Abstract
Within a large family of peroxidases, one member that catalyzes the reduction of organic peroxides to alcohols is known as alkyl hydroperoxide reductase, or AhpC. Gene disruption mutations in the gene encoding AhpC of Helicobacter pylori (ahpC) were generated by screening transformants under low-oxygen conditions. Two classes of mutants were obtained. Both types lack AhpC protein, but the major class (type I) isolated was found to synthesize increased levels (five times more than the wild type) of another proposed antioxidant protein, an iron-binding, neutrophil-activating protein (NapA). The other class of mutants, the minor class (type II), produced wild-type levels of NapA. The two types of AhpC mutants differed in their frequencies of spontaneous mutation to rifampin resistance and in their sensitivities to oxidative-stress chemicals, with the type I mutants exhibiting less sensitivity to organic hydroperoxides as well as having a lower mutation frequency. The napA promoter regions of the two types of AhpC mutants were identical, and primer extension analysis revealed their transcription start site to be the same as for the wild type. Gene disruption mutations were obtained in napA alone, and a double mutant strain (ahpC napA) was also created. All four of the oxidative-stress resistance mutants could be distinguished from the wild type in oxygen sensitivity or in some other oxidative-stress resistance phenotype (i.e., in sensitivity to stress-related chemicals and spontaneous mutation frequency). For example, growth of the NapA mutant was more sensitive to oxygen than that of the wild-type strain and both of the AhpC-type mutants were highly sensitive to paraquat and to cumene hydroperoxide. Of the four types of mutants, the double mutant was the most sensitive to growth inhibition by oxygen and by organic peroxides and it had the highest spontaneous mutation frequency. Notably, two-dimensional gel electrophoresis combined with protein sequence analysis identified another possible oxidative-stress resistance protein (HP0630) that was up-regulated in the double mutant. However, the transcription start site of the HP0630 gene was the same for the double mutant as for the wild type. It appears that H. pylori can readily modulate the expression of other resistance factors as a compensatory response to loss of a major oxidative-stress resistance component.
Helicobacter pylori is a spiral bacterium that colonizes the gastric mucosae of humans, leading to a variety of gastric inflammatory responses, including peptic ulcer disease and chronic gastritis. Prolonged infections and inflammation due to persistent H. pylori colonization lead to severe tissue damage and sometimes to adenocarcinomas of the stomach (4). The pathogenesis of H. pylori depends on its persistence, which in turn depends on its survival in a harsh environment, which can be characterized by acidity, peristalsis, and attack by phagocytes and their released oxygen species (19, 25). These and other host defenses are undoubtedly counteracted by the successful pathogen, and some of these host responses are even beneficial to further colonization (5). Even outside of the host, it is clear that a microaerobic environment is important for the survival of H. pylori. Consequently, there are prevailing views (12, 19, 24) that O2-mediated damage is commonly encountered by the bacterium and that protective pathways to deal with toxic O2-derived products may be key to the organism's ability to colonize the mucosal environment of the host.
Alkyl hydroperoxide reductase (AhpC) belongs to a family of peroxidases that are beginning to receive intense research focus due to their roles in dissipating damaging hydrogen peroxides (10) and related hydroperoxides. The specific role of AhpC in catalyzing the reduction of organic hydrogen peroxides to their respective alcohols and the roles of accessory enzymes in these reductions are being determined (13, 33, 34). In addition, AhpC of Salmonella enterica serovar Typhimurium was shown to confer resistance to reactive N intermediates (9). H. pylori AhpC is closely related to the peroxiredoxin of higher organisms, such as those found in Saccharomyces cerevisiae and Caenorhabditis elegans, but it also has considerable sequence identity to five bacterial AhpCs (2). The reductase system for pure H. pylori AhpC was identified and consists of thioredoxin and thioredoxin reductase; the flavoprotein reductant (AhpF) used by most bacteria to reduce AhpC was shown to be inactive for the H. pylori AhpC system (2). The complete system (AhpC with thiol-specific reductant) was capable of reducing a variety of hydroperoxide substrates that include both small aromatic and lipid hydroperoxides. It was concluded that H. pylori alkyl hydroperoxide reductase is essential for growth of the bacterium, since gene-disrupted deletion mutants could not be recovered (2, 8, 23).
Based on the genome sequence, H. pylori contains a battery of enzymes to combat oxidative stress, but the importance of these enzymes for the growth of the organism via targeted mutagenesis approaches has been addressed for only a few of these (24, 29; R. W. Seyler and R. J. Maier, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. D-185, 2001). To address the role of AhpC in H. pylori, we obtained strains with mutations in the ahpC gene, encoding AhpC, by screening the allelic-exchange transformants under 2% O2 conditions. Surprisingly, two types of mutants were recovered. Both classes lack AhpC, but the major class of mutant recovered was found to up-regulate another proposed antioxidant protein, an iron-binding (31), neutrophil-activating protein (NapA) (14). Consequently, we created single mutant strains with individual mutations in the gene for NapA and a double mutant strain with mutations in the genes for NapA and AhpC. Of the four types of mutants, the double mutant was the most sensitive to oxidative stress and by far had the highest frequency of spontaneous mutation. However, all the mutants could be distinguished from the wild type in some oxidative-stress resistance phenotype. The phenotypic results are consistent with the conclusion that H. pylori has multiple oxidative-stress-related proteins and that compensatory mutations occur to circumvent the loss of AhpC.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strain ATCC 43504 was used as the wild type. Cultures of H. pylori were grown microaerobically at 37°C in a 5% CO2 incubator (model 3130; Forma Scientific) under different continuously controlled levels of oxygen (between 2 and 12% partial pressure, as indicated). Plates coated with brucella agar (Difco) supplemented with 10% defibrinated sheep blood (Gibson Laboratories, Inc.) are called BA plates and were used for growth of H. pylori. Kanamycin (at 30 μg/ml) or chloramphenicol (at 20 μg/ml) was added as indicated. All strains were confirmed to be H. pylori because of the presence of urease activity and catalase activity and the helical morphology of the organisms as determined by phase-contrast microscopy (28). Genetic manipulations were performed with Escherichia coli strain DH5α. Luria-Bertani agar supplemented with ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), or kanamycin (30 μg/ml) was used for growing various E. coli strains, as noted. Strains and their sources are given in Table 1.
TABLE 1.
Strains and plasmids
| Strain, plasmid, or primer | Description | Source |
|---|---|---|
| Strains | ||
| H. pylori | ||
| ATCC 43504 | Parent strain for all H. pylori strains | ATCCa |
| ahpC::Kan type I | aphA3 insertion within ahpC | This study |
| ahpC::Kan type II | aphA3 insertion within ahpC | This study |
| napA::Cm | cat insertion within napA | This study |
| ahpC::napA | ahpC napA double mutant | This study |
| E. coli DH5α | Cloning strain | Lab stock |
| Plasmids | ||
| pBluescript KS+ | Cloning vector | Stratagene |
| pKSΔS | pBluescript KS+ with destroyed SacI site | This study |
| pAhpC | ahpC inserted into SmaI site of pBluescript KS+ | This study |
| pAhpCKan | aphA3 inserted into AflII site of pAhpC | This study |
| pNapA | napA cloned from ATCC 43504 into pBluescript KS+ | This study |
| pNapAT1 | napA cloned from ahpC::Kan type I mutant | This study |
| pNAΔS | napA cloned into pKSΔS | This study |
| pNapA::Cm | cat inserted in SacI site of pNAΔS | This study |
| Primers | ||
| ahpCF | 5′ AGCCACGCCCAATAACGATG 3′ | IDTb |
| ahpCR | 5′ TCGCCTTTTCTCCAACCTGCTG 3′ | IDT |
| NapAF | 5′ AACCACTAAATTAAAGGGTAACGGC 3′ | IDT |
| NapAR | 5′ CGCTGTAATCTCTCATGCTGGC 3′ | IDT |
| NapART | 5′ ACACGATCGCATCCGCTTGC 3′ | IDT |
| HP0630RT | 5′ AGAATGCCCGAACGCTTTGG 3′ | IDT |
ATCC, American Type Culture Collection.
IDT, Integrated DNA Technologies, Coralville, Coraville, Iowa.
Cloning of ahpC and construction of an ahpC::Kan mutant.
Oligonucleotide primers for cloning the ahpC gene (ahpCF and ahpCR) (Table 1) were designed from the sequenced strain J99 (1). PCR amplification was performed with Taq DNA polymerase (Fisher Scientific, Pittsburgh, Pa.) using chromosomal DNA from H. pylori strain ATCC 43504 as a template. The PCR product (851 bp) was then treated with T4 DNA polymerase (Promega) and phosphorylated with T4 polynucleotide kinase (Promega). The fragment was then cloned into a SmaI site of pBluescript KS+ (Stratagene) to give pAhpC. This construct was then sequenced by the University of Georgia Molecular Genetics Instrumentation Facility. The ahpC::Kan construct was created via insertion of kanamycin resistance cassette AphA3 (25) into the AflII site of pAhpC. Restriction analysis revealed that the Kanr cassette was in the orientation opposite that of the ahpC gene.
Cloning of napA and construction of pNapA::Cm.
A fragment of H. pylori DNA corresponding to the napA gene was amplified from genomic DNAs obtained from both H. pylori strain ATCC 43504 and the ahpC::Kan (type I) strain with the primers NapAF and NapAR (Table 1) and Taq DNA polymerase (Fisher Biotech). This 809-bp fragment contains the entire napA coding region as well as 339 bp upstream of the napA start codon. The PCR fragment was treated with T4 DNA polymerase to ensure blunt ends and phosphorylated with T4 polynucleotide kinase. The fragments were then ligated with pBluescript KS+ digested with SmaI. The resulting clones, pNapA (napA cloned from ATCC 43504) and pNapAT1 (napA cloned from the ahpC::Kan type I strain), were then sequenced at the University of Georgia Molecular Genetics Instrumentation Facility. In order to utilize the SacI unique restriction site within the napA coding region, it was necessary to excise the fragment with EcoRI and BamHI and clone it into a derivative of pBluescript KS+ in which the SacI site had been destroyed (pKSΔS), yielding pNAΔS. SacI-digested pNAΔS was then filled in with T4 DNA polymerase and ligated to the chloramphenicol resistance cassette (cat). The resulting plasmid, pNapA::Cm, was then electroporated into competent ATCC 43504 cells to yield the napA mutant napA::Cm and into competent ahpC::Kan type I cells to yield the ahpC napA double mutant. Insertion of the cassette was confirmed by isolation of chromosomal DNAs from the mutant strains and PCR amplification of the napA locus, followed by agarose gel electrophoresis, which was used to monitor the increase in size of napA due to the cat insertion.
Transformation of H. pylori.
Preparation of competent cells involved harvesting and washing cells four times in a solution of (ice-cold) sucrose-glycerol, followed by the immediate storage of the cells at −80°C. H. pylori cells were transformed with pAhpCKan by electroporation (27). During transformation, 2 μl of the plasmid was added to 50 μl of competent cells and electroporated with Transporator Plus (BTX, San Diego, Calif.) with a pulse of 2.5 kV. Immediately after the pulse, 50 μl of Mueller-Hinton broth (Difco) was added to the cuvette containing the competent cells. Aliquots of these cells were then immediately plated on nonselective BA plates, and the plates were incubated in the CO2 incubator under partial pressure (2% O2). After a 48-h incubation period, the cells from this nonselective medium were transferred to BA plates containing kanamycin. Kanr colonies were selected by incubation in a 2% O2 incubator. Genomic DNA isolated from Kanr strains were shown to have an increase in the size of the PCR product of ahpC from 851 bp to approximately 2,251 bp, as anticipated. Further, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and two-dimensional (2D) gels were used to confirm the lack of synthesis of AhpC protein in the gene-directed mutants.
Gel electrophoresis.
Plate-grown cells (2% O2) were harvested with a swab from the plate and resuspended in 50 mM Tris (pH 7.4) buffer containing 50 mM NaCl. The cells were collected by centrifugation (10,000 × g for 10 min), resuspended in Tris-NaCl buffer, and broken by two passages through a French pressure cell at 138,000 kPa (SLM Instruments, Inc.). Crude extracts were then cleared of unbroken cells by centrifugation at 10,000 × g for 10 min. The protein concentrations of the cell extracts were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). For SDS-PAGE, 5 μg of cell extract was placed into SDS buffer, boiled for 5 min, and applied to a denaturing 12.5% acrylamide gel as described previously (21). For 2D gel electrophoresis, 50 μg of total protein (crude extract) was added to 1.125× immobilized-pH-gradient (IPG) buffer (1× concentration: 8 M urea, 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 40 mM Tris base). The IPG strips (pHs 3 to 10; Bio-Rad, Hercules, Calif.) were then rehydrated overnight at room temperature with the IPG buffer-treated protein. Proteins were focused for 5 h at 3,000 V by using a Multiphore II isoelectric-focusing apparatus (Pharmacia Biotech, Piscataway, N.J.). The strips were then placed onto a 12.5% acrylamide gel, and the proteins were separated in the second dimension by SDS-PAGE, which was performed according to the recommendations of the manufacturer (Bio-Rad). Protein spots were visualized by silver staining as described previously (26).
Protein fractionation and sequencing.
Crude extracts of H. pylori wild-type cells were subjected to partial purification by fast-protein Q-Sepharose ion-exchange chromatography (Pharmacia Biotech). Proteins were prepared for sequencing by subjecting the starting material (partially purified ahpC::Kan type I crude extract for NapA, or the ahpC napA crude extract for HP0630) to SDS-PAGE (12.5% acrylamide), followed by electroblotting onto a polyvinylidene difluoride membrane (32). The appropriate bands were then excised and subjected to eight cycles of N-terminal protein sequencing. Protein sequencing was performed by the Molecular Genetics Instrumentation Facility at the University of Georgia.
RNA isolation and primer extension analysis.
Plates were streaked and incubated at 2% O2 for 3 days. The harvested cells of the wild type, napA single mutant, and ahpC napA double mutant were resuspended in 1 ml of phosphate-buffered saline (PBS). RNA was isolated as described previously (2). For the primer extension analysis, 10 pmol each of either primer NapART or HP0630RT (Table 1) was 5′-end labeled in the reaction with 5 μl of [γ-32P]ATP (6,000 Ci/mmol; Amersham) and 10 U of T4 polynucleotide kinase (Promega). An RNA sample (11 μl at a concentration of 200 μg/ml) was incubated with a 3-μl solution of the 32P-labeled primers and allowed to anneal at 70°C for 5 min, followed by slow cooling to 50°C for 3 h. A mixture containing 1 μl of avian myeloblastosis virus reverse transcriptase (Promega), 4 μl of 5× reverse transcriptase buffer (supplied with enzyme), and 1 μl of a 10 mM concentration of the deoxynucleoside triphosphates was added to the annealed primers for 1 h at 42°C. DNA-sequencing reactions of napA and HP0630 were performed simultaneously with the same primers and the fmol DNA-sequencing system (Promega). Aliquots of primer extension and DNA-sequencing reaction mixtures were heated prior to being loaded onto a 6% PAGE-7 M urea gel. Gels were run for 2 h at 65 W and dried, and bands were visualized by exposure to film.
Growth sensitivity to oxygen.
H. pylori mutant and wild-type strains were streaked for individual colony observation by use of three-way streaks onto BA plates. These plates were placed into the CO2 incubator under controlled O2 concentrations. The incubators were adjusted to different (but continuously controlled) oxygen levels (2, 3, 4, 5, 7.5, 10, or 12% partial pressure), and the plates were observed for the presence of isolated colonies in the area of the final streak after 3 days in the incubator. Individual colony sizes described in Table 2 are from the third streak, when individual colonies were the farthest apart.
TABLE 2.
Growth sensitivity to oxygen
| Strain | Growth sensitivity to oxygen concn (%)a:
|
||||||
|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 7.5 | 10 | 12 | |
| Wild type | ++ | +++ | +++ | +++ | +++ | +++ | +++ |
| ahpC::Kan type I | ++ | + | +/− | +/− | +/− | − | − |
| ahpC::Kan type II | ++ | + | +/− | +/− | +/− | − | − |
| napA::Cm | ++ | +++ | ++ | + | + | +/− | +/− |
| ahpC napA | + | + | +/− | +/− | − | − | − |
Score on day 3. Symbols: −, no growth; +/−, slight growth at the site of greatest inoculum, no individual colonies; +, significant growth at the site of greatest inoculum, no individual colonies; ++, significant growth and development of isolated colonies (pinpoint-like colonies, less than 0.5 mm in diameter); +++, healthy growth with individual colonies (more than 0.5 mm in diameter).
Tolerance to oxygen.
BA plates were heavily streaked and incubated at 2% O2 for 3 days to obtain confluent growth. ahpC::Kan and wild-type cells from these plates were harvested, and cell suspensions were prepared in PBS. The optical density of the cells was measured (at 600 nm) and adjusted to 0.1; 5-ml aliquots were exposed to either anaerobic or aerobic conditions at 37°C, as previously described (29). Undiluted samples from each set of conditions were plated hourly for a period of 6 h onto BA plates. CFU (survivors) were counted after 3 to 5 days of incubation with 2% O2.
Frequency of spontaneous mutations.
The frequency of spontaneous mutations was monitored by quantitating mutation to rifampin resistance (29). The strains were plated onto BA plates with or without rifampin (5 μg/ml). Cells were first harvested from the plates and then resuspended in PBS buffer. Optical density (at 600 nm) was measured, and dilutions were plated to determine the number of viable cells in the original (undiluted) suspension. The mutant was plated onto BA plates containing kanamycin, and the wild type was plated onto the same medium but without kanamycin. The original suspensions of wild-type and mutant cells were also plated onto BA plates supplemented with rifampin. The numbers of wild-type and mutant cells were then compared with the numbers of colonies identified on rifampin plates, and the mutation frequency was calculated (29).
Peroxide sensitivity.
Sensitivity to three different cytotoxic agents was evaluated by disk assays (29). Sterile filter paper disks 7.5 mm in diameter containing one of the agents in water (5% [vol/vol] cumene hydroperoxide, 5% [vol/vol] t-butyl hydroperoxide, or 2 mM paraquat) were placed on BA plates (100 by 15 mm, 25-ml volume) that had previously been streaked for confluent growth with either mutant or wild-type cells. The plates were then placed in the 2% O2 incubator. Following a 2-day incubation period, the clear zones surrounding the disks were measured. The data given represent the distances from the edge of the disk to the end of the clear zone, where growth begins. No growth inhibition was seen in disks containing H2O.
RESULTS
Sequencing and cloning of the ahpC gene.
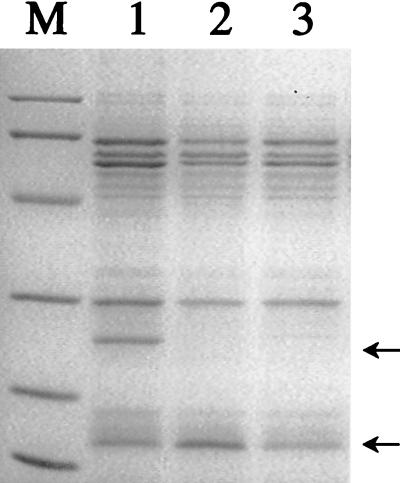
The H. pylori ahpC gene no. HP1563 (gene numbers are from reference 30) encodes an alkyl hydroperoxide reductase, known as AhpC (30). Our goal was to clone this gene to enable the generation of gene-directed mutants for use in physiological studies. The cloned gene was then used to make a Kanr ahpC-disrupted mutant by allelic exchange (see Materials and Methods). Importantly, the mutant strains were obtained by screening the transformants with O2 at low pressure and Kanr transformants were not recovered when the transformants were screened in 12% O2. SDS-PAGE was done to compare AhpC levels in wild-type and mutant cells. Alkyl hydroperoxide reductase was previously shown to be a predominant protein (third most abundant) in H. pylori, and it could be readily identified in 2D gels (18). In single-dimension gels of crude extracts of the wild type, we observed a predominant peptide that migrated at 26 kDa (Fig. 1); this peptide represented the putative AhpC absent in extracts from ahpC::Kan mutants.
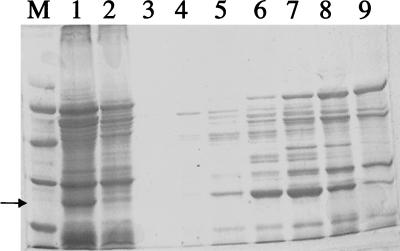
FIG. 1.
Fractionation of AhpC via Q-Sepharose chromatography. The arrow delineates the protein of interest, AhpC. Lane M, low-range standards composed of phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa); lane 1, H. pylori crude extract; lane 2, flowthrough eluent; lanes 3 through 9, fractions 20 through 26.
Fast protein Q-Sepharose ion-exchange chromatography was used to fractionate AhpC from extracts of the wild type (Fig. 1). Fraction 23 contained an abundance of the 26-kDa protein, which was well separated from other bands, so the polypeptides in this fraction were separated by SDS-PAGE and transblotted onto a polyvinylidene difluoride membrane. The excised 26-kDa band was subjected to eight cycles of N-terminal sequencing. The sequence result was in excellent agreement with that predicted for AhpC, with complete identity in the first 8 amino acids (MLVTKLAP). Based on SDS-PAGE analysis, all mutants chosen (as Kanr colonies) lacked the 26-kDa protein (i.e., AhpC). However, from the gel analysis, we also observed that the AhpC mutant (Fig. 2, lane 2) contained levels of another protein (of an approximate molecular mass of 17 kDa) higher than those in the wild type. This type of mutant was common (see below) and is referred to as type I. Sequencing of the first 8 amino acids of the up-regulated protein (by transblotting and excising) revealed 100% identity to H. pylori NapA (neutrophil-activating protein). Densitometric scanning of all the proteins in each lane of Fig. 2 showed that the NapA band (normalized to all protein bands) was fivefold greater in intensity than that determined for the wild type. Apparently, the loss of AhpC caused the concomitant overproduction of another oxygen stress-related protein, NapA. NapA in H. pylori is a ferritin-like iron-binding protein (31), and it may play a role in reducing toxic oxygen radical production via free-iron removal.
FIG. 2.
SDS-PAGE of H. pylori strains. Five micrograms of crude extract was loaded into each lane. Lane M, low-range markers (sizes are indicated in the legend to Fig. 1); lane 1, strain ATCC 43504; lane 2, ahpC::Kan type I strain; lane 3, ahpC::Kan type II strain. The arrows indicate proteins AhpC (26 kDa) and NapA (17 kDa).
Two types of mutant strains.
Upon further analysis of a series of transformant colonies, it was observed that we had obtained another type of AhpC mutant in which NapA was not up-regulated. Therefore, two types of mutants were discernible. To determine the prevalence of the two types of mutants, 36 individual colonies were chosen from three complete and independent transformation procedures (selecting for kanamycin insertions). The Kanr transformants from all three procedures were selected under the partial-pressure (2% O2) condition. Out of the 36 ahpC::Kan mutants, only 2 lacked the up-regulation of the NapA phenotype. These two isolates still made NapA, but they did so at the wild-type level. These isolates are referred to as type II mutants, and SDS gels of a representative of each of the two types of mutants are shown in Fig. 2. As a control, modA::Kan mutants were obtained by screening for transformants in the same manner (in 2% O2). In this case, no up-regulation of NapA was visible for any of the six mutants chosen; all modA mutants had normal (like wild-type) NapA levels. Therefore, the up-regulation phenotype was associated with the loss of AhpC, even though the second class of mutants was obtained in the same screening procedure. As further confirmation of the correlation between the lack of AhpC and a concomitant up-regulation of NapA, we prepared samples of the wild-type cells and both types of mutant cells and subjected them to 2D gel electrophoresis. The result (Fig. 3) confirmed the loss of AhpC in the ahpC gene-directed mutants and documented the up-regulation of NapA in the type I mutant and the normal levels of NapA in type II strains (data not shown).
FIG. 3.
Separation (2D) of crude extracts of the wild-type (ATCC 43504) (A), ahpC::Kan type I (B), napA::Cm (C), and ahpC napA (D) strains. The first dimension (isoelectric focusing) was carried out with the pH range between 3 (left) and 10 (right). The identified proteins are AhpC (within the ovals), NapA (within the rectangles), and HP0630 (circled).
The difference in NapA levels between the parent strain and the type I mutants was not due to a change in the napA promoter region. The DNA sequences of the intergenic region (209 bp) between HP0244 and the start of napA were sequenced from clones derived from both the wild-type strain ATCC 43504 and the ahpC::Kan type I mutant, and they were determined to be identical.
Due to the observed up-regulation of NapA and our desire to identify the roles of oxidative-stress resistance proteins, we created more mutant strains. These were a gene disruption mutant with a mutation in napA, designated the napA::Cm strain, and a double mutant with mutations in both napA and ahpC, designated the ahpC napA strain. Conferring additional stress on H. pylori via mutation of oxidative-stress resistance genes created another compensatory change: in the double mutant strains (ahpC napA mutant), the 2D gels revealed the induction of another protein of about 22 kDa in size. N-terminal sequencing of this protein showed 100% homology with HP0630 of H. pylori 26695 (30), a 21.8-kDa protein. Interestingly, a conserved domain search of the Pfam database (U.S. version 6.6) (3) revealed that this protein is part of a family of NAD(P)H dehydrogenases (pfam02525) which, in eukaryotic cells, catalyze the two-electron reduction of quinone and are involved in cellular protection against damage by free radicals and reactive oxygen species (22). The only bacterial member of the family to be characterized is the Mda66 protein of E. coli, which has been shown to be a NADPH quinone reductase but to which no physiological role has been assigned (16). Further analysis of this protein will bring a deeper understanding of its role in H. pylori, perhaps as an oxidative-stress resistance factor.
Primer extension analysis of napA and HP0630.
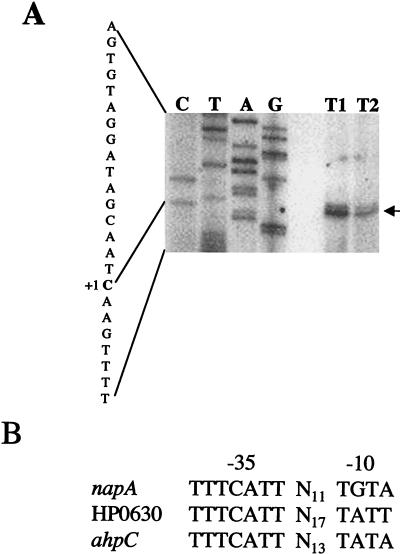
The transcription start sites of the napA and HP0630 genes were identified by primer extension analysis. Two possible transcription start sites of napA were identified 53 and 54 bp upstream of the translation initiation codon. The most prominent band indicating base C, located at 53 bp upstream, was designated +1 (Fig. 4A). All the strains analyzed (wild-type, ahpC::Kan type I, and ahpC::Kan type II strains) utilized the same transcription start site for napA. Similarly, the wild-type, ahpC::Kan type I, and ahpC napA strains had the same start site for the HP0630 gene, at 16 bp upstream of the translation start site (data not shown). The extended product for the ahpC::Kan type I mutant was more intense than that of the type II mutant (Fig. 4A), in agreement with the increased amount of protein observed for the type I mutant. Analysis of the promoter region for the two new genes of interest along with that of the previously determined ahpC promoter (2) revealed strong homology in the −35 regions among the three genes and some (but less) similarity in the location and sequence of the −10 regions among the three genes (Fig. 4B). The inverted repeat identified in the ahpC promoter (2) did not appear to be conserved in either napA or HP0630.
FIG. 4.
(A) Primer extension analysis of napA in ahpC::Kan type I and type II mutants. The extended product (designated by the arrow) is located 53 bp upstream from the translation initiation codon AUG and is indicated by “+1.” (B) Alignment of −35 and −10 promoter regions of napA and HP0630 with the previously identified ahpC regions (2). The −35 heptamers begin 33, 30, and 33 bp upstream from the transcription start sites of napA, HP0630, and ahpC, respectively.
Sensitivity to oxygen.
Oxidative-stress responses are usually related to an organism's ability to tolerate oxygen and to combat the toxic oxygen radicals and other related oxidative species that may arise. One way in which we sought to characterize mutants was to determine their sensitivity to oxygen. Initially, we measured the growth of the wild-type and mutant cells in environments having different levels of oxygen. BA plates were incubated at 2, 3, 4, 5, 7.5, 10, or 12% O2. The presence of isolated colonies, as well as growth at the site of inoculation (initial streak site) at each O2 level, was evaluated after 3 days (Table 2). In the wild-type strain of H. pylori, isolated colonies were visible at all concentrations of O2. On plates streaked with either of the AhpC-type mutants, small, isolated colonies were visible from the 2% O2 incubator. No isolated colonies were visible when the plates were incubated at pressures above 2% O2, and slight growth (but not individual colonies) was visible under partial-pressure conditions of 3, 4, 5, and 7.5% O2. Still, this growth was observed only at the site of greatest inoculum. No growth was visible (for any of the AhpC mutants) at 10% O2 or higher, O2 levels fully permissible for the growth of wild-type cells. NapA-deficient cells showed growth with the appearance of individual colonies under microaerobic conditions up to 3% O2, like wild-type cells, and this strain began to suffer growth inhibitions at 4% O2. The double mutant (ahpC napA) cells were highly oxygen sensitive. We observed no individual colonies of this strain at any O2 concentration; however, there was growth at the site of greatest inoculum. We previously reported that a sodB mutant was considerably more sensitive to oxygen killing than the wild-type strain in experiments measuring the viability loss of nongrowing cells exposed to air. Interestingly, unlike the sodB mutant (29), both types of ahpC::Kan mutants showed only slightly greater viability loss than the wild type when they were subjected to these non-growth-promoting, air exposure conditions (data not shown).
Sensitivity to cytotoxic agents.
A previously studied sodB mutant of H. pylori was shown to have increased susceptibility to hydrogen peroxide (29). To test whether our ahpC::Kan mutant showed increased sensitivity to an oxidative-stress reagent (such as peroxide exposure), a series of disk inhibition assays were performed. Inhibition zones were measured around agent-saturated disks (Table 3). Both types of AhpC mutants were more sensitive to cumene hydroperoxide than the wild type. However, with t-butyl hydroperoxide, the type I mutant was less sensitive than type II, as shown by a slight decrease in the size of the inhibition zone. This tolerance of t-butyl hydroperoxide by the type I mutant is probably related to the increased NapA level, since the type II mutant was considerably more sensitive to this peroxide than the wild type. Both types of AhpC mutants were more sensitive to paraquat than the wild-type strain. The napA::Cm mutant was similar to the wild type in its sensitivity to peroxides and paraquat, presumably because this strain contains abundant AhpC. The double mutant (ahpC napA) showed the greatest sensitivity of all the mutants to both t-butyl hydroperoxide and to paraquat.
TABLE 3.
Disk sensitivity assaya
| Strain | Disk sensitivity with the following treatmenta:
|
||
|---|---|---|---|
| 5% Cumene hydroperoxide | 5% t-Butyl hydroperoxide | 2 mM paraquat | |
| Wild type | 7.3 ± 1.5 | 13.5 ± 1.2 | 1.1 ± 0.1 |
| ahpC::Kan type I | 11.7 ± 2.4 | 9.0 ± 0.2 | 9.5 ± 0.5 |
| ahpC::Kan type II | 12.9 ± 2.3 | 19.8 ± 1.0 | 10.4 ± 1.1 |
| napA::Cm | 7.3 ± 1.3 | 14 ± 1.7 | 1.1 ± 0.1 |
| ahpC napA | 9.9 ± 1.3 | 20.5 ± 1.7 | 13.7 ± 0.9 |
Zones of inhibition were measured (in millimeters) around filter paper disks saturated with 10 μl of the indicated compounds. Water as a control did not yield any zones of growth inhibition. Results are the averages ± standard deviations from five independent experiments.
Spontaneous rifampin resistance.
Mutant strains of H. pylori lacking superoxide dismutase had higher frequencies of spontaneous mutation than that of the wild type (29). H. pylori deprived of functional AhpC may be more prone to DNA damage caused by reactive oxygen species as well. In three independent experiments on the AhpC mutants, we found that both of the mutants had significantly elevated spontaneous mutation frequencies compared with that of the wild-type strain. The average frequencies of Rifr mutation were 133 and 20 times greater for the type II (40 Rifr CFU per 108 cells) and type I (6 Rifr CFU per 108 cells) mutants, respectively, than that for the wild-type strain. The ahpC napA double mutant strain's frequency was 350-fold greater than that of the wild type, whereas the napA::Cm single mutant had a mutation frequency no greater than that of the wild type. Lipid hydroperoxides are known to undergo metal ion-mediated decomposition which results in modification of DNA and formation of so-called adducts. This type of alteration can cause frequent mutations (7) and could be one explanation for why the double mutant had a much higher mutation frequency than the other strains. The double mutant strain clearly has a deficiency in dissipating organic hydroperoxides (Table 3).
DISCUSSION
In order to combat reactive oxygen species and therefore persist in the gastric mucosa, pathogenic H. pylori is equipped with a number of putative detoxifying proteins (11, 24, 25). These include not only enzymes to detoxify oxygen-related radicals but N-containing reactive species as well. One of the enzymes observed in great abundance in H. pylori cells grown outside of the host is alkyl hydroperoxide reductase (18), and several groups have concluded that the organic-peroxide-detoxifying activity is important to H. pylori survival, since strains with mutations in ahpC could not be recovered (2, 8, 23). From our results, such mutants could be recovered under low-O2 conditions. AhpC clearly plays a role in oxidative-stress resistance, but whether this is due primarily to minimizing the level of organic peroxides (17) is unclear. For example, it was recently suggested that H2O2 may be the primary substrate for AhpC (10).
Two classes of mutants, both lacking AhpC, were obtained. The type I mutants contained increased levels of NapA protein, whereas the type II mutants did not. This increased NapA expression was associated with an increased resistance to t-butyl hydroperoxide and a lower spontaneous mutation frequency than that of the type II mutants. Previously, it was found that overexpressed levels of alkyl hydroperoxide reductase were able to overcome the high frequency of spontaneous mutations, which suggests that the reductase plays an important role in protecting against oxidative DNA damage (17). The same may be true for our (ahpC::Kan type I) mutants, where NapA appears to play a role in minimizing the spontaneous mutation frequency.
In other systems, a loss of oxidative-stress resistance function has been reported to cause a concomitant increase in levels of another oxidative-stress resistance function. Mutation of AhpCF of Bacillus subtilis led to increased expression of some factors that may help confer H2O2 resistance (6). Similarly, the antioxidant protein Dpr of Streptococcus mutans was identified due to its increased production upon mutation of other antioxidant-encoding genes (33). Dpr, a homologue of NapA, was proposed to help confer aerotolerance to S. mutans by keeping free-iron levels low, thus minimizing hydroxyl radical production (34).
Increased expression of an oxidative resistance factor appears to be a compensatory response used by oxidative-stress-deficient H. pylori to ensure that oxidative damage resistance is maintained. The peroxide sensitivity phenotype associated with AhpC mutants supports this compensatory up-regulation conclusion. Our results on the up-regulation phenomena prompted us to obtain additional mutants, namely, a napA::Cm mutant and a strain with a double gene disruption (ahpC napA). NapA is an iron-binding protein that may protect the cells by sequestering ferric iron released from iron sulfur clusters (20). The need for sequestering this type of iron is related to Fenton reactions that produce DNA-damaging hydroxyl radicals (15). Most of the results on the H. pylori napA::Cm mutants do not clearly define an oxidative-stress role for this protein, but the napA::Cm mutant strain was O2 sensitive compared to the wild type. However, the high spontaneous mutation frequency and marked sensitivity to organic peroxides of the double mutant are in agreement with the conclusion that both proteins (AhpC and NapA) play roles in oxidative agent detoxification. Further experiments on the role of the protein (HP0630) that is up-regulated in the ahpC napA mutant may prove to be useful in furthering our knowledge of oxidative-stress resistance in gastric pathogens.
Acknowledgments
This work was supported by NIH grant 1-RO1-DK60061-01.
We thank Richard Seyler and Matt Chenoweth for their expertise and assistance.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Baker, L. M., A. Raudonikiene, P. S. Hoffman, and L. B. Poole. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. Biomed. J. 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser, M. J., and J. Parsonnet. 1994. Parasitism by the “slow” bacterium Helicobacter pylori leads to altered gastric homeostasis and neoplasia. J. Clin. Investig. 94:4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bsat, N., L. Chen, and J. D. Helmann. 1996. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J. Bacteriol. 178:6579-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burcham, P. C. 1998. Genotoxic lipid peroxidation products: their DNA damaging properties and role in formation of endogenous DNA adducts. Mutagenesis 13:287-305. [DOI] [PubMed] [Google Scholar]
- 8.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., Q. W. Xie, and C. Nathan. 1998. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1:795-805. [DOI] [PubMed] [Google Scholar]
- 10.Costa Seaver, L., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doig, P., B. L. de Jonge, R. A. Alm, E. D. Brown, M. Uria-Nickelsen, B. Noonan, S. D. Mills, P. Tummino, G. Carmel, B. C. Guild, D. T. Moir, G. F. Vovis, and T. J. Trust. 1999. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev. 63:675-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, H. R., and L. B. Poole. 1997. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 36:13349-13356. [DOI] [PubMed] [Google Scholar]
- 14.Evans, D. J., Jr., D. G. Evans, T. Takemura, H. Nakano, H. C. Lampert, D. Y. Graham, D. N. Granger, and P. R. Kvietys. 1995. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 63:2213-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridovich, I. 1998. Oxygen toxicity: a radical explanation. J. Exp. Biol. 201:1203-1209. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, M., H. Ohzeki, H. Shimada, and T. Unemoto. 1996. NADPH-specific quinone reductase is induced by 2-methylene-4-butyrolactone in Escherichia coli. Biochim. Biophys. Acta 1273:165-170. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson, F. S., R. W. Morgan, M. F. Christman, and B. N. Ames. 1989. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J. Biol. Chem. 264:1488-1496. [PubMed] [Google Scholar]
- 18.Jungblut, P. R., D. Bumann, G. Haas, U. Zimny-Arndt, P. Holland, S. Lamer, F. Siejak, A. Aebischer, and T. F. Meyer. 2000. Comparative proteome analysis of Helicobacter pylori. Mol. Microbiol. 36:710-725. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, D. J. 1998. The physiology and metabolism of the human gastric pathogen Helicobacter pylori. Adv. Microb. Physiol. 40:137-189. [DOI] [PubMed] [Google Scholar]
- 20.Keyer, K., and J. A. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 93:13635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Li, R., M. A. Bianchet, P. Talalay, and L. M. Amzel. 1995. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. USA 92:8846-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundström, A. M., and I. Bölin. 2000. A 26 kDa protein of Helicobacter pylori shows alkyl hydroperoxide reductase (AhpC) activity and the mono-cistronic transcription of the gene is affected by pH. Microb. Pathog. 29:257-266. [DOI] [PubMed] [Google Scholar]
- 24.Marais, A., G. L. Mendz, S. L. Hazell, and F. Mégraud. 1999. Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol. Mol. Biol. Rev. 63:642-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGee, D. J., and H. L. T. Mobley. 1999. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr. Top. Microbiol. Immunol. 241:155-180. [DOI] [PubMed] [Google Scholar]
- 26.Morrissey, J. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 2:307-310. [DOI] [PubMed] [Google Scholar]
- 27.Olson, J. W., J. N. Agar, M. K. Johnson, and R. J. Maier. 2000. Characterization of the NifU and NifS Fe-S cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry 39:16213-16219. [DOI] [PubMed] [Google Scholar]
- 28.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 29.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 31.Tonello, F., W. G. Dundon, B. Satin, M. Molinari, G. Tognon, G. Grandi, G. Del Giudice, R. Rappuoli, and C. Montecucco. 1999. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 34:238-246. [DOI] [PubMed] [Google Scholar]
- 32.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Identification of a new gene responsible for the oxygen tolerance in aerobic life of Streptococcus mutans. Biosci. Biotechnol. Biochem. 64:1106-1109. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 182:3740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]