Abstract
Purpose
Methionine-sulfoxide reductases are unique, in that their ability to repair oxidized proteins and MsrA, which reduces S-methionine sulfoxide, can protect lens cells against oxidative stress damage. To date, the roles of MsrB1, -B2 and -B3 which reduce R-methionine sulfoxide have not been established for any mammalian system. The present study was undertaken to identify those MsrBs expressed by the lens and to evaluate the enzyme activities, expression patterns, and abilities of the identified genes to defend lens cells against oxidative stress damage.
Methods
Enzyme activities were determined with bovine lens extracts. The identities and spatial expression patterns of MsrB1, -B2, and -B3 transcripts were examined by RT-PCR in human lens and 21 other tissues. Oxidative stress resistance was measured using short interfering (si)RNA–mediated gene-silencing in conjunction with exposure to tert-butyl hydroperoxide (tBHP) and MTS viability measurements in SRA04/01 human lens epithelial cells.
Results.
Forty percent of the Msr enzyme activity present in the lens was MsrB, whereas the remaining enzyme activity was MsrA. MsrB1 (selenoprotein R, localized in the cytosol and nucleus), MsrB2 (CBS-1, localized in the mitochondria), and MsrB3 (localized in the endoplasmic reticulum and mitochondria) were all expressed by the lens. These genes exhibit asymmetric expression patterns between different human tissues and different lens sublocations, including lens fibers. All three genes are required for lens cell viability, and their silencing in lens cells results in increased oxidative-stress–induced cell death.
Conclusions.
The present data suggest important roles for both MsrA and -Bs in lens cell viability and oxidative stress protection. The differential tissue distribution and lens expression patterns of these genes, coupled with increased oxidative-stress–induced cell death on their deletion provides evidence that they are important for lens cell function, resistance to oxidative stress, and, potentially, cataractogenesis.
The eye lens consists of a single layer of epithelial cells that overlie concentric layers of differentiated and elongated fiber cells. Damage to lens cells and their components results in protein aggregation associated with age-related cataract, an opacity of the eye lens that is the major cause of blindness worldwide.1 Oxidative stress is believed to play a major role in cataract formation, since oxidation of proteins results in loss of protein function.2–5 One major modification resulting from oxidative stress is oxidation of methionine, which affects a multitude of biological functions through the direct inactivation of proteins.6 The content of oxidized methionine residues increases in the lens with age, and in cataracts as much as 60% of the total membrane-bound protein methionine is found as methionine sulfoxide (Met (O)), suggesting a possible link between methionine oxidation and age-related cataract.7–9
Reaction of methionine with reactive oxygen species (ROS) results in the formation of two epimers of methionine sulfoxide, referred to as Met-R-(O) and Met-S-(O). Unlike most protein modifications, these oxidations are reversible through the action of the methionine sulfoxide reductase (Msr) family of enzymes that exhibit two specific activities: MsrA and -B. MsrA is stereospecific for Met-S-(O), and MsrB is specific for Met-R-(O).10,11 To date, the only naturally occurring system for Met (O) reduction in cells is reduced thioredoxin.6 In mammals, there is only one gene encoding MsrA, but there are at least three MsrB genes called MsrB1 (selenoprotein R), MsrB2 (CBS-1) and MsrB3.12,13 Previous studies have indicated that MsrB1 is localized to the cell nucleus and cytoplasm, MsrB2 is localized to the mitochondria, and MsrB3 resides in the endoplasmic reticulum (ER).12 MsrA has been found in both mitochondria and cytoplasm.
The Msr family of genes is conserved throughout evolution and influences longevity in species, including Drosophila,14 mice,15 and yeast.16 For instance, overexpression of MsrA in transgenic flies renders them more resistant to oxidative stress and dramatically increases their lifespan.14 Overexpression of MsrA in yeast can extend lifespan and protect against oxidative stress in yeast,16,17 human T-lymphocytes17 and PC12 cells.18 By contrast, deletion of the MsrA gene in mice results in increased sensitivity to oxidative stress, a shortened lifespan, and neurologic impairment.15
In previous studies, we have demonstrated that overexpression of MsrA protects lens cells against oxidative stress, whereas deletion of MsrA renders them more resistant to oxidative stress and decreases cell viability in the absence of oxidative stress.19 In contrast to MsrA, no functional studies have been reported on any of the MsrB family of genes in lens or other mammalian systems.
In the present study, we examined the levels and spatial expression patterns of the MsrB gene family in the human lens and tested the viability of lens cells in which these genes were silenced when exposed to oxidative stress. The data demonstrate that the lens contains both MsrA and -B enzyme activity, show that all three MsrB genes are expressed by the human lens, and show that silencing of each of the three MsrB genes results in loss of lens cell viability and decreased resistance of lens cells to oxidative stress exposure. The varied expression of these genes in different tissues, lens sublocations, and cellular sublocations, coupled with their requirement for oxidative stress defense suggests that they play important roles in the repair of oxidized lens proteins and, potentially, in cataract formation.
Materials and Methods
SRA01/04 Cell Culture
All cells used in this work were transformed human lens epithelial cells (SRA01/04; HLE cells)20 grown and cultured in Dulbecco’s modified Eagle’s medium supplemented with 15% fetal bovine serum, gentamicin (50 U/mL), penicillin-streptomycin antibiotic mix (50 U/mL), and amphotericin B(5 μL/mL, Fungizone; all from Invitrogen, Gaithersburg, MD) at 36.5°C in the presence of 5% CO2.
Msr Activity in Bovine Lens Extracts
Fifty bovine eyes were obtained from Pel-Freeze (Rogers, AR) and were microdissected into lens epithelia, cortical fibers, and nuclear fibers. After microdissection, cell-free extracts were prepared from each separate section of the lens by sonication, centrifugation, and collection of the soluble fraction. Protein concentrations were determined by the Bradford assay, with concentrations of 3.85 mg/mL for the epithelial and 65 mg/mL for the fiber extracts. Msr activity was measured by a modification of a previously described procedure.21 The reactions (30 μL) contained 22 μL of cell extract, 100 mM Tris-HCl (pH 7.4), 15 mM dithiothreitol (DTT), and 5 nanomoles 3H-N-acetyl Met-R, S-(O) (130 cpm/picomole). N-acetylornithine (10 mM) was also added, to prevent deacetylation of the substrate. The 3H-N-acetyl methionine formed was assayed after extraction into ethyl acetate. To determine what percentage of the Msr activity in the cell was due to MsrA or -B, a competitive inhibitor of MsrA, S-methyl-p-tolyl sulfoxide (Sigma-Aldrich, St. Louis, MO), was added to incubations. In separate experiments, this inhibitor was determined to be greater than 95% effective at inhibiting purified MsrA at a ratio of 120:1 inhibitor-substrate, with no effect on MsrB activity (data not shown). The remaining Msr activity detected in the presence of the competitor was attributed to MsrB.
Msr Tissue Distribution
Tissue distribution of Msr mRNA was surveyed by semiquantitative RT-PCR, using total RNA from 21 different human tissues (BD Biosciences, Franklin Lakes, NJ). Clear human lenses were obtained 12 to 24 hours after death and microdissected or stored as whole lenses 24 to 36 hours after death. Microdissected lens components or whole lenses were stored at liquid nitrogen temperature before RNA extraction and RT-PCR analysis. The average age of lenses used in this study was 63 ± 10 years (SD). The lenses were approximately 55% female. RT-PCR was performed using 100 ng of total RNA. The primer sequences are shown in Table 1. GAPDH was used as a control gene and was amplified for 25 PCR cycles with an annealing temperature of 60°C. Msr transcripts were amplified for 33 PCR cycles with an annealing temperature of 56°C for MsrA and -B1 and 52°C for MsrB2 and -B3. All reactions were conducted with reagents contained in a commercial RT-PCR kit (One-Step; Invitrogen) according to the manufacturer’s protocol. Products were separated by gel electrophoresis and sequenced to ensure authenticity. Product formation was found to be linear over the number of PCR cycles indicated.
Table 1.
Primer Sequences
| RT-PCR Primer | Product Size (bp) | Primer Sequence |
|---|---|---|
| MsrA forward | 329 | 5′-AGTACCTGAGCAAGAACCCCA-3′ |
| MsrA reverse | 5′-TCACTCAGACCCCAGAAGACA-3′ | |
| MsrB1 forward | 328 | 5′-GACGTTACACCCTCACCTT-3′ |
| MsrB1 reverse | 5′-AGCTACTTCCGCACAGATT-3′ | |
| MsrB2 forward | 308 | 5′-CAAGGAAGCAGGAATGTATCA-3′ |
| MsrB2 reverse | 5′-ATGGTCAGTGTTTCCTTGGTTT-3′ | |
| MsrB3 forward | 328 | 5′-CCGGGTCGTGTAGGGATAAA-3′ |
| MsrB3 reverse | 5′-TGAGCACCACACTGAGAGCA-3′ | |
| GAPDH forward | 600 | 5′-CCACCCATGGCAAATTCCATGGCA-3′ |
| GAPDH reverse | 5′-TCTAGACGGCAGGTCAGGTCCACC-3′ | |
| siRNA | siRNA Sequence | |
| MsrA: siRNA A-8 | r(CAAAGUACAAAGGAAUUUAUU) and (UAAAUUCCUUUGUACUUUGUG) | |
| MsrB1/SelX: siRNA B1-1 | r(GCGUCCGGAGCACAAUAGA)d(TT) and r(UCUAUUGUGCUCCGGACGC)d(TT) | |
| MsrB2/CBS1: siRNA B2-1 | r(GUUCUACGUCACAAGAGAA)d(TT) and r(UUCUCUUGUGACGUAGAAC)d(TG) | |
| MsrB3: siRNA B3-1 | r(GUGCCUUUGAAGGAGAAUA)d(TT) and r(UAUUCUCCUUCAAAGGCAC)d(TT) |
RT-PCR primer sequences used for MsrA, -B1, -B2, and -B3 and GAPDH gene expression. SiRNA sequences used to generate MsrA, -B1, -B2, and -B3 gene silencing.
Analysis of Msr Transcript Levels in Microdissected Components of Whole Human Lenses
The relative levels of Msr transcripts were estimated between micro-dissected portions of adult human lenses by semiquantitative RT-PCR as just described. Eight clear human lenses were microdissected to remove the lens epithelium (central 8–9 mm) from the underlying fiber cells. RT-PCR was performed as described earlier for the tissue distribution, using the same primers for each gene. The products were separated by gel electrophoresis and were sequenced to ensure authenticity. Product formation was linear over the number of PCR cycles indicated.
Short Interfering RNA–Targeted Gene Silencing
Double-stranded short interfering (si)RNAs specific for each Msr were designed and manufactured (4-for-silencing system; Qiagen, Valencia, CA). HLE cells were then transfected with siRNA (Transmessenger Transfection Reagent kit; Qiagen) according to the manufacturer’s protocol. The cells were plated in six-well plates at a density of 500,000 cells per well and transfected with transfection reagent alone for mock samples and 4 μg of siRNA per well for Msr experimental samples. Four different double-stranded siRNA constructs were used. They are designated siRNAs A-8, B1-1, B2-1, and B3-1, targeted to MsrA, -B1, -B2, and -B3, respectively. Their sequences are listed in Table 1.
Cell Viability MTS Assays
A cell proliferation assay kit (Cell Titer 96 Aqueous One Solution; Promega, Madison, WI) containing the tetrazolium compound [3-(4,5-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (MTS) was used to monitor cell viability according to the manufacturer’s protocol. The final concentration of MTS added to the cells was 317 μg /μL, and the MTS color change at 490 nm was monitored with a universal plate reader (model ELX-800; Bio-Tek Instruments, Winooski, VT).
tBHP Sensitivity of Control and siRNA-Treated HLE Cells
HLE cells were plated in 96-well plates at a density of 20,000 cells per well and mock-transfected or transfected with 0.2 μg of siRNA per well 24 hours after seeding. At 48 hours after transfection, the cells were treated with increasing concentrations of tert-butyl hydroperoxide (tBHP) for 24 hours in serum-supplemented medium in sets of four identical treatments, and cell viability was monitored with the MTS assay. Mean absorbance and standard deviations for each treatment are indicated.
Results
MsrA and -B Enzyme Activity in Lens Epithelia and Fibers
Msr activity has been detected in the lens22; however, to date, the proportion of MsrA or -B enzyme activities in human lens has not been described. Figure 1 shows the time course of Msr activity in bovine lens epithelia and fiber extracts. As seen in Figure 1, significant Msr activity was present in both lens epithelia and fiber preparations, but the epithelial extracts exhibited a 10-fold increase in specific activity when compared with the fiber fraction. Individual contributions of MsrA and -B activities to the total Msr activity in these fractions were assayed by monitoring the reduction of 3H-N-acetyl met-R,S-(O), in the presence or absence of the specific competitive inhibitor of MsrA S-methyl-p-tolyl sulfoxide. As seen in Table 2, in both the lens epithelia and fiber fractions, approximately 60% of the total Msr activity was inhibited, suggesting that 40% of the Msr activity in these fractions is due to MsrB activity.
Figure 1.
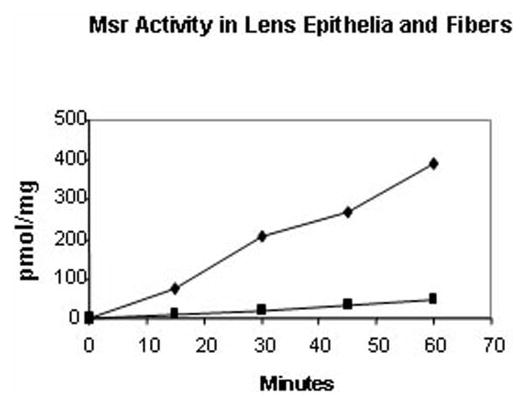
Msr activity in lens epithelia and fibers. Time course of Msr activity in lens epithelia and fiber extracts. Results are expressed as the average of two independent experiments, in which the results varied within a margin of ±5%. (▪) Lens cortical fiber extracts; (♦) lens epithelial extract.
Table 2.
MsrA and MsrB Enzyme Activity in Lens Fractions
| Enzyme | % Activity/Fiber Fraction | % Activity/Epithelial Fraction |
|---|---|---|
| MsrA | 63 | 60 |
| MsrB | 37 | 40 |
The percentage of each activity was determined by using S-methly-p-tolyl sulfoxide, a competitive inhibitor of MsrA.
Differential Distribution of MsrB Transcripts
Although the enzyme assays demonstrated that significant MsrA and -B activity was present in both lens epithelia and lens fibers, these assays cannot distinguish the activities or levels of individual MsrB gene products, because these are not substrate specific. To determine the relative levels of individual MsrBs in the lens and in other tissues, we used semiquantitative RT-PCR, to compare the transcript levels of MsrB1, -B2, and -B3 relative to MsrA in the lens and in 21 other human tissues (Fig. 2).
Figure 2.

Tissue distribution of Msrs transcripts detected by semiquantitative RT-PCR. Ethidium-bromide–stained gels showing the relative levels of indicated Msrs and GAPDH transcripts in 21 different human tissues: Co, colon; Ma, bone marrow; Ce, cerebellum; Si, small intestine; Fb, fetal brain; Fl, fetal liver; He, heart; Kd, kidney; Sc, spinal cord; Lu, lung; Pl, placenta; Pr, prostate; Sg, salivary gland; Sm, skeletal muscle; Sp, spleen; Te, testis; St, stomach; Th, thyroid; Tr, trachea; Ut, uterus; Wl, whole lens.
Consistent with previous studies,23 MsrA was present at relatively high and equal levels in the lens and most other human tissues (Fig. 2). By contrast, all MsrBs were expressed by the human lens but exhibited different levels of expression in the lens and in other human tissues. MsrB1 expression was highest in bone marrow, followed by kidney and skeletal muscle; MsrB2 expression was highest in the heart, followed by skeletal muscle and the cerebellum; and MsrB3 expression was highest in the heart followed by the colon and small intestine.
Differential Expression of MsrB Transcripts in Lens Epithelia and Fibers
To determine the relative levels of individual MsrB transcripts expressed in different parts of the lens, we prepared RNA from microdissected lens epithelium and fibers and analyzed the expression patterns of MsrB transcripts relative to MsrA by semiquantitative RT-PCR. As seen in Figure 3, MsrB1 and -B3 were approximately equally expressed between lens epithelium and fiber cells. By contrast, MsrA was expressed at significantly higher levels in the lens epithelium relative to fiber cells, whereas MsrB2 expression was greater in fiber cells. Based on densitometry measurements, approximately 73% of total Msr transcripts in lens epithelia was MsrA, 5% was MsrB1, 10% was MsrB2, and 11% was MsrB3, whereas only 13% of total Msr transcripts in lens fibers was MsrA, 6% was MsrB1, 65% was MsrB2, and 11% was MsrB3.
Figure 3.
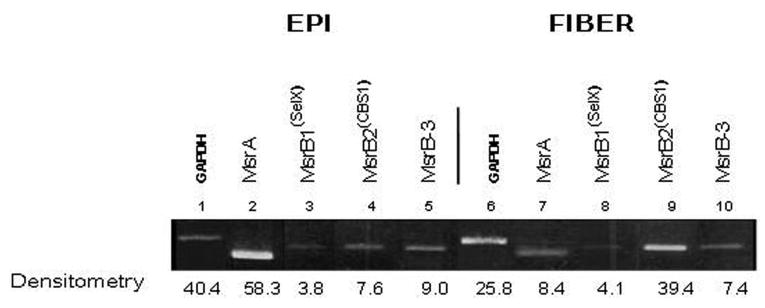
Spatial analysis of indicated Msr transcripts in microdissected human lenses. Ethidium-bromide–stained gels showing the relative levels of Msr transcripts and GAPDH between microdissected lens epithelial and fiber cells.
Effect of Silencing of Individual MsrB Genes in Lens Cells on Cell Viability and Resistance to Oxidative Stress
Previously, we showed that MsrA is necessary for human lens cell viability and provides resistance to oxidative stress in cultured lens cells.19 To examine the role of individual MsrB genes on lens cell viability and resistance to damage from oxidative stress, we used siRNA-mediated gene-silencing to suppress the levels of specific MsrB transcripts from lens cells, as shown in Figure 4A, and subsequently measured the viability of the gene-silenced cells in the presence or absence of increasing tBHP concentrations (Fig. 4B). Silencing of all Msr genes resulted in decreased lens cell viability, even in the absence of tBHP (compare untransfected [U] cells with gene-silenced cells not exposed to tBHP; 0; Fig. 4B). Silencing of the Msr genes individually resulted in increased tBHP-induced cell death, suggesting that these genes all provide oxidative stress protection to lens epithelial cells. The most dramatic effect was observed with MsrA consistent with oxidative stress producing a 50:50 mixture of S- and R-methionine sulfoxides and the fact that, in contrast to the functionally redundant MsrBs, all of which reduce R-methionine sulfoxides, only MsrA can reduce methionine-S-sulfoxides.
Figure 4.
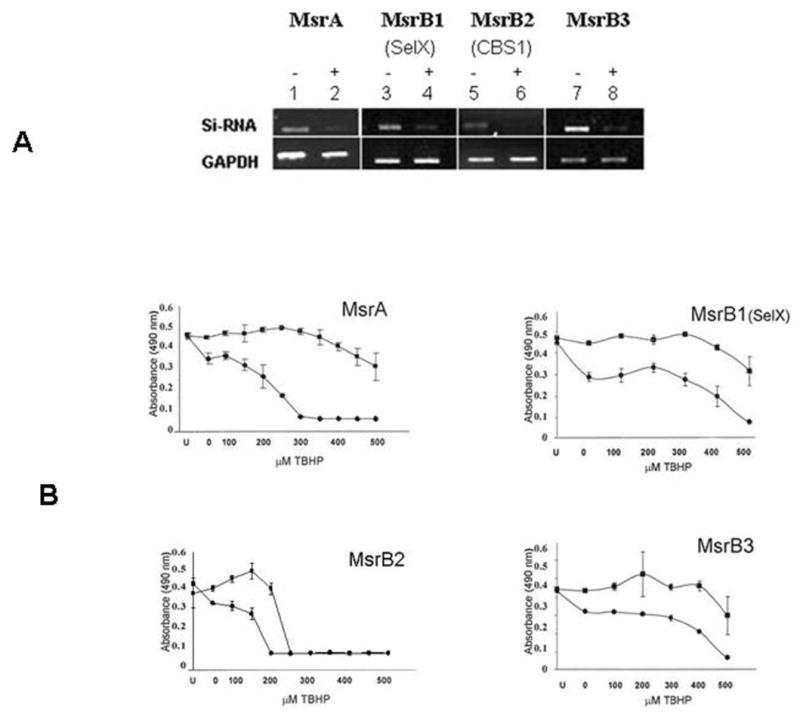
siRNA-mediated gene silencing of MsrA and -Bs in HLE cells. (A) Ethidium-bromide–stained gel showing MsrA- and -B-specific gene suppression 48 hours after transfection relative to mock transfected control cells. (B) Representative graphs depicting decreased resistance to tBHP stress 48 hours after transfection. U, untransfected cells; 0, siRNa-transfected control cells not exposed to tBHP. tBHP treatments were conducted for 24 hours in serum-supplemented medium and cell viability measured by MTS absorbance readings. (▪) Mock-transfected cells; (•) siRNA-transfected cells. Error bars represent standard deviations of four separate experiments.
Discussion
The results in the present study provide evidence that the lens contains both MsrA and -B enzyme activity and that all three MsrB genes are important for lens cell viability and oxidative stress resistance. We also found asymmetric distribution of MsrB transcripts between the lens and other human tissues, and we showed that MsrA is the predominant Msr expressed by the lens epithelium, whereas MsrB2 is the predominant Msr expressed by the lens fibers.
Previous studies have detected the presence of Msr activity in human and cow lenses22; however, in these studies, a substrate that does not distinguish between R-and S-methionine sulfoxide was used. In the present report, we used a stereo specific enzyme inhibitor to demonstrate that approximately 40% of lens Msr activity is due to MsrB activity. Although our enzyme assays cannot distinguish the individual activities of the separate MsrB enzymes, because they act on the same substrates, our semiquantitative RT-PCR data demonstrate that all three Msr genes are expressed by the lens, although they exhibit different abundances in different lens sublocations. In contrast to MsrA, which is evenly distributed in the different tissues examined, individual MsrB genes were differentially expressed between different tissues. This could indicate different roles for these genes in these tissues.
Antibodies for MsrBs are not available, and therefore corresponding protein levels could not be determined. Although we are confident that Msr protein levels parallel the mRNA levels, we cannot rule out the possibility that differences between mRNA and protein levels exist in the gene expression or gene-silencing studies.
Because only MsrA acts on the S-form of methionine sulfoxide, whereas all three MsrB genes act on the R-form, it is not surprising that MsrA would be ubiquitously expressed and the MsrB genes asymmetrically expressed. Previous studies have demonstrated that MsrA is expressed in multiple tissues, including the kidney, retinal pigmented epithelium, brain, blood, and alveolar macrophages.23 In the lens, MsrA transcripts were detected in the epithelium and nuclear fibers.19
MsrB1 and -B2 transcripts have been detected in liver kidney, heart skeletal muscle, and brain.24 Subcellular localization in mammalian cells indicated that MsrB1 was localized to the cytoplasm and nucleus. MsrB2 was localized to the mitochondria and MsrB3 to the ER, suggesting specialized subcellular roles for these proteins.12 Like MsrA,19 all three MsrB proteins were found in lens fibers, suggesting that both MsrA and -Bs participate in protein repair of both S- and R-methionine sulfoxides formed in lens fibers. MsrA has been shown to play a role in defense against oxidative stress in multiple cell systems, including human lens cells.19 In the present study, we provide evidence that all three MsrB genes are also important for cell viability and oxidative stress protection of lens cells.
MsrA and -B2 are localized to the mitochondria, and it is likely that these Msrs are important in the maintenance of mitochondrial function through direct scavenging of reactive oxygen species produced during mitochondrial respiration. Approximately 0.1% to 1% of respiratory oxygen is estimated to form reactive oxygen species during normal respiration25 and mitochondria are a major target for reactive oxygen species. It is interesting to note that both MsrA and -B2 were the predominant transcripts expressed in lens epithelia and lens fibers, respectively, suggesting that maintenance of mitochondrial function is important for these lens components.
MsrB1 and MsrB3 are localized to the cytosol and the ER,12 respectively, suggesting that they play specialized roles in these cellular compartments that could include repairing damage to cytosolic proteins and proteins in the ER, including newly synthesized proteins. These transcripts were detected at high levels in the lens and in bone marrow, intestine, heart, kidney, colon, and skeletal muscle, all of which are tissues with high levels of protein synthesis that could require coordinated repair from oxidative stress.
Although the targets for Msr action in the lens have yet to be defined, it has been shown that methionine oxidation of γ-crystallin results in loss of chaperone activity,2,26 which could play a role in cataract formation.27 Other likely targets for Msr repair are the γ-crystallins which are rich in methionine residues and are one of the first lens proteins to aggregate on cataract formation.28 MsrA function depends on the reducing system, and NADPH levels have been shown to decrease rapidly on cataract formation,29 which could reduce the ability of Msrs to repair proteins damaged through oxidation.
Regardless of the exact roles of the individual MsrB genes in lens cells, the present study provides evidence that these genes are important for the maintenance of lens cell viability and resistance to oxidative stress damage. These properties of Msrs, coupled with their presence in lens fibers, suggests that they play important roles in the repair of oxidized lens proteins and that loss of their normal activities is likely to contribute to cataract and other age-related diseases.
Acknowledgments
The authors thank Venkat Reddy for providing the SRA04/01 lens epithelia cells and the West Virginia Eye Bank and the Lions Eye Bank of Oregon for providing the human lenses used in the study.
Footnotes
Supported by National Eye Institute Grant EY13022 (MK) and Grant P200415 from the State of Florida Center of Excellence in Biomedical and Marine Biotechnology.
Disclosure: M.A. Marchetti, None; G.O. Pizarro, None; D. Sagher, None; C. DeAmicis, None; N. Brot, None; J.F. Hejtmancik, None; H. Weissbach, None; M. Kantorow, None
References
- 1.Kupfer C, Underwood B, Gillen T. Leading causes of visual impairment world wide. Principles and Practice of Ophthalmology Basic Science. Philadelphia: WB Saunders; 1994;1249–1255.
- 2.Smith JB, Jiang X, Abraham EC. Identification of hydrogen peroxide oxidation sites of alpha A- and alpha B-crystallins. Free Radic Res. 1997;26:103–111. doi: 10.3109/10715769709097789. [DOI] [PubMed] [Google Scholar]
- 3.McNamara M, Augusteyn RC. The effects of hydrogen peroxide on lens proteins: a possible model for nuclear cataract. Exp Eye Res. 1984;38:45–56. doi: 10.1016/0014-4835(84)90137-4. [DOI] [PubMed] [Google Scholar]
- 4.Bodaness RS, Leclair M, Zigler JS., Jr An analysis of the H2O2-mediated crosslinking of lens crystallins catalyzed by the heme-undecapeptide from cytochrome c. Arch Biochem Biophys. 1984;231:461–469. doi: 10.1016/0003-9861(84)90409-0. [DOI] [PubMed] [Google Scholar]
- 5.Zigler JS, Jr, Huang QL, Du XY. Oxidative modification of lens crystallins by H2O2 and chelated iron. Free Radic Biol Med. 1989;7:499–505. doi: 10.1016/0891-5849(89)90025-7. [DOI] [PubMed] [Google Scholar]
- 6.Weissbach H, Etienne F, Hoshi T, et al. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- 7.Spector A. Oxidative stress-induced cataract: mechanism of action. FASEB J. 1995;9:1173–1182. [PubMed] [Google Scholar]
- 8.Truscott RJ, Augusteyn RC. Oxidative changes in human lens proteins during senile nuclear cataract formation. Biochim Biophys Acta. 1977;492:43–52. doi: 10.1016/0005-2795(77)90212-4. [DOI] [PubMed] [Google Scholar]
- 9.Garner MH, Spector A. Selective oxidation of cysteine and methionine in normal and senile cataractous lenses. Proc Natl Acad Sci USA. 1980;77:1274–1277. doi: 10.1073/pnas.77.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskovitz J, Singh VK, Requena J, Wilkinson BJ, Jayaswal RK, Stadtman ER. Purification and characterization of methionine sulfoxide reductase from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochem Biophys Res Commun. 2002;290:62–65. doi: 10.1006/bbrc.2001.6171. [DOI] [PubMed] [Google Scholar]
- 11.Grimaud R, Ezraty B, Mitchell JK, et al. Repair of oxidized proteins: identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 12.Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Escribano J, Sarfarazi M, Coca-Prados M. Identification, expression and chromosome localization of a human gene encoding a novel protein with similarity to the pilB family of transcriptional factors (pilin) and to bacterial peptide methionine sulfoxide reductase. Gene. 1999;233:233–240. doi: 10.1016/s0378-1119(99)00131-6. [DOI] [PubMed] [Google Scholar]
- 14.Ruan H, Tang XD, Chen ML, et al. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koc A, Gasch AP, Rutherford JC, Kim HY, Gladyshev VN. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and-independent components of aging. Proc Natl Acad Sci USA. 2004;101:7999–8004. doi: 10.1073/pnas.0307929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci USA. 1998;95:14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yermolaieva O, Xu R, Schinstock C, et al. Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc Natl Acad Sci USA. 2004;101:1159–1164. doi: 10.1073/pnas.0308215100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kantorow M, Hawse JR, Cowell TL, et al. Methionine sulfoxide reductase A is important for lens cell viability and resistance to oxidative stress. Proc Natl Acad Sci USA. 2004;101:9654–9659. doi: 10.1073/pnas.0403532101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsui H, Lin LR, Ho YS, Reddy VN. The effects of up- and downregulation of MnSOD enzyme on oxidative stress in human lens epithelial cells. Invest Ophthalmol Vis Sci. 2003;44:3467–3475. doi: 10.1167/iovs.02-0830. [DOI] [PubMed] [Google Scholar]
- 21.Brot N, Werth J, Koster D, Weissbach H. Reduction of N-acetyl methionine sulfoxide: a simple assay for peptide methionine sulfoxide reductase. Anal Biochem. 1982;122:291–294. doi: 10.1016/0003-2697(82)90283-4. [DOI] [PubMed] [Google Scholar]
- 22.Spector A, Scotto R, Weissbach H, Brot N. Lens methionine sulfoxide reductase. Biochem Biophys Res Commun. 1982;108:429–434. doi: 10.1016/0006-291x(82)91884-8. [DOI] [PubMed] [Google Scholar]
- 23.Moskovitz J, Jenkins NA, Gilbert DJ, et al. Chromosomal localization of the mammalian peptide-methionine sulfoxide reductase gene and its differential expression in various tissues. Proc Natl Acad Sci USA. 1996;93:3205–3208. doi: 10.1073/pnas.93.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansel A, Jung S, Hoshi T, Heinemann SH. A second human methionine sulfoxide reductase (hMSRB2) reducing methionine-R-sulfoxide displays a tissue expression pattern distinct from hMSRB1. Redox Rep. 2003;8:384–388. doi: 10.1179/135100003225003429. [DOI] [PubMed] [Google Scholar]
- 25.Nicholls DG. Mitochondrial membrane potential and aging. Aging Cell. 2004;3:35–40. doi: 10.1111/j.1474-9728.2003.00079.x. [DOI] [PubMed] [Google Scholar]
- 26.Cherian M, Abraham EC. Diabetes affects alpha-crystallin chaperone function. Biochem Biophys Res Commun. 1995;208:675–679. doi: 10.1006/bbrc.1995.1954. [DOI] [PubMed] [Google Scholar]
- 27.Brady JP, Garland D, Duglas-Tabor Y, Robison WG, Jr, Groome A, Wawrousek EF. Targeted disruption of the mouse alpha A-crystallin gene induces cataract and cytoplasmic inclusion bodies containing the small heat shock protein alpha B-crystallin E. Proc Natl Acad Sci USA. 1997;94:884–889. doi: 10.1073/pnas.94.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown NP, Bron AJ. Lens Disorders: A Clinical Manual of Cataract Diagnosis. Oxford: Butterworth-Heineman; 1996.
- 29.Lee SM, Schade SZ, Doughty CC. Aldose reductase, NADPH and NADP+ in normal, galactose-fed and diabetic rat lens. Biochim Biophys Acta. 1985;841:247–253. doi: 10.1016/0304-4165(85)90065-0. [DOI] [PubMed] [Google Scholar]


