Abstract
Sitosterolaemia (phytosterolaemia) is an autosomal recessive disorder characterised by the presence of tendon xanthomas in the face of normal or mildly elevated plasma cholesterol levels, premature atherosclerotic disease and has diagnostically elevated plasma and tissue plant sterol concentrations. Affected individuals show an increased absorption of both cholesterol and sitosterol from the diet, decreased bile clearance of these sterols and their metabolites resulting in markedly expanded whole body cholesterol and sitosterol pools. The defective gene is therefore hypothesised to play a crucial role in regulating dietary cholesterol absorption, and its elucidation may shed light on these molecular processes. We have previously localised the defective gene to human chromosome 2p21, between microsatellite markers D2S1788 and D2S1352, a distance of approximately 15 cM. Recently, the disease locus interval has been narrowed to lie between D2S2294 and D2S2291/D2S2174. We have constructed a high-resolution YAC and BAC contigs by using known STSs and generating novel STSs from the minimal interval. Eight previously identified genes and 60 ESTs were mapped to these contigs. The BAC contig contains 60 BAC clones and 108 STSs and encompasses a physical distance of approximately 2.0 cM between microsatellite markers D2S2294 and D2S2291. These results will not only facilitate cloning of the sitosterolaemia gene, but also other disease genes located in this region, and accelerate sequencing of the corresponding genomic clones.
Keywords: BAC contig, mapping, positional cloning, atherosclerosis genes
Introduction
Sitosterolaemia (also known as phytosterolaemia, MIM number 210250) is a rare autosomal recessively inherited metabolic disorder, which was described in 1974 in two affected sisters.1 Sitosterolaemic patients develop tendon and tuberous xanthomas, haemolytic episodes, arthralgias and arthritis, and premature coronary and aortic atherosclerosis leading to cardiac fatalities.1–5 Affected individuals have very high levels of plasma plant sterols (sitosterol, campesterol, stigmasterol, avenosterol) and their 5α-saturated stanols, particularly sitostanol, but their blood cholesterol levels may be normal or only moderately increased.1,4 Increased intestinal absorption and decreased hepatic excretion of sitosterol (the major plant sterol) may be responsible for the accumulation of these non-cholesterol sterols in plasma and tissues of affected patients.4,6–10
In addition to the proposed defects of absorption and excretion of sitosterol, reduced whole body cholesterol synthesis has also been noted.2,11,12
Linkage analyses of 10 well-characterised pedigrees localised the genetic defect to human chromosome 2p21, between microsatellite markers D2S1788 and D2S1352.13 Recently, we have narrowed this interval to lie between microsatellite markers D2S2294 and D2S2291 (Lee et al, manuscript submitted). To refine the minimal critical region and isolate candidate gene(s), we have constructed high-resolution YAC and BAC contigs by using known STSs and by generating novel STSs from this region. We have mapped a number of ESTs to this interval, building a partial transcript map that should aid identification of the defective gene. Additionally, this may facilitate the identification of other disease loci mapped to this region, such as a QTL for serum leptin levels,14 as well as a locus for gingival fibromatosis.15
Materials and methods
Selection and STS contents of YAC clones
YAC clones were identified through the YAC databases developed by CEPH16,17 and the Whitehead Institute18 using all of the known markers and STSs in sitosterolaemia region (D2S2291, D2S2174, D2S1830, D2S1485, D2S2298, D2S119, D2S2294, D2S414). The YAC clones were purchased from Research Genetics, Inc (Birmingham, AL, USA). Single YAC colonies were grown at 30°C for 48 h in 15 ml of selective YPD medium. Total YAC DNA was prepared as described previously.19 The STS contents of the YACs were determined by using PCR amplifications.
Inter-Alu PCR
Inter-Alu PCR was performed using YAC DNA as template and the following primers: CL1, (5′TCCCAAAGTGCTGGGATTACA), CL2 (5′CTGCACTCCAGCCCTGGG) and used as CL2 alone or CL1 and CL2 combined primers.20,21 The PCR products were isolated and cloned into plasmid, pBluescript (Stratagene, La Jolla CA, USA) using TA cloning, as previously described,22 and sequenced using T3 and T7 primers. The sequences were scanned against the databases, using BLAST23 (http://www.ncbi.nlm.nih.gov/BLAST/) and the RepeatMasker program (http://ftp.genome.washington.edu/RM/webRepeatMasker.html). Unique sequences were used to design primers for further mapping (Table 1). Confirmation of chromosome 2 specific sequences was verified by PCR, using chromosome 2 specific humanhamster hybrid somatic cell line DNAs (Corell Cell Repository, Camden, NJ, USA).
Table 1.
Primer sequences used in YAC/BAC contigs
| aSTS | Forward primer (5′–3′) | Reverse primer (5′–3′) | Size (bp) | bAccession no. |
|---|---|---|---|---|
| C-506D15.F | CCAGTGGCATTTAGTACATTA | AATGCCACTGAATCACACAC | 207 | AZ051294 |
| C-506D15.R | AAAAAGGCTGCCCACCTTTA | GTCAAGAGGTAGATGAAATGC | 206 | AZ051295 |
| C-498C3.F | TCTTGTTTTCCGATTCTGTTC | GTCAATTTCTACAGTGTAGCT | 203 | AZ051296 |
| C-528A6.F | CCCAGCACCAAATACAGTGAA | CAGTACATCTTCCGGCTCTA | 218 | AZ051297 |
| C-528A6.R | CCCTGATATTTACCCAGCTC | CAAGAAGAATGAGATCTGGC | 266 | AZ051298 |
| C-569J16.F | TACCTGAGCTTCCTAGGATG | CAGACAGCCTCAGTCGCTA | 306 | AZ051299 |
| R-990A23.F | AACGCATGGCTCTATAGAGG | AGGACAGCAGTGTCATTTAC | 412 | AQ702225 |
| R-990A23.R | CTGGATGGAACGCTCACTA | CAGAAGCCTCAGGGTAAGA | 268 | AQ668383 |
| R-1081G2.F | GATAAAGTTTAAGGTTATCTC | GAAATAAGCTTAGCCTGTAC | 275 | AQ740126 |
| R-316B10.F | CACAACTATGACCACTTTGAC | CGTGAGGTTCTCCTTTCCC | 299 | AQ541175 |
| R-316B10.R | ACAGCTAAGGATAATGAGGCA | GTGTTTATCCCCCAAGCACT | 288 | AQ507714 |
| R-32814.R | GGTGAGAATTCTAGCAAGCTA | CTATGGATAGAACTCTCAGTG | 256 | AQ539167 |
| R-959M3.R | ACAACACTGACAGTCTATCCG | CCAGATAGATAGTATGAGTTCA | 215 | AQ667844 |
| R-646H10.R | AGGATAATGAGGCATGTGAAG | TCGTGGGTTCACATAGCACA | 372 | AQ516454 |
| R-72C11.R | CAGTGGTGCTTGTAGCAGG | CCCATAGTGATCAAGCACTA | 260 | AQ285023 |
| R-203O10.R | CACGTGCATTGAAGGCTAATA | TTGATCTAGCTAAGCTAGTCC | 214 | AQ418643 |
| C-520L5.F | AGAGTTTCTGCTCTCTATGG | AGTGATTCTTTGATGGGCAG | 301 | AZ051300 |
| C-520L5.R | ATTCTCCTCTAGGCCTCTAG | TCAGCCTCTGCCTCTTGGAA | 274 | AZ051301 |
| R-92L13.F | GCCTAACAGCCAATCTGAG | TAACCCTACATGTGTTCCCA | 285 | AQ322533 |
| R-92L13.R | TATAGGGATCCAACAGTACC | GAGCTACATGATGGCCTTCA | 288 | AQ322528 |
| R-2415.R | TGGTACTCGCATCTCCTTG | GTCAAGGAGTCTTCTTGGG | 243 | AQ013398 |
| Y888G9.L | CTACCTAAAAGGCTTGGTTATC | CTCTGCAGAGTATTGCCACA | 207 | AZ051302 |
| R-161J6.R | AACTGAAGTGGTACTGACAG | AAGACGGCAGATGTATCCTG | 185 | AQ376733 |
| C-535K2.F | AGGAAAGTCAAGCTCCAGAG | TTAAAGAGAGCCTTCAGCTTC | 233 | AZ051303 |
| C-535K2.R | CTTGTGCCACTACTGCACTC | TTGTGCTGTCCATTCCTAGAA | 244 | AZ051304 |
| C-325O15.F | AGGACATTCTTACAGGCTACA | CAGCTAGTTATCTGAAGCTGA | 253 | AZ051305 |
| C-441A7.F | TATTTGCTCATTTAATGAGCCTA | CCTTACATAGTTCTCATCCTC | 224 | AZ051306 |
| C-441A7.R | TGATGGGGGAAAGCCACAAA | CTGTGGCCTCCCAAATTTC | 306 | AZ051307 |
| C-2094M11.R | GGAAACTGTGCAAGTGAAGA | TTAACAACAGGAGTCCCGCT | 177 | AQ566340 |
| C-2117A16.R | TAGCAAATCCTGTGCCATTC | TATAAAGGAAGGTCCTGACC | 318 | AQ753530 |
| R-117H6.F | TCTGTGGACTGACCTAGTAG | TGGGCGCAAGATTTCTGAG | 257 | AQ350077 |
| C-285H9.F | CCCACTACTGTGCAAACTTC | ATAAGAGCCATCCGGATTGC | 208 | AZ051308 |
| 42C1 | GAACAAGATCTGTAAGGGGT | TTCAGTAACATTGCATATTTTTCT | 138 | AZ051309 |
| 42B2 | ATGGAAGAGGGTTGGATGTTG | TGAGTGTCTGCCGGTGTA | 163 | AZ051310 |
| 42 E7 | TGAGACCTTTCTGCTTCTATCC | CACTGGAGAGTTGGCTGTG | 314 | AZ051314 |
| 42B7 | GCTGAGAATATCACTTTACTCC | GATTTCCAAGGTTACAATGTGTA | 151 | AZ051315 |
| 45A12 | ACTTGCTTGGTTTTGGTAAT | ACAGTCTCTTTGTGATCTT | 217 | AZ051316 |
| 42F7 | GAAAGTAGGCTAAGAGAGTTAAT | GTGAGCCACTGCACCCAG | 158 | AZ051317 |
| 87A10 | GGTTCTGTTTCATGTGTATGG | CAACTAGAATTGGACTAGATACTC | 221 | AZ051318 |
| 45D2 | CACTGCTGAATGTGAACTGC | CCCATGGTTTGACAAATGATTTC | 262 | AZ051311 |
| 42C5 | CACTTCATCATGTAGAACAGG | AGGATGATAGAGGGATTGGTTT | 269 | AZ051313 |
| 45B4 | ACTGCTGAATGTGAACTGC | TGCTACTATTGCAGCCCT | 196 | AZ051319 |
| 42D12 | CTTACACATTGTTATGAAGTGCAC | GTCTCAGAGAAAGATGTCACA | 215 | AZ051323 |
| 42C9 | GTGTAGCCTATTCAGAGAAC | AGTCAGTCTTCACGGCCA | 181 | AZ051326 |
| 45D11 | GAACGTGGAATAATATAAGACC | TATCTCACCACCCACACTG | 187 | AZ051327 |
| 45E 11 | GTCAGCTTTATGGATAGGG | GAAATACTCAGAATCCAGAAAC | 214 | AZ051328 |
| 45B2 | CATTCTGAGGGCCAGATTT | AGATGTAATACTTGCAAGCC | 219 | AZ051329 |
| 45B10 | ACCAGAAAATGACACCTTC | CATAGTATGAGTGCTACTTGACTC | 242 | AZ051330 |
| 42G8 | GGCAAACTTTGGCTCATGG | GTGCTAGAATCATCAGTTTGTCAT | 272 | AZ051331 |
| 87A7 | CAGCCCTCAGAGACAATAGA | TGCTGCCAAGCCATCCAA | 222 | AZ051332 |
| 87A5 | TGACAGGGTGAGAGTCCATC | GCCTTACACTGACTGACAGAT | 300 | AZ051333 |
| 87A3 | CCTCAGTGGAGCAGATTGC | AAATTTCCTAGGAAAGTTGGG | 257 | AZ051334 |
| 87A2 | CACATTATCTCTGAGTAGAG | CTATGCTTCTGAATGCCAG | 178 | AZ051335 |
| 41HM9 | CCCACCAGCAGTGTATGAG | GTTCCACATCACTGGTCATC | 153 | AZ051344 |
| 42C2/T7 | CAGACCATAGCATCCTCTTT | TCACACTTCACACAAGGTC | 234 | AZ051337 |
| 42F10 | CAAGACTGGTTGCCATATGG | CATCTCTTTCCTCCCCTC | 201 | AZ051339 |
| 42 E2 | CCAGATTTGACCAAAAGCCC | AGATGTAATACTTGCAAGCC | 209 | AZ051340 |
| 42D8 | CCTACATGTGTTCCCATTGCA | TTGCCTTGATGCCTCCCA | 175 | AZ051341 |
| 42D3 | AACCACTCTTAACTCCAGGG | GCAAGCCTTCTTAAATAGGCATA | 237 | AZ051342 |
| 42B1 | AGGTGGATGTCTACAATGGTC | GGTTTGCATATAGCCAGTCAC | 187 | AZ051343 |
| 42H11 | GCACTCCAGCCTGGGCAA | AGAGGTGAAGCTTACTGGAA | 183 | AZ051338 |
| 87A1 | GATTACAGGCATCAGCCAC | CCAGTCCTCCAAAAATGGTC | 175 | AZ051336 |
| 42A3 | AGGCAATCTGGGTTACTAGG | CGACTGAACATACAGACACT | 210 | AZ051312 |
| 45F3 | CAAGTACTGTTCTAAGGGCT | TATGATAGAGGTATGCACTGG | 168 | AZ051320 |
| 45D2 | TGGCCACTATCATTATTAGAAA | CTCTTCAGAGAGTTTGGACC | 255 | AZ051321 |
| 42B3 | AACAGTCAGCTTCTCAAAGG | ATGGAGACTTCTTTAGGAGG | 217 | AZ051325 |
| 45A8 | CATCTTCATCATCAAGCAGTG | AAGTACTGTGCCAAGGCCTG | 240 | AZ051322 |
| D2S4009 | GATCCAGTGTCATTATGCATAC | GCCAGTTGTTAATATTTTGCC | 219 | G64673 |
| D2S4010 | CAGCGGTAGTCTCTATGATA | TCAGAAGGTTCCTTATACAAGGC | 172 | G64671 |
| D2S4014 | TGCAGACTGTAATTGTGGGCT | GACTCCAGATGAGATCTATGACTG | 297 | G64669 |
| D2S4015 | CTCAAATCTCTGACTCCAGATC | GGCTATCCACTCAATAATTC | 297 | G64672 |
| D2S4016 | GATAAGCAAGCTGGTCACACTC | ATTTGAGCTTCAGAGGTCAA | 253 | G64670 |
| D2S4019 | ATGATCTGCATGAGGGTCAAGG | GAGTATATTTAGAAATTTCCATAA | 102 | G64675 |
| D2S4020 | TAGTCTTAATGTTTCCCTTGG | GAGACTAGTTTTCTGACTCAAG | 189 | G64676 |
| D2S4023 | GAGATTCTTTTTATTCTGATTTTTTGAG | ATGATCTGCATGAGGGTCAA | 127 | G64677 |
Table shows the primer sets for all the unique STS and microsatellite repeats identified in this study and not available in the public databases.
Prefix C is CITB-SHP-C BAC library; R is RPCI-11 BAC library; Y is CEPH YAC library; D2S is a microsatellite marker.
Prefixes AZ and G are from this study.
BAC clone screening
PCR-based library screening
The CITB-SHP-C Human BAC library,24 (Research Genetics, Inc., Huntsville, AL, USA) was screened by a PCR-based assay of DNA super-pools and plates according to the vendor recommended procedures. Positive clones were obtained from Research Genetics, Inc., plated on agar plates containing 12.5 μg/ml chloramphenicol and colonies screened by PCR for STS content verification.
Hybridisation-based library screening
High-density gridded filters of BAC libraries (RPCI-11) were obtained from Roswell Park Cancer Institute (Dr. Peter de Jong's laboratory, Buffalo NY, USA), and screened with radioactive probes from the IMAGE cDNA clones of ESTs mapped to the YAC contig. Positive clones were obtained from the Roswell Park Cancer Institute.
Selection from database
All known STS, EST and Alu PCR sequences were checked by a Basic BLAST against the Alu database (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) and masked by RepeatMasker23 and unique sequences thus identified were used as probes. Sequenced BACs in the public databases were identified by a BLAST 2.0 alignment search of the HTGS database25,26 (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) and the complete BAC sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/query_form.html). BACs with known end-sequence information were determined by searching the BAC End Sequence Database at TIGR (http://www.tigr.org/tdb/hum-gen/). The overlapped BACs or BAC contigs were obtained by searching the Washington University Human mapping database (http://genome.wustl.edu/gsc/cgibin/fpchuman.-single.pl) for likely matches to specified clones.
Sequencing of YAC and BAC ends
Isolation of YAC ends was performed using a modified vectorette method, using primers as previously described.27 YAC DNA (0.1 μg) was digested with 10 units of RsaI and AluI in 30 μl reaction buffer. Five microlitres of digested YAC DNA was ligated to vectorette adapters using 10 units of T4 DNA ligase in a total volume of 50 μl by incubation overnight at room temperature. The YAC end fragments were purified by Qiagen PCR column kit and directly sequenced using the left or right internal primers.
To obtain BAC end-sequences, BAC plasmid DNA was prepared using alkaline lysis procedure and tip-500 columns (Qiagen).28 The quality and quantity of DNA samples were tested by HindIII digestion pattern on agarose gels, as well as by the presence of expected STS markers. Direct BAC end sequencing was performed using an automated ABI 373 DNA sequencer. Three micrograms of BAC DNA and 50 pmoles of primer were used in a total volume of 40 μl. The following primers were used: T7 (5′-TAATACGACTCACTATAGGG-3′) and SP6 (5′ATTTAGGTGACACTATAG-3′). PCR reactions were carried out under the following cycle conditions: initial denaturation at 96°C for 4 min; 100 cycles of 96°C for 10 s, 50°C for 10 s, 60°C for 4 min. The end sequences of some BAC clones were obtained by searching BAC End Sequence Database at TIGR.
Transcript map
To identify candidate genes, known genes and ESTs previously mapped to the region between D2S177 and D2S337 were selected from the Human Transcript Map.29 The selected ESTs and genes were tested by PCR amplification against our YAC and BAC contigs and positive clones further characterised, as described above.
Results
Construction of a YAC contig
The initial goal was to construct an extensive YAC contig spanning the sitosterolaemia candidate region on chromosome 2, between markers D2S2174 and D2S2294. Based on the publicly available contig maps (contig WC2.4) from Whitehead Institute/MIT Center for Genome Research (WI/MIT) (http://carbon.wi.mit.edu) and the CEPH-Généthon (CEPH) (http://www.cephb.fr/infoclone.html), 30 YAC clones were identified, using following microsatellite and STSs markers (D2S414, D2S2294, D2S119, D2S2298, D2S1484, D2S1486, D2S1485, D2S1830, D2S2174 and D2S229). Additionally, information from a published partial YAC contig was also available.30 All YAC clones were screened for the markers and confirmed by testing three colonies of each clone for their STS contents (Figure 1). Two gaps were identified at the telomeric ends, and no further YACs were identified, despite additional library screening. However, we subsequently identified a BAC, R-489G24, positive for markers D2S2259 and an EST T71978, that linked YACs 888g9 and 899b1, thus closing one gap (Figure 1). Since the sitosterolaemia locus is located towards the centromeric end, no further attempts were made to close the more distal gap. Using the YAC contig, new markers were generated, employing a combination of YAC end-sequence analyses and inter-Alu PCR. To confirm that the identified STSs were from chromosome 2, all of these markers were screened for their presence in human chromosome 2-specific somatic cell hybrid cell-lines. Sixty ESTs from the databases (Unigene and GeneMap98, see Materials and methods) were screened by PCR against the YAC contig. Six of these were positive for the YAC contig (Figure 1), of which five mapped to within the region of interest. The sixth EST (T71978) was found to be positive on a linking BAC (see above). By performing inter-Alu PCR using YACs 919c10, 930a1 and 761e1 as templates, we generated 35 additional unique sequences from Alu PCR clones for obtaining STS markers and eight microsatellite repeat markers that allowed for further fine-mapping of the sitosterolaemia locus (Lee et al, manuscript submitted). A total of 76 STSs, including 17 new STSs generated from YAC insert-end sequences and inter-Alu PCR products and nine EST markers, were used to order the clones (Table 1 and Figure 1) and span an approximate distance of 5 cM. This physical map provided a resource for the construction of a BAC contig.
Figure 1.
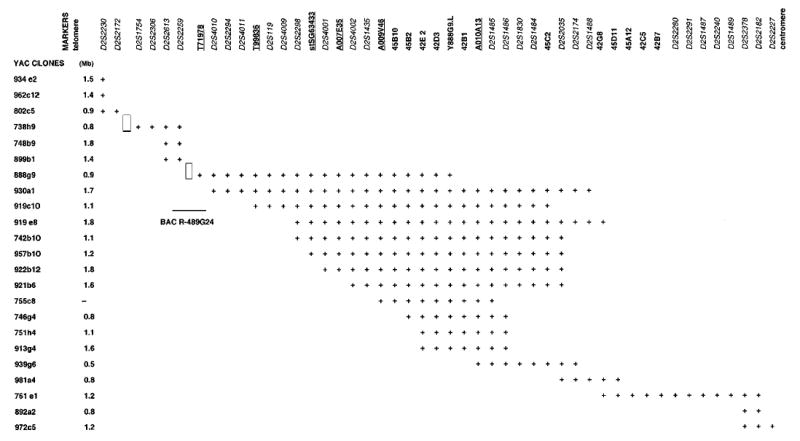
YAC clone contig encompassing the sitosterolaemia locus. The markers are oriented along the X-axis from telomeric end at the left to the centromeric end at the right, and the YAC clones are indicated along the Y-axis. There are two gaps in the contig (boxed areas, see Text). The distal centromeric gap is closed by a linking BAC, R-489G24, giving a contiguous contig from D2S1754 to D2S2227. ESTs mapped to the YAC contig are underlined, microsatellite markers are italicised and STSs are in normal font.
Note also that our YAC contig is a telomeric extension of a published adjoining YAC contig, thus providing a continuous map that spans chromosome 2p15–2p21 (D2S1364–D2S1754, ~14 Mb) and contains several human disease loci.31
Construction of a BAC contig
To construct a BAC map, we used the following strategy; (1) identify BAC clones using both a PCR-based and filter hybridisation-based BAC library screenings, (2) screen all positive BACs for STS content by PCR, (3) search BAC-related databases for updated information, and (4) perform chromosome-walks using selected STSs generated from BAC ends. Initially, PCR was used to screen the CITB-978SK-B human BAC library using six repeat polymorphic markers (D2S2294, D2S119, D2S2298, D2S1830, D2S2174, and D2S2291), a YAC end-sequence (from 888g9L) and an EST marker (T71978). Twelve BAC clones, positive for D2S2294, D2S119, D2S2291, 888g9L and T71978, were identified. For hybridisation-based BAC library screening, high-density filters were hybridised with a mixture of five probes consisting of ESTs T99836, T71978, A007E35, stSG63433, A010A13, previously mapped to the YAC contig. Eight more positive BAC clones were obtained from RPCI-11 BAC library. The BAC end sequences of identified clones were determined by direct automated sequencing or by searching the BAC end sequence database at TIGR. BAC end sequences of the inserts of BAC clones were used to develop further STS markers. All STS markers were tested by PCR amplification against all identified BAC clones, to verify true positives. By searching the databases in an iterative manner, we identified 18 sequenced BACs. In total, we used 118 markers, composed of 29 microsatellite markers, 53 new STSs from BAC/YAC end sequences and inter-Alu PCR sequences, and 36 EST markers. The constructed BAC contig contains 60 BAC clones, which contains a high density of STS markers, at an average of about 20 kb for each marker, and covers a physical distance of about 2.0 Mb (Figure 2). A significant number of these BACs have been sequenced (boxed, Figure 2), but about 500 kb sequence is not publicly available.
Figure 2.

BAC clone contig encompassing the sitosterolaemia locus. The markers are oriented along the X-axis from telomeric end at the left to the centromeric end at the right, and the BAC clones are indicated along the Y-axis. Prefixes of the BAC clones are as follows, R; from RPCI-11 BAC library; and C; from CITB-SHP-C BAC library. ESTs are underlined, microsatellite markers are shown in italics and STSs are in normal font. A box indicates BACs, for which almost complete sequence information is available in the genome databases. Four gaps (indicates by vertical lines) were identified, but these BACs are indicated, as they contain markers placed on the YAC contig framework (Figure 1). Of these the more centromeric is spanned by YAC 888g9. BAC R-436K12 (not shown) is linked to the contig published by Kirchener et al,31 and links our YAC contig at the centromeric end.
Mapping of known genes and ESTs to the YAC/BAC contigs
We have constructed a transcript map (Table 2) of the BAC contig using two methods. From GeneMap'99, based upon two radiation hybrid panels,32,33 we selected 80 genes and ESTs between anchor markers D2S177 and D2S2291. All ESTs were verified by PCR against the BAC contig. Of the 80 markers, only eight known genes and 30 ESTs mapped unambiguously to our BAC contig. The eight known genes are KIAA0544 protein,34 ERF2 protein,35 3-hydroxyanthranilic acid dioxygenase,36 CGI 60 protein,37 leucinerich protein,38 protein phosphatase 1B (formerly PP2C),39 Na+-independent neutral and basic amino acid transporter (solute carrier family 3, SLC3A1),40 and KIAA0436.41 In the second approach, using the known human genomic sequences from STSs and sequenced BACs between D2S2294 and D2S2291, we identified a further 30 ESTs by a BLASTN search of the EST databases. All of these 30 identified ESTs contain unique sequences, >95% matched to genomic sequences, and have not been previously mapped to a chromosome. A summary of the mapped ESTs to our BAC contig is shown in Table 2. We computed the expression patterns for many of these ESTs (Table 3). Additionally, we screened each of the mapped ESTs against the databases, looking for homologous ESTs/genes identified in other species, on the assumption that highly conserved expressed sequences may reflect proteins that have highly conserved and critical functions, such as selective sterol absorption. Only sequences that had >100 bp of sequence identity and >70% homology are reported (Table 3). Although such analysis is limited by the lack of depth of the EST databases for the other species, we identified 11 ESTs that appear to have homologues in non-human sequence databases (Table 3), although none from the Drosophila database were identified.
Table 2.
ESTs and genes mapped to the YAC/BAC contigs in this study
| NCBI No (GeneMap '99) | Aliases or synonyms | UniGene No. | Genbank Accession No. | Image Clone ID | Known genes | Mapping data |
|---|---|---|---|---|---|---|
| WI-20996 | stSG41980, T48876 | Hs.19280 | R26389 | 132199 | KIAA0544 protein | R-501O7 |
| SGC33875 | T98917 | 122669 | R-78I14 | |||
| stSG48396 | Hs.98023 | AA854974 | 1394041 | R-459K11 | ||
| SGC34340 | WI-6575, SGC34340 | Hs.78909 | X78992 | ERF-2 protein | R-339H12 | |
| STSG16054 | A009V10 | Hs.17711 | R98822 | 207006 | R-339H12 | |
| WI-14187 | G21943 | Hs.16063 | AA515534 | 925219 | R-339H12 | |
| stSG52431 | Hs.165571 | AI566776 | 2168475 | R-489G24 | ||
| T89476 | sts-T89476 | Hs.16587 | AA934036 | 1551421 | R-489G24 | |
| stSG15818 | R83265 | 194194 | R-489G24 | |||
| WI-8407 | Hs.108441 | Z29481 | 3-Hydroxyanthranilic acid dioxygenase | R-489G24 | ||
| BCD1971 | M79071 | no image clone | R-489G24 | |||
| T71978 | sts-T71978 | Hs.168439 | AA534545 | 925906 | R-489G24 | |
| stSG58568 | Hs.58598 | AI359618 | 2013757 | R-489G24 | ||
| stSG30561 | Hs.58598 | AA169121 | 594556 | 506D15 | ||
| H96893 | stSG21270 | Hs.32241 | AI274775 | 1986682 | R-489K21 | |
| stSG21136 | H58682 | 205857 | R-489K21 | |||
| AF151818 | CGI 60 protein | R-489K21 | ||||
| T99836 | Hs.18176 | T99836 | 123200 | R-489K21 | ||
| WI16988 | A007E35 | Hs.142718 | AA034046 | 429916 | R-1081G2 | |
| stSG63433 | Hs.190354 | AA700586 | 433330 | R-1081G2 | ||
| stSG32054 | stSG1757, SHGC-8019 T17102 | Hs.182490 | M92439 | Leucine-rich protein mRNA | R-1081G2 | |
| Hs.128293 | AI223013 | 1838809 | R-559M23 | |||
| Hs.225721 | AI873444 | 2362159 | Trans-prenyltransferase (TPT) | R-559M23 | ||
| Hs.225721 | AA889371 | 1471263 | R-559M23 | |||
| AA457390 | 838194 | R-559M23 | ||||
| AA828868 | 1374287 | R-559M23 | ||||
| Hs.187945 | AA937699 | 1491139 | R-559M23 | |||
| A004I37/H99661 | stSG51096 | Hs.169652/Hs.5687 | AA164383/AA565932 | PP2C | Protein phosphatase 2C | R-24I5 |
| stSG3387 | A003R48 | R11895 | 25315 | R-24I5 | ||
| stSG52154 | Hs.112916 | AA620873 | 1049335 | R-24I5 | ||
| M95548* | SHGC-9884, stSG4626 | Hs.198294/Hs.154834 | D82326/M95548 | Amino acid transporter, SLC3A1 | R-24I5 | |
| M95548* | SHGC-9884, stSG4626 | Hs.110 | AB007896 | KIAA0436 mRNA | R-24I5 | |
| A009V46 | Hs.174862, Hs.220859 | H95593 | 242930 | R-194L1 | ||
| A010A13 | WI-18144 | Hs.124990 | H58934 | 207758 | R-194L1 | |
| D29089 | D29089 | no image clone | R-194L1 | |||
| stSG8383 | H60063 | 205767 | R-194L1 | |||
| Hs.132799 | AA922097 | 1543611 | R-194L1 | |||
| Hs.129473 | AA994134 | 1628550 | R-194L1 | |||
| Hs.213492 | AI928677 | 2466254 | R-194L1 | |||
| Hs.124990 | H60592 | 207898 | R-194L1 | |||
| W80452 | 415494 | R-194L1 | ||||
| stSG26329 | H57813 | 205424 | R289E6 | |||
| Hs.136519 | AA601487 | 1100969 | R-442O5 | |||
| T87425 | 115418 | R-442O5 | ||||
| WI-3495 | G02557 | Hs.188588 | AA583683 | 1088083 | R-442O5 | |
| AA835723 | 1372934 | R-442O5 | ||||
| H64341 | 210718 | R-442O5 | ||||
| AA838139 | 1385549 | R-442O5 | ||||
| Hs.170428 | AI459058 | 2149952 | R-442O5 | |||
| Hs.170428 | AW206717 | 2722480 | R-442O5 | |||
| Hs.233172 | AW022706 | 2486137 | R-442O5 | |||
| stSG46410 | Hs.97696 | AA399659 | 729207 | R-89K21 | ||
| N24094 | N24095 | 266792 | R-576F1 | |||
| WI-3976 | SHGC-17237 | Hs.246042 | N75945 | 295200 | R-576F1 | |
| stSG49702 | Hs.167640 | H87795 | 220658 | R-436K12 | ||
| WI-18791 | U03911, SHGC-2762, SHGC-10660 | Hs.78934 | HSU03911 | (hMSH2) | Mismatch repair protein (MSH2) mRNA | R-436K12 |
| stSG60189 | Hs.122384 | AI015254 | 1641212 | R-436K12 | ||
| embl-AA007353 | sts-AA007353 | Hs.256042 | AA007353 | 429281 | R-436K12 | |
| SGC34683 | SHGC-34683, stSG28638, stSG9035 | Hs.117085 | AA677756 | 430606 | R-436K12 |
All ESTs and genes that were mapped to the YAC and BAC contigs (Figures 1 and 2) are shown. For clarity, only the BAC ID is shown in the far right column. BAC R-436K12 is not indicated on the BAC contig (Figure 2), but is contiguous with the centromeric end. Only a representative EST or Image clone is indicated, where multiple clones were identified. The asterisk indicates a GeneMap ID, M95548, which identifies two separate genes that share the 3′ UTR (see text). Additionally, there are two GeneMap98 IDs for the same gene (PP2C) that have been consolidated.
Table 3.
Expression pattern of ESTs and genes
| NCBI No (GeneMap'99) | GenBank Accession No. | Expression pattern | Known gene | Human | Mouse | Rat | Bovine | Porcine | Zebrafish | Chicken |
|---|---|---|---|---|---|---|---|---|---|---|
| WI-20996/KIAA0544 | R26389 | Multiple tissues | KIAA0544 protein | 57 | 4 | 0 | 0 | 1 | 0 | 0 |
| SGC33875 | T98917 | Fetal liver, spleen | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG48396 | AA854974 | Testis | 12 | 0 | 0 | 0 | 0 | 0 | 0 | |
| SGC34340 | X78992 | Multiple tissues | ERF-2 protein | >75 | 16 | 15 | 6 | 0 | 1 | 0 |
| STSG16054 | R98822 | Fetal liver, spleen | 5 | 0 | 0 | 0 | 0 | 0 | 0 | |
| WI-14187 | AA515534 | Multiple tissues | 30 | 0 | 1 | 0 | 0 | 0 | 0 | |
| stSG52431 | AI566776 | Brain, eye, heart, pancreas, uterus, thymus | 18 | 0 | 0 | 0 | 0 | 0 | 0 | |
| embl-T89476 | AA934036 | Bone, germ cell, prostate | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG15818 | R83265 | Fetal liver, spleen | 40 | 40 | 7 | 1 | 0 | 0 | 0 | |
| WI-8407 | Z29481 | Colon, kidney, lung, placenta, spleen, uterus | 3-hydroxyanthranilic acid dioxygenase | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| BCD1971 | M79071 | Brain | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| embl-T71978 | AA534545 | Colon, kidney, liver, lung | 22 | 23 | 1 | 0 | 1 | 0 | 0 | |
| stSG58568 | AI359618 | Multiple tissues | 5 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG30561 | AA169121 | Multiple tissues | 6 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H96893 | AI274775 | Multiple tissues | 32 | 0 | 1 | 0 | 0 | 0 | 0 | |
| stSG21136 | H58682 | Fetal liver, spleen | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| AF151818 | Multiple tissues | CGI 60 protein | 72 | 8 | 6 | 0 | 0 | 0 | 0 | |
| T99836 | T99836 | Fetal liver, spleen | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| WI16988 | AA034046 | Fetal liver, spleen | 6 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG63433 | AA700586 | Fetal liver, spleen | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG32054 | M92439 | Multiple tissues | Leucine-rich protein mRNA | >100 | 5 | 0 | 0 | 0 | 0 | 0 |
| AI223013 | Testis | 4 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| AI873444 | Ovary | Trans-prenyltransferase (TPT) | 3 | 2 | 0 | 0 | 0 | 1 | 0 | |
| AA889371 | Ovary | 3 | 2 | 0 | 0 | 0 | 1 | 0 | ||
| AA457390 | Retina | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AA828868 | Ovary | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AA937699 | Skin | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| A004I37/H99661 | AA164383/AA565932 | Multiple tissues | Protein phosphatase 2C | 48 | 26 | 8 | 0 | 0 | 0 | 0 |
| stSG3387 | R11895 | Brain | 4 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG52154 | AA620873 | Testis | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| M95548* | D82326/M95548 | Brain, kidney, pancreas, uterus, colon | Amino acid transporter, SLC3A1 | 36 | 38 | 2 | 1 | 1 | 0 | 0 |
| M95548* | AB007896 | Multiple tissues | KIAA0436 mRNA | 100 | 16 | 3 | 0 | 0 | 0 | 0 |
| A009V46 | H95593 | Fetal liver, spleen | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A010A13 | H58934 | Fetal liver, spleen | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| D29089 | D29089 | Epidermis, keratinocyte | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG8383 | H60063 | Fetal liver, spleen | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| AA922097 | Testis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AA922097 | Testis | 3 | 5 | 0 | 1 | 0 | 0 | 0 | ||
| AA994134 | Tonsil | 5 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AI928677 | Brain | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| H60592 | Fetal liver, spleen | 3 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| W80452 | Fetal liver, spleen | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| stSG26329 | H57813 | Fetal liver, spleen | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| AA601487 | Adrenal gland | 1 | 2 | 0 | 0 | 0 | 0 | 0 | ||
| T87425 | Fetal liver, spleen | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| WI-3495 | AA583683 | Kidney, nose | 5 | 0 | 0 | 0 | 0 | 0 | 0 | |
| AA835723 | Germinal center B cell | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| H64341 | Fetal liver, spleen | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AA838139 | Ovary | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AI459058 | Lung | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AW206717 | Lung | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AW022706 | Ear | 1 | 3 | 0 | 0 | 0 | 0 | 0 | ||
| stSG46410 | AA399659 | Testis | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| N24095 | N24095 | Melanocyte | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| WI-3976 | N75945 | Whole blood | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stSG49702 | H87795 | Retina, colon | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| WI-18791 | HSU03911 | Multiple tissues | Mismatch repair protein (MSH2) | 63 | 13 | 1 | 1 | 0 | 0 | 3 |
| stSG60189 | AI015254 | Testis | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| AA007353 | AA007353 | Lung | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| SGC34683 | AA677756 | Fetal liver, spleen, neuroepithelium | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
Expression profiles were determined for the ESTs and genes, based upon the identification of the EST or gene transcript in various cDNA libraries. Thus this profile is a minimal expression pattern. Additionally, homologues for the ESTs and genes were searched for (see Materials and methods) and the number of ESTs thus identified are indicated in the columns on the right. No homologues (based upon parameters specified in the text) were found in the C. elegans or D. melanogaster databases.
Discussion
Positional cloning techniques, combined with computer-assisted data analyses of the sequence rich databases generated by human genome projects,42,43 has considerably facilitated the identification of disease genes. The availability of complete and detailed clone contigs of candidate regions make for efficient positional cloning projects. We first constructed a YAC contig of this region and used it as a resource for the construction of a deep BAC contig. At the centromeric end of our YAC contig, there is a YAC, 972c5, which contains markers D2S2182 and D2S2227, which are also located in a published adjoining YAC contig.31 Thus combined with this published YAC contig, this provides a continuous map that spans chromosome 2p15-2p21 (D2S1364–D2S1754, ~14 Mb) and contains several human disease loci.31
Sixty-seven new STSs were identified by inter-Alu PCR and YAC/BAC end sequencing. The high-resolution physical map generated in this study spans ~2 Mb with complete coverage of the minimal region of sitosterolemia. The data presented here have been parsed for multiple ESTs for single genes represented in the databases and we have attempted to summarise data that are found scattered in a number of different databases, increasing the utility of this information. A summary of the results is provided in Figure 3. Based upon the radiation hybrid mapping databases, our initial YAC contig spans approximately 10 cM. However, this area appears to span only 5 Mb in physical length, suggesting a lower than expected recombination frequency (Figure 3). Assuming that all the non-redundant ESTs mapped to the BAC contig are unique transcripts and taking into account the small number of genes known to map into the BAC contig, we estimate that the gene density is approximately 1 gene per 50 kb of genomic DNA (Figure 3, 40 ESTs and genes mapped with the 2 Mb area).
Figure 3.
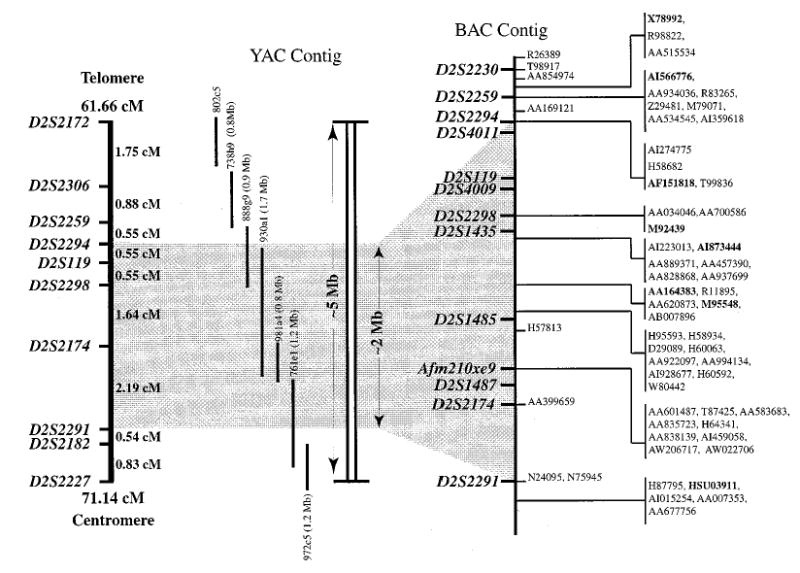
Summary of the YAC, BAC and EST mapping data. The figure shows a summary of the data presented in this study, indicating the genetic distance, physical distance and Genbank Accession numbers for mapped ESTs and genes located in the region of interest. Note that the genetic distance, based upon publicly available databases, spans ~10 cM, but spans ~5 Mb. Although only the Genbank IDs are shown, all identified EST can be obtained by utilising the Unigene or the GeneMap'99 identifiers shown in Table 2. Accession numbers in bold represent known genes, the remaining represent putative ESTs. The exact order of the ESTs at any given map location can not be determined at present and are thus grouped, indicated by the vertical lines.
One of our findings is the mis-assignment of BAC R-35M22. This BAC was previously assigned to chromosome 4 (Genbank accession number AC016338, Birren et al, direct submission), but is positive to DNA sequences from BACs R-24I5 and R-194L1. Additionally, it also contains ESTs A004I37, H99661, stSG3387, stSG52154, M95548, and M95548, 9 of 14 exons of KIAA0436 protein and exon 2 of Na+-independent neutral and basic amino acid transporter, thus placing it firmly on chromosome 2, in the interval D2S119-D2S2291.
Our integrated BAC contig allows for more accurate placement of genes and ESTs than the corresponding region in Genemap'99. In the D2S119-D2S2291 interval from GeneMap'99, 43 ESTs listed, 39 of which are unique. However, only nine of these 39 ESTs actually map to the D2S119-D2S2291 interval into our BAC contig, 30 of 39 map outside of this region. Of the 40 ESTs we have physically mapped to the D2S119-D2S2291 interval of our BAC contig, 31 of these were previously assigned to lie outside of this region. Therefore, the accuracy of GeneMap'99 for the D2S119-D2S2291 interval is only 25%, which is similar to the 30% reported by Kirschener et al for the D2S123-D2S2251 interval, but much lower than 75% in the D2S2291-D2S123 interval reported by the same authors.31
In summary, we have developed 67 new STSs, constructed an integrated YAC and BAC contigs for sitosterolaemia region and mapped eight known genes and 48 ESTs to the contig. These results will facilitate the identification of the sitosterolaemia gene and other disease genes located in this region. Additionally, this information may be useful in ordering some of the sequenced BAC contigs and accelerate sequencing of the corresponding genomic clones.
Acknowledgments
We are grateful to Dr Anand Srivastava for expert advice and critical review of our work, to Starr Hazard and the BioMolecular Computing Resource for assistance with the software. This work was funded by a Scientist Development Award from the American Heart Association grant 9730087N (SB Patel) and by the National Institutes of Health, NHLBI Grant HL60616 (SB Patel) and MO1 RR01070-25 (MUSC GCRC), and by an intramural award from the University Research Committee, Medical University of South Carolina (SB Patel).
References
- 1.Bhattacharyya AK, Connor WE. Beta-sitosterolemia and xanthomatosis. A newly described lipid storage disease in two sisters. J Clin Invest. 1974;53:1033–1043. doi: 10.1172/JCI107640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miettinen TA. Phytosterolaemia, xanthomatosis and premature atherosclerotic arterial disease: a case with high plant sterol absorption, impaired sterol elimination and low cholesterol synthesis. Eur J Clin Invest. 1980;10:27–35. doi: 10.1111/j.1365-2362.1980.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 3.Kwiterovich P, Jr, Bachorik PS, Smith HH, et al. Hyperapobeta-lipoproteinaemia in two families with xanthomas and phytosterolaemia. Lancet. 1981;1:466–469. doi: 10.1016/s0140-6736(81)91850-x. [DOI] [PubMed] [Google Scholar]
- 4.Salen G, Shefer S, Nguyen L, et al. Sitosterolemia. [Review] J Lipid Res. 1992;33:945–955. [PubMed] [Google Scholar]
- 5.Shefer S, Salen G, Bullock J, et al. The effect of increased hepatic sitosterol on the regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase and cholesterol 7 alpha-hydroxylase in the rat and sitosterolemic homozygotes. Hepatology. 1994;20:213–219. doi: 10.1016/0270-9139(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 6.Salen G, Ahrens E, Jr, Grindy SM. Metabolism of beta-sitosterol in man. J Clin Invest. 1970;49:952–967. doi: 10.1172/JCI106315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregg RE, Connor WE, Lin DS, Brewer H., Jr Abnormal metabolism of shellfish sterols in a patient with sitosterolemia and xanthomatosis. J Clin Invest. 1986;77:1864–1872. doi: 10.1172/JCI112513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salen G, Shore V, Tint GS, et al. Increased sitosterol absorption, decreased removal, and expanded body pools compensate for reduced cholesterol synthesis in sitosterolemia with xanthomatosis. J Lipid Res. 1989;30:1319–1330. [PubMed] [Google Scholar]
- 9.Nguyen LB, Salen G, Shefer S, et al. Decreased cholesterol biosynthesis in sitosterolemia with xanthomatosis: diminished mononuclear leukocyte 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and enzyme protein associated with increased low-density lipoprotein receptor function. Metab Lin Exp. 1990;39:436–443. doi: 10.1016/0026-0495(90)90260-j. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharyya AK, Connor WE, Lin DS, McMurry MM, Shulman RS. Sluggish sitosterol turnover and hepatic failure to excrete sitosterol into bile cause expansion of body pool of sitosterol in patients with sitosterolemia and xanthomatosis. Arterio Throm. 1991;11:1287–1294. doi: 10.1161/01.atv.11.5.1287. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen LB, Shefer S, Salen G, et al. A molecular defect in hepatic cholesterol biosynthesis in sitosterolemia with xanthomatosis. J Clin Invest. 1990;86:923–931. doi: 10.1172/JCI114794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel SB, Honda A, Salen G. Sitosterolemia: exclusion of genes involved in reduced cholesterol biosynthesis. J Lipid Res. 1998;39:1055–1061. [PubMed] [Google Scholar]
- 13.Patel SB, Salen G, Hidaka H, et al. Mapping a gene involved in regulating dietary cholesterol absorption. The sitosterolemia locus is found at chromosome 2p21. J Lipid Res. 1998;102:1041–1044. doi: 10.1172/JCI3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comuzzie AG, Hixson JE, Almasy L, et al. A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nat Genet. 1997;15:273–276. doi: 10.1038/ng0397-273. [DOI] [PubMed] [Google Scholar]
- 15.Hart TC, Pallos D, Bowden DW, et al. Genetic linkage of hereditary gingival fibromatosis to chromosome 2p21. Am J Hum Genet. 1998;62:876–883. doi: 10.1086/301797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen D, Chumakov I, Weissenbach J. A first-generation physical map of the human genome. Nature. 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- 17.Chumakov IM, Rigault P, Le Gall I, et al. A YAC contig map of the human genome. Nature. 1995;377:175–297. doi: 10.1038/377175a0. [DOI] [PubMed] [Google Scholar]
- 18.Hudson TJ, Stein LD, Gerety SS, et al. An STS-based map of the human genome [see comments] Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- 19.Cruts M, Backhovens H, Wang SY, et al. Molecular genetic analysis of familial early-onset Alzheimer's disease linked to chromosome 14q24.3. Hum Mol Genet. 1995;4:2363–2371. doi: 10.1093/hmg/4.12.2363. [DOI] [PubMed] [Google Scholar]
- 20.Strong TV, Tagle DA, Valdes JM, et al. Widespread expression of the human and rt Huntington's disease gene in brain and nonneural tissues. Nat Genet. 1993;5:259–265. doi: 10.1038/ng1193-259. [DOI] [PubMed] [Google Scholar]
- 21.Qin S, Nowark NJ, Zhang J, et al. A high-resolution physical map of human chromosome 11. Proc Natl Acad Sci USA. 1996;93:3149–3154. doi: 10.1073/pnas.93.7.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemeth-Slany A, Talmud P, Grundy SM, Patel SB. Activation of a cryptic splice-site in intron 24 leads to the formation of apolipoprotein B-27. Atherosclerosis. 1997;133:163–170. doi: 10.1016/s0021-9150(97)00105-6. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Kim UJ, Birren BW, Slepak T, et al. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 25.Ouellette BF, Boguski MS. Database divisions and homolgy search files: a guide for the perplexed. Genome Res. 1997;7:952–955. doi: 10.1101/gr.7.10.952. [DOI] [PubMed] [Google Scholar]
- 26.Center TSCaTWUGS. Towards a complete human genomic sequence. Genome Res. 1998;8:1097–1108. doi: 10.1101/gr.8.11.1097. [DOI] [PubMed] [Google Scholar]
- 27.Riley J, Butler R, Ogilvie D, et al. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirschner LS, Stratakis CA. Large-scale preparation of sequence-ready bacterial artificial DNA using QIAGEN columns. Biotechniques. 1999;27:72–74. [PubMed] [Google Scholar]
- 29.Deloukas P, Schuler GD, Gyapay G, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- 30.Schell U, Wienberg J, Kohler A, et al. Molecular characterization of breakpoints in patients with holoprosencephaly and definition of the HPE2 critical region 2p21. Hum Mol Genet. 1996;5:223–229. doi: 10.1093/hmg/5.2.223. [DOI] [PubMed] [Google Scholar]
- 31.Kirschner LS, Taymans SE, Pack S, et al. Genomic mapping of chromosomal region 2p15–p21 (D2S378–D2S391): integration of Genemap'98 within a framework of yeast and bacterial artificial chromosomes. Genomics. 1999;62:21–33. doi: 10.1006/geno.1999.5957. [DOI] [PubMed] [Google Scholar]
- 32.Stewart EA, McKusick KB, Aggarwal A, et al. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 33.Gyapay G, Schmitt K, Fizames C, et al. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- 34.Nagase T, Ishikawa K, Miyajima N, et al. Predicition of the coding sequence of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 35.Nie XF, Maclean KN, Kumar V, McKay IA, Bustin SA. ERF-2, the human homologue of the murine Tis11d early response gene. Gene. 1995;152:285–286. doi: 10.1016/0378-1119(94)00696-p. [DOI] [PubMed] [Google Scholar]
- 36.Malherbe P, Kohler C, Da Prada M, et al. Molecular cloning and functional expression of human 3-hydroxyanthranilic-acid dioxygenase. J Biol Chem. 1994;269:13792–13797. [PubMed] [Google Scholar]
- 37.Lai CH, Chou CY, Ch'ang LY, Lui CS, Lin W. Identification of Novel Human Genes Evolutionarily Conserved in Caenorhabditis elegans by Comparative Proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou J, Wang F, McKeehan WL. Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HepG2) In Vitro Cell Dev Biol Anim. 1994;30A:111–114. doi: 10.1007/BF02631402. [DOI] [PubMed] [Google Scholar]
- 39.Marley AE, Kline A, Crabtree G, Sullivan JE, Beri RK. The cloning expression and tissue distribution of human PP2Cbeta. FEBS Lett. 1998;431:121–124. doi: 10.1016/s0014-5793(98)00708-x. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto K, Segawa H, Tatsumi S, et al. Effects of truncation of the COOH-terminal region of a Na+-independent neutral and basic amino acid transporter on amino acid transport in Xenopus oocytes. J Biol Chem. 1996;271:16758–16763. doi: 10.1074/jbc.271.28.16758. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa K, Nagase T, Makajima D, et al. Prediction of the coding sequences of unidentified human genes. VIII.78 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1997;4:307–313. doi: 10.1093/dnares/4.5.307. [DOI] [PubMed] [Google Scholar]
- 42.Collins FS. Positional cloning moves from perditional to traditional [published erratum appears in Nat Genet 1995 Sep;11(1):104] Nat Genet. 1995;9:347–350. doi: 10.1038/ng0495-347. [DOI] [PubMed] [Google Scholar]
- 43.Boehm T. Positional cloning and gene identification. Methods. 1998;14:152–158. doi: 10.1006/meth.1997.0574. [DOI] [PubMed] [Google Scholar]


