Abstract
Ammonium transport (Amt) proteins appear to be bidirectional channels for NH3. The amt genes of the hyperthermophiles Aquifex aeolicus and Methanococcus jannaschii complement enteric amtB mutants for growth at 25 nM NH3 at 37°C. To our knowledge, Amt proteins are the first hyperthermophilic membrane transport proteins shown to be active in a mesophilic bacterium. Despite low expression levels, His-tagged Aquifex Amt could be purified by heating and nickel chelate affinity chromatography. It could be studied genetically in Escherichia coli.
Hyperthermophilic Amt proteins are active at mesophilic temperatures.
Previous studies of Amt proteins in enteric bacteria and their homologues in Saccharomyces cerevisiae provided evidence that they facilitate diffusion of the uncharged species NH3 across the cytoplasmic membrane (10-12). We amplified the amtB gene of the hyperthermophilic bacterium Aquifex aeolicus (GenBank accession number AAC06478; 423 residues) and the two genes (amt and amtB) of the hyperthermophilic archaeon Methanococcus jannaschii (GenBank accession numbers AAB98038 [MJ0058; 391 residues] and AAB99352 [MJ1343; 420 residues], respectively) by PCR and cloned them under the control of the tac promoter in plasmid pJES1242, which also codes for a C-terminal six-His tag. (pJES1242 was derived from pJES1130, which carries Escherichia coli amtB under the control of the tac promoter [10], and pJES1139, a derivative of pET21a [Novagen, Inc.], which codes for an E. coli AmtB-His fusion protein under the control of the T7 promoter. Cloning into pJES1242 entailed removal of the ribosome binding site of E. coli amtB.) All three genes from hyperthermophiles (amtB from A. aeolicus and amt and amtB from M. jannaschii, which we call amt1 and amt2 in conformity with nomenclature used for other organisms) complemented an E. coli amtB mutant for growth at 0.5 mM ammonium at pH 5 (25 nM NH3) at 37°C (Table 1), although the amt2 gene of M. jannaschii worked least well. (The E. coli amtB mutant has no growth defect at 5 mM ammonium at pH 5 or 0.5 mM ammonium at pH 7 [10].) Thus, the hyperthermophilic proteins not only are active but also are successfully inserted into the cytoplasmic membrane and folded at mesophilic temperatures. They are apparently tolerant of the lipid composition of enteric membranes.
TABLE 1.
The amt genes of A. aeolicus and M. jannaschii complement an E. coli amtB mutant for growth at low ammonium and [14C]methylammonium uptake
| Straina | Doubling timeb (min) at IPTG concn (μM)
|
MA uptakec (pmol/[ml × OD600 × min]) | ||
|---|---|---|---|---|
| 0 | 10 | 100d | ||
| NCM1458 (10) (wild type) | 80 | NAe | NA | 80 |
| NCM2019 (10) (amtB) | 600 | NA | NA | ≤3 |
| NCM3404 (amtB/pJES1335 [A. aeolicus AmtB-His6]) | 160 | 100 | 270 | 100 |
| NCM3421 (amtB glnA/pJES1335 [A. aeolicus AmtB-His6]) | NA | NA | NA | ≤3 |
| NCM3452 (amtB/pJES1348 [M. jannaschii Amt1-His6]) | 110 | 110 | No growth | 155 |
| NCM3453 (amtB/pJES1349 [M. jannaschii Amt2-His6]) | 320 | 250 | No growth | 13 |
| NCM3896 (amtB/pJES1452 [E. coli AmtB-His6])f | 300 | No growth | No growth | 200 |
Plasmids pJES1335, pJES1348, and pJES1349 were derived by subcloning from pJES1331, pJES1343, and pJES1344, respectively, in which the amt genes are under the control of a T7 promoter and ribosome binding site. The latter were used for overexpression. All plasmids yield C-terminally His-tagged proteins.
Cells were grown in N- and C-free medium at pH 5 with 0.2% glucose as the carbon source and 0.5 mM NH4Cl as the nitrogen source (10) and with IPTG as indicated.
Transport of [14C]methylammonium (MA). Cells were grown in N- and C-free medium at pH 7 with 0.4% glucose as the carbon source and 2 mM glutamine as the nitrogen source and with 10 μM IPTG. Growth on glutamine induces the synthesis of glutamine synthetase and of AmtB in the wild-type strain.
Slowing of growth at high IPTG concentrations is due to toxicity of the Amt proteins. It is also observed with the E. coli protein, whether or not it carries a His tag, under similar circumstances.
NA, not applicable.
pJES1452 carries the gene that codes for E. coli AmtB-His6 in the same vector used for the genes from hyperthermophiles. Plasmid pJES1452 is toxic even on enriched medium at pH 7, probably because the E. coli protein is highly expressed. The E. coli gene carries only five rare codons, whereas the genes from hyperthermophiles carry between 21 and 36 rare codons (mostly ATA for isoleucine). Vectors from which the E. coli AmtB-His6 protein is apparently less well expressed are less toxic. They complement fully for growth and less well for [14C]methylammonium uptake (data not shown).
As expected from their effect on growth, all three amt genes also complemented for accumulation of 14C label from the ammonium analogue [14C]methylammonium (Table 1) (10). In the strain carrying the Aquifex amt gene, which was the only one tested, the analogue was quantitatively converted to [14C]methylglutamine in the ATP-dependent reaction catalyzed by glutamine synthetase (not shown). This occurs in E. coli itself (10), and hence, apparent concentration of [14C]methylammonium by Amt proteins is actually due to an energy-dependent metabolic conversion to [14C]methylglutamine. Indeed, the Aquifex Amt protein (called A. aeolicus AmtB) failed to restore accumulation of 14C label in a mutant background (glnA amtB) in which glutamine synthetase was inactive (Table 1). Like E. coli AmtB, all three hyperthermophilic Amt proteins allowed accumulation of 14C label from [14C]methylammonium down to temperatures of 4°C (not shown), an observation that is easily rationalized if they are NH3 channels. Accumulation at lower temperatures also depended on residual activity of E. coli glutamine synthetase and peaked earlier and at lower levels than at 37°C.
A. aeolicus AmtB can be partially purified.
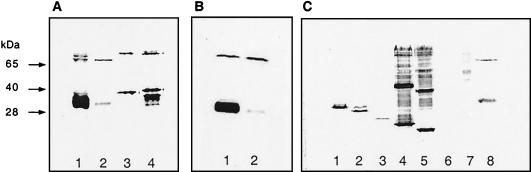
Hyperthermophilic amt genes were placed under the control of the T7 promoter and ribosome binding site in pET21a, which also codes for a C-terminal six-His tag. When induced cell extracts were run on sodium dodecyl sulfate (SDS)-polyacrylamide gels and immunoblotted, each hyperthermophilic protein gave at least two His-tagged bands, as was the case for the E. coli protein (Fig. 1A). The relative amounts of upper and lower bands varied with many aspects of handling, including freezing and dilution. Further study of the Aquifex protein provided preliminary evidence that the upper band was an oligomer (perhaps a dimer), whereas the lower band was a monomer. (Similar behavior was seen for the AqpZ protein of E. coli [1], a member of the aquaporin family.) First, we never saw bands of higher mobility than the lower band and the lower band was seen whether or not protease inhibitors were present during preparation of extracts. Second, doubly N- and C-terminally tagged A. aeolicus AmtB protein yielded lower and upper bands of the same mobility as C-terminally His-tagged protein (not shown). Third, some missense mutant forms of the protein (see below and Table 2) yielded far more lower than upper band, as if the functional oligomer was less stable (Fig. 1B). Finally, some N-terminal-deletion-containing proteins and C-terminally truncated proteins (see below) yielded only single bands of higher mobility than the lower band (Fig. 1C and data not shown). Again, these may have oligomerized less stably than the full-length protein. We have not ruled out other explanations for the two bands of A. aeolicus AmtB, including the possibility that lower bands are proteolytic cleavage fragments and upper bands are intact monomers.
FIG. 1.
Immunoblots of C-terminally His-tagged Amt proteins in cell extracts. All proteins were tagged with six histidine residues. Cell extracts were subjected to SDS-10% (A and B) or 12% (C) polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and blotted with a specific anti-His antibody (INDIA HisProbe-HRP; Pierce). Plasmids for panels A and C were derived from vector pET21a (Novagen). The host strain was BL21(DE3)(pLysE). Plasmids for the experiment in panel B were derived from pJES1335. The host strain was NCM2019 (10). Expression of Amt proteins was induced as described in the text for purification of A. aeolicus AmtB. (A) Lane 1, E. coli AmtB (pJES1139); lane 2, A. aeolicus AmtB (pJES1331); lane 3, M. jannaschii Amt1 (pJES1345); lane 4, M. jannaschii Amt2 (pJES1346). (B) Lane 1, A. aeolicus AmtBT253I, T259M, S404F (pJES1378); lane 2, A. aeolicus AmtB (pJES1335). (C) Lane 1, A. aeolicus AmtBΔ2-51 (pJES1432); lane 2, AmtBΔ2-87 (pJES1433): lane 3, AmtBΔ2-143 (pJES1434); lane 4, AmtBΔ2-170 (pJES1435); lane 5, AmtBΔ2-209 (pJES1436); lane 6, AmtBΔ398-423 (pJES1437); lane 7, molecular weight standards; lane 8, A. aeolicus AmtB (pJES1331).
TABLE 2.
Effects of mutations on the ability of the A. aeolicus amtB gene to complement for growth and [14C]methylammonium uptake and on the toxicity of its producta
| Plasmid | Genotype | Substitution or deletionb | Complementation
|
Toxicityc | |
|---|---|---|---|---|---|
| MA uptaked (pmol/[ml × OD600 × min]) | Doubling time (min)e | ||||
| pJES1335 | Wild type (423 aa) | 55 | 105 | Toxic | |
| pJES1377 | G314 → A | Gly105 → Asp | 40 | 120 | Toxic |
| pJES1376 | G388 → A | Ala130 → Thr | 45 | 145 | Toxic |
| pJES1378 | C758 → T | Thr253 → Ile | 55 | 100 | Toxic |
| C776 → T | Thr259 → Met | ||||
| C1211 → T | Ser404 → Phe | ||||
| pJES1379 | G1129 → A | Val377 → Ile | 55 | 120 | Toxic |
| pJES1389 | G96 → A | Trp32 → stopf | ≤3 | No growth | Not toxic |
| pJES1393 | G236 → A | Trp79 → stop | ≤3 | No growth | Not toxic |
| pJES1391 | C301 → T | Gln101 → stop | ≤3 | No growth | Not toxic |
| pJES1392 | G503 → A | Trp168 → stop | ≤3 | No growth | Weakly toxic |
| pJES1395 | G509 → A | Trp170 → stop | ≤3 | No growth | Toxic |
| pJES1394 | G791 → A | Trp264 → stop | ≤3 | No growth | Toxic |
| pJES1390 | G806 → A | Trp269 → stop | ≤3 | No growth | Toxic |
| pJES1388 | C1105 → T | Gln369 → stop | ≤3 | No growth | Toxic |
| pJES1426 | Δ2-51 (ΔTMf1) | ≤3 | No growth | Toxic | |
| pJES1427 | Δ2-87 (ΔTM1, 2) | ≤3 | No growth | Toxic | |
| pJES1428 | Δ2-143 (ΔTM1-3) | ≤3 | No growth | Toxic | |
| pJES1429 | Δ2-170 (ΔTM1-4) | ≤3 | No growth | Toxic | |
| pJES1430 | Δ2-209 (ΔTM1-5) | ≤3 | No growth | Toxic | |
| pJES1431 | Δ398-423 (ΔC terminus) | 9 | Slow growth | Toxic | |
The host strain was NCM2019 (amtB) and all plasmids were derived from pJES1335, which codes for C-terminally His-tagged A. aeolicus AmtB. All proteins carry two extra residues at the N terminus and eight at the C terminus. Positions of nucleotide bases and amino acid residues are for the native protein.
aa, amino acids. Transmembrane spanning regions predicted by the PHDhtm topology program (http://maple.bioc.columbia.edu/predictprotein/): TM1 (32-50), TM2 (64-85), TM3 (120-137), TM4 (142-165), TM5 (186-203), TM6 (219-237), TM7 (248-266), TM8 (291-315), TM9 (334-351), and TM10 (376-396).
Toxicity was assessed as described in the text.
Transport of [14C]methylammonium. Cells were grown as described in footnote c of Table 1.
Growth was assessed in liquid culture or on plates and was compared to that of NCM2019 (amtB) and NCM3404 (amtB/pJES1335). The medium was as described in footnote b of Table 1 with 10 μM IPTG.
The mutation Trp32 → stop results in reinitiation of translation at Met33 (see text).
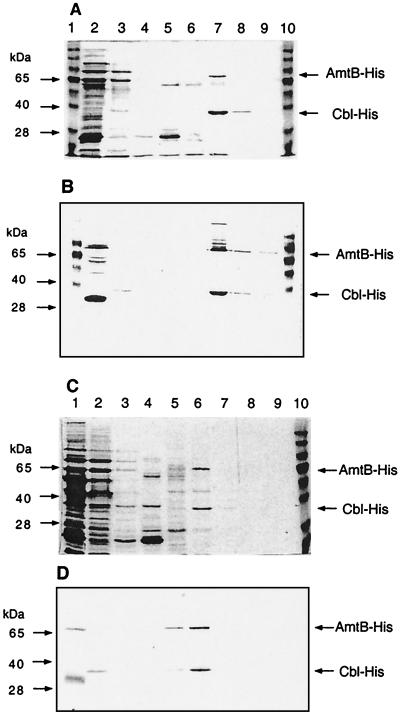
To purify A. aeolicus AmtB, a culture of strain NCM3404 (amtB/pJES1331) was grown in maximal induction medium (8) to an optical density at 600 nm (OD600) of 0.6 and induced with 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. Bacterial cells were harvested, concentrated 400-fold in breakage buffer (50 mM sodium phosphate, pH 8.0; 0.3 M NaCl; 5 mM β-mercaptoethanol; 0.1 mM phenylmethylsulfonyl fluoride; 1% n-octyl-β-d-glucopyranoside [octylglucoside]), and disrupted by being passed through a French pressure cell twice at 12,000 lb/in2. Adding octylglucoside before disruption of cells was critical. After centrifugation at 8,000 × g for 15 min at 4°C, Triton X-100 was added to the supernatant to 0.5%, and it was heated for 20 min at 70°C and centrifuged at 6,000 × g for 5 min at room temperature. All of the A. aeolicus AmtB protein remained in the heated supernatant (Fig. 2, lane 2), whereas 90% of the other protein in the extract was precipitated. (The Aquifex protein also remained in the supernatant after heating for 30 min at 85°C.) The results indicated that a membrane protein from a hyperthermophile was heat tolerant even in the absence of lipid from the organism in which it naturally occurred.
FIG. 2.
Metal affinity purification of A. aeolicus AmtB-His6 and mutant form W32 → stop (see the text). Samples taken during purification (see the text) were subjected to SDS-10% polyacrylamide gel electrophoresis. (A and C) Coomassie blue-stained gels; (B and D) Western blots with anti-His antibody (see the legend to Fig. 1). (A and B) Lane 1, molecular weight standards; lane 2, crude extract after heat treatment; lane 3, flowthrough of Ni-NTA affinity column; lane 4, first wash (in breakage buffer modified to contain 0.1 mM phenylmethylsulfonyl fluoride and 0.1% octylglucoside); lane 5, second wash (in breakage buffer adjusted to pH 6, modified as described above, and containing 10% glycerol); lane 6, eluate with 50 mM imidazole added to the second wash buffer; lane 7, eluate with 200 mM imidazole; lane 8, eluate with 500 mM imidazole; lane 9, eluate with 1,000 mM imidazole; lane 10, molecular weight standards. (C and D) Lane 1, crude extract after heat treatment; lane 2, flowthrough of Ni-NTA affinity column; lane 3, first wash (pH 8.0); lane 4, second wash (pH 6.0); lanes 5 to 8, eluates with 50, 200, 500, and 1,000 mM imidazole, respectively; lane 9, wash with 0.2 M acetic acid; lane 10, molecular weight standards.
After dialysis of the heated supernatant against a buffer that did not contain Triton X-100, a soluble His-tagged “carrier” protein, CysB-like (Cbl)-His6 (6, 14), was added to it (0.5 mg of carrier/10 mg of protein), and it was applied to a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (Qiagen). Both His-tagged proteins bound (Fig. 2A and B). (When Cbl-His6 was omitted, AmtB-His6 did not bind to the column [data not shown].) The column was washed essentially as recommended by the manufacturer (see the legend to Fig. 2), and His-tagged proteins were eluted in steps with increasing imidazole concentrations (50 to 1,000 μM at pH 6.0). Only the upper band of A. aeolicus AmtB-His6 was recovered, even after the column was stripped with acetic acid (data not shown and Fig. 2D; see below). We verified by mass spectrometry (2) that the upper band contained AmtB (a C-terminal hydrophilic tryptic peptide [VSEEEELELDSSLHGEK] was detected) and that the lower band contained Cbl (14 tryptic peptides were detected). When the eluate from the Ni-NTA affinity column was heated at 70°C for 20 min, A. aeolicus AmtB remained in the supernatant (Fig. 3, lane 2). Although some of the His-tagged Cbl carrier protein also remained in the supernatant, much of it precipitated and could be removed along with several prominent contaminants (Fig. 3, lane 3). About 50 μg of His-tagged A. aeolicus AmtB protein was obtained from 1.5 liters of culture. Hence, purification of A. aeolicus AmtB for biochemical and structural studies is promising.
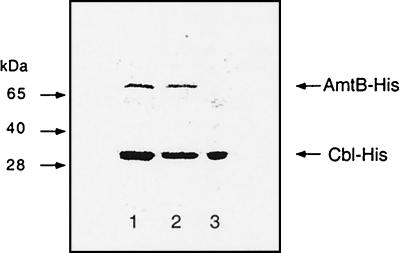
FIG. 3.
Heat inactivation of the Cbl-His6 carrier used for purification of A. aeolicus AmtB-His6. Samples were treated as described for Fig. 2, and the resulting SDS gel was stained with Coomassie blue. Lane 1, eluate from Ni-NTA affinity column with 200 mM imidazole (sample in lane 7 of Fig. 2A and B); lane 2, supernatant of sample in lane 1 after heat treatment at 70°C for 20 min; lane 3, pellet.
A. aeolicus AmtB can be studied genetically in E. coli.
A. aeolicus AmtB is active in E. coli and, when overexpressed, is toxic. (Toxicity is also observed with the E. coli protein.) To see whether it was feasible to explore determinants for activity and toxicity of A. aeolicus AmtB genetically, we mutagenized plasmid pJES1335 with 2 M hydroxylamine and 1 mM EDTA at 50°C for 2 h. Mutagenized plasmids were recovered from a silica gel column and transformed into E. coli amtB strain NCM2019. We screened 1,000 transformants for a growth defect specifically at 0.5 mM ammonium at pH 5 (using 10 μM IPTG, a low concentration, to induce synthesis of A. aeolicus AmtB). This yielded 12 transformants that appeared to have a growth defect and that also had at least one mutation in the Aquifex amtB gene. Mutant forms of the gene were recloned and plasmids were again introduced into NCM2019. After recloning, plasmids bearing all eight nonsense mutations failed to complement for growth on 0.5 mM ammonium at pH 5 and resulted in complete loss of [14C]methylammonium uptake activity (Table 2). By contrast, the occurrence of missense mutations (a total of three in one instance) had little effect on growth or uptake. In the case of at least one missense mutation (Val377 → Ile), we confirmed that an additional mutation in the vector contributed to poor growth at low ammonium in the initial screen. Though we have characterized only small numbers of mutant forms, these early results hint that it may be difficult to obtain inactive Amt proteins with single amino acid substitutions.
Toxicity of mutant A. aeolicus AmtB proteins was assessed by their effect on growth at pH 7 with 1 mM ammonium as the nitrogen source (Table 2), a condition under which the function of Amt as an NH3 channel is not required. A high concentration of IPTG (100 μM) was used for induction. Transformants were compared to NCM3404 (amtB/pJES1335), which produces the toxic intact A. aeolicus AmtB protein and therefore grows poorly, and to NCM2019 (amtB), which grows well. The four plasmids carrying missense mutations in amtB yielded proteins that remained toxic at high concentrations, whereas the four plasmids coding for the shortest nonsense fragments yielded nontoxic or weakly toxic proteins (Table 2). The remaining four plasmids carrying nonsense mutations yielded proteins that remained toxic despite their loss of activity. Thus, toxicity of Amt is apparently not a function of its activity as an NH3 channel.
Surprisingly, upon SDS electrophoresis and immunoblotting for the His tag, one of the C-terminally His-tagged nonsense variants, the W32 → stop variant, yielded bands essentially the same as those from intact AmtB (Fig. 2D). As expected, none of the other nonsense variants was detected in this way. The W32 → stop variant, which was not detected with an N-terminal His tag (see below), appears to have reinitiated at methionine 33. The PHDhtm topology program (Table 2) predicts that Met33 lies at or near the beginning of the first transmembrane spanning segment of the AmtB protein. Although the Δ1-32 mutant protein was not toxic, it was nevertheless found in the membrane fraction (160,000 × g pellet) rather than the soluble fraction or inclusion bodies (inferred to be in the 8,000 × g pellet). It could be purified by exactly the same procedure used for the full-length protein (Fig. 2C and D). Attempts to select suppressor mutations that restore the activity of Δ1-32 are under way.
To detect the other nonsense variants, all genes carrying nonsense mutations were subcloned into pET28a, which codes for both N- and C-terminal six-His tags. Properties of the resulting proteins are summarized in Table 2.
Finally, to determine whether N-terminally truncated Aquifex proteins that lacked more than the first 32 residues were active and/or nontoxic, we used PCR to delete residues from 2 through 51, 87, 143, 170, or 209. The resulting peptides are predicted by the PHDhtm topology program to lack transmembrane spanning segments 1, 1 and 2, 1 to 3, 1 to 4, and 1 to 5, respectively. Like the peptide beginning with Met33, all were inactive, but, unlike it, all were toxic. All peptides were readily detected by immunoblotting of crude cell extracts (Fig. 1C; also, see above). In addition to a lower band with higher mobility than that from intact AmtB, which is likely to be a monomer, the shortest two peptides yielded many additional bands of lower mobility, which may be aggregates. Deletion of the C terminus of AmtB from residue 398 to the end greatly decreased the activity of the protein and left it toxic. The peptide was not readily detected (Fig. 1C). At present there is no easy way to summarize the basis for toxicity of A. aeolicus AmtB.
Implications.
Upon first consideration, it seems odd that hyperthermophiles would require protein channels for NH3 gas. Perhaps the lipid compositions of their cytoplasmic membranes, which allow them to withstand high temperatures (reviewed in reference 9), also restrict passive movement of NH3. We have noted previously that the membranes of Saccharomyces cerevisiae may be more restrictive to passive diffusion of NH3 than their counterparts in enteric bacteria (12).
Despite the fact that the Amt proteins of A. aeolicus, M. jannaschii, and E. coli are each predicted to have different numbers of membrane-spanning segments (i.e., 10 to 12) (3, 7, 13, 15) and have very different C termini, their substrate specificities appear to be the same. There is no evidence that the Amt proteins of the autotrophs, which were identified by homology to those of other organisms, function in diffusion of carbon dioxide or methane rather than NH3. We have speculated that the Rhesus proteins, the only known homologues of Amt, are channels for CO2 (12). These proteins are notably absent in both the archaea and the bacteria (4, 5), which may be too small to need them.
Acknowledgments
We are grateful to Monika M. Hryniewicz for the generous gift of plasmid pMH243 (Cbl-His6) and to Hiroshi Nikaido for critical review of the manuscript.
This work was supported by National Institutes of Health grant GM37537 to D.F.H. and by National Science Foundation grant MCB 9874443 to S.K.
REFERENCES
- 1.Borgnia, M. J., D. Kozono, G. Calamita, P. C. Maloney, and P. Agre. 1999. Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J. Mol. Biol. 291:1169-1179. [DOI] [PubMed] [Google Scholar]
- 2.Hanss, B., E. Leal-Pinto, A. Teixeira, R. E. Christian, J. Shabanowitz, D. F. Hunt, and P. E. Klotman. 2002. Cytosolic malate dehydrogenase confers selectivity of the nucleic acid-conducting channel. Proc. Natl. Acad. Sci. USA 99:1707-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howitt, S. M., and M. K. Udvardi. 2000. Structure, function and regulation of ammonium transporters in plants. Biochim. Biophys. Acta 1465:152-170. [DOI] [PubMed] [Google Scholar]
- 4.Huang, C.-H., P. Z. Liu, and J. G. Cheng. 2000. Molecular biology and genetics of the Rh blood group system. Semin. Hematol. 37:150-165. [DOI] [PubMed] [Google Scholar]
- 5.Huang, C. H., and P. Z. Liu. 2001. New insights into the Rh superfamily of genes and proteins in erythroid cells and nonerythroid tissues. Blood Cells Mol. Dis. 27:90-101. [DOI] [PubMed] [Google Scholar]
- 6.Iwanicka-Nowicka, R., and M. M. Hryniewicz. 1995. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene 166:11-17. [DOI] [PubMed] [Google Scholar]
- 7.Marini, A. M., and B. André. 2000. In vivo N-glycosylation of the Mep2 high-affinity ammonium transporter of Saccharomyces cerevisiae reveals an extracytosolic N-terminus. Mol. Microbiol. 38:552-564. [DOI] [PubMed] [Google Scholar]
- 8.Popham, D., J. Keener, and S. Kustu. 1991. Purification of the alternative sigma factor, sigma 54, from Salmonella typhimurium and characterization of sigma 54-holoenzyme. J. Biol. Chem. 266:19510-19518. [PubMed] [Google Scholar]
- 9.Schouten, S., E. C. Hopmans, R. D. Pancost, and J. S. S. Damste. 2000. Widespread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc. Natl. Acad. Sci. USA 97:14421-14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soupene, E., L. He, D. Yan, and S. Kustu. 1998. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc. Natl. Acad. Sci. USA 95:7030-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soupene, E., H. Lee, and S. Kustu. 2002. Ammonium/methylammonium transport (Amt) proteins facilitate diffusion of NH3 bidirectionally. Proc. Natl. Acad. Sci. USA 99:3926-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soupene, E., R. M. Ramirez, and S. Kustu. 2001. Evidence that fungal MEP proteins mediate diffusion of the uncharged species NH3 across the cytoplasmic membrane. Mol. Cell. Biol. 21:5733-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas, G. H., J. G. L. Mullins, and M. Merrick. 2000. Membrane topology of the Mep/Amt family of ammonium transporters. Mol. Microbiol. 37:331-344. [DOI] [PubMed] [Google Scholar]
- 14.van der Ploeg, J. R., R. Iwanicka-Nowicka, M. A. Kertesz, T. Leisinger, and M. M. Hryniewicz. 1997. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J. Bacteriol. 179:7671-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Wiren, N., S. Gazzarrini, A. Gojon, and W. B. Frommer. 2000. The molecular physiology of ammonium uptake and retrieval. Curr. Opin. Plant Biol. 3:254-261. [PubMed] [Google Scholar]