Abstract
Replacement of the recBCD genes of Escherichia coli with the red recombination genes of bacteriophage lambda results in a strain in which adaptive mutation occurs at an elevated frequency. Like RecBCD-dependent adaptive mutation, Red-mediated adaptive mutation is dependent upon recA and ruvABC functions.
Adaptive mutation is a process by which an organism under nonlethal selective pressure produces mutations that relieve the selective pressure (see reference 5 for a review). It is illustrated particularly well by Escherichia coli strain FC40, a strain bearing an F′ episome carrying a lac allele with a frameshift mutation that makes it phenotypically Lac−. When FC40 is plated on lactose-minimal medium, Lac+ revertants appear at a high rate over several days, largely in the absence of cell growth, death, or turnover (2, 4). The formation of these adaptive mutations, unlike mutations that occur during growth under nonselective conditions, depends on the RecA-RecBCD recombination pathway (2, 8).
E. coli strains in which the RecBCD function is replaced by the Red recombination system of bacteriophage λ exhibit a hyperrecombination phenotype (13, 14). The possibility that the activity of the cell's recombination system might be a rate-limiting factor in the production of adaptive revertants led us to test whether Red+ bacteria might produce such revertants at an elevated rate. We report here that they do.
Bacterial strains.
P1 transduction was used to replace the recC-ptr-recB-recD gene cluster in FC40 and FC691 with a sequence bearing both Ptac-gam-bet-exo and the cat gene. (The Ptac-gam-bet-exo sequence is designated red.) Recombinants were selected for chloramphenicol resistance, producing strains TP694 and TP730. Strains TP705 and TP732 were made by transforming TP694 and TP730 with linear DNA resulting from the digestion of plasmid pTP822 (16). Recombinants, in which ΔrecBCD::red-cat was replaced by ΔrecBCD::red-pae-cI822, were selected for immunity to phage λ and screened for sensitivity to chloramphenicol and kanamycin. These and other strains employed in this study are described in Table 1.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Reference or construction |
|---|---|---|
| FC29 | F′ Δ(lacI lacZ) | 2 |
| FC40 | F′ Φ(lacI33-lacZ) | 2 |
| FC691 | F− Φ(lacI33-lacZ) | 18 |
| JC15329 | Δ(srl-recA)306::Tn10 | A. J. Clark |
| KM32 | AB1157 recBCDΔ::red-cat | 16 |
| N4454 | ΔruvABC::cat | 12 |
| TP538 | ΔrecG::tet6200 | 15 |
| TP547 | KM32 lacZ::tet805 | KM32 × linear DNA |
| TP694 | FC40 ΔrecBCD::red-cat | FC40 × P1(TP547) |
| TP705 | FC40 ΔrecBCD::red-pae-cI822 | TP694 × linear DNA |
| TP709 | TP705 ΔrecG::tet6200 | TP705 × P1(TP538) |
| TP710 | TP705 Δ(srl-recA)306::Tn10 | TP705 × P1(JC15329) |
| TP724 | TP705 ΔruvABC::cat | TP705 × P1(N4454) |
| TP725 | FC40 ΔruvABC::cat | FC40 × P1(N4454) |
| TP732 | FC691 ΔrecBCD::red-pae- cI822 | FC691 × linear DNA |
| TP734 | TP732 ΔrecG::tet6200 | TP732 × P1(TP538) |
Media.
The lysogeny broth (LB) used contained, per liter, 10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, and 1 ml of 1 M NaOH. It was solidified with 15 g of agar per liter and supplemented as necessary with 25 μg of tetracycline/ml, 20 μg of chloramphenicol/ml, 0.1 mM isopropylthiogalactopyranoside (IPTG), or 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml.
M9 salt solution contained, per liter, 6.3 g of Na2HPO4, 3.0 g of KH2PO4, 0.5 g of NaCl, and 1.0 g of NH4Cl. M9 glycerol-minimal medium consisted of M9 salt solution plus 1 mM MgSO4, 5 μg of thiamine/ml, 0.1 mM CaCl2, 10 μg of gelatin/ml, and 1 mg of glycerol/ml. M9 lactose-minimal agar consisted of M9 salt solution plus 1 mM MgSO4, 5 μg of thiamine/ml, 1 mg of lactose/ml, and 15 mg of agar/ml.
Measurement of stationary-phase reversion to Lac+.
The strains to be tested were scraped from frozen cultures in storage vials and streaked on LB plates supplemented with appropriate antibiotics. A single colony was used to inoculate 5 ml of LB broth, and the culture was grown with aeration at 30 or 37°C to apparent saturation. The LB culture was diluted 105-fold with M9 glycerol-minimal medium, with a final 100-fold dilution into three to six tubes containing 1 ml each. These cultures in M9 glycerol-minimal medium were grown with aeration at 30 or 37°C to apparent saturation, and 0.01 to 0.5 ml of each culture was plated on one M9 lactose-minimal plate with approximately 2 × 109 scavenger cells. Scavenger cells (FC29) were grown to saturation in M9 glycerol-minimal medium and concentrated by centrifugation and resuspension in 0.1 volume of M9 salt solution; thereafter, 0.1 ml was plated. The plates were incubated at 37°C; colony counts were recorded on days 2 through 6 or 7. (Lac+ cells present in the culture at the time of plating take 2 days to form colonies.) The few plates with large numbers of preexisting revertants were ignored. Titers of the tested cultures were determined by plating on LB agar at 37°C.
Red-mediated reversion of episomal lac.
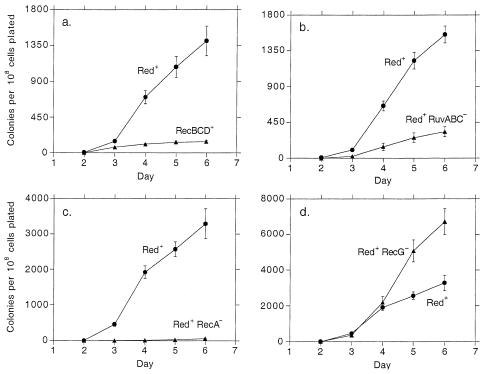
A derivative of the F′ Φ(lacI33-lacZ)-bearing E. coli strain FC40, in which the recBCD gene cluster is replaced by the red system of phage λ, exhibits an elevated rate of adaptive mutation to Lac+ relative to its recBCD+ parent (Fig. 1a). As in recBCD+ bacteria (7, 9), the rate is significantly lower in the absence of the ruv genes (Fig. 1b) and lower still in the absence of recA (Fig. 1c). Deletion of recG from the red-substituted strain mildly increases the rate of reversion (Fig. 1d). The red+ recG strain and its recBCD+ recG counterpart exhibit approximately equal rates of reversion (data not shown).
FIG. 1.
Rates of reversion to Lac+. Points represent the means of the numbers of independent cultures specified below. Error bars (some of which are smaller than the symbols) represent the standard errors of the means. (a) Six cultures each of TP705 (circles) and FC40 (triangles); (b) six cultures of TP705 (circles) and five of TP724 (triangles); (c) three cultures of TP705 (circles) and six of TP710 (triangles); (d) three cultures of TP705 (circles) and six of TP709 (triangles).
Although Red stimulates the rate of adaptive reversion, it apparently has little effect on the rate of mutation occurring during nonselective growth. Revertants arising during the growth of the cultures form visible colonies in 2 days. The average numbers of colonies appearing on day 2 were 3.5 per 108 cells plated for the wild type (average of 8 experiments) and 3.9 per 108 cells plated for the red-substituted strain (average of 10 experiments) (data not shown).
Red-mediated adaptive reversion appears to depend on the same E. coli recombination functions as does Red-mediated recombination, and like Red-mediated recombination, it is apparently mildly inhibited by RecG function (15). RecBCD-mediated adaptive mutation, however, differs from RecBCD-mediated recombination in that it is greatly inhibited by RecG function while RecBCD-mediated recombination is mildly stimulated by RecG function (7, 9).
The DNA segment replacing recBCD in the strains used in these experiments includes genes encoding the PaeR7 restriction-modification system and the λ repressor, in addition to the λ red system. These additional functions apparently do not contribute to the elevated rate of reversion; TP694, another red-substituted strain, lacking pae and cI, reverts at an equally high rate (data not shown).
Red-mediated reversion of chromosomal lac.
E. coli FC691 is F− and carries the Φ(lacI33-lacZ) allele on its chromosome (18). A derivative of FC691 in which the recBCD gene cluster is replaced by the red system of phage λ also exhibits an increased rate of stationary-phase mutation to the Lac+ phenotype, as shown in Table 2. The observed increase was approximately fourfold. The deletion of recG did not appear to increase the rate of reversion further (Table 2).
TABLE 2.
Reversion of chromosomal lac mutants
| Strain | Relevant genotype | Rate of reversiona |
|---|---|---|
| FC691 | F− Φ(lacI33-lacZ) | 0.63 ± 0.22 |
| TP732 | FC691 ΔrecBCD::red-pae-cI822 | 2.47 ± 0.34 |
| TP734 | TP732 ΔrecG::tet6200 | 2.39 ± 0.69 |
Rates are given as mean numbers of revertants per day per 109 cells plated ± standard errors of the means for 5 to 12 cultures. Newly arising colonies were counted between 4 and 7 days after plating. Plating conditions were the same as those for the F′ Φ(lacI33-lacZ)-bearing strains, except that greater numbers of cells were plated (1 × 109 to 2 × 109 per plate), and no scavenger cells were used.
Stability of the Red-generated revertants.
If Lac+ revertants of the episomal lac allele are generated by amplification, they form sectored colonies when streaked on nonselective medium containing X-Gal (1, 10). To determine whether amplification underlies the high rate of Red-mediated stationary-phase reversion, 10 late-arising Lac+ revertants of strain TP705 and 2 of FC40 were streaked on LB plates supplemented with X-Ga1 and IPTG and incubated at 37°C. The great majority of the colonies in all cases were solid blue. A few pure-white colonies were observed as well, possibly representing the scavenger strain. Of approximately 100 well-isolated TP705 revertant colonies, only one appeared to have a mixed phenotype, but its appearance could be accounted for by a pure-white colony and a pure-blue colony formed in close proximity. By the sectoring test, therefore, the great majority of the late-arising Lac+ colonies of the red+ F′ F(lacI33-lacZ) strain appear to represent true reversion events rather than amplifications of the mutant allele. In contrast, when this test was repeated with Red-mediated revertants of the chromosomal lac allele, 7 of 12 late-arising colonies tested gave rise to numerous sectored colonies. Thus, Red increases stable amplification on the chromosome but not on the episome.
Amplification is involved in the reversion of the episomal lac allele, although its true role is debated (4, 10, 11, 17). Since adaptive reversion of the chromosomal lac allele in FC691 is not dependent on RecA and therefore does not involve amplification (6), it appears that Red may induce an entirely new pathway for adaptive change on the chromosome. Alternatively, the high frequency of sectored colonies among Red-mediated chromosomal Lac revertants may reflect not a greater frequency of formation but a greater stability of amplifications on the chromosome in the Red+ background. Although the chromosomal amplifications have not been characterized, based on previous reports, they are expected to be the same type of tandem repeats as are the episomal amplifications (3).
Acknowledgments
This research was supported by the University of Massachusetts and by grant NSF MCB-9996308 from the National Science Foundation (to P.L.F.).
REFERENCES
- 1.Anderson, D. I., E. S. Slechta, and J. R. Roth. 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282:1133-1135. [DOI] [PubMed] [Google Scholar]
- 2.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edlund, T., and S. Normark. 1981. Recombination between short DNA homologies causes tandem duplication. Nature 292:269-271. [DOI] [PubMed] [Google Scholar]
- 4.Foster, P. L. 1994. Population dynamics of a Lac− strain of Escherichia coli during selection for lactose utilization. Genetics 138:253-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster, P. L. 1999. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu. Rev. Genet. 33:57-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster, P. L., and J. M. Trimarchi. 1995. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc. Natl. Acad. Sci. USA 92:5487-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster, P. L., J. M. Trimarchi, and R. A. Maurer. 1996. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics 142:25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264:258-260. [DOI] [PubMed] [Google Scholar]
- 9.Harris, R. S., K. J. Ross, and S. M. Rosenberg. 1996. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics 142:681-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings, P. J., H. J. Bull, J. R. Klump, and S. M. Rosenberg. 2000. Adaptive amplification: an inducible chromosomal instability mechanism. Cell 103:723-731. [DOI] [PubMed] [Google Scholar]
- 11.Hendrickson, H., E. S. Slechta, U. Bergthorsson, D. I. Anderson, and J. R. Roth. 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. USA 99:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35-45. [DOI] [PubMed] [Google Scholar]
- 13.Murphy, K. C. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poteete, A. R., and A. C. Fenton. 1993. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages λ and P22. Genetics 134:1013-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poteete, A. R., and A. C. Fenton. 2000. Genetic requirements of phage λ Red-mediated gene replacement in Escherichia coli K-12. J. Bacteriol. 182:2336-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poteete, A. R., A. C. Fenton, and K. C. Murphy. 1999. Roles of RuvC and RecG in phage λ Red-mediated recombination. J. Bacteriol. 181:5402-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell, S. C., and R. M. Wartell. 2001. Different characteristics distinguish early versus late arising adaptive mutations in Escherichia coli FC40. Mutat. Res. 473:219-228. [DOI] [PubMed] [Google Scholar]
- 18.Rosche, W. A., and P. L. Foster. 1999. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96:6862-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]