Abstract
ATP-dependent proteases degrade denatured or misfolded proteins and are recruited for the controlled removal of proteins that block activation of regulatory pathways. Among the ATP-dependent proteases, those of the Clp family are particularly important for the growth and development of Bacillus subtilis. Proteolytic subunit ClpP, together with regulatory ATPase subunit ClpC or ClpX, is required for the normal response to stress, for development of genetic competence, and for sporulation. The spx (formally yjbD) gene was previously identified as a site of mutations that suppress defects in competence conferred by clpP and clpX. The level of Spx in wild-type cells grown in competence medium is low, and that in clpP mutants is high. This suggests that the Spx protein is a substrate for ClpP-containing proteases and that accumulation of Spx might be partly responsible for the observed pleiotropic phenotype resulting from the clpP mutation. In this study we examined, both in vivo and in vitro, which ClpP protease is responsible for degradation of Spx. Western blot analysis showed that Spx accumulated in clpX mutant to the same level as that observed in the clpP mutant. In contrast, a very low concentration of Spx was detected in a clpC mutant. An in vitro proteolysis experiment using purified proteins demonstrated that Spx was degraded by ClpCP but only in the presence of one of the ClpC adapter proteins, MecA or YpbH. However, ClpXP, either in the presence or in the absence of MecA and YpbH, was unable to degrade Spx. Transcription of spx, as measured by expression of spx-lacZ, was slightly increased by the clpX mutation. To exclude a possible effect of clpX and clpP on spx transcription, the spx gene was placed under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Pspac promoter. In this strain, Spx accumulated when ClpX or ClpP was absent, suggesting that ClpX and ClpP are required for degradation of Spx. Taken together, these results suggest that Spx is degraded by both ClpCP and ClpXP. The putative proteolysis by ClpXP might require another adapter protein. Spx probably is degraded by ClpCP under as yet unidentified conditions. This study suggests that the level of Spx is tightly controlled by two different ClpP proteases.
ATP-dependent proteases play an important role in many cellular processes in both prokaryotes and eukaryotes (reviewed in reference 47). They function in the cell's response to stress by eliminating damaged or misfolded proteins and degrading truncated products of aborted translation. They are also involved in degradation of specific, short-lived regulatory proteins, many of which function in developmental processes. Members of one class of ATP-dependent proteases, the Clp proteases, are composed of two subunits, a regulatory ATPase component and the peptidase, ClpP (reviewed in reference 37). While ClpP alone exhibits peptidase activity, its association with the ATPase imparts substrate specificity and is required for the degradation of polypeptides longer than six amino acids (41). The Clp ATPase hexamers bind to and unfold substrates and then translocate them to the two rings of ClpP heptamers (16, 40). Clp ATPases, in the presence or absence of ClpP, also function as molecular chaperones (reviewed in reference 12). For example, ClpX chaperone activity is necessary for the dissociation of the MuA complex following strand transfer during Mu transposition (23). In Escherichia coli, ClpAP degrades RepA (46), bacteriophage P1 protein (22), and the MazE protein (1), which is involved in programmed cell death. ClpXP degrades MuA transposase (23), λ O (49), and the stationary-phase σ factor RpoS (39). Proteins from the translation of truncated mRNA are targeted for the addition of 11-residue peptides to the carboxy termini, a process which is mediated by SsrA RNA (also called 10Sa RNA or tm RNA) (42). The SsrA-tagged proteins are degraded by ClpAP and ClpXP (13).
In Bacillus subtilis, Clp proteases include those bearing the ClpC, ClpE, and ClpX ATPase subunits (reviewed in reference 38). Unlike E. coli clpP and clpX mutants, which grow normally (39), the B. subtilis clp mutants are highly pleiotropic and exhibit poor growth phenotypes on certain media such as minimal medium and Luria-Bertani (LB) medium even under nonstress conditions (10, 26, 31). Immunocytochemical experiments showed that ClpCP and ClpXP are directly involved in degradation of misfolded or denatured proteins after either heat shock or treatment with puromycin (20). These treatments also induce transcription of clpC, clpP, and clpX (10, 11, 19, 26, 27) and lead to increased ClpC and ClpP protein levels (20). ClpC, ClpP, and ClpX are essential for growth at high temperature following heat shock induction (10, 19, 26, 27). Other common features observed in the clpC, clpP, and clpX mutants include a salt-sensitive and nonmotile phenotype (10, 19, 26). No obvious phenotype which is associated with the clpE mutation has been found so far (5). Recently, ClpXP was shown to be responsible for degradation of SsrA-tagged proteins in B. subtilis cells (48).
In addition to their growth phenotype and sensitivity to stress, mutants lacking ClpC, ClpP, or ClpX show defects in developmental pathways including sporulation and competence (10, 26, 31, 32). A clpP null mutation confers the most severe sporulation defect, followed, in order of decreasing severity, by the clpC and clpX mutations. The sporulation phenotype of the clpC mutation appeared more obvious at high temperature (32), while a mutation in clpE, encoding another ATPase subunit, produces no sporulation phenotype (5). Since ClpP is thought only to function with Clp ATPases, it is not clear why mutations in clpX, -C, and -E have little or no sporulation phenotype. Perhaps the ATPases perform redundant functions during the sporulation process. That ClpP is necessary for entry into sporulation is supported by the finding that Spo0A-dependent expression of spoIIA and spoIIG operons, which encode σF and σE, respectively, was adversely affected by a clpP mutation (26). This result suggests that the phosphorelay leading to the phosphorylation of Spo0A is defective in the clpP mutant. In fact, a spo0E mutation partially suppresses clpP; that is, Spo0A-dependent gene expression is observed in a spo0E clpP mutant background but sporulation was not restored (33). The clpC mutation did not affect Spo0A-dependent expression of spoIIA and spoIIE; however, the mutation caused an impairment of σF-dependent gene expression (35). Apparently ClpCP is required after phosphorylation of Spo0A and before activation of σF. A recent study demonstrated that ClpCP increased σF activity by directly degrading SpoIIAB and anti-σF factor and by indirectly promoting polar septum formation, which is a prerequisite for the activation of σF (35). ClpCP is also involved in the degradation of σH, an RNA polymerase sigma subunit required for sporulation initiation, at high temperature and might be responsible for the degradation of σH after it accomplishes its task in early sporulation (32). Finally ClpX, but not ClpP, is required for σH-RNA polymerase activity in vitro and in vivo (25). Thus the role of the ClpXP protease in sporulation, if any, is elusive.
Although ClpC, ClpP, and ClpX are involved in competence development, the effect of clpC is opposite to that of clpP and clpX. Both clpP and clpX mutants are defective in competence (26, 31), while the clpC mutant shows a mec (medium-independent expression of competence) phenotype (27), indicating a negative role for ClpC. ClpC forms a complex with MecA (18) and ComK (45) in noncompetent cells; this complex sequesters transcription factor ComK in an inactive state (44). Thus, ClpC negatively regulates comK-dependent expression of comK and late competence genes. In addition, ComK is degraded by ClpCP, and the proteolysis requires adapter protein MecA (43). The small peptide ComS (7, 14), whose synthesis is activated by a quorum-sensing pathway, accumulates in stationary phase and releases ComK from the inhibitory complex (44). Liberated ComK is no longer a target of ClpCP; instead MecA and ComS are degraded by ClpCP (43). Two additional roles for ClpP in competence have been found. ClpP, together with ClpX, is required for srf (comS) expression (31). Amino acid substitutions in the carboxy-terminal domain of the α subunit of RNA polymerase suppress either clpX or clpP with respect to comS expression (31). Although the exact role of ClpXP in comS expression remains to be elucidated, a direct involvement of ClpXP in comS transcription was suggested. Furthermore ClpP is also needed to degrade Spx (YjbD), the accumulation of which negatively affects competence and sporulation (28). The clpP spx double mutant completely and partially restored competence and sporulation, respectively. In this study, we examined how the level of Spx is regulated by ClpP proteases and found that both ClpCP and ClpXP are involved in degrading Spx, probably under different conditions.
MATERIALS AND METHODS
Bacterial strains and media.
All B. subtilis strains listed in Table 1 are derivatives of JH642. Construction of LAB2876 (clpX::spc) (25), LAB2972 (clpP::erm) (34), and ORB3834 (spx::neo) (28) was described previously. ORB3976 was constructed by transforming JH642 with chromosomal DNA prepared from QPB418 (clpC::tet) (35).
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain | Relevant genotype or properties | Source and/or reference |
|---|---|---|
| B. subtilis strains | ||
| JH642 | trpC2 pheA1 | J. A. Hoch |
| QPB418 | clpC::tet | 35 |
| LAB2876 | trpC2 pheA1 clpX::spc | 25 |
| LAB2972 | trpC2 pheA1 clpP::erm | 34 |
| ORB3673 | trpC2 pheA1 clpX::spc yjbFG::pPS34 | 28 |
| ORB3834 | trpC2 pheA1 yjbD::neo | 28 |
| ORB3976 | trpC2 pheA1 clpC::tet | This study |
| ORB4059 | trpC2 pheA1 yjbD-lacZ cat | This study |
| ORB4065 | trpC2 pheA1 clpX::spc yjbD-lacZ cat | This study |
| ORB4075 | trpC2 pheA1 amyE::Pspac-yjbD cat | This study |
| ORB4078 | trpC2 pheA1 yjbD::neo amyE::Pspac-yjbD | This study |
| ORB4079 | trpC2 pheA1 yjbD::neo clpX::spc amyE::Pspac-yjbD | This study |
| ORB4080 | trpC2 pheA1 yjbD::neo clpP::erm amyE::Pspac-yjbD | This study |
| ORB4081 | trpC2 pheA1 yjbD::neo clpC::tet amyE::Pspac-yjbD | This study |
| Plasmids | ||
| pDR66 | Plasmid allowing IPTG-dependent gene expression | A. Grossman, 15 |
| pLysS | Plasmid to produce T7 lysozyme | Stratagene |
| pPS34 | Integrative plasmid | P. Serror and A. L. Sonenshein, unpublished |
| pTKlac | Plasmid for construction of lacZ transcriptional fusion | 17 |
| pTYB1 | Cloning vector for IMPACT T7 system | New England BioLabs |
| pTYB2 | Cloning vector for IMPACT T7 system | New England BioLabs |
| pClpC | pTYB2 with clpC | This study |
| pClpP | pTYB1 with clpP | This study |
| pMMN464 | pPS34 carrying oppB to yjbF | This study |
| pMMN470 | pTYB4 with yjbD | 28 |
| pMMN497 | pDR66 with yjbD under control of Pspac | This study |
| pSN3 | pTYB1 with mecA | This study |
| pSN5 | pTYB1 with ypbH | This study |
| pSN16 | pTKlac carrying yjbD-lacZ | This study |
A strain which carries the spx coding sequence downstream of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Pspac promoter was constructed as follows. Primers oMN01-173 (5′-CGAGGAAGCTTAGATGTTCATCCTACTA-3′) and oMN01-174 (5′-TACCAGCAGGTCGACAAATAAAAGAAGG-3′) were used to amplify the spx gene by using JH642 chromosomal DNA as the template. The PCR product was digested with HindIII and SalI and ligated with pDR66 digested with the same enzymes. After the spx sequence was verified, the resultant plasmid, pMMN497, was used to transform JH642 to generate ORB4075. ORB4075 was generated by integration of pMMN497 at the amyE locus by double-crossover recombination and screening for the Amy− (amylase-negative) phenotype. ORB4078 was constructed by transforming OEB4075 with chromosomal DNA prepared from LAB3834 (spx::neo). The insertion of the neo marker into spx located at the original locus and not into Pspac-spx located at amyE was confirmed by testing the genetic linkage between the chloramphenicol resistance (Cmr; derived from pMMN497) and neomycin resistance (Neor) markers. ORB4079, ORB4080, and ORB4081 were constructed by transforming ORB4078 with LAB2876 (clpX::spc), LAB2972 (clpP::erm), and ORB3976 (clpC::tet) DNA, respectively.
B. subtilis cells were grown in rich media, 2× yeast extract-tryptone (YT) (29) and LB medium, Difco sporulation medium (DSM) (29), and one-step competence medium (CM) (8). Antibiotics were added to the following concentrations: chloramphenicol, 5 μg/ml; erythromycin plus lincomycin, 1 and 25 μg/ml, respectively; tetracycline, 12.5 μg/ml; spectinomycin, 75 μg/ml; neomycin, 5 μg/ml; ampicillin, 25 μg/ml.
Purification of proteins.
For production of proteins used in this study, the IMPACT system (New England BioLabs) (4), which utilizes the inducible self-cleaving intein tag, was used. Overproduction and purification of Spx (28) and ClpX (25) were previously described.
The clpP gene was amplified by PCR using primers oclpPex1 (5′-GGAGGAGCCATATGAATTTAAT-3′) and oclpPex2 (5′-ATTAGGAAGAGCCCTTTTTGTCTTCTGTGTGA-3′) and chromosomal DNA prepared from JH642 as the template. The PCR product digested with NdeI and SapI was cloned into pTYB1 (New England BioLabs), which had undergone digestion with the same enzymes. The resultant plasmid was digested with SapI, treated with T4 DNA polymerase in the presence of deoxynucleoside triphosphate to fill in the single-stranded end, and self-ligated to generate pClpP.
The clpC gene was amplified by PCR using primers oClpCex1 (5′-GGATGAATCCATATGATGTTTGGAAGATTT-3′) and oClpCex2 (5′-TCCCCCGGGATTCGTTTTAGCAGTCGTTTT-3′) and JH642 chromosomal DNA as the template. The amplified fragment, after being digested with NdeI and SmaI, was cloned into pTYB2 (New England BioLabs) digested with the same enzymes to generate pClpC.
The mecA gene was amplified by PCR using JH642 chromosomal DNA and two primers, oMN01-156 (5′-GGAAGGTTGGCATATGGAAATTG-3′) and oMN01-157 (5′-GACTTAGCTCTTCCGCATGATGCAAAGTGTTTT-3′). The amplified fragment, after being digested with NdeI and SapI, was inserted into pTYB1 (digested with the same enzymes) to construct plasmid pSN3.
The ypbH gene was amplified by PCR using JH642 chromosomal DNA and two primers, oMN01-164 (5′-GAAGATCATATGCGGCTTGAGCGT-3′) and oMN01-165 (5′-GCCGCCGCTCTTCCGCATGAAAAATGAGTTTGTA-3′). The amplified fragment was inserted into pTYB1 by using the same enzymes as those used for cloning the mecA gene to generate pSN5.
The sequences of all the genes cloned above were verified by DNA sequencing. The clpC gene in pClpC has a mutation in codon 795; however, the amino acid (Asp) was not affected by the change, GAT to GAC. E. coli ER2566 (New England BioLabs) carrying pClpP, pClpC, or pSN3 was used to produce ClpP, ClpC, and MecA, respectively. E. coli BL21(DE3) carrying pLysS (Stratagene) and pSN5 was used for the production of YpbH. The proteins were purified by using a chitin column as recommended by the manufacturer. Purified MecA and YpbH produced by this construct have no extra amino acids. ClpP and ClpC have extra amino acids at their carboxy ends (Gly-Ser-Ser-Tyr for ClpP and Pro-Gly for ClpC).
Purification of the Spx protein in the presence of CuSO4.
E. coli ER2566 carrying pMMN470 was cultured in 2× YT at 37°C. When the optical density at 600 nm reached 0.4, CuSO4 was added to the culture at the final concentration of 500 μM. The procedure for further purification was described previously (28).
In vitro degradation assay.
An in vitro degradation assay was carried out as described previously (43) with slight modification. ClpP (4 μM) and Spx (4 μM) were incubated at 37°C for 0 or 30 min in the presence or absence of MecA (2.5 μM), YpbH (2.5 μM), ClpX (2.5 μM), and ClpC (2.5 μM) in a final volume of 50 μl of reaction buffer (25 mM MOPS [morpholinepropanesulfonic acid]-KOH [pH 7.0], 100 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 4 mM ATP, 2 mM phosphoenol pyruvate, 0.93 μM pyruvate kinase [Sigma]). Phosphoenol pyruvate and pyruvate kinase were used to regenerate ATP. After the incubation, 10-μl portions of the samples were transferred to 5 μl of stop solution. The proteins were analyzed on a 15% polyacrylamide-sodium dodecyl sulfate (SDS) gel, followed by staining with Coomassie blue.
Western blot analysis.
Cells were cultured in CM and harvested at T2 (2 h after the onset of the stationary phase). Cells were broken by passage through a French press, and whole-cell extracts were used in Western analysis or cell debris was first removed by centrifugation (17,000 × g for 15 min) to generate a cleared extract. The protein concentration of each sample was measured with a Bio-Rad protein assay kit. Thirty micrograms of each protein was applied to a 15% polyacrylamide-SDS gel. After electrophoresis, the proteins were transferred to a nitrocellulose membrane. Immunodetection was performed with the anti-Spx antibody (28) followed by incubation with the secondary antibody conjugated to alkaline phosphatase.
Construction of a transcriptional spx-lacZ fusion.
B. subtilis strain ORB3673, which has plasmid pPS34 inserted into the yjbFG region, was constructed in a previous study (28). Chromosomal DNA prepared from ORB3673 was digested with ApaI and self-ligated. The ligation mixture was used to transform E. coli DH5α with selection for ampicillin resistance (Ampr). Plasmid pMMN464, isolated from the Ampr transformant, carries 9.3 kbp of B. subtilis chromosomal DNA containing the 3′ ends of oppB, oppCDE, yjbBCDE, and mecA and the 5′ end of yjbF. pMMN464 was cut with BglII, and the resulting 4.6-kbp fragment was recovered from a low-melting-point agarose gel. The fragment was then digested with BclI and HaeIII sequentially, which produces a 1.4-kbp fragment containing the 3′ end of yjbB, the entire yjbC gene, and the 5′ end of spx (yjbD). The fragment was inserted into promoter-probe vector pTKlac (17), which was digested with SmaI and BamHI to generate pSN16. JH642 was transformed with pSN16, and a Cmr transformant was selected as ORB4059. ORB4065 was isolated by transforming ORB4059 with chromosomal DNA prepared from LAB2876 (clpX::spc) with selection for Cmr and Spcr.
Assay of β-galactosidase activity.
Cells were grown in CM, DSM, or 2× YT, and samples were withdrawn at 30-min or 1-h time intervals during growth. β-Galactosidase activity in each sample was determined as described previously (29) and is presented as Miller units.
RESULTS
Spx accumulates in clpP and clpX mutants but not in a clpC mutant.
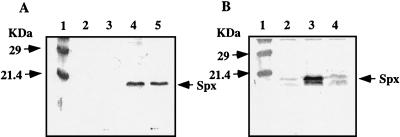
We have previously shown that Spx concentration was low in wild-type cells grown in CM but markedly higher in a clpP mutant (28). This suggests that Spx is a substrate of ClpP protease(s). In B. subtilis ATPase subunits ClpC, ClpX, and ClpE are known or thought to function with catalytic subunit ClpP (reviewed in reference 38). To identify which regulatory subunit is involved in degradation of Spx, we first examined the amount of Spx in a clpX mutant. The result showed that Spx accumulated in the clpX mutant to a level similar to that observed in a clpP strain (Fig. 1A). In contrast, Spx concentration in a clpC mutant was as low as that in the wild type (Fig. 1B). As was the case for the clpC mutant, no increase in Spx concentration was observed in either the mecA or ypbH mutant. This result suggested that degradation of Spx is dependent primarily on ClpXP.
FIG. 1.
Effect of mutations on Spx level as measured by Western blot analysis using anti-Spx antiserum. Samples were taken at T2 (2 h after the onset of the stationary phase) from cells grown in CM. (A) Lane 1, molecular mass markers; lane 2, JH642 (wild type); lane 3, ORB3834 (spx::neo); lane 4, LAB2972 (clpP::erm); lane 5, LAB2876 (clpX::spc). (B) Lane 1, molecular mass markers; lane 2, JH642; lane 3, LAB2972; lane 4, ORB3976 (clpC::tet).
Spx is degraded in vitro by ClpCP but not ClpXP.
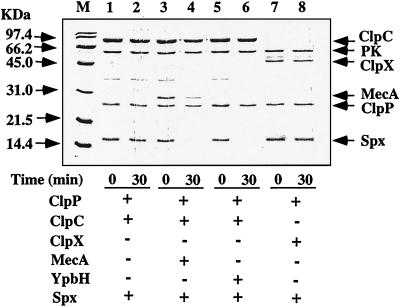
Although the result described above suggests that Spx is a substrate of ClpXP, alternative interpretations, such as the possibility that a regulator of spx expression is a target of ClpXP protease, could not be excluded. To investigate whether Spx is degraded by ClpXP, in vitro proteolysis experiments using purified proteins were carried out. We have previously found that Spx binds to copper (30), and Fig. 2 shows the data from experiments using copper-bound Spx, prepared as described in Materials and Methods. However, a similar pattern of proteolysis was observed by using the copper-free Spx protein (data not shown). We also examined the effect of ClpCP protease on Spx degradation, since we observed that Spx could form a complex with the ClpCP adapter, MecA, and could contribute to ComK inhibition (30). Degradation of Spx was observed in the presence of ClpP, ClpC, and MecA (Fig. 2, lanes 3 and 4). MecA, to a lesser extent, was also degraded by ClpCP, as previously reported (43). In the absence of MecA (lanes 1 and 2) or ATP (data not shown), Spx was stable after incubation for 30 min. This indicates that MecA acts as the adapter for ClpCP-catalyzed proteolysis of Spx, as well as for MecA-dependent ComK and ComS proteolysis by ClpCP (43). Under these same conditions, we observe that ComK is degraded by MecA-dependent, ClpCP-catalyzed proteolysis (data not shown), as was shown previously. YpbH is similar in amino acid sequence to MecA and is a possible MecA paralog. Therefore we examined whether YpbH can act as the adapter protein for ClpC. The result showed that Spx was degraded by ClpCP in the presence of YpbH (lanes 5 and 6), indicating that YpbH acts as the adapter for Spx proteolysis by ClpCP. We could not discern whether YpbH itself was degraded by ClpCP because YpbH and ClpP comigrated during SDS-polyacrylamide gel electrophoresis. Surprisingly, Spx was not degraded in vitro in the presence of ClpX, ClpP, and ATP (lanes 7 and 8). Addition of MecA and YpbH did not promote ClpXP-dependent proteolysis (data not shown).
FIG. 2.
Degradation of Spx in vitro. Spx and ClpP were incubated at 37°C in the presence of ATP and an ATP-generating system with (+) or without (−) ClpC, ClpX, MecA, and YpbH as described in Materials and Methods. Samples were taken at the times indicated and were analyzed by SDS-polyacrylamide gel electrophoresis, followed by staining with Coomassie blue. Lane M, molecular mass markers. PK, pyruvate kinase, used as an ATP-regenerating system.
Spx accumulates less in clpX and clpP cells grown in DSM than in cells grown in CM or 2× YT.
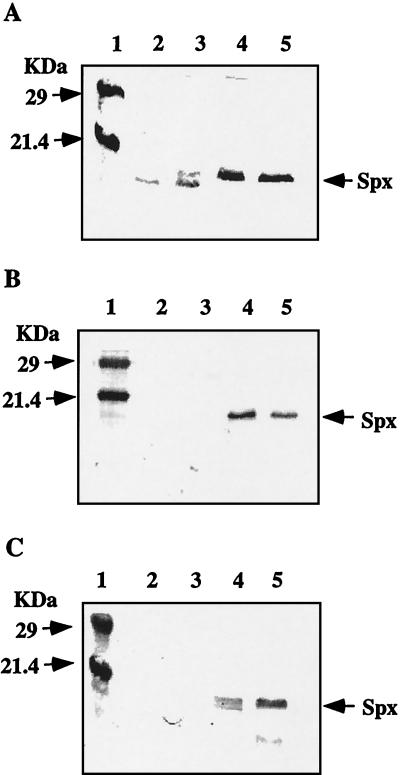
Although Spx was degraded by ClpCP in vitro, Spx did not accumulate in the clpC mutant grown in CM. However, the clpC mutation might affect the concentration of Spx under different conditions. First we examined whether Spx accumulates in clpC cells cultured in DSM. The result showed that the level of Spx in the clpC strain was as low as that in the wild type (data not shown). In addition, Spx accumulates in the clpX and clpP mutants, but at a level much lower than those in the same strains grown in CM (Fig. 3A) or 2× YT (data not shown). The band with a molecular weight lower than that of Spx, which may be a degradation product of Spx, was sometimes detected (as shown in lanes 2 and 3).
FIG. 3.
Western blot analysis using anti-Spx antiserum. (A) Effect of culture media on Spx level as measured by Western blot analysis. Samples were taken at T2 from cells grown in DSM and in CM. Lane 1, molecular mass markers; lane 2, LAB2876 (clpX::spc) grown in DSM; lane 3, LAB2972 (clpP::erm) grown in DSM; lane 4, LAB2876 grown in CM; lane 5, LAB2972 grown in CM. (B) Cells carrying Pspac-spx were grown in CM in the presence of 1 mM IPTG. Samples were taken at T2. Lane 1, molecular mass markers; lane 2, ORB4078 (wild type); lane 3, ORB4081 (clpC::tet); lane 4, ORB4079 (clpX::spc); lane 5, ORB4080 (clpP::erm). (C) The wild type and the clpX mutant carrying Pspac-spx were grown in DSM or CM in the presence of 1 mM IPTG. Samples were taken at T2. Lane 1, molecular mass markers; lane 2, ORB4078 in DSM; lane 3, ORB4078 in CM; lane 4, ORB4079 in DSM; lane 5, ORB4079 in CM.
The clpX mutation has no significant effect on spx transcription.
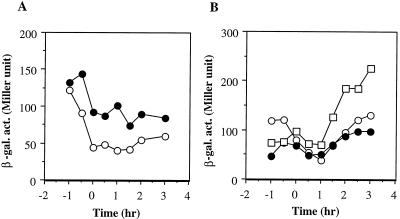
That Spx is abundant in the clpX mutant whereas ClpXP was unable to degrade Spx in vitro might be explained if clpX negatively regulates production of Spx at the level of spx transcription. To test this possibility, a transcriptional spx-lacZ fusion was constructed as described in Materials and Methods and its expression in wild-type and clpX cells was examined. β-Galactosidase activity from spx-lacZ was expressed constitutively in wild-type cells during exponential and stationary phases of growth in CM. Transcription of spx in the clpX mutant was slightly affected by the clpX mutation (Fig. 4A). However, the increase in the spx transcription in the clpX mutant was two- to threefold at the most, which cannot fully explain the large difference in Spx level between the wild type and the clpX mutant.
FIG. 4.
Expression of spx-lacZ. (A) Cells were grown in CM. Time zero, onset of the stationary phase. ○, ORB4059 (wild type); •, ORB4065 (clpX::spc). (B) ORB4059 (wild type) was grown in CM (○), DSM (•), or 2× YT (□). β-gal. act., β-galactosidase activity.
We also examined whether the expression of spx is medium dependent. Levels of expression of spx when cells were grown in CM and DSM were similar, and the expression is higher in stationary-phase cultures grown in 2× YT (Fig. 4B). This result suggests that the smaller amount of Spx in the clpX and clpP mutant cells grown in DSM than in those grown in CM is attributable to posttranscriptional regulation.
ClpXP is responsible for low levels of Spx in Pspac-spx cells.
The results described above suggested that higher concentrations of Spx in the clpX and clpP mutants are probably due to loss of proteolysis. To confirm this, spx was placed under the control of IPTG-inducible promoter Pspac (50) and the effect of the clpX, clpP, and clpC mutations on the amount of Spx was examined. In these strains, Pspac-spx was integrated at the amyE locus and the original spx gene was mutated by insertion of a Neor gene cassette. When Pspac-spx strains with or without the clp mutations were grown in CM in the presence of 1 mM IPTG, Spx proteins were hardly detected in the wild type and the clpC mutant (Fig. 3B). In contrast, the amounts of Spx in the clpX and clpP mutant strains were dramatically increased. From these results we concluded that ClpXP is required for Spx degradation. The Spx level in the Pspac-spx clpX mutant was higher when cells were grown in CM than when they were grown in DSM (Fig. 3C). However, the Spx concentration was significantly elevated even in the DSM cultures in the absence of clpX. This result suggests that both ClpP and another protease(s) are responsible for the degradation of Spx in B. subtilis cells grown in DSM.
DISCUSSION
Spx is highly conserved in gram-positive bacteria and was identified as the site of clpP suppressor mutations in Lactococcus lactis (9) and of clpX (and clpP) mutations in B. subtilis (28). The accumulated Spx in B. subtilis is harmful to cell growth on certain media and to developmental pathways such as genetic competence and sporulation. This study indicated that the concentration of Spx is controlled by multiple pathways of ClpP-dependent proteolysis.
Western blot analysis in a previous paper (28) and in this study indicated that Spx markedly accumulated in the clpP and clpX mutants grown in CM or 2× YT. Using the Pspac-spx construct, we demonstrated that the amount of Spx is regulated either by ClpXP-dependent proteolysis or, less likely, by an effect on spx translation that is indirectly influenced by ClpXP. The poor growth of the clpX and clpP mutants on minimal media (such as CM) and rich media (such as LB medium and 2× YT) is caused by the increased amount of Spx under these growth conditions. In contrast, the clpX and clpP mutants grow relatively well on DSM, where Spx accumulates to considerably lower levels. The growth of clpP spx and clpX spx mutants on CM and LB medium is similar to that of the wild type. Furthermore the clpX and clpP mutants carrying the Pspac-spx construct grew well on CM and LB medium in the absence of IPTG; however, growth was severely reduced in the presence of IPTG (data not shown). The growth of the Pspac-spx clpC strain was not affected by IPTG. Taken together, these results are consistent with the hypothesis that accumulation of Spx is unfavorable to normal cell growth. It also indicates that the level of Spx in the clpP strain is not high enough to affect growth in DSM but is sufficient to inhibit sporulation because the spx mutation in the clpP background only partially restored sporulation (28). Our results also suggested that both ClpP and another protease(s) are involved in the degradation of Spx in DSM cultures.
Although Western blot analysis clearly showed that both clpX and clpP are responsible for maintaining low Spx concentrations, Spx is not degraded by ClpXP in vitro. In fact, at present, there is no experimental evidence demonstrating that B. subtilis ClpXP is proteolytically active in vitro, although in vivo studies have suggested various proteins as substrates. Comparison of proteins by two-dimensional gel electrophoresis showed that several proteins, such as GroEL, PpiB, PyrK, SucD, YhfP, YqkF, YugJ, and YvyD, were more abundant in clpP and clpX mutants than in the wild-type cells, suggesting that some of them might be the substrates for ClpXP (10). CtsR, the negative regulator of class III heat-inducible genes such as clpP, clpE, and clpC, is degraded by ClpXP under nonstress conditions (6). Finally evidence that SsrA-tagged proteins in B. subtilis are degraded by ClpXP was reported (48). We assume that the degradation of Spx by ClpXP requires another protein, which may work as an adapter or a stimulator. In E. coli, ribosome-associated protein SspB, by binding to SsrA-tagged proteins, enhances proteolysis by ClpXP but not by ClpAP (24). In the degradation of the stationary-phase sigma subunit of E. coli, RpoS, RssB acts as an adapter for ClpXP-dependent RpoS proteolysis (3, 51). We are currently searching for a protein that may be involved in ClpXP-dependent proteolysis of Spx in B. subtilis.
The in vivo studies clearly indicated that ClpXP, not ClpCP, is responsible for maintaining low intracellular levels of Spx. In contrast, in vitro proteolysis experiments using purified proteins revealed that ClpCP, but not ClpXP, degraded Spx. The degradation of Spx by ClpCP requires adapter protein MecA or YpbH, a putative paralog of MecA. Although several ClpCP target proteins were suggested by in vivo studies (see the introduction), only two cases have been analyzed in vitro. ComK and ComS are the best-studied targets of ClpCP proteolysis in B. subtilis (43). The degradation of ComK and ComS by ClpCP also requires MecA. The other substrate of ClpCP is CtsR. As mentioned above, CtsR is degraded by ClpXP under nonstress conditions; however, it becomes a target of ClpCP upon heat shock (21). In vitro proteolysis of CtsR by ClpCP was observed in the absence of MecA. It has yet to be determined how MecA presents ComK, ComS, and Spx to ClpCP and how CtsR is directly recognized by ClpCP. There is a similar example from studies of ClpXP proteolysis in E. coli. The E. coli stationary sigma factor, σs (39), and λ O (49) are substrates for ClpXP. Whereas the proteolysis of σs requires response regulator RssB both in vivo and in vitro, the ClpXP-dependent proteolysis of λ O does not require RssB (51). Interestingly, RssB can also act like an anti-σs factor when the cellular RssB/σs ratio is elevated and proteolysis is reduced (3). This is analogous to the ternary complex formed by MecA, ClpC, and ComK, which sequesters ComK. We have recently shown that Spx interacts with MecA, thus forming the quaternary complex and enhancing ComK binding to ClpC-MecA (30).
The contradictory in vitro and in vivo results of studies aimed at determining the role of ClpCP in Spx proteolysis are puzzling. One possibility is that the result of the in vitro proteolysis is an artifact. However, we think that this is unlikely because the proteolysis requires either MecA or YpbH. Another plausible explanation is that an unidentified protein is negatively involved in ClpCP-dependent proteolysis of Spx in vivo. The protein could be an inhibitor that specifically binds to Spx and protects it from proteolysis by ClpCP or could be the putative adapter for ClpXP that binds to Spx. This interaction could render Spx resistant to proteolysis by ClpCP.
Although the increased amount of Spx is detrimental to cells, Spx is likely needed under certain conditions. The spx gene resides downstream of yjbC, and these two genes probably constitute an operon (2). The transcription of the yjbC and spx genes was induced by heat, salt, and ethyl alcohol stress (36). A yjbC mutant and, to a lesser extent, an spx mutant exhibited a salt-sensitive phenotype (36). It is possible that Spx is produced under conditions of extreme stress as one way to suppress developmental processes and allow the cell to devote its energy to dealing with the cellular damage caused by harsh conditions. It is apparent from our studies and those of others that Spx is involved, positively and negatively, in various cellular functions in gram-positive bacteria.
Acknowledgments
We thank Alan Grossman and Rich Losick for their kind gift of a plasmid and a strain.
This work was supported by grant GM45898 from the National Institutes of Health.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal ‘addiction module' regulated by 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, G., E. Klauck, and R. Hengge-Aronis. 2000. The response regulator RssB, a recognition factor for σs proteolysis in Escherichia coli, can act like an anti-σs factor. Mol. Microbiol. 35:657-666. [DOI] [PubMed] [Google Scholar]
- 4.Chong, S., F. B. Mersha, D. G. Comb, M. E. Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus, and M. Q. Xu. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192:271-281. [DOI] [PubMed] [Google Scholar]
- 5.Derré, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 6.Derré, I., G. Rapoport, and T. Msadek. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37οC. Mol. Microbiol. 38:335-347. [DOI] [PubMed] [Google Scholar]
- 7.D'Souza, C., M. M. Nakano, and P. Zuber. 1994. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 91:9397-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 9.Frees, D., P. Varmanen, and H. Ingmer. 2001. Inactivation of a gene that is highly conserved in gram-positive bacteria stimulates degradation of non-native proteins and concomitantly increases stress tolerance in Lactococcus lactis. Mol. Microbiol. 41:93-103. [DOI] [PubMed] [Google Scholar]
- 10.Gerth, U., E. Krüger, I. Derré, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 11.Gerth, U., A. Wipat, C. R. Harwood, N. Carter, P. T. Emmerson, and M. Hecker. 1996. Sequence and transcriptional analysis of clpX, a class-III heat-shock gene of Bacillus subtilis. Gene 181:77-83. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman, M. E., and W. A. Hendrickson. 2000. Protein folding and unfolding by Escherichia coli chaperones and chaperonins. Curr. Opin. Microbiol. 3:197-202. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamoen, L. W., H. Eshuis, J. Jongbloed, G. Venema, and D. van Sinderen. 1995. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 15:55-63. [DOI] [PubMed] [Google Scholar]
- 15.Härtl, B., W. Wehrl, T. Wiegert, G. Homuth, and W. Schumann. 2001. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J. Bacteriol. 183:2696-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoskins, J. R., S. K. Singh, M. R. Maurizi, and S. Wickner. 2000. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. USA 97:8892-8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny, T. J., and C. P. Moran, Jr. 1991. Genetic evidence for interaction of σA with two promoters in Bacillus subtilis. J. Bacteriol. 173:3282-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong, L., K. J. Siranosian, A. D. Grossman, and D. Dubnau. 1993. Sequence and properties of mecA, a negative regulator of genetic competence in Bacillus subtilis. Mol. Microbiol. 9:365-373. [DOI] [PubMed] [Google Scholar]
- 19.Krüger, E., U. Völker, and M. Hecker. 1994. Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance. J. Bacteriol. 176:3360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krüger, E., E. Witt, S. Ohlmeier, R. Hanschke, and M. Hecker. 2000. The Clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 182:3259-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krüger, E., D. Zühlke, E. Witt, H. Ludwig, and M. Hecker. 2001. Clp-mediated proteolysis in gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehnherr, H., and M. B. Yarmolinsky. 1995. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levchenko, I., L. Luo, and T. A. Baker. 1995. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 9:2399-2408. [DOI] [PubMed] [Google Scholar]
- 24.Levchenko, I., M. Seidel, R. T. Sauer, and T. A. Baker. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 289:2354-2356. [DOI] [PubMed] [Google Scholar]
- 25.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation of Bacillus subtilis. Mol. Microbiol. 33:415-428. [DOI] [PubMed] [Google Scholar]
- 26.Msadek, T., V. Dartois, F. Kunst, M.-L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 27.Msadek, T., F. Kunst, and G. Rapoport. 1994. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc. Natl. Acad. Sci. USA 91:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano, M. M., F. Hajarizadeh, Y. Zhu, and P. Zuber. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383-394. [DOI] [PubMed] [Google Scholar]
- 29.Nakano, M. M., M. A. Marahiel, and P. Zuber. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano, M. M., S. Nakano, and P. Zuber. Spx (YjbD), a negative effector of competence in Bacillus subtilis, enhances ClpC-MecA-ComK interaction. Mol. Microbiol., in press. [DOI] [PubMed]
- 31.Nakano, M. M., Y. Zhu, J. Liu, D. Y. Reyes, H. Yoshikawa, and P. Zuber. 2000. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase α can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol. Microbiol. 37:869-884. [DOI] [PubMed] [Google Scholar]
- 32.Nanamiya, H., Y. Ohashi, K. Asai, S. Moriya, N. Ogasawara, M. Fujita, Y. Sadaie, and F. Kawamura. 1998. ClpC regulates the fate of a sporulation initiation sigma factor, σH protein, in Bacillus subtilis at elevated temperatures. Mol. Microbiol. 29:505-513. [DOI] [PubMed] [Google Scholar]
- 33.Nanamiya, H., K. Takahashi, M. Fujita, and F. Kawamura. 2000. Deficiency of the initiation events of sporulation in Bacillus subtilis clpP mutant can be suppressed by a lack of the Spo0E protein phosphatase. Biochem. Biophys. Res. Commun. 279:229-233. [DOI] [PubMed] [Google Scholar]
- 34.Ogura, M., L. Liu, M. Lacelle, M. M. Nakano, and P. Zuber. 1999. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol. Microbiol. 32:799-812. [DOI] [PubMed] [Google Scholar]
- 35.Pan, Q., D. A. Garsin, and R. Losick. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-σ factor in B. subtilis. Mol. Cell 8:873-883. [DOI] [PubMed] [Google Scholar]
- 36.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp proteases: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 38.Schumann, W., M. Hecker, and T. Msadek. 2001. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 39.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Marin. 1996. Regulation of Escherichia coli starvation sigma factor (σs) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, S. K., R. Grimaud, J. R. Hoskins, S. Wickner, and M. R. Maurizi. 2000. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. USA 97:8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, M. W., S. K. Singh, and M. R. Maurizi. 1994. Processive degradation of proteins by the ATP-dependent Clp proteases from Escherichia coli: requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 269:18209-18215. [PubMed] [Google Scholar]
- 42.Tu, G.-F., G. E. Reid, J.-G. Zhang, R. L. Moritz, and R. J. Simpson. 1995. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem. 270:9322-9326. [DOI] [PubMed] [Google Scholar]
- 43.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turgay, K., L. W. Hamoen, G. Venema, and D. Dubnau. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11:119-128. [DOI] [PubMed] [Google Scholar]
- 45.van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. ComK encodes the competence transcription factor, the key regulator protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 46.Wickner, S., S. Gottesman, D. Skowyra, J. Hoskins, K. McKenney, and M. R. Maurizi. 1994. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 91:12218-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1892. [DOI] [PubMed] [Google Scholar]
- 48.Wiegert, T., and W. Schumann. 2001. SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol. 183:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wojtkowiak, D., C. Georgopoulos, and M. Zylicz. 1993. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J. Biol. Chem. 268:22609-22617. [PubMed] [Google Scholar]
- 50.Yansura, D. G., and D. J. Henner. 1984. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 81:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets σs for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]