes distinct DNA and/or protein substrates. TraG-like proteins (named for the protein of IncP plasmid RP4 [31]) are present in all conjugative DNA transfer systems and in several type IV secretion systems. Although TraG-like proteins are essential components in these secretion systems (12), their function remains poorly understood.
The Dtr systems of conjugative plasmids are best characterized at the initiation stage of DNA processing. The relaxosome (20) is a complex of several Dtr proteins (relaxosomal proteins) bound to a specific DNA sequence, the origin of transfer (oriT) of the conjugative plasmid. This complex initiates DNA transfer by producing a single-stranded scission at the nic cleavage site within oriT. In this reaction, the catalytic key component, called the relaxase, becomes transiently attached to the 5′ end of the DNA single strand via a phosphodiester bound with its active-site tyrosine residue (42). By acting as DNA chaperones, auxiliary DNA binding proteins facilitate this reaction. The Mpf system comprises the subunits for pilus biogenesis and a set of membrane-associated proteins which mediate intimate contact between donor and recipient cells (48).
A TraG-like protein is the only RP4 component needed in addition to the Mpf system for mobilization of non-self-transmissible IncQ plasmid RSF1010 (6, 9, 30). TraG proteins have been named coupling proteins since they were said to mediate specific interaction between the Dtr and the Mpf functions (9, 10, 23, 30). Physical interactions between relaxosomal proteins and TraG-like proteins are suggested by genetic experiments (9, 10, 23, 30) and biochemical data (17, 25, 43, 54). In contrast, no direct evidence for an interaction between a TraG-like protein and an Mpf component has been reported so far.
TraG-like proteins have two common motifs resembling the type A and B nucleotide binding motifs of NTP hydrolases (NTPases) and ABC transporters (31, 32, 50). These motifs were shown to be essential for transfer activity (4) and nucleotide binding activity (type A motif) (37). Two TraG-like proteins have been biochemically analyzed so far: TraD of F (IncF) and TrwB of R388 (IncW). TraD was characterized as an inner membrane protein with potential DNA binding ability and ATPase activity (40). However, no ATPase activity has been detected for a soluble fragment of TrwB lacking the transmembrane part (TrwBΔN70) (37), and the ATPase activity present in TraD was assigned to an impurity (this study). TraD was shown to interact with TraM of F (17), a relaxosomal component binding to three different sites within the oriT region of F (16). Membrane topology analysis of TraD revealed that TraD has a short cytoplasmic N terminus, followed by a periplasmic domain of about 60 residues and a long cytoplasmic C-terminal tail (29). A similar topology was determined for the related VirD4 protein of the Ti plasmid of A. tumefaciens (15). The crystal structure of R388 TrwBΔN70 has recently been solved (22). It consists of a ring-shaped hexamer of identical subunits, strongly resembling the structure of ring helicases and of F1 ATPase (21). Apart from binding to nucleotides, TrwBΔN70 was reported to bind to double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) (37). Furthermore, TrwBΔN70 was demonstrated to enhance relaxase activity of R388 TrwC and topoisomerase activity of E. coli topoisomerase I (37). The genome sequence of H. pylori (57) revealed a coding sequence for a TraG-like protein, HP0524, which is encoded by a gene of the cag pathogenicity island (PAI). The cag PAI is responsible for virulence of H. pylori (1, 11) and contains a set of genes with homology to virulence genes of type IV secretion systems and transfer genes of conjugative transfer systems (13). HP0524 is essential for CagA export (52) but is dispensable for interleukin-8 (IL-8) induction in gastric epithelial cells (18). In another study, deletion of hp0524 was seen to cause an increase of IL-8 induction, implying that HP0524 downregulates IL-8 induction (14), although this result could not be confirmed (18).
Here, three members of the family of TraG-like proteins, TraG (RP4), TraD (F), and HP0524 (H. pylori), were biochemically characterized in detail. We showed that these proteins form oligomers, bind to DNA, and do not hydrolyze NTPs. By the use of surface plasmon resonance (SPR), RP4 TraG was demonstrated to specifically interact with relaxase TraI. The membrane topology of TraG was determined, revealing that TraG has cytosolic N and C termini, which are separated by a short periplasmic domain close to the N terminus. We use these data to propose a model of the RP4 relaxosome's architecture. In this model, the relaxosome is attached to the postulated TraG inner membrane pore by TraG-TraI and TraG-DNA interactions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Escherichia coli strains (Table 1) were grown in yeast extract-tryptone (YT) medium (36) or YT medium with additives (YT+) (26). NZCYM medium (47) was buffered with 25 mM 3(N-morpholino)propanesulfonic acid (MOPS), pH 8.0. When appropriate, antibiotics were added as follows: ampicillin (sodium salt; 100 μg/ml), chloramphenicol (10 μg/ml), kanamycin sulfate (40 μg/ml), nalidixic acid (30 μg/ml), epicillin (dihydroampicillin; 100 μg/ml), and tetracycline-HCl (15 μg/ml). Plasmids are listed in Table 2.
TABLE 1.
E. coli strains used in this study
| E. coli strain | Source, reference, or genotype | Growth mediuma |
|---|---|---|
| SCS1 | Stratagene | YT+ |
| HB101 | 7 | YT+ |
| HB101 Nxr | Spontaneous, nalidixic acid-resistant derivative of HB101 | YT+ |
| BL21(DE3)pLysS | Novagen | NZCYM |
| XL1-Blue | Stratagene | YT |
| XK1200 | 38 | YT |
| SG13109 | 39 | YT |
| CC191 | F128 lac1q Δ(lacZ)m15 traD/Δ(ara-leu)7697 araD139 Δ(lac)X74 galE galK thi rpsE phoA20 rpoB argE(am) recA1 | YT |
Compositions are in Materials and Methods.
TABLE 2.
Plasmids and phages used in this study
| Plasmid | Description | Relevant genotype | Selective marker(s)a | Reference or source |
|---|---|---|---|---|
| M13mJF182 | M13mp18Ω(pJF142 BamHI-BamHI, 0.8-kb fragment) | oriT | 63 | |
| pBS140 | pJF119HEΔ(HincII-SphI)Ω(RP4 48710-46495b) | traG+ | Ap | 61 |
| pDB127 | pDB126Δ(RP4 SfiI-SspI 48374-46670) | (trbB-trbM)+(traF-traM)+traG0 | Cm | 4 |
| pET14b | Vector for production of His6 fusion proteins | Ap | Novagen | |
| pFS141 | pMS470Δ8Δ(NdeI-HindIII)Ω(RP4 48495-46588 his6c) | traG-his6+ | Ap | This work |
| pFS241 | pMS470Δ8Δ(NdeI-HindIII)Ω(his6 RP4 48495-46588) | his6-traG+ | Ap | This work |
| pHY524Δ1 | pET14bΔ(NdeI-BamHI)Ω(Hp 550217-551951d) | his6-hp0524Δ1+ | Ap | This work |
| pJF142 | pBR329Ω(BamHI, BamHI-XmaIII linker, RP4 XmaIII-XmaIII 50995-51770, XmaIII-BamHI linker) | oriT | Ap, Cm | 20 |
| pJF143 | pBR329Ω(BamHI, BamHI-XmaIII linker, RP4 XmaIII-AccI 50995-51269, AccI-BamHI linker) | oriT | Ap, Cm | 20 |
| pKI410 | HpaI-BamHI fragment from pMP1 in pSPORT | traD+ | Ap | 33 |
| pMS119EH | Vector, Plac/lacIq | Ap | 53 | |
| pMS470Δ8 | Vector, pMS119EHΔ(XbaI-PstI)Ω(pT7-7 XbaI-NdeI, 40-bp fragment, R751 traC AvaI-SphI, 1.4 kb), Plac/lacIq | traC+ | Ap | 5 |
| pOX38traD411 | In vivo recombinant of pOX38 × pK1411 | traD0 | Km | 33 |
| pSK410 | pMS470Δ8Δ(NdeI-HindIII)Ω(F 23779-25937e) | traD+ | Ap | This work |
| pSK410CH | pMS470Δ8Δ(NdeI-HindIII)Ω(F 23779-25937 his6) | traD-his6+ | Ap | This work |
| pSK410NH | pMS470Δ8Δ(NdeI-HindIII)Ω(his6 F 23779-25937) | his6-traD+ | Ap | This work |
| pSK470 | pMS470Δ8Δ(NdeI-HindIII)Ω(RP4 48495-46588) | traG+ | Ap | This work |
| pSK470ΔB | pSK470Δ(BamHI)Ω(GATC) | traG+ | Ap | This work |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin.
his6 sequence: CACCATCACCATCACCAT.
Reagents were obtained from the following suppliers: Ni-nitrilotriacetic acid (NTA) Superflow, Qiagen; Superdex 200 columns and radioactive nucleotides, Amersham Pharmacia Biotech; nucleotides, Roche Molecular Biochemicals; enzymes, New England Biolabs; Brij 58, Zwittergent 3-14, and Zwittergent 3-16, Fluka; Triton X-100, Sigma.
Buffers.
Buffers (compositions are in parentheses) were as follows: buffer A (100 mM Tris-HCl [pH 7.6], 40 mM NaCl, 8% [wt/vol] sucrose, 0.44 mg of lysozyme/ml, 0.15% [wt/vol] Brij 58), buffer B (50 mM Tris-HCl [pH 7.6], 1 M NaCl, 0.25% Brij 58), buffer C (20 mM Tris-HCl [pH 7.6], 100 mM NaCl), buffer D (50 mM 2-[N-cyclohexylamino]ethanesulfonic acid [CHES]-NaOH [pH 9.5], 1 M NaCl, 5 mM MgCl2, 1% [vol/vol] Triton X-100), buffer E (50 mM CHES-NaOH [pH 9.5], 500 mM NaCl), buffer F (50 mM Tris-HCl [pH 8.7], 1 M NaCl, 10 mM Zwittergent 3-14, 1 mM dithiothreitol [DTT], 0.1 mM EDTA, 10% [wt/vol] glycerol), buffer G (50 mM Tris-HCl [pH 7.6], 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 10% [wt/vol] glycerol), buffer H (20 mM Tris-HCl [pH 7.6], 500 mM NaCl, 2 mM DTT, 0.1% [wt/vol] Brij 58, 0.1 mM EDTA), and buffer I (20 mM Tris-HCl [pH 7.6], 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.05% [wt/vol] Brij 58, 50 μg of bovine serum albumin [BSA]/ml).
DNA techniques.
Standard molecular cloning techniques were performed as described previously (47). PCR fragments were generated with DeepVent DNA polymerase (New England Biolabs) or, for his6-hp0524Δ1, Pfu DNA polymerase (Stratagene). Nucleotide sequences of PCR fragments were verified. RP4 traG (accession no. X54459) in pSK470 was obtained as a full-length gene by PCR by using pBS140 as a template. Primers were CGACGACTCATATGAAGAACCGAAACAACG and GCCTACGAAGCTTGGTGAGGCGCTGGAAGC (nucleotides corresponding to the RP4 sequence are italicized). his6-traG and traG-his6 in pFS241 and pFS141, respectively, were generated by PCR with pBS140 as the template and primer pair GCATTCCCATATGCACCATCACCATCACCATAAGAACCGAAACAACG and GCCTACGAAGCTTGGTGAGGCGCTGGAAGC and primer pair CGACGACTCATATGAAGAACCGAAACAACG andGCTAATAAGCTTGCTCAATGGTGATGGTGATGGTGTATCGTGATCCCCTCCC (deviations from the original sequence are underlined). Full-length F traD (accession no. NC002483) in pSK410 was obtained as an NdeI/HindIII PCR fragment with pKI410 as the template and primers CGACGACTCATATGAGTTTTAACGC and GCCTACGAAGCTTCATCAGAAATCATC. Primers for his6 fusions to traD were GCATTCCCATATGCACCATCACCATCACCATAGTTTTAACGCAAAGG, GCCTACGAAGCTTCATCAGAAATCATC (his6-traD in pSK410NH), CGACGACTCATATGAGTTTTAACGC, and GCTAATAAGCTTCATCAATGGTGATGGTGATGGTGGAAATCATCTCCCGG(traD-his6 in pSK410CH). H. pylori hp0524 (accession no. AE000566) was generated as a truncated his6 fusion (his6-hp0524Δ1) by PCR on H. pylori DNA with primers GGCCCCATATGCGGACTAGAGATATAGGAGCG and CCGGATCCTCACAGTTCGCTTGAACCCACAGG. pHY524Δ1 was constructed by inserting this NdeI/BamHI PCR fragment into pET-14b. For construction of pSK470ΔB, the unique BamHI site in pSK470 was disrupted by filling in its 5′ overhangs with T4 DNA polymerase. ssDNA was isolated from phage M13mp18 (63) or from phage M13mJF182, which contains the T strand of RP4 oriT (Table 2) (35).
Isolation of the traG insertion mutants.
E. coli strain CC191 carrying pSK470ΔB was infected with replication-deficient λ phages containing either TnlacZ/in or TnphoA/in transposons, as described previously (29). Transpositions of the ISlacZ/in or ISphoA/in element onto the plasmid were isolated by selecting for plasmid-borne chloramphenicol resistance (Cmr). About 85,000 Cmr colonies were screened, of which 20 (11 from the screen for lacZ fusions and 9 from the screen for phoA fusions) were identified as different in-frame translational fusions to traG after DNA sequence analysis (see Table 6). The IS elements were removed from the traG sequence by in vitro manipulation of the plasmids with BamHI digestion and religation. This resulted in a 31-codon insertion left in the traG gene at the site of the original transposition event. For all of these mutations, 27 of the 31 inserted codons were the same, with variation at each end caused by duplication of target sequences during the transposition event: XDSYTQVASWTEPFPFSIQGDPRSDQETXXX. The plasmids generated were named ptraG::i31X, and the resulting TraGi31 insertion mutant proteins were named TraGiX, with “X” designating the last unaltered residue of TraG before the insertion sequence (Table 6).
TABLE 6.
Transfer activity and trypsin sensitivity of TraG insertion mutants
| traG plasmid | TraG protein | No. of transconjugants/donor | Transfer activitya | Trypsin sensitivityb |
|---|---|---|---|---|
| pSK470 | TraG | 40 | ++ | − |
| pMS119 | None | <10−6 | − | ND |
| ptraG::i31Ser30 | TraGiS30 | 4 × 10−4 | +/− | ND |
| ptraG::i31Gly32 | TraGiG32 | l × 10−4 | +/− | ND |
| ptraG::i31Gly53 | TraGiG53 | 10 | ++ | + |
| ptraG::i31Gly74 | TraGiG74 | 2 × 10−2 | + | + |
| ptraG::i31Gly87 | TraGiG87 | 8 × 10−5 | +/− | + |
| ptraG::i31Ala119 | TraGiA119 | 1 × 10−3 | + | − |
| ptraG::i31Glu141 | TraGiE141 | 3 | ++ | ND |
| ptraG::i31Ser153 | TraGiS153 | 2 | ++ | − |
| ptraG::i31Glu175 | TraGiE175 | 4 × 10−3 | + | ND |
| ptraG::i31Asn243 | TraGiN243 | 7 × 10−1 | ++ | − |
| ptraG::i31Glu255 | TraGiE255 | 4 × 10−2 | + | ND |
| ptraG::i31Ala344 | TraGiA344 | 5 × 10−3 | + | − |
| ptraG::i31Ser397 | TraGiS397 | 74 | ++ | − |
| ptraG::i31Gly455 | TraGiG455 | <10−6 | − | ND |
| ptraG::i31Lys473 | TraGiK473 | <10−6 | − | ND |
| ptraG::i31Asn482 | TraGiN482 | 6 × 10−5 | +/− | − |
| ptraG::i31Glu516 | TraGiE516 | 2 × 10−4 | +/− | − |
| ptraG::i31His517 | TraGiH517 | <10−6 | − | ND |
| ptraG::i31Glu552 | TraGiE552 | <10−6 | − | ND |
| ptraG::i31Val612 | TraGiV612 | 61 | ++ | − |
Transfer activity is defined as positive (++) with wt TraG (pSK470) or negative (−) without TraG (pMS119) expressed from the complementary plasmid in the mating experiment. + and +/−, intermediate and weak activities, respectively.
ND, not determined.
Characterization of the TraGi31 mutant proteins.
The stability and steady-state expression levels of the TraG insertion mutant proteins were examined by Western blot analysis, using an antiserum specific for the insertion epitope (34). Of the 20 different mutant proteins, only 3 did not accumulate to significant levels in the cells. The membrane topology of TraG was characterized by a trypsinization assay of spheroplast cells expressing stable TraG mutants, as previously described (29). Briefly, cells carrying ptraG::i31 expression plasmids were converted to spheroplasts and then incubated with trypsin. Spheroplasts were collected in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and proteins were examined on Western blots with the 31-residue-specific antiserum. The ability of the TraGi31 insertion mutant proteins to participate in RP4-mediated conjugation was examined in conjugation experiments (see below).
Conjugations.
Mating experiments to determine transfer frequencies of RP4-mediated conjugation were carried out on filters (4). Conjugation of the F plasmid was as described previously except that mating was permitted for 45 min (28).
Protein purification.
For overproduction, broth cultures of E. coli strains carrying the appropriate plasmid were grown at 30°C. IPTG (isopropyl-β-d-thiogalactopyranoside)-mediated induction of expression, cell harvest, resuspension, and freezing of cells were performed as described previously (26). Further steps were carried out at 4°C or on ice unless noted otherwise. The RP4 TraI protein was purified as described previously (41). RP4 TraG, F TraD, and HP0524 were purified as N-terminally hexahistidine (His6)-tagged or truncated (Δ) derivatives His6-TraG, His6-TraD, and His6-HP0524Δ1. These derivatives were used in all assays and were referred to as TraG, TraD, and HP0524Δ1, respectively. Protein purity and concentrations were determined by laser-densitometric quantification of Coomassie blue (Serva)-stained gels using serial concentrations of BSA as a reference with ImageQuant software, version 5.0 (Molecular Dynamics). Western blot analysis of RP4 TraG was performed with TraG-specific antiserum (66). Purified proteins were stored at −20°C in buffer containing 50% (wt/vol) glycerol.
Purification of RP4 TraG.
SCS1 (pFS241) cells (35 g, resuspended in 200 ml) were thawed, supplemented with 400 ml of buffer A, and stirred for 90 min at room temperature. After centrifugation at 100,000 × g for 60 min, the supernatant was kept and the pellet was resuspended in 100 ml of buffer B with a Dounce homogenizer. The suspension was centrifuged as before, and the supernatants of both centrifugation steps were combined (fraction I, 660 ml). Proteins were precipitated by addition of (NH4)2SO4 (60% saturation), collected by centrifugation at 25,000 × g for 60 min, and resuspended in 100 ml of buffer C (fraction II, 100 ml). Fraction II was dialyzed against buffer C and applied to a Ni-NTA column. The column was washed with buffer C and buffer C-20 mM imidazole [pH 7.6]. Proteins were eluted with buffer C-250 mM imidazole. Fractions containing TraG were pooled (fraction III, 85 ml) and concentrated by dialysis against buffer C-20% (wt/vol) polyethylene glycol 20000.
Purification of F TraD.
SCS1(pSK410NH) cells (18 g, resuspended in 90 ml) were thawed, supplemented with 180 ml of buffer A, and stirred and centrifuged as described for RP4 TraG. The supernatant was discarded, and the pellet was resuspended with a homogenizer in 28 ml of buffer D. The suspension was centrifuged as before, and the supernatant was collected (fraction I, 28 ml). Fraction I was applied to a Ni-NTA column equilibrated with buffer E. The column was washed stepwise with buffer E and with buffer E-35 mM imidazole [pH 7.6]. The TraD protein was eluted with buffer E-250 mM imidazole. Fractions containing TraD were pooled (fraction II, 38 ml) and concentrated by (NH4)2SO4 precipitation (40% saturation). The precipitate was resuspended in 13 ml of buffer D and dialyzed against the same buffer.
Purification of HP0524Δ1.
BL21DE3(pLysS) cells carrying pHY524Δ1 (6 g, resuspended in 30 ml) were thawed and supplemented with 30 ml of lysis buffer (0.5% [wt/vol] Brij 58, 1.8 M NaCl, 0.1 M Tris-HCl [pH 7.6], 1.34 mg of lysozyme/ml, 10% [wt/vol] sucrose). The mixture was stirred for 80 min and centrifuged for 45 min at 81,000 × g. The supernatant was discarded, and the pellet was washed twice by thorough resuspension with a homogenizer, first with 38 ml of wash buffer 1 (0.25% [wt/vol] Brij 58, 1 M NaCl, 50 mM Tris-HCl [pH 7.6], 10 mM spermidine·3HCl, 1 mM EDTA) and second with 38 ml of wash buffer 2 (10 mM Zwittergent 3-16, 1 M NaCl, 50 mM Tris-HCl [pH 8.7]) and centrifuged as before. The remaining pellet was resuspended in 250 ml of wash buffer 2 containing Zwittergent 3-14 instead of Zwittergent 3-16. The suspension was sonicated (Sonifier; Branson), warmed briefly to 25°C, and centrifuged as before. The supernatant (fraction I, 250 ml) was concentrated by (NH4)2SO4 precipitation (40% saturation) and resuspension of the precipitate in 18 ml of buffer F (fraction II, 18 ml). Fraction II was dialyzed against buffer F and subjected to gel filtration on a HiLoad 16/60 Superdex 200-pg column. The purest fractions were pooled and concentrated by dialysis against buffer F-20% (wt/vol) polyethylene glycol 20000 (fraction III, 12 ml).
Gel filtration.
A Superdex 200 HR 10/30 column was calibrated with a gel filtration standard (Bio-Rad) consisting of four globular proteins of 670, 158, 44, and 17 kDa and vitamin B12 (1.4 kDa). The column was run with buffer G or with buffer F at a flow rate of 0.4 ml/min. Protein elution was monitored at 280 nm. Elution volumes of reference proteins were identical for both buffer systems. TraG, TraD, and HP0524Δ1 were subjected to gel filtration under the same conditions with buffer G for TraG and TraD and buffer F for HP0524Δ1. A trend line correlating the elution volumes of the gel filtration standards to the corresponding Mr was used to obtain an estimate of the Mrs of TraG, TraD, and HP0524Δ1.
Glycerol gradient centrifugation.
Purified TraG (630 μg) or TraD (372 μg) in 150 μl of buffer H was layered on a 3.7-ml, 15 to 35% (wt/vol) linear glycerol gradient. Centrifugation was at 270,000 × g for 38 h at 3°C. Fractions of 250 μl were collected, analyzed by SDS-PAGE, and tested for ATPase activity (see below). Three proteins were centrifuged in a parallel experiment for reference: aldolase (158 kDa, s20,w = 7.8), BSA (67 kDa, s20,w = 4.4), and ovalbumin (43.5 kDa, s20,w = 3.6).
Sequence alignment of TraG-like proteins.
A BLAST search (2) for TraG-like proteins was performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/), with the RP4 TraG sequence as the query. From the search results, 19 representative proteins with significant similarity to RP4 TraG were chosen. The sequences were aligned with Clustal W software (55), with the BLOSUM series as the protein weight matrix. Profile alignments were used for alignments of more distantly related sequences to a set of aligned TraG-like sequences.
NTPase assay.
NTPase activity was determined by monitoring hydrolysis of [γ-32P]-labeled nucleoside triphosphates (NTPs) as described previously (26) except for using 50 mM Tris-HCl (pH 7.6) instead of 50 mM CHES (pH 9.5). When appropriate, 50 ng of ssDNA/μl was added. P4α protein (0.25 μM) and HP0525 protein (0.25 μM) with known NTPase activity (26, 67) served as positive controls in the experiments. The measured ATPase activity was expressed as the rate of conversion of ATP (in micromolar per minute) in the presence of 1 μM protein and 50 ng of ssDNA/μl.
Fragment retardation assay.
pJF143 (Table 2) was digested with EcoRI and BamHI, yielding an oriT-containing fragment of 287 bp and additional fragments of 608 and 3,543 bp. After 5′ labeling with [γ-32P]ATP and T4 polynucleotide kinase, the fragments (0.75 nM each) were incubated for 30 min at 37°C with increasing amounts of protein in a total volume of 20 μl of buffer I. In competition experiments, the previously incubated protein-DNA mixture was supplemented with increasing amounts of ssDNA and incubated for 30 min at 37°C. Samples were electrophoresed on nondenaturing polyacrylamide gels as described previously (67). The 32P-labeled DNA bands were visualized by the storage phosphor method (24) and analyzed with ImageQuant software (Molecular Dynamics).
Transmission electron microscopy.
dsDNA (25 fmol of pJF143 digested with EcoRI and BamHI) and ssDNA (25 fmol of M13mp18) were incubated for 10 min at room temperature with TraG or TraD (0.48 pmol each). Following fixation with 0.2% (wt/vol) glutaraldehyde for 10 min, the samples were prepared for electron microscopy (Philips; EM400) by adsorption to mica as described previously (51).
Protein interaction analysis by SPR.
The interaction between RP4 TraG and TraI was observed with the BIAcore system (Pharmacia Biosensor AB). The TraI protein (571 resonance units [RU]) was covalently coupled to flow cell 3 of Pioneer chip B1 with the BIAcore amine coupling kit. Second and third flow cells served as controls and were loaded with 231 RU of BSA (flow cell 2) or without protein (flow cell 1; saturated with ethanolamine). HBS-EP buffer (BIAcore) was used as the eluant and dilution buffer. TraG was injected at increasing concentrations (10 to 200 nM) at a rate of 100 μl/min for 2 min. Dissociation was monitored for 5 min. Between injections, flow cells were washed with 10 μl of 0.2 M Na2CO3 and 10 μl of HBS-EP-0.01% (wt/vol) SDS. The association rate constant (ka) was determined by using the Langmuir binding model for a 1:1 interaction between analyte and ligand.
RESULTS
RP4 TraG, F TraD, and cag HP0524 were purified after N-terminal modification.
Since several attempts to purify native TraG and native TraD failed, the proteins were N-terminally modified for affinity purification on Ni-NTA. The full-length traG and traD genes were fused to six histidine codons (his6) at either end by PCR amplification, yielding pFS241, pFS141, pSK410NH, and pSK410CH (Table 2). To verify the in vivo activity of the modified proteins, the plasmids were assayed for complementation in mating experiments (Table 3). Both N-terminal His6 fusions (proteins encoded by pFS241 and pSK410NH) complemented the transfer-defective phenotype conferred by plasmids pDB127 (ΔtraG) and pOX38 traD411 (ΔtraD) as efficiently as the wild-type (wt) alleles in trans. These constructions were therefore selected for protein purification and further experiments. A C-terminal His6 fusion to TraG did not alter the ability of the protein to promote DNA transfer, while the C-terminal fusion to TraD resulted in a reduction of the transfer frequency of 3 orders of magnitude, compared to that for wt proteins.
TABLE 3.
Transfer activity of modified TraG and TraD proteins
| Donor strain (transfer deficient)a | Recipient straina | Plasmid tested for complementation | Description | Transfer rateb |
|---|---|---|---|---|
| HB101(pDB127) (Cm) | HB101 Nxr (Nx) | pFS241 | his6-traG | 5.0 × 10−1 |
| pFS141 | traG-his6 | 3.0 × 10−1 | ||
| pSK470 | wt traG (overproducer) | 3.0 × 10−1 | ||
| pBS140 | wt traG | 5.0 × 10−1 | ||
| pMS119EH | Vector alone | <1.0 × 10−6 | ||
| XK1200(pOX38traD411) (Km) | SG13109 (Tc) | pSK410NH | his6-traD | 3.3 × 100 |
| pSK410CH | traD-his6 | 2.0 × 10−3 | ||
| pSK410 | wt traD (overproducer) | 7.6 × 100 | ||
| pK1410 | wt traD | 3.7 × 100 | ||
| pMS119EH | Vector alone | <1.0 × 10−7 |
Antibiotic resistances are in parentheses. Nx, nalidixic acid; Tc, tetracycline.
The transfer rate is given by the number of transconjugants per donor cell. Transconjugants were selected for expression of Cm and Nx (HB101 Nxr) resistance or Km and Tet (SG13109) resistance.
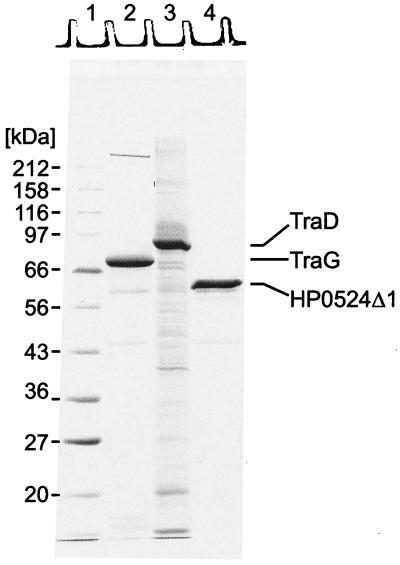
TraG was overproduced in E. coli cells harboring plasmid pFS241 (Table 2). Proteins from the crude cell extract were precipitated by addition of (NH4)2SO4 (60% saturation) and were applied to Ni-NTA. Elution from the affinity matrix yielded a TraG preparation with a purity of 55% (Table 4 and Fig. 1). TraG migrates as a 70-kDa protein on SDS-polyacrylamide gels. Its identity was confirmed by N-terminal sequencing. The TraG preparation was further analyzed by Western blotting using polyclonal antibodies against TraG (data not shown). The major impurity, a 59-kDa protein, was identified by N-terminal sequencing as a C-terminally truncated His6-TraG derivative.
TABLE 4.
Purification of TraG, TraD, and HP0524Δ1
| Protein | Fraction | Purification step | Amt of protein (mg) | Yield (%) | Purity (%)a |
|---|---|---|---|---|---|
| TraG | I | Crude extract | 2,017 | 100 | 22 |
| II | (NH4)2SO4 precipitation | 1,139 | 57 | 18 | |
| III | Ni-NTA | 87 | 4.3 | 55 | |
| TraD | I | Crude extract | 179 | 100 | 35 |
| II | Ni-NTA | 34 | 19.2 | 42 | |
| HP0524Δ1 | I | Crude extract | 107 | 100 | 50 |
| II | (NH4)2SO4 precipitation | 81 | 76 | 50 | |
| III | Gel filtration | 48 | 45 | 72 |
Laser-densitometric evaluation of Coomassie blue-stained gels.
FIG. 1.
The N-terminally modified forms of TraG, TraD, and HP0524 were solubilized and purified as described in Materials and Methods. Purification steps and yields are listed in Table 4. For each protein 7 μg was loaded onto an SDS-15% polyacrylamide gel, resolved by electrophoresis, and stained with Coomassie blue R. Lane 1, marker proteins; lane 2, purified TraG (His6-TraG); lane 3, purified TraD (His6-TraD); lane 4, purified HP0524Δ1 (His6-HP0524Δ1).
TraD was overproduced in E. coli cells harboring plasmid pSK410NH. The protein was solubilized in a buffer containing 1% Triton X-100 as described previously (40). It was then purified by affinity chromatography on Ni-NTA, yielding a His6-TraD preparation of only 42% purity but containing no major impurities (Table 4; Fig. 1). By SDS-PAGE, His6-TraD has a size of 81 kDa.
Attempts to overproduce the full-length HP0524 failed. However, overproduction of an N-terminally truncated protein lacking the first 170 residues (HP0524Δ1) was achieved with plasmid pHY524Δ1 (Table 2). The deletion of the first 170 residues of HP0524 removes three potential transmembrane helices predicted by the PHDhtm program (http://cubic.bioc.columbia.edu/predictprotein). The primary difficulty in the purification of HP0524Δ1 was the solubilization of the protein. Extracts of induced E. coli(pHY524Δ1) cells contained large amounts of HP0524Δ1, but these remained insoluble under nondenaturing conditions when Brij 58, Triton X-100, CHAPS (3-[{3-cholamidopropyl}-dimethylammonio]-1-propanesulfo-nate), Zwittergent 3-16, and other detergents were used. Solubilization of HP0524Δ1 was achieved by the use of Zwittergent 3-14, a zwitterionic detergent containing a hydrophobic chain of 14 carbon atoms. These solubility properties were of great advantage for the purification of HP0524Δ1, yielding a crude protein extract of 50% purity; the extract was further purified by gel filtration (Table 4). HP0524Δ1 has a size of 60 kDa by SDS-PAGE (Fig. 1).
TraG, TraD, and HP0524Δ1 have common biophysical properties.
Cells grown at 37 or 30°C yielded equal amounts of overproduced proteins, as seen on SDS-PAGE of complete cell extracts (not shown). However, solubilization experiments under nondenaturing conditions showed that the overproduced proteins were insoluble when cells were grown at 37°C but could be solubilized when cells had been grown at 30°C. This indicates that the overproduced TraG-like proteins might form insoluble aggregates in more rapidly growing cells.
In vitro formation of multimers or aggregates was deduced from several observations. Upon gel filtration, TraG and TraD eluted within the void volume of the column. The values for the elution volume of TraG remained unaltered when gel filtration was run with buffer containing 1 M NaCl and 0.1% (wt/vol) Brij 58 or when samples of TraG were sonicated prior to gel filtration. Given that the gel filtration matrix has an exclusion limit of 1,200 kDa, TraG and TraD probably form multimers of at least 18 and 16 subunits, respectively (Table 5). A molecular mass of approximately 320 kDa was calculated from the elution volume of HP0524Δ1. From this we propose that HP0524Δ1 forms multimers of four or five subunits. Glycerol gradient centrifugation of TraG and TraD resulted in very broad sedimentation profiles (Fig. 2). Failure of both proteins to sediment as discrete bands indicates that TraG and TraD are polydisperse under the conditions applied. TraG and TraD did not significantly interact with chromatographic matrices including DEAE-Sephacell, heparin-Sepharose, and ATP-agarose. This property could not be verified for HP0524Δ1 since the high-salt conditions, which were needed to keep the protein in solution, were incompatible with chromatography on these matrices.
TABLE 5.
Molecular masses and oligomeric states of purified TraG, TraD, and HP0524Δ1 as evaluated by gel filtration
| Protein | Molecular mass of the monomera(kDa) | Mr as evaluated by gel filtrationb | Calculated oligomeric state |
|---|---|---|---|
| TraG | 70.7 | >1,300 | >18 |
| TraD | 82.2 | >1,300 | >16 |
| HP0524Δ1 | 68.0 | 321 | 4-5 |
Calculated from the predicted amino acid sequence.
Gel filtration on a column calibrated with globular proteins.
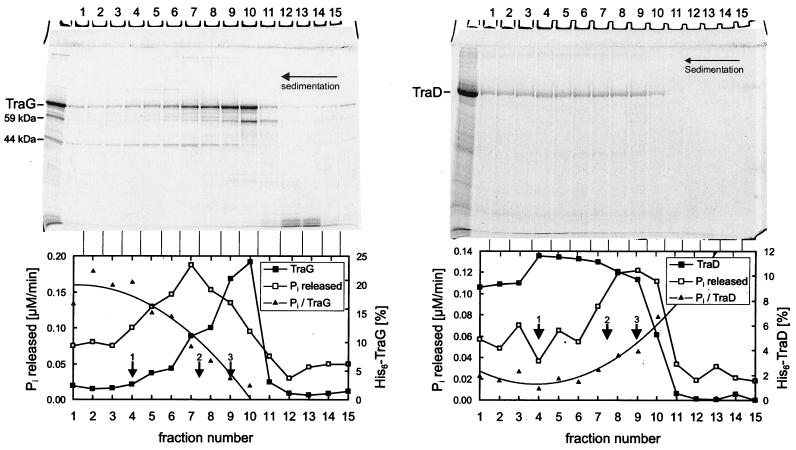
FIG. 2.
TraG and TraD preparations contained ATP-hydrolyzing impurities that were removed by glycerol gradient centrifugation. Purified TraG or TraD was layered on a 15 to 35% (wt/vol) linear glycerol gradient. Centrifugation was at 270,000 × g for 38 h. Aliquots of fractions were analyzed on an SDS-15% polyacrylamide gel (top) and were assayed for ATPase activity. The graphs on the bottom show the TraG and TraD contents (in percentage of total TraG or TraD) and the ATPase activity (expressed as time-dependent release of Pi) in each fraction. The ratio of ATPase activity to protein content (Pi/TraG or TraD) is also shown. Reference proteins 1 to 3 migrated as indicated by arrows. 1, aldolase, 158 kDa, s20,w = 7.8; 2, BSA, 67 kDa, s20,w = 4.4; 3, ovalbumin, 43.5 kDa, s20,w = 3.6. The very broad sedimentation profiles of TraG and TraD indicate the presence of several oligomeric states of these proteins.
The topology of integral membrane protein TraG resembles the topology of F TraD and Ti VirD4.
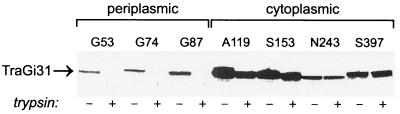
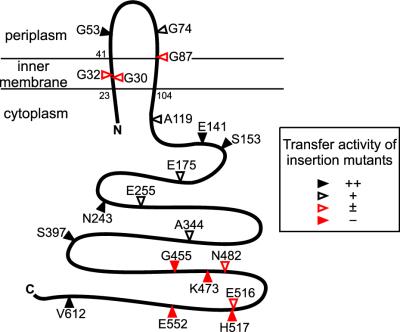
TraG contains two significant clusters of hydrophobic residues (from Val23 to Gly70 and from Ala83 to Val104), consistent with the protein potentially possessing two or three transmembrane domains. To evaluate the membrane topology of TraG, in-frame insertion mutants containing 31-residue insertion sequences were created (Table 6). When cells are converted to spheroplasts, periplasmic domains of membrane proteins can be exposed to proteases. The 31-residue insertion sequence contains a single Arg residue, which enables cleavage by trypsin when the insertion is exposed in periplasmic domains. In contrast, insertions in cytoplasmic domains are protected by the inner membrane. This method has been used previously to confirm the membrane topologies of LacY and Tsr and to determine the membrane topology of F TraD (29). The periplasmic domain of TraG is normally trypsin resistant, as is seen for many cytoplasmic membrane proteins with substantial periplasmic domains (such as Tsr and TraD). The collection of mutants tested consists of several that retain a high level of transfer activity in mating assays along with a few that are conjugation deficient and that still express stable proteins (see below). TraGiG53, TraGiG74, and TraGiG87 were sensitive to trypsin, while the remaining mutants were resistant to trypsinization of spheroplasts (Fig. 3). This observation suggests that the insertions after residues 53, 74, and 87 are translocated to the periplasmic space. The insertions in the remaining mutants are likely retained in the cytoplasm. We propose that the N and C termini of TraG are localized to the cytoplasm and that the protein possesses two transmembrane sequences and a periplasmic domain of ca. 40 residues, spanning approximately from residue His44 to Arg82 (Fig. 4). Thus, the topology of RP4 TraG is very similar to the topology determined for F TraD (29) and Ti VirD4 (15).
FIG. 3.
Trypsin sensitivity of TraG insertion mutants. Cells expressing TraG insertion mutants (TraGi31) were converted to spheroplasts and treated with (+) or without (−) trypsin prior to immunoblot analysis. Antibodies directed against the trypsin-sensitive insertion sequence were used for detection of the different mutant proteins (29).
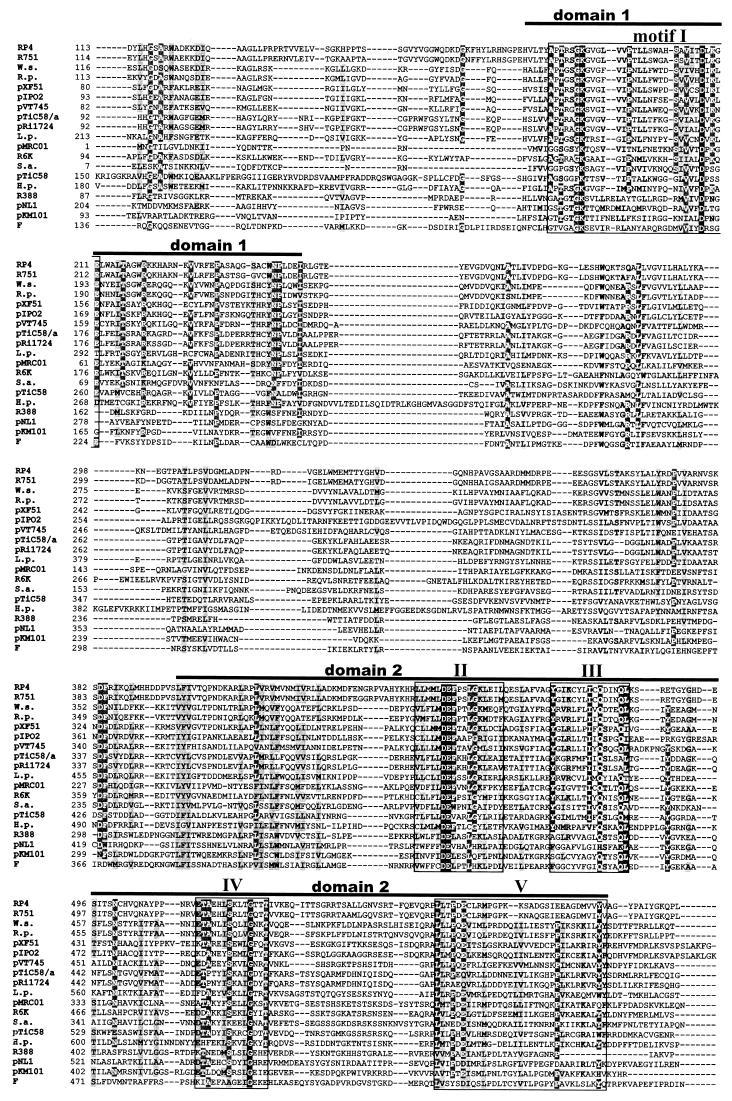
TraG contains two functionally essential domains.
Sequence comparison of 19 representative TraG-like proteins of conjugative DNA transfer systems (such as RP4, R388, F, Ti, and pKM101 plasmids) and of other type IV secretion systems involved in pathogenicity (H. pylori, Wolbachia spp., and Rickettsia prowazeckii) shows that these proteins contain two conserved domains (domains 1 and 2; see Fig. 5). These two domains also map the nucleotide binding domains (NBD) of R388 TrwB, as identified from the crystal structure of TrwBΔN70 (22). A previous study identified motifs I and II (see Fig. 5) as essential motifs for transfer activity of RP4 TraG (4). Three amino acid substitutions in motif I (K187T, K209T, and E211Q) and one substitution in motif II (D449N) resulted in reduced or abolished transfer activity (4). Motifs I and II are similar to the Walker A and B nucleotide binding motifs of NTPases (59) and ABC transporters (50). The Walker A consensus motif (G/AxxxxGKS/T), the phosphate-binding loop (P loop), is present in R388 TrwB and F TraD but contains a substitution in RP4 TraG and HP0524, where the sequence is AptrsGKG (the derivation for the consensus motif is underlined). This sequence almost perfectly matches the P-loop sequence of adenylate kinases (49), GxxxsGKG, and may therefore be considered a functional P-loop motif. The 20 TraG insertion mutants (Table 6) were analyzed for their ability to support conjugative transfer. The different pTraG::i31 plasmids were provided in trans for complementation of pDB127 (ΔtraG)-mediated conjugation. Among the 20 mutants tested, four insertion mutants had a transfer-defective phenotype and another five insertion mutants had substantially decreased transfer (listed in Table 6; Fig. 4, red). Strikingly, all of the mutations leading to a complete loss of transfer activity are located inside domain 2, downstream of motif II (Fig. 5). We conclude that conserved motifs I to V play essential roles in TraG function in conjugative transfer. Three mutations severely affecting transfer activity are located inside or adjacent to the proposed transmembrane domains of TraG (residues 23 to 41 and 87 to 104). Insertion of the 31-codon sequence into a transmembrane sequence is likely to prevent the insertion of this domain into the membrane. We propose that insertions at residues 30 and 32 (and perhaps 87) inhibit proper translocalization of the transmembrane domains of TraG into the inner membrane, indicating that membrane association is critical for TraG activity.
FIG. 5.
TraG-like proteins of conjugal transfer systems and of other type IV secretion systems contain two conserved domains probably forming a functional NBD. Amino acid sequences of 19 different TraG-like proteins were aligned. Bars, domains 1 and 2. Conserved motifs I to V inside these domains are boxed. The origin of each protein (plasmid or strain) is indicated to the left of the sequence. Black background, amino acid positions that are conserved throughout; dark shading, identical residues present in at least 68% of the sequences; light shading, similar residues. Accession numbers are as follows: TraG (RP4), S22999; TraG (R751), S22992; VirD4 (Wolbachia sp. strain wKueYO [W.s.]), BAA97443; VirD4 (R. prowazeckii [R.p.]), H71684; VirD4 (pXF51), NP_061672; TraN (pIPO2), AJ297913 (delimited DNA sequence); MagB12 (pVT745), NP_067572; VirD4 (pTiC58/a), P18594; VirD4 (pRi1724), NP_066751; LvhD4 (L. pneumophila [L.p.]), CAB60062; TrsK (pMRC01), NP_047302; TaxB (R6K), CAA71845; TrsK (pGO1 [S.a.]), C56976; TraG (pTiC58), Q44346; HP0524 (H. pylori [H.p.]), NP_207320; TrwB (R388), S43877; TraD (pNL1), NP_049138; TraJ (pKM101), T30861; TraD (F), NP_061481.
FIG. 4.
Proposed topology of RP4 TraG. In frame TraG insertion mutants were generated for identification of periplasmic and cytoplasmic domains (Fig. 3) and for detection of nonpermissible insertion sites. Arrowheads, insertion sites. The last unaltered amino acid and its position are given. The transfer frequency of each mutant was determined in mating assays (Table 6).
TraG, TraD, and HP0524Δ1 do not hydrolyze NTPs.
NTPase activity was tested by incubating the purified proteins with [γ-32P]NTPs and measuring the release of radioactive phosphate (Pi). Although the TraG preparation contained detectable ATP- and GTP-hydrolyzing activity, which could be stimulated by addition of ssDNA (specific ATPase activity: 0.39 ± 0.05 μM/min), the majority of this activity was separable from TraG by glycerol gradient centrifugation (Fig. 2, left). The ATPase activity present in the collected fractions did not correlate with the amount of TraG (the plot of Pi versus TraG in Fig. 2 is not a constant value). The ATPase activity measured in the TraD preparation (3.5 ± 0.5 μM/min) also failed to correlate with the amount of TraD (Fig. 2, right). Thus the NTPase activity present in these protein preparations appears to be due to impurities. No ATPase activity was detected in the HP0524Δ1 preparation under the conditions tested. We propose that NTP-hydrolyzing activity is not intrinsic to the TraG, TraD, and HP0524 proteins.
TraG, TraD, and HP0524Δ1 bind to DNA.
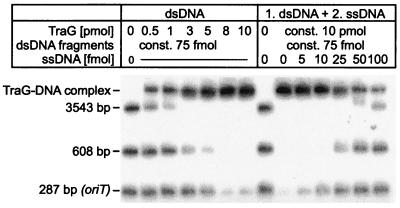
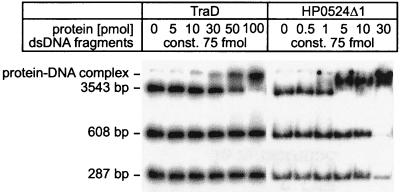
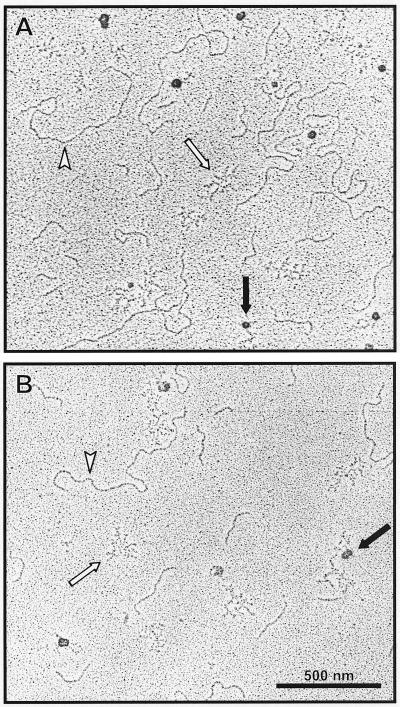
TraG, TraD, and HP0524Δ1 were tested for DNA binding ability and possible specificity for oriT sequences. Fragment retardation on nondenaturing gels revealed that all three proteins possess dsDNA binding ability. When increasing amounts of protein were added to radiolabeled dsDNA, a reduced mobility of the DNA was observed, indicating the formation of dsDNA-protein complexes (Fig. 6 and 7). TraG did not preferentially bind RP4 oriT-containing DNA compared to other DNA sequences (Fig. 6), nor were F oriT-containing fragments preferentially bound by TraD (data not shown). In competition assays, the addition of unlabeled ssDNA to a previously incubated protein-dsDNA mixture showed that ssDNA is the preferred binding substrate for TraG and TraD (as shown for TraG in Fig. 6). In contrast, HP0524Δ1-dsDNA complexes remained intact after addition of competitor ssDNA (not shown), indicating that dsDNA is the preferred substrate for HP0524Δ1. Complexes of TraG and TraD with dsDNA or ssDNA were also visualized by electron microscopy. When a mixture of equal amounts of ssDNA and dsDNA was incubated with the proteins, complex formation was seen to occur only with ssDNA (Fig. 8).
FIG. 6.
RP4 TraG binds to both dsDNA and ssDNA with preference for ssDNA. (Left) 32P-labeled dsDNA fragments were incubated with increasing amounts of TraG and electrophoresed as described in Materials and Methods. Protein-DNA complexes were retained in the gel slots. The smallest of the dsDNA fragments (287 bp) contains the core sequence of oriT, which is the sequence specifically recognized by TraI and TraJ transfer proteins during relaxation of RP4. No binding preference for this sequence was detected for TraG. (Right) Increasing amounts of ssDNA were added as competitor to a previously incubated dsDNA-TraG mixture. dsDNA was released from TraG in the presence of small amounts of ssDNA, indicating a preference for ssDNA binding.
FIG. 7.
TraD and HP0524Δ1 bind to dsDNA. 32P-labeled dsDNA fragments were incubated with increasing amounts of TraD or HP0524Δ1 and electrophoresed as described in Materials and Methods. Protein-DNA complexes were retained in the gel slots.
FIG. 8.
Complexes of TraG (A) and TraD (B) with ssDNA visualized by transmission electron microscopy. Equal amounts of dsDNA and ssDNA were incubated with TraG or TraD. Proteins appear as large dark spots (black arrows) and are located exclusively near ssDNA (bundles of short, thin strokes [white arrows]). dsDNA strands (white arrowheads) remain unbound in the presence of ssDNA, demonstrating the preference of TraG and TraD to bind to ssDNA.
TraG binds tightly and specifically to the relaxase TraI.
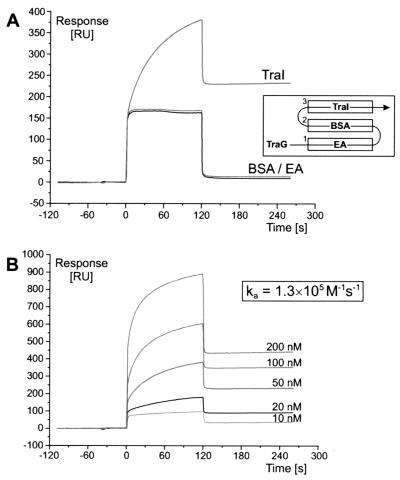
SPR was used to monitor the binding of TraG to TraI. The purified TraI protein was immobilized to a sensor chip in a flow cell, and BSA was immobilized in a reference unit. When the TraG protein was injected, tight binding to TraI was observed, while BSA and the bald chip surface remained unbound (Fig. 9A), demonstrating that the binding of TraG to TraI is specific. The association rate constant was determined to be 1.3 × 105 M−1 s−1 (Fig. 9B).
FIG. 9.
TraG binds tightly and specifically to relaxase TraI of RP4. Real-time monitoring of complex formation between TraG and TraI is demonstrated by SPR. The TraI protein was covalently attached to the sensor chip by amide coupling. Immobilized ethanolamine (EA) and BSA (flow cells 1 and 2) served as a control for specificity. After injection, TraG protein solution flowed through flow cells 1, 2, and 3 successively (A, inset). (A) Specific binding of 30 nM TraG to immobilized TraI expressed in RU per second. The “binding curves” of nonspecific interaction with EA and BSA are displayed for comparison. At injection start (0 s) and end (120 s), the abrupt change in RU in all flow cells is due to the contribution of bulk solution on the surface layer of the sensor chip. (B) Binding of TraG to immobilized TraI at five different ligand concentrations. The association rate constant (ka) was determined by using the Langmuir binding model for 1:1 interaction between analyte and ligand.
DISCUSSION
TraG-like proteins contribute in an essential manner to the translocation of protein and DNA substrates in conjugation and the related type IV secretion systems. Despite their significance for these processes, the nature of their activity remains obscure. To provide insights into the function of TraG-like proteins, biochemical properties of three members of this family originating from different type IV secretion systems, TraG, TraD, and HP0524, were determined. These proteins belong to conjugative DNA transfer systems of plasmids RP4 and F and to the pathogenicity-related type IV secretion system of gastric pathogen H. pylori, respectively. TraG and TraD were purified as His-tagged full-length proteins (His6-TraG and His6-TraD), and HP0524 was purified as a truncated derivative lacking the predicted transmembrane domain (His6-HP0524Δ1) (Fig. 1). A pronounced tendency to form oligomers or aggregates was observed for each of these proteins (Fig. 2 and Table 5). TraG, TraD, and HP0524Δ1 were found to bind to DNA without sequence specificity (Fig. 6 and 7) and did not possess an intrinsic NTPase activity under the conditions tested (Fig. 2). Specific and strong binding of RP4 TraG to relaxase TraI was demonstrated by SPR (Fig. 9). Functionally essential domains and the membrane topology of TraG were determined by analysis of in-frame insertion mutants (Fig. 3 and 4). Thus, TraG was shown to be a membrane-anchored protein with a topology corresponding to that of its F and Ti plasmid analogues, TraD and VirD4 (15, 29).
The crystal structure of a truncated derivative of the TraG-like protein of plasmid R388 lacking the transmembrane part (TrwBΔN70) has recently been solved (22). TrwB is proposed to form a hexameric, pore-like structure, strongly resembling the structure of F1 ATPase and ring helicases. The identification of functionally essential regions of TraG enables comparison with the related structural domains of TrwB. The core of the TrwB structure, consisting of the NBD, is apparently conserved in TraG (Fig. 5) and is critical for TraG activity in mating experiments (4) (Table 6).
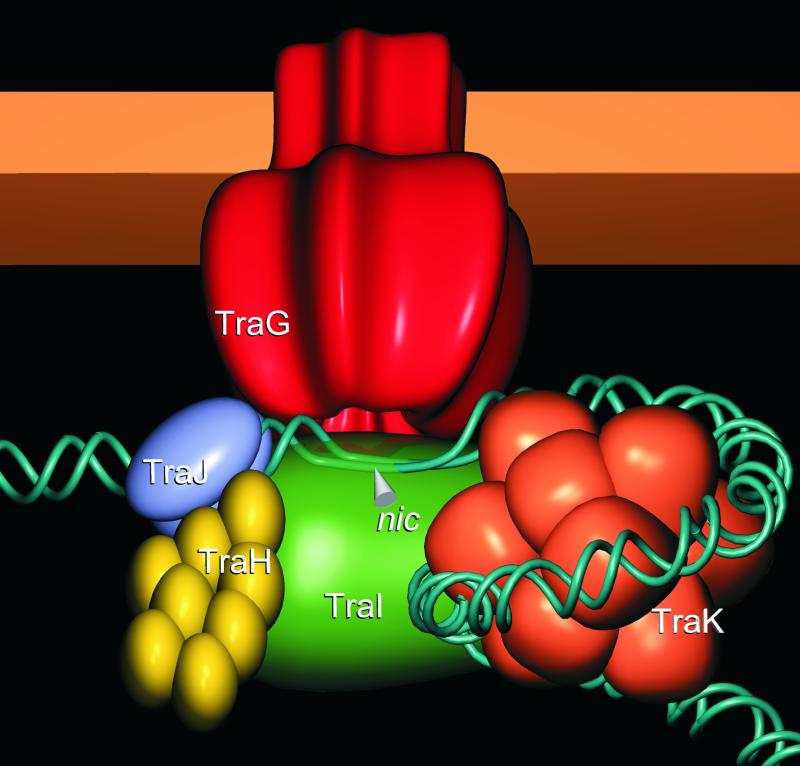
Taken together, the biochemical characteristics of the three TraG-like proteins presented in this study and the properties of TrwBΔN70 published earlier support the view that TraG-like proteins assemble as multimeric forms that are anchored in the inner membrane of the gram-negative bacterial cell envelope. Genetic and biochemical evidence clearly indicates a physical association between TraG-like proteins and components of the relaxosome via protein-protein and protein-DNA interactions. These findings lead to the hypothesis that TraG-like proteins form an inner membrane pore, which is specifically recognized by the secreted substrates. A model illustrating the proposed architecture of the TraG-associated RP4 relaxosome is shown (Fig. 10). TraG is depicted as a homohexamer based on the structural data for TrwBΔN70 (22). The relaxosome consists of relaxase TraI, which cleaves the T strand of oriT DNA at the nic site, along with accessory proteins contributing to relaxase activity (TraJ), relaxosome stability (TraH), and oriT accessibility (TraK) (41, 65). Here, we propose that this complex is linked to the potential TraG inner membrane pore via TraG-TraI and TraG-DNA interactions.
FIG. 10.
Proposed model for the RP4 relaxosome. TraG (red) is a membrane-anchored, multimeric protein probably forming a pore-like structure that could serve as a channel for translocation of the transferred ssDNA (T-DNA). The relaxase TraI (green) and the plasmid DNA both bind to this TraG pore. TraI cleaves the oriT sequence of RP4 at the nic site and is covalently attached to the 5′ end of the DNA single strand. TraJ (blue) binds to the sequence upstream of the nic site (srj) and is required for relaxase activity. TraH (yellow) is a homomultimer that stabilizes the TraI-TraJ-DNA complex, probably by bridging TraJ and TraI. TraK (orange) binds to a sequence downstream of the nic site and functions as a DNA chaperone, facilitating the formation of the TraI-DNA adduct. A helicase for displacement of the transferred strand has not been identified in the RP4 system.
While this overall assembly is well supported by the present data, the role of TraG-like proteins in the macromolecular transport process remains speculative. The interaction of TraG-like proteins with DNA and with proteins that prepare substrate DNA for transfer implies a direct role for these proteins in the translocation mechanism itself or in its regulation. The inner membrane localization and postulated multimeric pore-like structure of the TraG-like proteins are highly suggestive of a translocation portal or passageway for exported substrates. Less consistent with this view is the observation that TraG-like proteins do not seem to exhibit NTP-hydrolyzing activity under the applied conditions (reference 37 and this work). The possibility that an accessory factor (or several factors) may be required for these proteins to function as active NTPases cannot be excluded.
Genetic analysis has revealed that hp0524 is absolutely required for infectivity of H. pylori (13, 18). Here, comparison of the biochemical properties of HP0524Δ1 to those of other TraG-like proteins of conjugative transfer systems revealed close similarities. In view of the fact that the 145-kDa CagA protein is the only known substrate for the type IV secretion system of H. pylori, our finding that HP0524Δ1 binds to DNA is remarkable. A conjugation-like mechanism for DNA transfer between Helicobacter strains was suggested earlier (27), although the genetic determinants have not been identified. Two predicted relaxases encoded by open reading frames hp0996 and hp1004 (3, 56) are possible candidates for interaction with HP0524. Thus, involvement of HP0524 in a DNA transfer system cannot be excluded. On the other hand, type IV secretion systems of pathogens have most probably evolved from conjugative DNA transfer systems (62). Thus the DNA binding activity of HP0524 may be a residual activity from a TraG ancestor of a conjugative transfer system. Whether DNA is still actively transported by this secretion system or whether it is even involved in pathogenicity remains to be elucidated.
In type IV secretion systems, the transported substrates consist of a protein (such as CagA of H. pylori cag) and/or of a protein complexed to DNA (such as TraI-oriT of RP4). The A. tumefaciens Ti plasmid secretion system transports the VirD2-T-DNA complex, along with virulence-associated proteins VirE2 and VirF, into plant cells. VirE2 and VirF translocation depends on the VirB/VirD4 transport system (related to the Mpf/TraG system of RP4) but does not require DNA transfer (58). Secretion of CagA by H. pylori is equally dependent on the corresponding VirB/VirD4 system of cag PAI (18). It is conceivable that analogous Mpf/TraG-dependent protein secretion exists in the conjugative transfer system of RP4. TraC, a functional analogue of VirE2, is transferred into recipient cells during RP4-mediated conjugation (45). Since TraC and VirE2 are cytoplasmic proteins lacking a signal sequence for secretion by the sec system (GSP system), TraG and VirD4 may possibly mediate their transport through the inner membrane.
The putative translocation activity of TraG-like proteins appears to be limited to crossing the inner membrane. It is notable that the only type IV secretion system clearly lacking a TraG homologue is found in B. pertussis. The pertussis toxin liberation system, Ptl, employs a two-step mechanism for secretion. Pertussis toxin subunits rely on the sec system for translocation through the inner membrane, and the Ptl system is responsible for transition of the holotoxin across the outer membrane barrier (8). The activity of the Ptl transport system is limited to the outer membrane and is therefore distinct from that of the VirB prototype of type IV transporters, which convey substrates across both the inner and outer membranes. The notable absence of a traG homologue in B. pertussis implies that the putative ancestral TraG-like protein of this secretion system has been replaced by the sec system of the host during evolution.
In type IV secretion systems the functional connection between TraG-like proteins and the envelope-spanning Mpf components remains uncertain. Physical interactions between the TraG-like proteins and Mpf proteins have been postulated (10, 23), but biochemical evidence for this remains to be supplied. The periplasmic domain of TraG could possibly mediate interactions of this type. During the present study we tested the possibility that TrbB, a membrane-associated cytosolic ATPase of the Mpf system of RP4 (31), might provide an interface with TraG. However, a TrbB-TraG interaction was not detected by affinity chromatography of TrbB on a His6-TraG-loaded Ni-NTA column, whereas the TraI-TraG interaction was detected by the same technique (25). The most promising approaches for the detection of protein-protein interactions, SPR and affinity chromatography, are hampered by the limited solubility of Mpf proteins. Thus, direct evidence for a linkage between TraG-like proteins and components of the Mpf transfer machinery is still lacking. Resolution of this challenging aspect of TraG function will probably prove decisive to unraveling the mechanism of type IV secretion.
Acknowledgments
E. Lanka, G. Schröder and S. Krause thank Hans Lehrach for generous support. The expert technical assistance of Marianne Schlicht is greatly appreciated. We thank Florian Sack and Vanessa Csitkovits for their contributions to this study. The project was stimulated by discussions within the EU-BIOTECH concerted action BIO4-CT-0099, Mobile Genetic Elements' Contribution to Bacterial Adaptability and Diversity (MECBAD).
Work in E. Lanka's laboratory was supported by the Deutsche Forschungsgemeinschaft. E. L. Zechner and B. Traxler were supported by the Austrian FWF P13277 and the NSF, respectively. Work in G. Waksman's laboratory was supported by the ASAP.
REFERENCES
- 1.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backert, S., E. Nickisch-Rosenegk, and T. F. Meyer. 1998. Potential role of two Helicobacter pylori relaxases in DNA transfer? Mol. Microbiol. 30:673-674. [DOI] [PubMed] [Google Scholar]
- 4.Balzer, D., W. Pansegrau, and E. Lanka. 1994. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J. Bacteriol. 176:4285-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzer, D., G. Ziegelin, W. Pansegrau, V. Kruft, and E. Lanka. 1992. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 20:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beijersbergen, A., A. Den Dulk-Ras, R. A. Schilperoort, and P. J. J. Hooykaas. 1992. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256:1324-1327. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 8.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 2:25-29. [DOI] [PubMed] [Google Scholar]
- 9.Cabezón, E., E. Lanka, and F. de la Cruz. 1994. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 176:4455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabezón, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 11.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 14.Crabtree, J. E., D. Kersulyte, S. D. Li, I. J. Lindley, and D. E. Berg. 1999. Modulation of Helicobacter pylori induced interleukin-8 synthesis in gastric epithelial cells mediated by cag PAI encoded VirD4 homologue. J. Clin. Pathol. 52:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, A., and Y. H. Xie. 1998. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol. Microbiol. 27:405-414. [DOI] [PubMed] [Google Scholar]
- 16.Di Laurenzio, L., L. S. Frost, and W. Paranchych. 1992. The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol. Microbiol. 6:2951-2959. [DOI] [PubMed] [Google Scholar]
- 17.Disqué-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, W., J. Püls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 19.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fürste, J. P., W. Pansegrau, G. Ziegelin, M. Kröger, and E. Lanka. 1989. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. USA 86:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomis-Rüth, F. X., and M. Coll. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 33:839-843. [DOI] [PubMed] [Google Scholar]
- 22.Gomis-Rüth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezón, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1- ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton, C. M., H. Lee, P. L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, R. F., S. C. Pickett, and D. L. Barker. 1990. Autoradiography using storage phosphor technology. Electrophoresis 11:355-360. [DOI] [PubMed] [Google Scholar]
- 25.Krause, S. 1999. Die Transferproteine TraG und TrbB des konjugativen Plasmides RP4: Strukturelle und funktionelle Gemeinsamkeiten zu analogen Komponenten anderer Transportsysteme. Ph.D. thesis. Freie Universität Berlin, Berlin, Germany.
- 26.Krause, S., W. Pansegrau, R. Lurz, F. de la Cruz, and E. Lanka. 2000. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182:2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuipers, E. J., D. A. Israel, J. G. Kusters, and M. J. Blaser. 1998. Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori. J. Bacteriol. 180:2901-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupelwieser, G., M. Schwab, G. Högenauer, G. Koraimann, and E. L. Zechner. 1998. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 275:81-94. [DOI] [PubMed] [Google Scholar]
- 29.Lee, M. H., N. Kosuk, J. Bailey, B. Traxler, and C. Manoil. 1999. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J. Bacteriol. 181:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessl, M., D. Balzer, K. Weyrauch, and E. Lanka. 1993. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J. Bacteriol. 175:6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessl, M., W. Pansegrau, and E. Lanka. 1992. Relationship of DNA-transfer-systems: essential transfer factors of plasmids RP4, Ti and F share common sequences. Nucleic Acids Res. 20:6099-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llosa, M., S. Bolland, and F. de la Cruz. 1994. Genetic organization of the conjugal DNA processing region of the Incw plasmid R388. J. Mol. Biol. 235:448-464. [DOI] [PubMed] [Google Scholar]
- 33.Maneewannakul, K., P. Kathir, S. Endley, D. Moore, J. Manchak, L. Frost, and K. Ippen-Ihler. 1996. Construction of derivatives of the F plasmid pOX-tra715: characterization of traY and traD mutants that can be complemented in trans. Mol. Microbiol. 22:197-205. [DOI] [PubMed] [Google Scholar]
- 34.Manoil, C., and J. Bailey. 1997. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J. Mol. Biol. 267:250-263. [DOI] [PubMed] [Google Scholar]
- 35.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics, p. 431-433. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Moncalián, G., E. Cabezón, I. Alkorta, M. Valle, F. Moro, J. M. Valpuesta, F. M. Goni, and F. de la Cruz. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274:36117-36124. [DOI] [PubMed] [Google Scholar]
- 38.Moore, D., B. A. Sowa, and K. Ippen-Ihler. 1981. The effect of tra mutations on the synthesis of the F-pilin membrane polypeptide. Mol. Gen. Genet. 184:260-264. [DOI] [PubMed] [Google Scholar]
- 39.Nurse, P., K. H. Zavitz, and K. J. Marians. 1991. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J. Bacteriol. 173:6686-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panicker, M. M., and E. G. Minkley, Jr. 1992. Purification and properties of the F sex factor TraD protein, an inner membrane conjugal transfer protein. J. Biol. Chem. 267:12761-12766. [PubMed] [Google Scholar]
- 41.Pansegrau, W., D. Balzer, V. Kruft, R. Lurz, and E. Lanka. 1990. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl. Acad. Sci. USA 87:6555-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 54:197-251. [DOI] [PubMed] [Google Scholar]
- 43.Pansegrau, W., and E. Lanka. 1996. Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPα plasmid RP4. J. Biol. Chem. 271:13068-13076. [DOI] [PubMed] [Google Scholar]
- 44.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 45.Rees, C. E., and B. M. Wilkins. 1990. Protein transfer into the recipient cell during bacterial conjugation: studies with F and RP4. Mol. Microbiol. 4:1199-1205. [DOI] [PubMed] [Google Scholar]
- 46.Salmond, G. P. C. 1994. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 32:181-200. [Google Scholar]
- 47.Sambrook, J., E. T. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Samuels, A. L., E. Lanka, and J. E. Davies. 2000. Conjugative junctions in RP4-mediated mating of Escherichia coli. J. Bacteriol. 182:2709-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15:430-434. [DOI] [PubMed] [Google Scholar]
- 50.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 51.Spiess, E., and R. Lurz. 2001. Electron microscopic analysis of nucleic acids and nucleic acid-protein complexes. Methods Microbiol. 20:293-323. [Google Scholar]
- 52.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA 97:1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strack, B., M. Lessl, R. Calendar, and E. Lanka. 1992. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the α protein of the Escherichia coli satellite phage P4. J. Biol. Chem. 267:13062-13072. [PubMed] [Google Scholar]
- 54.Szpirer, C. Y., M. Faelen, and M. Couturier. 2000. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol. Microbiol. 37:1283-1292. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorsted, P. A., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. N. Thomas. 1998. Complete sequence of the IncP-β plasmid R751—implications for evolution and organization of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 57.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 58.Vergunst, A. C., B. Schrammeijer, A. Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 59.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waters, V. L. 1999. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 4:D433-D456. [DOI] [PubMed] [Google Scholar]
- 61.Waters, V. L., B. Strack, W. Pansegrau, E. Lanka, and D. G. Guiney. 1992. Mutational analysis of essential IncPα plasmid transfer genes traF and traG and involvement of traF in phage sensitivity. J. Bacteriol. 174:6666-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winans, S. C., D. L. Burns, and P. J. Christie. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 64.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative DNA transfer processes, p. 87-173. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 65.Ziegelin, G., W. Pansegrau, R. Lurz, and E. Lanka. 1992. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J. Biol. Chem. 267:17279-17286. [PubMed] [Google Scholar]
- 66.Ziegelin, G., W. Pansegrau, B. Strack, D. Balzer, M. Kröger, V. Kruft, and E. Lanka. 1991. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Sequence 1:303-327. [DOI] [PubMed] [Google Scholar]
- 67.Ziegelin, G., E. Scherzinger, R. Lurz, and E. Lanka. 1993. Phage P4 α protein is multifunctional with origin recognition, helicase and primase activities. EMBO J. 12:3703-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]