Abstract
The proteins required for adherence of the pathogen Mycoplasma pneumoniae to host respiratory epithelial cells are localized to a polar structure, the attachment organelle. A number of these proteins have been characterized functionally by analysis of noncytadhering mutants, and many are components of the mycoplasma cytoskeleton. Mutations in some cytadherence-associated proteins have pleiotropic effects, including decreased stability of other proteins, loss of adherence and motility, and abnormal morphology. The function of protein HMW3, a component of the attachment organelle, has been difficult to discern due to lack of an appropriate mutant. In this paper, we report that loss of HMW3 resulted in decreased levels and more diffuse localization of cytoskeletal protein P65, subtle changes in morphology, inability to cluster the adhesin P1 consistently at the terminal organelle, reduced cytadherence, and, in some cells, an atypical electron-dense core in the attachment organelle. This phenotype suggests a role for HMW3 in the architecture and stability of the attachment organelle.
Mycoplasma pneumoniae is a bacterial pathogen of the human respiratory tract. In general, infections are mild, target children and young adults, and manifest as tracheobronchitis or primary atypical pneumonia (9). M. pneumoniae cells have no cell wall and therefore are pleomorphic. Nevertheless, a single morphology is predominant and is characterized by spindle-shaped cells with a distinct asymmetry due to the presence of a polar terminal structure, the attachment organelle (5). The major adhesin protein P1 is densely clustered in the membrane of wild-type Mycoplasma cells at the attachment organelle (3, 11, 18). As its name suggests, this structure mediates the adherence of M. pneumoniae cells to host epithelium, an activity that is essential for successful infection of the respiratory tract (30). In addition, the attachment organelle is the leading pole as mycoplasma cells travel by gliding motility (7). Moreover, M. pneumoniae cells divide by binary fission, and bifurcation of the attachment organelle is thought to be an early step in this process (5, 32). Examination of thin sections of the attachment organelle by electron microscopy reveals a membrane-bound, laterally oriented, electron-dense core that ends distally in a bulbous knob (terminal button). The electron-dense core is a major component of the Triton X-100-insoluble M. pneumoniae cytoskeleton (13), as reviewed by us recently (24).
Since correct assembly of the attachment organelle is paramount for attachment, considerable research has focused on the isolation and characterization of noncytadhering mutants (16, 22) and resulted in the identification of several cytadherence accessory proteins, including HMW1, HMW2, HMW3, and P65 (16, 22, 26, 29). Each is associated with the attachment organelle (20, 35, 36), and mounting evidence reveals a complex circuit of interactions among these proteins. For example, loss of HMW1 results in an inability to cluster P1 at the attachment organelle (15) and reduced levels of HMW2 (M. J. Willby et al., unpublished data) and P65 (20). Somewhat conversely, loss of HMW2 results in accelerated turnover of HMW1, HMW3, and P65 (2, 20, 27) in addition to the inability to localize P1 properly (3). However, additional studies regarding the function of these cytadherence proteins and the chronology of their incorporation into the attachment organelle have been hampered by the lack of genetic tools available for use with M. pneumoniae. For example, homologous recombination in M. pneumoniae has not been described, preventing targeted mutations in proteins of interest. Instead, the generation of new mutants must rely on identifiable spontaneous mutations or serendipitous transposition events.
HMW3 is a cytoskeletal protein that is thought to be peripherally associated with the inner surface of the M. pneumoniae membrane (29, 36). Analysis by immunoelectron microscopy suggests that polymers of HMW3 wrap around the electron-dense core and terminal button of the attachment organelle in a linear pattern (36). This protein has a deduced mass of 73,725 Da but a relative mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of 140,000 (27). The major structural feature of HMW3 is an acidic, proline-rich (APR) domain (residues 88 to 488) that probably contributes to the anomalous migration on SDS-PAGE (27). While steady-state levels of HMW3 (along with HMW1 and P65) are reduced in hmw2 mutants due to accelerated protein turnover (2, 20, 28), before this study, no hmw3 mutant had been isolated. In the present report, we describe an hmw3 transposon insertion mutant. Loss of HMW3 resulted in alterations of mycoplasma cellular morphology, localization of P1 and P65, stability of P65, hemadsorption capacity, and the ultrastructure of the electron-dense core, suggesting a role for HMW3 in its stabilization.
MATERIALS AND METHODS
Organisms and growth conditions.
Wild-type, cytadhering M. pneumoniae strain M129-B18 (25) was used in this study. Mycoplasma cells were cultured in Hayflick medium or on PPLO agar as previously described (14). Gentamicin (18 μg/ml) was included in cultures of transformants except where indicated otherwise. The hmw3::Tn4001 transformant was filter cloned as previously described (37) to obtain a clonal population.
Mapping of the transposon insertion.
The following program was used for all PCRs: heating at 95°C for 5 min, denaturing at 95°C for 2 min, annealing at 50°C for 1 min, extension at 72°C for 1 min, repetition of the last three steps 40 times, and extension at 72°C for 10 min. Sequencing of PCR products was carried out by the University of Georgia Molecular Genetics Instrumentation Facility as described elsewhere (10). Sequences were analyzed with Genetics Computer Group software (Wisconsin Package, version 10.1; Genetics Computer Group, Madison, Wis.).
Protein analysis by SDS-PAGE and Western immunoblotting.
M. pneumoniae cultures were harvested as described elsewhere (14). Total protein was quantitated for SDS-PAGE by the bicinchoninic acid assay (Pierce, Rockford, Ill.). SDS-PAGE and Western blotting were performed as previously described (14) with the following antibodies at the dilutions indicated: anti-HMW3, 1:5,000 (36); anti-HMW1, 1:10,000 (35); anti-HMW2, 1:7,500 (23); anti-P30, 1:3,000 (20); anti-B, 1:500 (34); anti-P1, 1:1,000 (MAB134P; Maine Biotechnology Services, Inc.); anti-P65, 1:1,000 (29), anti-P43, 1:100.
Hemadsorption assay.
M. pneumoniae cultures were assessed qualitatively for hemadsorption as detailed elsewhere (14). Quantitative hemadsorption assays were performed as described by Fisseha et al. (12) with modifications. M. pneumoniae cultures were inoculated from frozen stocks into 15 ml of Hayflick broth containing 200 μCi of [3H]thymidine (6.7 Ci/mmol; Dupont NEN, Boston, Mass.) and incubated at 37°C to mid-log phase. Cells were harvested, washed twice in cold phosphate-buffered saline (PBS; pH 7.2), and suspended in 3 ml of Hayflick broth by repeated passage through a 25-gauge needle. Suspensions were centrifuged for 5 min at 123 × g in a clinical centrifuge (International Equipment Company, Needham Heights, Mass.) to remove any remaining aggregates. Six 100-μl aliquots were removed; three were incubated at 4°C, and three were incubated at 37°C, each for 30 min. A 1:1 suspension of fresh chicken erythrocytes in Alsever's solution was washed twice with PBS (pH 7.2), and then the erythrocytes were suspended in PBS (pH 7.2) to 4% (vol/vol). Fifty microliters of the 4% red blood cell suspension was added to each 100-μl aliquot of M. pneumoniae cells and incubated at 4 or 37°C for an additional 30 min. The M. pneumoniae-erythrocyte mixtures were then overlaid onto 150 μl of 40% sucrose and centrifuged at 1,690 × g for 90 s. The resulting pellets were suspended in 100 μl of PBS, 10 μl of 10% SDS was added, and the incubation was continued overnight at room temperature. On the following day, 5 μl of H2O2 was added and samples were incubated for an additional 2 h at 37°C, after which scintillation fluid was added and radioactivity was measured.
Microscopy.
Samples were prepared for scanning electron microscopy and immunoelectron microscopy as described elsewhere (13), except that samples intended for immunoelectron microscopy were incubated in blocking buffer overnight at 4°C; mouse anti-P1 monoclonal antibody was used at a dilution of 1:150, 1:200, or 1:250; and samples were not stained. Scanning electron microscopy was performed with a LEO 982 scanning electron microscope, and transmission electron microscopy was performed with a JEOL 100CX II transmission electron microscope.
Cells were prepared for fluorescence microscopy as previously described (20) and examined with a Nikon TE300 epifluorescence microscope with a tetramethylrhodamine isothiocyanate filter cube (528 to 552 nm) and equipped with phase-contrast optics. Samples were probed with P65-specific antibodies diluted 1:100 and indocarbocyanine (Cy3)-conjugated donkey anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.) diluted 1:75, and images were digitized with a Micromax charge-coupled device camera (Princeton Scientific Instruments, Monmouth Junction, N.J.) with an exposure time of 0.1 s for phase-contrast images and 0.5 s for fluorescent images.
For ultrastructural examination, 50-ml mid-log-phase cultures were harvested by gently scraping adherent cells into the spent medium and centrifuging them at 20,000 × g for 20 min at 4°C. After three washes in cold PBS, the cells were divided into two 1.5-ml Eppendorf tubes. Subsequent washes were done with Sorensen's phosphate buffer (pH 7.2) (6). Samples were fixed for 1 h at 4°C in 2% glutaraldehyde-2% paraformaldehyde, washed overnight at 4°C, and washed two more times for 30 min each on the following day. Samples were then postfixed in 1% OsO4 for 1 h at room temperature, washed twice for 3 min each time, dehydrated by transfer through a series of ethanol concentrations (30, 50, 70, 85, 95, 100, and 100%) for 10 min each time, and then subjected to two 15-min incubations in propylene oxide. Samples were incubated overnight in 1:1 propylene oxide-Epon 812 at room temperature, followed by 2 h in 100% Epon 812 and embedding in Epon 812 for 24 h at 60°C. Thin sections were cut with a diamond knife and collected on Formvar-coated nickel grids. Grids were stained in 2% aqueous uranyl acetate-4.4% lead citrate before examination.
Isolation of excision revertants.
Strain hmw3::Tn4001 was passaged five times in 5 ml of Hayflick broth in the absence of gentamicin in 25-cm2 tissue culture flasks. For each passage, cultures were grown to mid-log phase as indicated by the phenol red pH indicator, the spent medium was decanted, and the monolayer was washed three times with 10 ml of cold PBS and gently scraped into 10 ml of PBS. Twenty-five to 50 μl of the cell suspension was then used to inoculate 5 ml of fresh Hayflick broth without gentamicin. Following the fifth passage, cells were washed and scraped as described above and dilutions were plated on PPLO agar and incubated at 37°C for 7 to 9 days. Colonies were tested for hemadsorption, and 10 hemadsorption-positive colonies were picked as described previously (17) and inoculated into 1 ml of Hayflick broth. The resulting cultures were replated and tested for a hemadsorption-positive phenotype. Gentamicin sensitivity was tested by growth in Hayflick broth with or without gentamicin in 24-well tissue culture dishes.
RESULTS
Identification of an hmw3 insertion mutant.
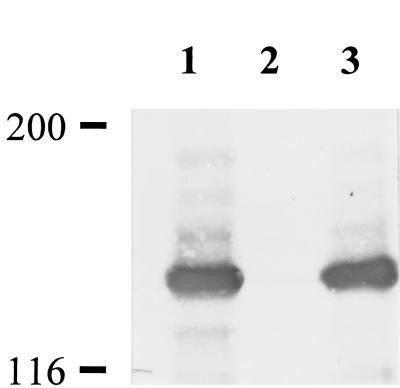
Wild-type M. pneumoniae was transformed with pKV201, a derivative of pISM2062 (21) that contains a modified Tn4001 transposon into which a gene fusion encoding mouse dihydrofolate reductase and a very small fragment of hmw1 had been cloned as part of a separate study. Gentamicin-resistant transformants were isolated from agar plates and inoculated into Hayflick broth. With one transformant, we observed cells floating in the growth medium, as well as attached to the flask, suggesting that cytadherence was impaired. Significantly, this phenotype was unique to this particular transformant; all of the other transformants examined attached normally to plastic, suggesting that the loss of attachment was insertion site specific. Western blots of this transformant probed individually with antisera against various cytadherence-associated proteins indicated that HMW3 was completely absent (Fig. 1), raising the possibility of a fortuitous transposon insertion within or near the hmw3 gene. The original focus of this transformation, the dihydrofolate reductase fusion protein, was not detected in Western blots (data not shown).
FIG. 1.
Western immunoblot analysis of M. pneumoniae with anti-HMW3 serum. Equal amounts of total protein were electrophoresed on an SDS-4.5% polyacrylamide gel, transferred to nitrocellulose, and probed with antisera (1:10,000). Lanes: 1, wild-type M. pneumoniae; 2, hmw3::Tn4001 transformant; 3, transformant control (modified Tn4001 transposon inserted elsewhere). Protein size standards are indicated in kilodaltons.
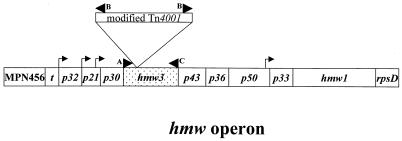
Following filter cloning of this transformant to ensure a clonal population, we verified the site of transposon insertion by PCR amplification and nucleotide sequencing. Mycoplasma cells were harvested, and DNA was isolated as described elsewhere (39). The transposon insertion was localized to the 5′ end of hmw3 by PCR on genomic DNA with three primers that we designed for this purpose (Fig. 2). Primer A was complementary to a sequence immediately upstream of hmw3, primer C was complementary to the 3′ end of hmw3, and primer B was complementary to the inverted repeat of the insertion sequence (IS) element of Tn4001 (8). The estimated size of the PCR products generated with primer combinations A-B and B-C and analyzed by agarose gel electrophoresis approximated that of the insertion within hmw3. Sequencing of a PCR product generated with primer pair A-B localized the insertion precisely between nucleotides 267 and 268 in the hmw3 coding region. Analysis of the composite sequence hmw3::Tn4001 revealed an in-frame stop codon 13 bp into the IS element of the transposon. We did not detect a truncated HMW3 protein on Western blots probed with polyclonal antibodies prepared against full-length HMW3 (data not shown).
FIG. 2.
Schematic of the hmw operon (10, 37) illustrating the point of insertion of the transposon into hmw3 (shaded for emphasis). Arrowheads indicate the positions of the primers used for PCR analysis. Arrows indicate the positions of the promoters within the operon. Insertion of the transposon immediately downstream of nucleotide 267 was determined by sequencing the PCR product generated using primer pair A-B. The diagram is not drawn to scale.
Only the steady-state level of P65 is reduced in the absence of HMW3.
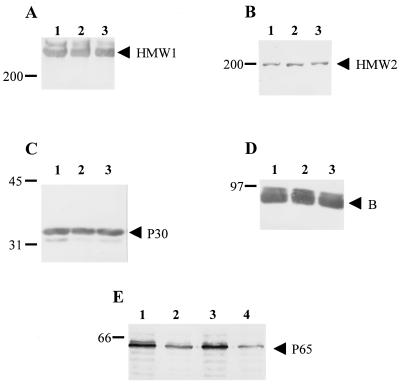
We examined the consequences of loss of HMW3 on the steady-state levels of other cytadherence-related proteins. Western blots were prepared and probed with antisera directed against cytadherence-associated proteins HMW1, HMW2, B, P65, and P30 (Fig. 3), as well as P1 and P28 (data not shown). Examination of these blots revealed wild-type levels of all of the proteins tested except P65, which was decreased in the hmw3::Tn4001 transformant to nearly the same level as that seen in hmw2 mutant I-2. The levels of all of the proteins examined, including P65, were indistinguishable in wild-type M. pneumoniae and a transformant thereof with the modified Tn4001 transposon inserted elsewhere (Fig. 3).
FIG. 3.
Western immunoblot analysis of M. pneumoniae to assess steady-state levels of certain cytadherence-associated proteins. Equal amounts of protein were electrophoresed on either a 4.5% (A, B, and D) or a 10% (C and E) polyacrylamide gel, transferred to nitrocellulose, and probed with the following antisera: A, anti-HMW1; B, anti-HMW2; C, anti-P30; D, anti-B; E, anti-P65. Lanes: 1, wild-type M. pneumoniae; 2, hmw3::Tn4001 transformant; 3, transformant control; 4 (panel E only), mutant I-2. Protein size markers are given in kilodaltons to the left of each blot, and arrowheads designate proteins of interest.
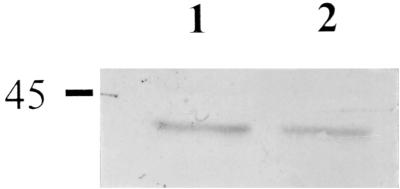
The gene for HMW3 is part of a large transcriptional unit (38; Fig. 2) and is immediately followed by and possibly transcriptionally linked to the gene for P43 (10). To determine whether transposon insertion in hmw3 might have polar consequences, we compared P43 levels in wild-type M. pneumoniae and the hmw3::Tn4001 transformant by Western immunoblotting. No decline in P43 levels was evident (Fig. 4), indicating that the gene for P43 is expressed in this mutant, probably from an outward-reading promoter in the IS element of Tn4001 (8).
FIG. 4.
Western immunoblot analysis of M. pneumoniae to compare levels of protein P43. Equal amounts of protein were electrophoresed on a 12% polyacrylamide gel, transferred to nitrocellulose, and probed with antisera (1:100). Lanes: 1, wild-type M. pneumoniae; 2, hmw3::Tn4001 transformant. The protein size standard is indicated in kilodaltons.
Cytadherence.
Adsorption to erythrocytes by M. pneumoniae (hemadsorption) correlates strongly with adherence to the respiratory epithelium (33) and was assessed qualitatively and quantitatively in the hmw3 mutant. Qualitative examination revealed that the majority of hmw3::Tn4001 transformant colonies had erythrocytes attached only around their periphery, whereas wild-type and transformant control colonies were uniformly coated with erythrocytes and colonies of the nonadherent mutant II-3 had no attached erythrocytes (data not shown). Results of the quantitative assay indicated an intermediate level of hemadsorption by the hmw3::Tn4001 transformant (22% ± 5.3% of the wild-type level), compared with 0% ± 3.3% of the wild-type level for the negative control (mutant II-3) and 108% ± 6.1% of the wild-type level for the transformant control. The intermediate hemadsorption phenotype of hmw3::Tn4001 is consistent with the qualitative hemadsorption findings, as well as the original observation that these cells adhered only poorly to the plastic tissue culture flask.
Morphology and ultrastructure of hmw3::Tn4001.
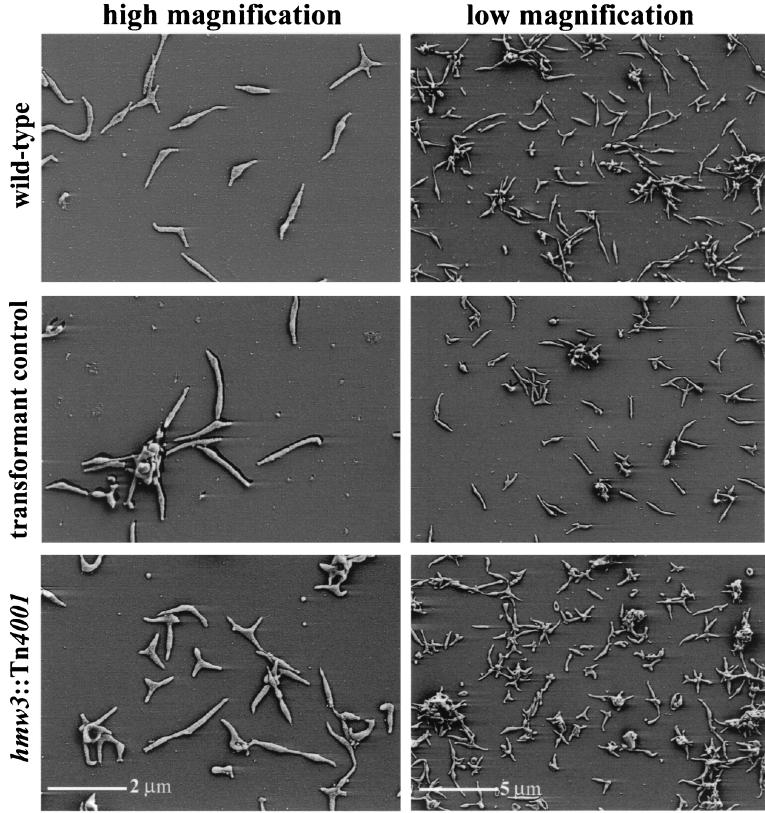
Wild-type M. pneumoniae cultures are typically pleomorphic, with the predominant morphology being an asymmetric spindle with a polar attachment organelle (Fig. 5). The spindle shape results from the presence of a distended area (cell body) adjacent to the attachment organelle, with a width approximately two to three times that of the leading and trailing filaments. Cells of the hmw3::Tn4001 transformant were also pleomorphic (Fig. 5) but distinct from wild-type populations. In particular, the prototypical, filamentous wild-type morphology was much less common in the hmw3 mutant population. The most striking morphological feature of this mutant was the unusually large number of cells having multiple branches (filaments) or more than one distended area. The transformant control containing the modified Tn4001 transposon at another site on the chromosome was morphologically indistinguishable from wild-type M. pneumoniae (Fig. 5).
FIG. 5.
Morphological analysis of wild-type (top row), transformant control (middle row), and hmw3::Tn4001 (bottom row) M. pneumoniae by scanning electron microscopy. A high-magnification image is shown on the left, and a lower-magnification survey shot is shown on the right so that individual cells, as well as a population of cells, can be examined.
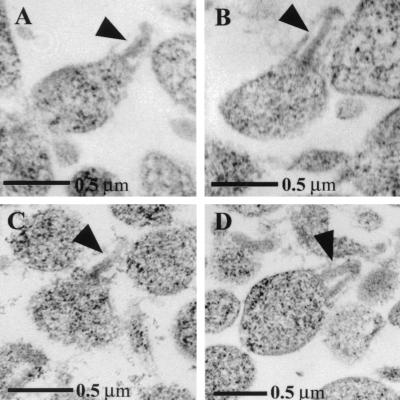
The characteristic ultrastructural feature of wild-type M. pneumoniae cells is the electron-dense core that defines the attachment organelle (Fig. 6A and B). This core structure terminates in a bulbous knob and can be easily identified in thin sections of wild-type cells (4). Immunoelectron microscopy studies localized HMW3 to this knob and along the shaft of the electron-dense core (36). Somewhat surprisingly, despite the loss of HMW3 in the mutant, many cells contained wild-type-like cores, with a single electron-dense rod (Fig. 6C). However, in at least 20% of the cells, the core was not seen as a single electron-dense rod, as it was in 100% of the wild-type cells, but rather was V shaped with the vertex distal, suggesting spreading at the base of the core and a potential role for HMW3 in holding the core together (Fig. 6D, arrowhead).
FIG. 6.
Transmission electron microscopic analysis of thin sections of M. pneumoniae cells showing the electron-dense core (arrowheads). A, wild-type M. pneumoniae; B, transformant control; C, hmw3::Tn4001 transformant with wild-type core; D, hmw3::Tn4001 transformant with altered core.
Localization of cytadherence-associated proteins P1 and P65 in the absence of HMW3.
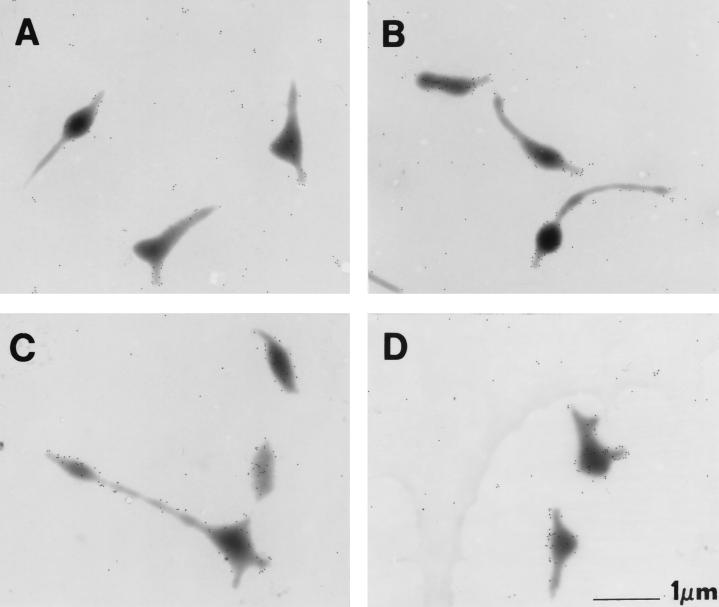
Adhesin protein P1 localizes primarily to the attachment organelle in wild-type M. pneumoniae (3, 11, 18; Fig. 7A). We used immunogold labeling to examine P1 localization in the hmw3::Tn4001 transformant. Due to the abnormal morphology of the mutant, it was often difficult to identify attachment organelles with complete certainty, yet some cells exhibited a local concentration of P1 on one or two branches in addition to the P1 scattered on the rest of the cell (Fig. 7D). However, this pattern was the exception and was only seen in approximately 10% of hmw3::Tn4001 cells, compared with approximately 60% of wild-type cells. In a majority of hmw3::Tn4001 cells, the labeling was dispersed along the cell surface (Fig. 7C). No obvious correlation was apparent between the presence or absence of P1 clustering and cell morphology. Some cells morphologically similar to prototypical wild-type cells exhibited no attachment organelle-localized P1, while other cells that were morphologically irregular exhibited P1 clustering. The transformant control was indistinguishable from wild-type cells (Fig. 7B).
FIG. 7.
Protein P1 distribution on wild-type and transformant M. pneumoniae cells. A, wild-type M. pneumoniae; B, transformant control; C and D, hmw3::Tn4001 transformant.
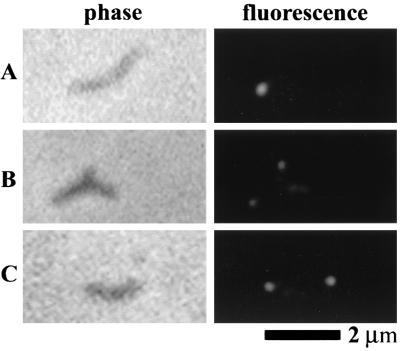
Recent reports have described the localization of P65 to the attachment organelle of wild-type M. pneumoniae with immunofluorescent labeling (20, 32). P65 is predicted to be incorporated late in the assembly of this structure (24). Additionally, our data revealing reduced levels of P65 concurrent with loss of HMW3 suggest an interaction between these two proteins. Therefore, we examined whether loss of HMW3 has consequences for the localization of P65 (Fig. 8). Cells were labeled with anti-P65 antibody and a fluorescent secondary antibody and examined by fluorescence-phase-contrast microscopy. hmw3::Tn4001 transformant cells generally had a minimum of one fluorescent focus (usually polar) with patchy fluorescence on the rest of the cell similar to the patchy labeling pattern of mutant I-2 cells (20). This is in contrast to the exclusively polar labeling observed with wild-type cells and the transformant control.
FIG. 8.
Localization of P65 protein in wild-type M. pneumoniae (A), mutant I-2 (B), and an hmw3::Tn4001 transformant (C). On the left are individual cells viewed by phase-contrast microscopy, and on the right are the corresponding fluorescent images. The contrast was adjusted to compensate for protein level differences among the strains (see Fig. 3E).
Isolation and characterization of an hmw3::Tn4001 excision revertant.
The hmw3::Tn4001 transformant was passaged five times in the absence of gentamicin; resulting colonies that were gentamicin sensitive regained the ability to adsorb erythrocytes, implying excision of the transposon. Gentamicin-sensitive, hemadsorption-positive colonies were selected and cultured in Hayflick broth. All putative excision revertants analyzed by Western immunoblotting regained wild-type levels of full-length HMW3 (data not shown). One revertant was characterized further. DNA sequencing of the hmw3 gene confirmed precise excision of the transposon, consistent with reacquisition of HMW3. Likewise, this revertant exhibited a characteristic wild-type phenotype with respect to protein profile, hemadsorption, cell morphology, and P1 and P65 localization (data not shown), establishing a clear correlation between insertional inactivation of hmw3 and the altered phenotype described here.
DISCUSSION
Several scenarios could account for the failure to isolate an hmw3 mutant previously (17, 22). The most likely explanation, however, is that mutations affecting cytadherence were identified previously by screening for complete loss of attachment (22), in some cases including enrichment steps for nonadherent cells (17). Loss of HMW3 resulted in only a partial reduction in cytadherence; hence, hmw3 mutants might have been overlooked in previous studies. Significantly, a 1999 study in which M. pneumoniae and Mycoplasma genitalium were both subjected to global transposon mutagenesis yielded a transposon insertion in the M. genitalium hmw3 homologue (19), hinting that this protein is dispensable in vitro, at least in M. genitalium and by correlation in M. pneumoniae.
Many of the M. pneumoniae cytadherence-associated proteins identified to date exhibit a stabilizing interdependency, most likely reflecting a complex web of interactions (24). For example, loss of HMW2 results in accelerated turnover of several proteins, including P65, which is encoded by the gene immediately upstream of hmw2 (23), and HMW1 and HMW3 (28), proteins encoded by the hmw operon, which is quite some distance from hmw2. Similarly, loss of HMW1 is associated with reduced levels of HMW3, HMW2 (Willby et al., unpublished), and P65 (20). However, loss of HMW3 in the hmw3::Tn4001 transformant affected only the level of P65. Significantly, reduction of the level of HMW3 was the single common denominator of decreased levels of P65 in hmw1, hmw2, and hmw3 mutants, suggesting a role for HMW3 in P65 stabilization and expanding the functional relationship between the protein products of the two operons. However, HMW3 is not sufficient to stabilize P65, as defects involving protein P30, which is encoded by the gene preceding hmw3, do not affect levels of HMW3 but result in diminished P65 levels (20, 24). Therefore, both HMW3 and P30, which are encoded by adjacent genes in the hmw operon, are likely required for stabilization of P65. Furthermore, this correlation between the levels of HMW3 and P65 may indicate a physical interaction between these two proteins that stabilizes P65 within the attachment organelle.
It is reasonable to assume that the interactions among cytadherence-associated proteins not only serve to stabilize one another but also assist in achieving and maintaining the appropriate distribution of these proteins within the cell. Recent fluorescence microscopy studies have shown that P65 is localized to a single pole in wild-type M. pneumoniae (20, 32) cells, the same pole at which P1 is found (32). These studies also showed wild-type localization of P65 in mutants in cytadherence proteins A, B, C, P1, and P30 but patchy distribution of P65 in mutants in which HMW1, HMW2, and HMW3 are absent or present at reduced levels (mutants I-2 and M6) (20). In the present study, in the absence of HMW3, P65 was not confined to a single focus. Patches of P65 were dispersed throughout the cell, although often with a dense cluster still identifiable at a pole (Fig. 8). Therefore, wild-type levels of HMW1 and HMW2 alone are not sufficient for wild-type localization of P65; HMW3 is also required. In contrast, preliminary studies have revealed HMW1 consolidated in a discrete focus in the hmw3::Tn4001 transformant (data not shown), as in wild-type cells (32, 35).
The major adhesin P1 is concentrated at the attachment organelle but present to a lesser extent along the rest of the wild-type M. pneumoniae cell (3, 11, 18). The clustering of P1 presumably enhances attachment of the bacterium to host cells. The loss of cytadherence accessory protein HMW1 or HMW2 affects the ability of P1 to localize properly (3, 15). Likewise, while some cells of the hmw3::Tn4001 transformant showed clustered P1 on branches, most did not (Fig. 7), a finding that is consistent with poor adherence to plastic and an intermediate hemadsorption phenotype. It is difficult to know whether these rare clusters of P1 are significant, since it is possible that in cells with randomly distributed P1, local concentrations of the adhesin could occur by chance along the length of the cell. HMW3 is thought to associate with the cytoplasmic surface of the cell membrane of the attachment organelle with no exposure on the mycoplasma cell exterior (36). Therefore, while HMW3 does not appear to play a direct role in attachment, loss of HMW3 results in defective P1 localization and, consequently, the dramatic decline in attachment efficiency revealed by qualitative and quantitative hemadsorption assays.
The hmw3 mutant is striking morphologically and ultrastructurally. While the classic wild-type spindle morphology was not completely absent in hmw3::Tn4001 cultures (Fig. 5), most cells had multiple branches and numerous distended areas. Branched cells are a common feature of cell division abnormalities in E. coli and other bacteria (1) and might indicate a similar consequence of loss of HMW3. Significantly, defects in P30, the gene for which precedes the gene for HMW3, likewise result in a branched morphology (31), as does loss of other cytadherence proteins (15, 31, 32). The electron-dense cores observed in thin sections of hmw3::Tn4001 cells in many cases appeared similar to those of wild-type cells (Fig. 6). Due to the nature of sectioning such small organisms, it is difficult to determine in cells with multiple branches whether more than one branch contains a core, so we could not assess whether the increased number of branches in the HMW3 mutant correlates with an increased number of cores. However, V-shaped cores were also identified in the mutant. These might actually represent a cross section of a cone. V-shaped cores accounted for approximately 20% of the cores seen in the hmw3::Tn4001 transformant. It is likely that this number is an underestimation of the actual number of abnormal cores present in the mutant since the sectioning process is arbitrary and the ability to distinguish V-shaped cores depends on the plane through which the cell is sectioned. Regardless, cores with this appearance have not been previously reported in wild-type cells or other mutants, suggesting that loss of HMW3 promotes a defect either in core structure or in the regulation of core construction. Previous studies indicated that polymers of HMW3 might surround the core and the terminal button in a linear pattern (36), possibly serving to stabilize this structure. In the absence of HMW3, the core may be less stable, leading to the V or, possibly, cone shape. Alternatively, this shape may reflect a change in the frequency at which new cores form prior to cell division, as formation of a second attachment organelle is thought to be an early step in this process (5, 32).
Studies based on this hmw3 mutant have advanced our understanding of M. pneumoniae on two related fronts. First, we examined more directly the function of HMW3, and secondly, we were able to lend weight to the model of attachment organelle assembly proposed by Krause and Balish (24). According to this model, proteins are incorporated into the attachment organelle via one of two pathways. Proteins HMW2, HMW1, HMW3, P30, and P65 follow one pathway and stabilize one another as they associate with the nascent attachment organelle. In this study, we showed that HMW1 is stable in the absence of HMW3, suggesting that it is one of the first proteins incorporated into the nascent attachment organelle. P65 is neither stable nor properly localized in the absence of HMW3, supporting the theory that P65 is incorporated into the attachment organelle after both HMW1 and HMW3. P1, on the other hand, was only occasionally found in polar clusters in hmw3::Tn4001 cells. Krause and Balish have proposed that P1 is incorporated into the attachment organelle via a pathway separate from HMW1 and HMW3 (24). The two pathways intersect in the attachment organelle, where proteins from each pathway likely interact, resulting in a functional structure. Its association with the attachment organelle, cytoskeleton, and cell membrane positions HMW3 to secure interactions with proteins found in each of these sites, for instance, maintaining localization of P1 to the attachment organelle and holding together the electron-dense core. Consistent with this hypothesis is the apparent ability of HMW3 to exist in multimeric form or as part of a complex of proteins (36), as HMW3 released from cells extracted sequentially with Triton X-100 and potassium iodide takes the form of clusters and chains (36). Furthermore, while HMW3 is a peripheral membrane protein, it pellets with alkali-treated cell membranes (Balish and Krause, unpublished data), suggesting that it is highly insoluble, which is perhaps attributable to its presence within a complex of proteins.
We hypothesize that HMW3 is not essential for formation of the attachment organelle but may be required for proper timing of its duplication or for stabilization or maintenance of its structure. Additionally, the attachment organelle is believed to function in adherence, motility, and cell division. Absence of HMW3 resulted in an intermediate hemadsorption phenotype. The colonial morphology of M. pneumoniae strains grown on soft agar with a liquid overlay has been correlated with the presence or absence of motility in M. pneumoniae strains (J. Jordan et al., unpublished data). We observed the hmw3::Tn4001 transformant under these conditions and, on the basis of preliminary observations, believe that this mutant has very limited motility in the absence of HMW3. Furthermore, the ultrastructural and morphological abnormalities associated with loss of HMW3 suggest a defect in cell division, a conclusion supported by the fact that HMW3 is located in the region of the cell where early steps in cell division are thought to occur. Thus, loss of HMW3 has effects on all of the functions thought to be associated with the attachment organelle, underscoring its fundamental importance.
Acknowledgments
This work was supported in part by Public Health Service research grant AI22362 from the National Institute of Allergy and Infectious Diseases (D.C.K.) and a National Science Foundation Research Training Grant in Prokaryotic Diversity (NSF BIR9413235) (M.J.W.).
We are very grateful to Richard Herrmann and Jarrat Jordan for generously sharing antisera. We also thank Mitchell Balish for critical reading of the manuscript and helpful suggestions.
REFERENCES
- 1.Ackerlund, T., K. Nordstrom, and R. Bernander. 1993. Branched Escherichia coli cells. Mol. Microbiol. 10:849-858. [DOI] [PubMed] [Google Scholar]
- 2.Balish, M. F., T.-W. Hahn, P. L. Popham, and D. C. Krause. 2001. Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the triton shell. J. Bacteriol. 183:3680-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baseman, J. B., R. M. Cole, D. C. Krause, and D. K. Leith. 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 151:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biberfeld, G., and P. Biberfeld. 1970. Ultrastructural features of Mycoplasma pneumoniae. J. Bacteriol. 102:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boatman, E. S. 1979. Morphology and ultrastructure of the mycoplasmatales, p. 63-102. In M. F. Barile and S. Razin (ed.), The Mycoplasmas, vol 1. Academic Press, Washington, D.C. [Google Scholar]
- 6.Bozzola, J. J., and L. D. Russell. 1992. Electron microscopy: principles and techniques for biologists. Jones and Bartlett Publishers, Boston, Mass.
- 7.Bredt, W. 1979. Motility, p. 141-155. In M. F. Barile and S. Razin (ed.), The mycoplasmas. vol 1. Academic Press, Washington, D.C. [Google Scholar]
- 8.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 9.Collier, A. M. 1972. Pathogenesis of Mycoplasma pneumoniae infection as studied in the human fetal trachea in organ culture Ciba Found. Symp., p. 307-327.
- 10.Dirksen, L. B., T. Proft, H. Hilbert, H. Plagens, R. Herrmann, and D. C. Krause. 1996. Sequence analysis and characterization of the hmw gene cluster of Mycoplasma pneumoniae. Gene 171:19-25. [DOI] [PubMed] [Google Scholar]
- 11.Feldner, J., U. Göbel, and W. Bredt. 1982. Mycoplasma pneumoniae adhesin localized to the tip structure by monoclonal antibody. Nature (London) 298:765-767. [DOI] [PubMed] [Google Scholar]
- 12.Fisseha, M., H. W. H. Göhlmann, R. Herrmann, and D. C. Krause. 1999. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J. Bacteriol. 181:4404-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Göbel, U., V. Speth, and W. Bredt. 1981. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J. Cell Biol. 91:537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, T.-W., K. A. Krebes, and D. C. Krause. 1996. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol. Microbiol. 19:1085-1093. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, T.-W., M. W. Willby, and D. C. Krause. 1998. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J. Bacteriol. 180:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, E. J., R. M. Wilson, and J. B. Baseman. 1979. Isolation of mutants of Mycoplasma pneumoniae defective in hemadsorption. Infect. Immun. 23:903-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedreyda, C. T., and D. C. Krause. 1995. Identification of a possible cytadherence regulatory locus in Mycoplasma pneumoniae. Infect. Immun. 63:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, P.-C., R. M. Cole, Y.-S. Huang, J. A. Graham, D. E. Gardner, A. M. Collier, and W. A. Clyde, Jr. 1982. Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216:313-315. [DOI] [PubMed] [Google Scholar]
- 19.Hutchison, C. A., III, S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 2000. Global transposon mutagenesis and a minimal mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 20.Jordan, J. L., K. M. Berry, M. F. Balish, and D. C. Krause. 2001. Stability and subcellular localization of cytadherence-associated protein P65 in Mycoplasma pneumoniae. J. Bacteriol. 183:7387-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knudtson, K. L., and F. C. Minion. 1993. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene 137:217-222. [DOI] [PubMed] [Google Scholar]
- 22.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause, D. C., T. Proft, C. T. Hedreyda, H. Hilbert, H. Plagens, and R. Herrmann. 1997. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J. Bacteriol. 179:2668-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krause, D. C., and M. F. Balish. 2001. Structure, function and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 198:1-7. [DOI] [PubMed] [Google Scholar]
- 25.Lipman, R. P., and W. A. Clyde, Jr. 1969. The interrelationship of virulence, cytadsorption, and peroxide formation in Mycoplasma pneumoniae. Proc. Soc. Exp. Biol. Med. 131:1163-1167. [DOI] [PubMed] [Google Scholar]
- 26.Morrison-Plummer, J., D. K. Leith, and J. B. Baseman. 1986. Biological effects of anti-lipid and anti-protein monoclonal antibodies on Mycoplasma pneumoniae. Infect. Immun. 53:398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogle, K. L., K. K. Lee, and D. C. Krause. 1992. Nucleotide sequence analysis reveals novel features of the phase-variable cytadherence accessory protein HMW3 of Mycoplasma pneumoniae. Infect. Immun. 60:1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popham, P. L., T.-W. Hahn, K. A. Krebes, and D. C. Krause. 1997. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs posttranslationally. Proc. Natl. Acad. Sci. USA 94:13979-13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proft, T., and R. Herrmann. 1994. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol. Microbiol. 13:337-348. [DOI] [PubMed] [Google Scholar]
- 30.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 31.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 5:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobeslavsky, O., B. Prescott, and R. M. Chanock. 1968. Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cell and possible role in virulence. J. Bacteriol. 96:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sperker, B., P.-C. Hu, and R. Herrmann. 1991. Identification of gene products of the P1 operon of Mycoplasma pneumoniae. Mol. Microbiol. 5:299-306. [DOI] [PubMed] [Google Scholar]
- 35.Stevens, M. K., and D. C. Krause. 1991. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike triton shell. J. Bacteriol. 173:1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens, M. K., and D. C. Krause. 1992. Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J. Bacteriol. 174:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tully, J. G. 1983. Cloning and filtration techniques for mycoplasmas. Methods Mycoplasmol. 1:173-177. [Google Scholar]
- 38.Waldo, R. H., III, P. L. Popham, C. E. Romero-Arroyo, E. A. Mothershed, K. K. Lee, and D. C. Krause. 1999. Transcriptional analysis of the hmw gene cluster of Mycoplasma pneumoniae. J. Bacteriol. 181:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wenzel, R., and R. Herrmann. 1988. Physical mapping of the Mycoplasma pneumoniae genome. Nucleic Acids Res. 16:8323-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]