Abstract
Benzoyl coenzyme A (benzoyl-CoA) reductase is a key enzyme in the anaerobic metabolism of aromatic compounds catalyzing the ATP-driven reductive dearomatization of benzoyl-CoA. The enzyme from Thauera aromatica uses a reduced 2[4Fe-4S] ferredoxin as electron donor. In this work, we identified 2-oxoglutarate:ferredoxin oxidoreductase (KGOR) as the ferredoxin reducing enzyme. KGOR activity was increased 10- to 50-fold in T. aromatica cells grown under denitrifying conditions on an aromatic substrate compared to that of cells grown on nonaromatic substrates. The enzyme was purified from soluble extracts by a 60-fold enrichment with a specific activity of 4.8 μmol min−1 mg−1. The native enzyme had a molecular mass of 200 ± 20 kDa (mean ± standard deviation) and consisted of two subunits with molecular masses of 66 and 34 kDa, suggesting an (αβ)2 composition. The UV/visible spectrum was characteristic for an iron-sulfur protein; the enzyme contained 8.3 ± 0.5 mol of Fe, 7.2 ± 0.5 mol of acid-labile sulfur, and 1.6 ± 0.2 mol of thiamine diphosphate (TPP) per mol of protein. The high specificity for 2-oxoglutarate and the low Km for ferredoxin (∼10 μM) indicated that both are the in vivo substrates of the enzyme. KGOR catalyzed the isotope exchange between 14CO2 and C1 of 2-oxoglutarate, representing a typical reversible partial reaction of 2-oxoacid oxidoreductases. The two genes coding for the two subunits of KGOR were found adjacent to the gene cluster coding for enzymes and ferredoxin of the catabolic benzoyl-CoA pathway. Sequence comparisons with other 2-oxoacid oxidoreductases indicated that KGOR from T. aromatica belongs to the Halobacterium type of 2-oxoacid oxidoreductases, which lack a ferredoxin-like module which contains two additional [4Fe-4S]1+/2+ clusters/monomer. Using purified KGOR, ferredoxin, and benzoyl-CoA reductase, the 2-oxoglutarate-driven reduction of benzoyl-CoA was shown in vitro. This demonstrates that ferredoxin acts as an electron shuttle between the citric acid cycle and benzoyl-CoA reductase by coupling the oxidation of the end product of the benzoyl-CoA pathway, acetyl-CoA, to the reduction of the aromatic ring.
An increasing number of bacteria that metabolize various aromatic compounds completely to CO2 in the absence of molecular oxygen have been discovered in the past decades. Most of the known pathways of the anaerobic aromatic metabolism lead to a common central intermediate, benzoyl coenzyme A (benzoyl-CoA), which becomes reductively dearomatized by benzoyl-CoA reductase (23, 25). This key enzyme catalyzes the ATP-driven transfer of two electrons from reduced ferredoxin to benzoyl-CoA, yielding a nonaromatic conjugated cyclic diene (see Fig. 5) (4, 7). The reduction of the benzene ring is an energetically and mechanistically difficult process which requires an extremely low redox potential (∼−1.9 V, as determined for the first electron transfer to the model compound S-ethyl thiobenzoate) and probably involves radical intermediates (7, 10, 16). Benzoyl-CoA reductase overcomes energetic limitations by using a low-potential electron donor (ferredoxin) (6), by coupling the electron transfer to a stoichiometric ATP hydrolysis (two ATP/two electrons) (4, 5), and by an appropriate binding of the substrate (35). The process of ATP-driven electron transfer in benzoyl-CoA reductase can be considered analogous to the well-studied enzymatic dinitrogen reduction (27).
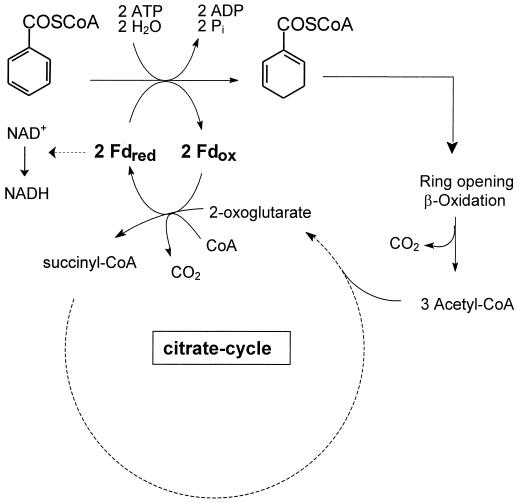
FIG. 5.
Proposed role of KGOR in the anaerobic metabolism of T. aromatica. Note that three molecules of 2-oxoglutarate are formed per molecule of benzoyl-CoA reduced. Oxidation of one molecule of acetyl-CoA would be sufficient to provide the two electrons required for the reduction of one molecule of benzoyl-CoA. The surplus reduced ferredoxin which is due to the oxidation of residual acetyl-CoA is suggested to be channeled to NAD+, catalyzed by ferredoxin:NAD+ oxidoreductase.
The biochemistry of benzoyl-CoA reductase has been studied only for the denitrifying bacterium Thauera aromatica so far. The 163-kDa enzyme consists of four different subunits with molecular masses of 49, 48, 44, and 30 kDa, resulting in an αβγδ composition (4). It contains three cysteine-ligated [4Fe-4S]1+/2+ clusters with redox potentials more negative than −500 mV (8). Based on amino acid sequence comparisons with similar enzymes (activase and 2-hydroxyglutaryl-CoA dehydratase from glutamate-fermenting Clostridium [22]), two functionally and structurally different modules can be distinguished in benzoyl-CoA reductase: (i) the 49- and 30-kDa subunits are highly similar to each other and coordinate one [4Fe-4S]1+/2+ cluster in the interface, resulting in an architecture similar to that of the Fe protein of nitrogenase, and (ii) the 48- and 44-kDa subunits form the substrate reduction module harboring two other [4Fe-4S]1+/2+ clusters. In spectroscopic studies, growing evidence was gained that ATP hydrolysis is coupled to the electron activation process, which is required for the electron transfer to the aromatic ring. During ATP hydrolysis, a [4Fe-4S]1+ cluster reversibly changed its spin state from the low-spin (S = 1/2) to the high-spin (S = 7/2) state (5, 8). Very recently, it has been demonstrated that a phosphorylated enzyme is transiently formed during catalysis (43).
In T. aromatica, a 2[4Fe-4S]1+/2+ cluster containing ferredoxin serves as the in vivo electron donor for benzoyl-CoA reductase (6). Evidence has been provided that ferredoxin is the natural electron donor for this enzyme (Km = ∼10 μM; cellular concentration of ferredoxin = ∼80 μM [6]). The gene coding for ferredoxin is located adjacent to the four genes coding for the four subunits of benzoyl-CoA reductase. Together with other genes, they form the bad gene cluster (benzoic acid degradation), as has been shown for the denitrifying T. aromatica (13) and the phototrophic bacterium Rhodopseudomonas palustris (20). Ferredoxin from T. aromatica belongs to the Chromatium vinosum type of 2[4Fe-4S] ferredoxins. This type differs from the clostridial type by an unusually low redox potential of one of the two [4Fe-4S]1+/2+ clusters (e.g., −587 mV for the ferredoxin from T. aromatica [9] and −660 mV for the ferredoxin from C. vinosum [29]).
The aim of this work was to identify and characterize the reaction that generates reduced ferredoxin for enzymatic aromatic ring reduction, to purify and characterize the enzyme catalyzing this reaction, and finally, to reconstitute purified benzoyl-CoA reductase and ferredoxin with the ferredoxin-reducing system to accomplish the reduction of the aromatic ring. No evidence for the presence of hydrogenase, formate dehydrogenase, or CO dehydrogenase activities in this bacterium was obtained. These enzymes are known to generate electrons of a potential low enough to reduce at least one of the two [4Fe-4S]1+/2+ clusters of the ferredoxin from T. aromatica. Instead, we identified 2-oxoglutarate:ferredoxin oxidoreductase (KGOR) as the ferredoxin-reducing enzyme in T. aromatica. KGOR was purified and the genes coding for the two structural subunits were cloned and sequenced. In an anaerobic assay, the electron transfer chain from 2-oxoglutarate to the aromatic ring was shown.
MATERIALS AND METHODS
Growth of bacterial cells.
T. aromatica (DSM 6984, formerly termed strain K172) was isolated in our laboratory and has been deposited in the Deutsche Sammlung von Mikroorganismen (Braunschweig, Germany) (1). It was grown anoxically at 28°C in a mineral salt medium in a 200-liter fermentor; 4-hydroxybenzoate and nitrate in a molar ratio of 1:3.5 or acetate and nitrate in a molar ratio of 1:1 served as sole sources of energy and cell carbon. Continuous feeding of the substrates, cell harvesting, and anaerobic preparation of cell extracts were carried out as described previously (11).
Cells of Escherichia coli strain XL1 Blue, MRF′ {Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)]}, and E. coli strain XLOLR, Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIqZΔM15 Tn10r)] Su− λr, were grown at 37°C in Luria-Bertani (LB) medium (40). Antibiotics were added to E. coli cultures as follows (final concentrations): ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 20 μg/ml.
Protein purification and storage.
For the preparation of cell extracts for large-scale purifications, 200 g (wet mass) of cells of T. aromatica grown on 4-hydroxybenzoate and nitrate were anaerobically suspended in 20 mM triethanolamine-HCl (pH 7.8), 4 mM MgCl2, and 1 mM dithioerythritol (DTE). French press treatment and ultracentrifugation were performed under strictly anaerobic conditions as described earlier (11). The purification included four chromatographic steps using DEAE-Sepharose and Hi Load Q-Sepharose (both from Amersham Biosciences, Inc.), Reactive Red agarose (Sigma), and Superdex-200 gel filtration (Amersham Biosciences, Inc.). If not otherwise indicated, all purification steps were performed in an anaerobic glove box with an N2/H2 (95%-5% [vol/vol]) atmosphere. Buffer A is defined as 20 mM triethanolamine-HCl (pH 7.8), 4 mM MgCl2, 10% glycerol, 0.2 mM thiamine diphosphate (TPP), and 1 mM DTE; all buffers used in the glove box were made anaerobic by flushing with nitrogen gas before use (100%, 0.2 bar, 10 min). All columns and protein fractions were cooled to 6 to 10°C during the purification. For the first chromatographic step, 250 ml of the crude extract was applied on a DEAE-Sepharose column which had been equilibrated with buffer A (for DEAE-Sepharose chromatography, all buffers additionally contained 0.5 mM sodium dithionite; column volume, 660 ml; column diameter, 5.1 cm; flow rate, 10 ml min−1). The column was washed with two bed volumes of buffer A and with two bed volumes of buffer A plus 90 mM KCl. The washing steps were followed by a 2-liter gradient from buffer A plus 90 mM KCl to buffer A plus 200 mM KCl. KGOR activity eluted at the beginning of the gradient at ∼95 mM KCl in 400 ml. For the next purification steps, a 70-ml fraction of the KGOR activity containing the DEAE pool was applied on a Hi Load Q-Sepharose column (volume, 65 ml; diameter, 2.6 cm; flow rate, 3 ml min−1) which had been equilibrated with buffer A plus 90 mM KCl. As this chromatography step was finished within less than 3 h, it could be performed under aerobic conditions without a loss of KGOR activity due to oxygen damage. KGOR was only weakly bound to Q-Sepharose under the conditions used and was eluted by continuous washing of the column with buffer A plus 90 mM KCl (∼4 bed volumes). The KGOR-activity-containing fractions were pooled and made anaerobic by flushing with N2. The third purification step was performed on a Reactive-Red-agarose column in the glove box which had been equilibrated with anoxic buffer B (same as buffer A but without DTE; volume, 70 ml; diameter, 2.2 cm; flow rate, 3 ml min−1). The column was washed with two bed volumes of buffer B, two bed volumes of buffer B plus 2 mM 2-oxoglutarate-500 mM KCl, and two bed volumes of buffer B plus 2 mM 2-oxoglutarate-1.2 M KCl. KGOR activity eluted in the last washing step (total volume, 53 ml). Prior to the next purification step, the pooled KGOR-activity-containing fractions were concentrated in microconcentrators (exclusion limit, 50 kDa; Filtron) by centrifugation. The concentrated protein (1 to 2 ml) was applied on a Superdex 200 gel filtration column which was run with buffer A plus 100 mM KCl (volume, 300 ml; diameter, 2.6 ml; flow rate, 1.5 ml min−1). The KGOR-activity-containing enzyme fractions eluted between 150 and 170 ml and were concentrated again to a final volume of 1 to 2 ml. Purified KGOR was anaerobically stored in gas-tight sealed glass tubes at −20°C for several weeks without significant loss of activity.
Oxidoreductase assays.
KGOR activity was routinely determined in a continuous spectrophotometric assay following the enzyme-, substrate-, and time-dependent reduction of oxidized benzyl viologen at λ = 600 nm (ɛBVred(600 nm) = 10 mM−1 cm−1 [21]) at 30°C. The typical assay (total volume, 420 to 470 μl) was performed in anaerobic gas-tight sealed glass cuvettes (gas phase: 95% N2-5% H2, by volume) and contained 100 mM potassium phosphate (pH 7.8), 1 mM benzyl viologen, 2.5 mM 2-oxoglutarate, 0.5 mM coenzyme A, 4 mM MgCl2, 0.025 mM sodium dithionite, and 5 to 50 μl of KGOR-activity-containing protein fraction. Additions of the substrates were made by using gas-tight glass syringes, and the reaction was usually started by adding 2-oxoglutarate. For the determination of the Km values of KGOR for 2-oxoglutarate and CoA, all substrates except the one to be tested were added in saturating concentrations and the initial rates were plotted against the substrate concentration by using the Lineweaver-Burk transformation. For testing the substrate specificity, various 2-oxoacids were added at 4 mM. Electron acceptor specificity (1 mM) was tested in continuous anaerobic spectrophotometric assays following the reduction of the acceptor (in 100 mM potassium phosphate [pH 7.8]). The following molar absorption coefficients were used: benzyl viologen, ɛ600 = 10 mM cm−1 (21); methyl viologen, ɛ600 = 13 mM−1 cm−1 (17); NADH, ɛ365 = 3.4 mM−1 cm−1; and NADPH, ɛ365 = 3.5 mM−1 cm−1. Ferredoxin from T. aromatica cells grown anaerobically on an aromatic substrate was a kind gift of M. Unciuleac (University of Freiburg); purification was as previously described (6). Aliquots of a 50 to 100 μM ferredoxin stock solution were added to the enzyme assay, resulting in a final concentration of 22 to 30 μM. Full oxidation of the ferredoxin was accomplished by titration with air by using a gas-tight syringe which was monitored by taking UV/visible (VIS) spectra (ratio of A280/A390 = 1.57 for the fully oxidized ferredoxin [5]). The time-, enzyme-, 2-oxoglutarate-, and CoA-dependent reduction of oxidized ferredoxin was recorded in the continuous spectrophotometric assay at 415 nm (ɛ(Fdox-Fdred) = 8,200 M−1 cm−1) at 30°C as previously described (6). Donor:NAD(P)+ oxidoreductase activities were determined in the buffer described above in this section. In this continuous spectrophotometric assay, the time- and enzyme-dependent oxidation of reduced methyl viologen (0.5 mM) by NAD(P)+ (1 mM) was recorded at 600 nm at 30°C.
Isotope exchange assay.
The isotope exchange reaction between 14CO2 and the C-1 carboxyl group of the 2-oxoacid substrate is a common partial reaction of 2-oxoacid oxidoreductases. Here this partial reaction was followed by monitoring the time- and KGOR-dependent formation of 14C-labeled 2-oxoglutarate. The typical assay was carried out under anaerobic conditions (95% N2-5% H2, vol/vol) in gas-tight sealed glass tubes at 30°C as described for the spectrophotometric assay (KGOR concentration, 50 μg ml−1). The assay (total volume, 0.5 ml) additionally contained 20 mM NaHCO3-3.6 kBq of [14C]Na2CO3 (specific activity, 2 MBq/μmol), whereas CoA and an electron acceptor were omitted. The reaction was started by addition of 2-oxoglutarate; samples were taken at 0, 2, 10, and 20 min and transferred to scintillation vials containing 10 μl of 3 M perchloric acid. For the removal of [12/14C]CO2, the open vials were shaken for 2 h at room temperature. Finally, the radioactivity in the samples was measured by scintillation counting.
Assay for electron transfer from 2-oxoglutarate to benzoyl-CoA.
For testing the electron transfer from 2-oxoglutarate to benzoyl-CoA, purified KGOR, ferredoxin and benzoyl-CoA reductase were passed over desalting columns (Sephadex G-25, Amersham Biosciences, Inc.; volume, ∼1 ml) which had been equilibrated with 100 mM MOPS (morpholinepropanesulfonic acid) buffer containing 10 mM MgCl2 in an anaerobic glove box (95% N2-5% H2, vol/vol). The purification of benzoyl-CoA reductase and ferredoxin was as described previously (4, 6). Due to the removal of the reductant in the desalting step, KGOR, ferredoxin, and benzoyl-CoA reductase were mostly in the oxidized state, as tested by taking UV/VIS spectra of each of the proteins. The assay (total volume, 0.5 ml) was performed in a gas-tight sealed glass tube at 37°C and contained the following components: 100 mM MOPS-KOH (pH 7.3), 10 mM MgCl2, 10 mM ATP, 1 mM coenzyme A, 2.5 mM 2-oxoglutarate, 0.16 mM benzoyl-CoA, 0.2 mg benzoyl-CoA reductase (specific activity, 0.3 μmol min−1 mg−1), 0.02 mg of ferredoxin, and 0.0325 mg of KGOR (specific activity, 4.8 μmol min−1 mg−1). Samples (50 μl) were taken at different time points, and the reaction was quenched by addition of 5 μl of 50% H2SO4. After centrifugation (10,000 × g, 10 min), the supernatants were analyzed by C18 reversed-phase high-performance liquid chromatography (HPLC) as previously described (4). In this analysis, benzoyl-CoA and the products formed (cyclohexa-1,5-diene-1-carbonyl-CoA and the hydration product 6-hydroxycyclohex-1-ene-1-carbonyl-CoA) were separated and quantitatively analyzed using a UV spectroscopic (260 nm) flow cell.
UV/VIS spectroscopy.
For recording the UV/VIS spectrum of KGOR, a fraction obtained after gel filtration was anaerobically concentrated to ∼20 μM (in the glove box as described above) and subsequently passed over a Sephadex G-25 desalting column (diameter, 1 cm; volume, ∼10 ml; Amersham Biosciences, Inc.) which had been equilibrated with anaerobic 100 mM potassium phosphate (pH 7.5). The spectra were recorded in gas-tight sealed quartz cuvettes (sample volume, 500 μl; final KGOR concentration, 2.1 μM). Reduction was carried out by addition of 5 μl of an anaerobically prepared 10 mM sodium dithionite stock solution (final concentration of dithionite in the cuvette was 0.1 mM).
Cloning and sequencing of DNA containing genes coding for KGOR.
In general, standard protocols were used for DNA isolation, cloning, transformation, amplification, and purification (40). The genes involved in benzoic acid degradation in T. aromatica are organized in the bad gene cluster and have been sequenced recently (13). From the 5′ region of this gene cluster (containing the beginning of an open reading frame [ORF] coding for a putative 2-oxoacid oxidoreductase in the opposite direction of the other genes of this cluster [13]), a 500-bp digoxigenin-11-dUTP-labeled (Roche Diagnostics) gene probe was amplified by PCR. The DNA fragment obtained was used for screening a λ-ZAP gene library of Sau3A-digested genomic DNA from T. aromatica. Genomic gene libraries were prepared according to the ZAP Express Cloning Kit instruction manual (Stratagene, Amsterdam, The Netherlands). Probes were detected with alkaline phosphatase (AP)-conjugated anti-digoxigenin Fab fragments and CSPD (Roche Diagnostics) in Southern blot analysis, followed by a 30-min exposure to an X-ray film (Hyperfilm MP; Amersham). For screening of hybridizing λ clones, labeled probe was detected with anti-digoxigenin-AP antibody, nitroblue tetrazolium chloride, and X-Phosphate (5-chloro-4-bromo-3-indolyl-phosphate toluidine salt) (Biomol, Hamburg, Germany). Using standard hybridization (60°C) and washing conditions, a 2.54-kb phagemid was obtained. DNA sequences were obtained with an ALFexpress automated sequencer. Sequence regions were extended by primer walking. Sequence comparisons were carried out using the BLAST network at the National Center for Biotechnology Information (Bethesda, Md.).
Analytical methods.
The native molecular mass of KGOR was determined by gel filtration using a Superdex 200 Hi Load FPLC column (diameter, 1.6 cm; volume, 120 ml; flow rate, 0.5 ml min−1; Pharmacia). For this purpose, KGOR (1.5 mg in 0.5 ml) was applied to the column; bovine serum albumin (67 kDa), alcohol dehydrogenase (150 kDa), catalase (232 kDa), and ferritin (440 kDa) were used as molecular mass standard proteins. N-terminal amino acid sequencing was performed by automatized Edman degradation as described elsewhere (26). Protein was determined by the method of Bradford (12) using bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (15% acrylamide) was performed as described by Laemmli (30), and visualization of the proteins was by Coomassie staining (45). The following mass standards were used: phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic acid anhydrase (30 kDa), and lysozyme (14 kDa). Iron was determined according to the method described by Lovenberg et al. (33) by using ferrous ammonium sulfate as the standard; determination of acid-labile sulfur was according to Beinert (2) using the 2[4Fe-4S] ferredoxin from T. aromatica as the standard. The determination of thiamine derivatives was performed according to the method described by Penttinen (37) and proceeded via the oxidation of TPP to thiochrome, which was quantitatively determined by fluorescence spectroscopy. The excitation wavelength was 365 nm, and the emission was recorded at 430 nm. Benzoyl-CoA was synthesized from benzoic acid anhydride and CoA (41).
Nucleotide sequence accession number.
The sequence data reported here were submitted to the GenBank database under accession no. AJ224959, which was updated on this occasion. The sequences of the DNA coding for the two subunits of T. aromatica KGOR were added to DNA sequences coding for enzymes involved in anaerobic aromatic metabolism which have been submitted previously (13).
RESULTS
Results are summarized as follows. (i) KGOR activity in extracts of T. aromatica.
The presence of KGOR activity in extracts of T. aromatica cells grown on aromatic and nonaromatic substrates was investigated. KGOR activity was determined in an anoxic spectrophotometric assay following the 2-oxoglutarate (2.5 mM)- and coenzyme A (0.5 mM)-dependent reduction of oxidized benzyl viologen (1 mM). The specific activity was 80 nmol of 2-oxoglutarate oxidized min−1 (mg of protein) −1 in cells grown anaerobically on benzoate (5 mM) and nitrate (17.5 mM). All KGOR activity was found in the water-soluble protein fraction (100,000 × g supernatant). In cells grown on nonaromatic substrates under denitrifying conditions (20 mM nitrate), KGOR activity was only 9% (when grown on 5 mM glutarate) or 2% (when grown on 20 mM acetate) of the activity determined in extracts of cells grown on benzoate. Thus, KGOR activity was induced 10- to 50-fold in T. aromatica cells during anaerobic growth on an aromatic substrate. Neither a pyruvate:ferredoxin oxidoreductase activity nor an NAD(P)+-dependent 2-oxoglutarate dehydrogenase activity was present in extracts of cells grown under denitrifying conditions on benzoate. In contrast, in cell extracts of T. aromatica grown aerobically on benzoate, the latter activity was ∼100 nmol min−1 mg−1 (32).
(ii) Purification of KGOR.
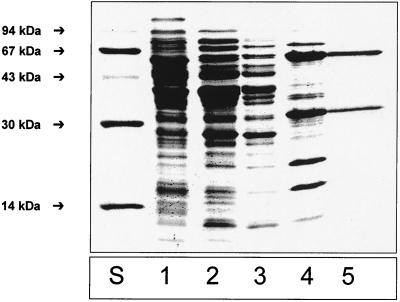
KGOR was purified from extracts of T. aromatica grown anaerobically on 4-hydroxybenzoate and nitrate with a 60-fold enrichment and a yield of 39% (Table 1). Purified KGOR was brownish and exhibited a weak sensitivity towards oxygen; approximately 50% of the activity was recovered after aerobic storage for 1 day at 4°C compared to the anaerobically stored enzyme (100% nitrogen atmosphere). Therefore, all purification steps except the rapidly performed chromatography on Q-Sepharose (exposure to oxygen was less than three hours) were performed in an anaerobic glove box under a nitrogen-hydrogen (95%-5%, by volume) atmosphere. All buffers contained 4 mM MgCl2, 1 mM DTE (expect for chromatography on Reactive Red agarose), 10% glycerol, and 0.2 mM TPP, as these additions exhibited a stabilizing effect on KGOR. A representative SDS-PAGE gel of the KGOR-activity-containing protein fractions obtained after each purification step is shown in Fig. 1. After the last purification step (gel filtration), two protein bands with a 1:1 stoichiometry (after normalization for the different molecular masses) were obtained. Approximately 50 mg of purified KGOR was obtained from 200 g (wet mass) of cells, corresponding to 1.7% of the water-soluble protein fraction in T. aromatica cell extracts.
TABLE 1.
Purification of KGOR from T. aromaticaa
| Purification step | Amt of protein (mg) | Total activity (U) | Sp act (U mg−1) | Enrichment n-fold | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 7,440 | 595 | 0.08 | 1 | 97 |
| DEAE-Sepharose | 2,280 | 615 | 0.27 | 3.3 | 100 |
| Q-Sepharose | 1,002 | 450 | 0.45 | 5.6 | 73 |
| Reactive Red | 100 | 290 | 2.9 | 36 | 49 |
| Gel filtration | 50 | 240 | 4.8 | 60 | 39 |
KGOR was purified from 200 g of cells of T. aromatica grown anaerobically on benzoate under denitrifying conditions. One unit corresponds to 1 μmol of 2-oxoglutarate oxidized to succinyl-CoA per min.
FIG. 1.
Results of SDS-PAGE (15% acrylamide) of enzyme fractions (5 to 20 μg of protein) obtained during purification of KGOR. Lanes: S, molecular mass standards; 1, supernatant (100,000 × g centrifugation) of cell extract; 2, DEAE-Sepharose fraction; 3, Q-Sepharose fraction; 4, Reactive-Red-agarose fraction; 5, gel filtration fraction (5 μg of protein).
(iii) Molecular and UV/VIS spectroscopic properties of KGOR.
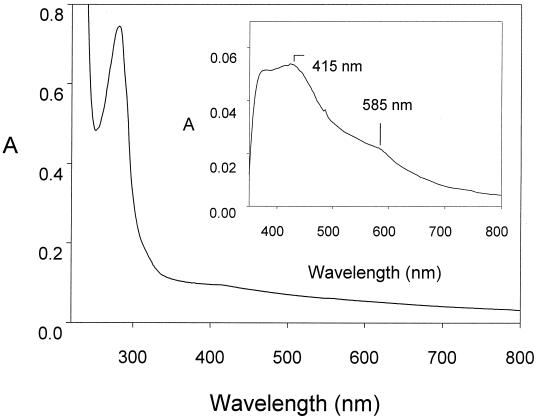
The molecular mass of KGOR was estimated to be approximately 200 ± 20 kDa by Superdex-200 chromatography (not shown). After SDS-PAGE analysis, purified KGOR consisted of two subunits with masses of 66 and 34 kDa, suggesting an (αβ)2 composition (Fig. 1). One mole of enzyme contained 8.3 ± 0.5 mol of nonheme iron, 7.5 ± 0.5 mol of acid-labile sulfur, and 1.6 ± 0.2 nmol of TPP. The numbers represent mean values of three independent determinations, and the range of deviations is indicated. The UV/VIS spectrum of KGOR isolated in the absence of a reducing agent was typical for an iron-sulfur protein, with a broad shoulder around 420 nm, and had a more unusual feature, an additional slight shoulder at 585 nm (Fig. 2). The estimated molar absorption coefficients were ɛ281 = 350,000 M−1 cm−1 and ɛ415 = 45,000 M−1 cm−1. Reduction of the enzyme by either dithionite or 2-oxoglutarate resulted in a loss of absorption from 370 to 700 nm. The difference spectra between the oxidized and the reduced spectra were highly similar for both reductants; the one obtained with dithionite is shown in the inset of Fig. 2. No evidence for the presence of a flavin cofactor was given by this difference spectrum. The difference absorption coefficient, ɛ415(oxidized-reduced KGOR), was 6,500 M−1 cm−1. It has been established that the molar difference absorption coefficient between the oxidized and reduced form of a cysteine-coordinated single [4Fe-4S]1+/2+ cluster is ∼4,000 M−1 cm−1 (33). Thus, as the iron content of KGOR indicates the presence of two [4Fe-4S]1+/2+ clusters, approximately 85% of these clusters were reduced by dithionite or 2-oxoglutarate at pH 7.8.
FIG. 2.
UV/VIS spectrum of KGOR (2.1 μM). The oxidized enzyme as isolated in 100 mM potassium phosphate (pH 7.8) is shown. The inset shows a part of the difference spectrum of oxidized KGOR minus dithionite-reduced KGOR.
(iv) Catalytic properties of KGOR.
The maximal specific activity of KGOR was 4.8 μmol min−1 mg−1 (30°C), yielding a catalytic number of 17 s−1, assuming a molecular mass of 200 kDa. The pH optimum was at 8.3 when using glycylglycine buffer. However, as the activity was threefold higher in phosphate buffer than in glycylglycine or Tris buffer (at pH 7.5 to 7.8), activity was routinely measured in 100 mM potassium phosphate buffer (pH 7.8). To test the substrate specificity of purified KGOR, several 2-oxoacids (4 mM each) and electron acceptors (0.028 mM for T. aromatica ferredoxin and 1 mM for viologens and nicotinamide adenine dinucleotides) were tested. KGOR was highly specific for 2-oxoglutarate and virtually no conversion was obtained with oxaloacetate, pyruvate, 2-oxoisovalerate, and phenylpyruvate. The enzyme used oxidized ferredoxin from T. aromatica (for details, see below) and oxidized benzyl viologen most efficiently as electron-accepting substrates, whereas oxidized methyl viologen was only reduced with half of the maximal activity. Neither NAD+ nor NADP+ served as electron acceptor for KGOR. Purified KGOR was shown to catalyze an isotope exchange between the 14CO2 and the C-1 carboxyl group of the substrate 2-oxoglutarate in the absence of CoA. The rate was 320 nmol min−1 (mg of protein)−1, which was 7% of the 2-oxoglutarate-dependent benzyl viologen reduction rate. The apparent Km values for 2-oxoglutarate and CoA were determined by varying their substrate concentration (whereas all other substrates were present in saturated concentrations). The Km values were 110 μM for 2-oxoglutarate and 290 μM for CoA (for a summary of the catalytic properties of KGOR, see Table 3). For ferredoxin, the Km could not be determined precisely by this technique, as the molar absorption coefficient (ɛFdox(415 nm) = 40,500 M−1 cm−1 [6]) enabled only the use of 10 to 30 μM Fd in the assay. Therefore, we estimated the Km for reduced ferredoxin as the concentration of reduced ferredoxin at which the half-maximal rate was observed in continuous spectrophotometric assay following the reduction of oxidized ferredoxin (30 μM). The half- maximal rate for KGOR was at approximately 10 μM Fdox and 20 μM Fdred, as estimated from the different absorption coefficients of the reduced and oxidized forms (6). It is of note that this method considers neither possible competitive effects of reduced ferredoxin nor the change of the redox potential during the reaction. Both effects could contribute to a lowering of the rate in the assay. Thus, the Km value of KGOR for ferredoxin is expected to be rather lower than the observed one.
TABLE 3.
General properties of KGOR from T. aromatica
| Property | Value |
|---|---|
| Reaction catalyzed | 2-oxoglutarate + coenzyme A + 2 Fdox → succinyl-CoA + CO2 + 2Fdred + 2 H+ |
| Electron donor | 2-oxoglutarate |
| Electron acceptors | Ferredoxin (100%), benzyl viologen (97%), methyl viologen (46%) |
| Apparent Km values | 2-oxoglutarate, 110 μM; coenzyme A, 290 μM; ferredoxin, ∼10 μM |
| Specific activity | 4.8 μmol of 2-oxoglutarate min−1 mg−1 |
| Catalytic number | 17 s−1 |
| pH optimum | 8.3 (determined in glyclglycine buffer) |
| Molecular mass | 200 ± 20 kDa (determined by gel filtration) |
| Subunits | 66 kDa (α), 34 kDa (β) (determined by SDS-PAGE) |
| Suggested composition | (αβ)2 |
| Iron | 8.3 ± 0.5 mol/mol |
| Acid-labile sulfur | 7.5 ± 0.5 mol/mol |
| TPP | 1.6 ± 0.2 mol/mol |
| Absorption coeffcients | ɛ281 = 354,000 M−1 cm−1; shoulder at 415 nm: ɛ415 = 45000 M−1 cm−1 |
| Stabilization by: | KCl (500 mM), glycerol (10%), DTE (1 mM) |
(v) Electron transfer from 2-oxoglutarate to benzoyl-CoA catalyzed by KGOR and benzoyl-CoA reductase.
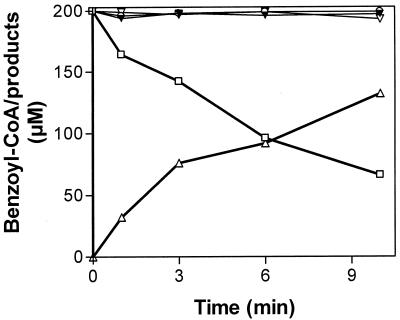
The main goal of this work was to identify the electron-generating reaction for enzymatic ring reduction. Since KGOR and benzoyl-CoA reductase used the same ferredoxin as electron acceptor/donor, an electron transfer chain from 2-oxoglutarate to benzoyl-CoA was assumed. To demonstrate this unambiguously, a discontinuous anaerobic assay was developed following the 2-oxoglutarate-, KGOR-, and ferredoxin-dependent reduction of benzoyl-CoA catalyzed by benzoyl-CoA reductase in the presence of the cosubstrates MgATP (benzoyl-CoA reductase) and coenzyme A (KGOR). HPLC analysis of the time-, enzyme-, and substrate-dependent conversion of benzoyl-CoA into the product(s) was as described recently (7); a quantitative analysis is shown in Fig. 3. Only in the presence of all components was benzoyl-CoA time dependently reduced to the product cyclohexa-1,5-diene-1-carbonyl-CoA, which was further hydrated to 6-hydroxycyclohex-1-ene-1-carbonyl-CoA. The latter product was formed by traces of a highly active dienoyl hydratase which was present as a contaminant in the benzoyl-CoA reductase preparation (7). The initial activity of benzoyl-CoA reduction was estimated to be approximately 200 nmol min−1 mg−1, which was 75% of benzoyl-CoA reductase activity with reduced methyl viologen as artificial electron donor under the conditions used. In control experiments in which 2-oxoglutarate, ferredoxin, or KGOR was omitted, virtually no benzoyl-CoA conversion was observed. This clearly indicates that the reaction measured depended on the components added and excludes the presence of alternative electron donating compounds, such as traces of dithionite.
FIG. 3.
Conversion of benzoyl-CoA to nonaromatic products by benzoyl-CoA reductase in the presence of 2-oxoglutarate/KGOR/ferredoxin as the electron-donating system. The anaerobic assay mixture (0.5 ml) contained 2-oxoglutarate, CoA, benzoyl-CoA, and MgATP, all in saturating concentrations; furthermore, ferredoxin (0.02 mg), KGOR (0.325 mg), and benzoyl-CoA reductase (0.2 mg) were added. Consumption of benzoyl-CoA (□) and formation of Δ cyclohexadiene-1-carbonyl-CoA plus 6-hydroxycyclohex-1-ene-1-carbonyl-CoA were in the presence of all components listed above. For control experiments, ferredoxin (▾), KGOR (○), or 2-oxoglutarate (▿) was omitted. Analysis of benzoyl-CoA and the products formed was by HPLC using UV/VIS (260 nm) detection. Note that the product of benzoyl-CoA reduction, cyclohexa-1,5-diene-1-carbonyl-CoA, was further converted by traces of a highly active hydratase impurity to the corresponding hydrated species. The amounts of both compounds were added up to determine the total amount of products formed from benzoyl-CoA.
(vi) Donor:NAD(P)+-oxidoreductase activities in extracts of T. aromatica grown on different substrates.
When growth was on benzoate, benzoyl-CoA reductase was considered the only catabolic enzyme using reduced ferredoxin as the electron donor. However, one benzoyl-CoA is converted to three acetyl-CoA and one CO2 by β-oxidation, yielding three 2-oxoglutarate in the citric acid cycle. Consequently, reduced ferredoxin would accumulate and KGOR activity would be blocked due to depletion of the oxidized electron acceptor. Therefore, besides benzoyl-CoA reductase, additional ferredoxin-oxidizing activities were expected in T. aromatica. To test this possibility, we determined ferredoxin:NAD(P)+ oxidoreductase activities in a spectrophotometric assay following the NAD(P)+-dependent oxidation of reduced methyl viologen. When T. aromatica cells were grown on different substrates, significant differences in the corresponding enzyme activities were found. For cells grown on benzoate, the donor:NAD+ oxidoreductase activity was five times higher than that in acetate-grown cells; a similar result was obtained when NADP+ replaced NAD+. In both cases, the activity was 10 times higher with NAD+ than with NADP+ as the electron acceptor.
(vii) N-terminal amino acid sequence and genes coding for subunits of KGOR.
To identify the genes coding for the two subunits of KGOR, a gene probe deduced from the 5′region of the bad gene cluster (accession number AJ224959) was amplified by PCR. By the use of this probe, a phagemid (2.54 kDa) was obtained which contained the DNA with the ORFs referred to as korA and korB. They were located adjacent to the bad gene cluster in the opposite direction, separated by a 0.4-kb intergenic segment (Fig. 4). The sequence for korB was completed to the C terminus by primer walking. The N-terminal amino acid sequences of the subunits of KGOR were identical to those deduced from the korA and korB genes (Table 2; the cysteine in KorB which was identified as valine by Edman degradation is usually not detectable by this technique). No further adjacent ORF was found up to 0.3 kb in the 5′ direction of korB, confirming that KGOR consists of two subunits only. The theoretical molecular masses of the gene products deduced from the sequences of korA and korB were 60.5 and 29.8 kDa, which are in the range of the molecular masses of the subunits of KGOR as determined by SDS analysis (66 and 34 kDa, respectively).
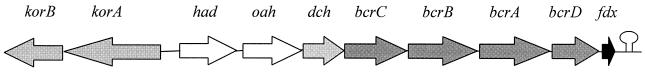
FIG. 4.
Organization of the genes involved in the benzoyl-CoA pathway of benzoate degradation (bad operon) and the two genes coding for the subunits of KGOR. korAB code for the two subunits of KGOR, bcrABCD code for the four structural genes of benzoyl-CoA reductase, and fdx codes for ferredoxin. The other genes of the bad operon code for an alcohol dehydrogenase (had), a ring hydrolase (oah), and a dienoyl-CoA dehydratase (dch), which are all involved in the benzoyl-CoA pathway. A stem-loop structure forms at the 3′ end of fdx, suggesting that transcription is terminated at this site.
TABLE 2.
Comparison of chemically determined amino acid sequences of two subunits of KGOR from T. aromatica with amino acid sequences deduced from the corresponding genes
| Subunit | Source of sequence | Amino acid sequence |
|---|---|---|
| KorA (66 kDa) | Experimentally determined | TARSVSITFAGSGGAGVMTA |
| Deduced from the gene | MTARSVSITFAGSGGAGVMTA | |
| KorB (34 kDa) | Experimentally determined | MDATSTVPSYSARDYKSEVK |
| Deduced from the gene | MDATSTCPSYSARDYKSEVK |
Amino acid sequence comparisons with similar enzymes yielded evidence that KGOR from T. aromatica belongs to the Halobacterium type of 2-oxoacid oxidoreductases (for details, see Discussion).
DISCUSSION
(i) General properties of KGOR from T. aromatica.
The general properties of KGOR are summarized in Table 3. The native molecular mass, the subunit composition, and the amount of iron, acid-labile sulfur, and TPP suggests an (αβ)2 architecture with two [4Fe-4S] clusters and two TPP per dimer. In addition to the oxidative decarboxylation of 2-oxoglutarate, KGOR also catalyzes the isotope exchange between 14CO2 and the C-1 carboxyl group of 2-oxoglutarate. The specific activity of KGOR in cell extracts is 80 nmol min−1 mg−1 (∼300 U/100 g [wet mass]), which is approximately double the activity of benzoyl-CoA reductase and meets the requirements of cells growing on benzoate at 30°C. KGOR from T. aromatica is highly specific for 2-oxoglutarate as the 2-oxoacid substrate; the apparent Km values for all substrates are in the physiological range. As expected, KGOR used only one-electron accepting substrates as viologens and the ferredoxin from T. aromatica (ferredoxins from other organisms were not tested). KGOR was unable to catalyze a hydride transfer to NAD(P)+. This is in accordance with the lack of a flavin cofactor which is usually required for electron transfer from a [4Fe-4S]1+ cluster to NAD(P)+. An example of a flavin cofactor containing 2-oxoacid oxidoreductase is phenylglyoxylate:NAD+ oxidoreductase (CoA benzoylating) from the denitrifying bacterium Azoarcus evansii, which is involved in anaerobic phenylalanine metabolism (26). This enzyme has the typical composition of a 2-oxoacid:ferredoxin oxidoreductase but in addition contains a flavin cofactor harboring subunit.
(ii) Novel function for 2-oxoacid:acceptor oxidoreductase.
In anaerobic bacteria using aromatic compounds as sources for energy and cell carbon, the strategy is to reduce the benzene ring first before it is oxidized to three acetyl-CoA and one CO2 in a modified β-oxidation (23, 25). The 2[4Fe-4S]1+/2+ cluster containing ferredoxin which has been shown to act as electron donor for benzoyl-CoA reductase in T. aromatica is now identified as the electron acceptor for KGOR. The in vivo operation of an electron transfer chain from 2-oxoglutarate to benzoyl-CoA by means of ferredoxin is supported in several respects. (a) In a coupled enzyme assay, the electron transfer from 2-oxoglutarate to benzoyl-CoA was followed in this work. (b) KGOR activity was increased 10 to 50 times in cells grown anaerobically on an aromatic substrate, indicating that KGOR is linked to the anaerobic aromatic metabolism. In contrast, NAD+-dependent 2-oxoglutarate dehydrogenase activity was abolished under anaerobic conditions. (c) The genes coding for the two subunits of KGOR are located adjacent to genes of the bad gene cluster (separated by a 0.4-kbp intergenic segment), providing the molecular basis of the observed coexpression of the proteins encoded in the bad gene cluster and by korA and korB.
The role of KGOR in the aromatic metabolism of T. aromatica is depicted in Fig. 5. It couples the complete acetyl-CoA oxidation to aromatic ring reduction by the use of the low-potential electron shuttle ferredoxin. Three acetyl-CoA (yielding three 2-oxoglutarate) and one CO2 are formed per benzoyl-CoA metabolized. The data obtained support the notion that excess reduced ferredoxin is regenerated by an NAD(P)H:ferredoxin oxidoreductase. Corresponding activities were clearly induced in T. aromatica when grown anaerobically on an aromatic substrate.
The role of KGOR in T. aromatica differs from the known functions of other 2-oxoacid oxidoreductases: in strictly anaerobic bacteria they are involved in substrate-level phosphorylation (e.g., Clostridium, sulfate reducers, hyperthermophiles, methanogenic archaea [references 17 and 18 and references therein]); moreover, they play a role in CO2 fixation in bacteria using the reductive tricarbonic acid cycle (15, 42), in acetyl-CoA assimilation (many obligate anaerobes (3, 19, 34), or as a source for low-potential electron equivalents for nitrogen fixation (14, 44).
In the denitrifying bacterium A. evansii, a 2-oxoglutarate:NADP+ oxidoreductase is induced when the bacterium is anaerobically grown on an aromatic substrate (32). It is conceivable that in A. evansii the electron donor for enzymatic ring reduction ferredoxin is not directly reduced by a 2-oxoacid:ferredoxin oxidoreductase as in T. aromatica but rather by the combined action of 2-oxoacid:NADP+ oxidoreductase and NADPH:ferredoxin oxidoreductase. Nothing is known about the electron transfer to the aromatic ring in other anaerobic bacteria using aromatic compounds.
(iii) Comparison of subunit architecture with other 2-ketoacid:acceptor oxidoreductases.
Based on molecular and phylogenetic homologies, 2-oxoacid oxidoreductases are grouped in a single enzyme family. In general, enzymes of this family are composed of four modules (a, b, c, and d) with molecular masses between 10 and 50 kDa. The b module binds one TPP and a single [4Fe-4S] cluster; the c module is involved in CoA binding. The d module is highly similar to a ferredoxin and usually contains two further [4Fe-4S] clusters, whereas a distinct function has not been ascribed to the a module. Depending on the organization of the four modules, different types are distinguished in the 2-oxoacid oxidoreductase family which have been described in detail previously (28, 46). The “Halobacterium type” of 2-oxoacid oxidoreductases is distinguished from all other types by the lack of the two [4Fe-4S] clusters containing the d module and is usually referred to as the (αβ)2 type. The α subunit of this type contains the a and c modules on a single polypeptide.
The amino acid sequences deduced from the sequences of of korA and korB showed similarities with several other 2-oxoacid:ferredoxin oxidoreductases. Highest similarity values were found with enzymes of the (αβ)2 architecture consisting of two subunits with masses of approximately 60 to 70 kDa (α subunit) and 30 to 35 kDa (β subunit). The amino acid sequence deduced from korA (containing the a and c modules) exhibited highest similarities (28 to 32% identity and 45 to 50% similarity) with the corresponding subunits of 2-oxoacid:ferredoxin oxidoreductases from halophilic bacteria/archaea (2-oxoacid:ferredoxin oxidoreductases from several Halobacterium species and Bacillus halodurans), of a putative 2-oxoacid oxidoreductase from Staphylococcus aureus, and of oxidoreductases from several thermophilic bacteria (e.g., Thermotoga maritima, Sulfolobus tokodaii, Hydrogenobacter thermophilus, Thermoplasma acidophilum; see accession numbers [Table 4]). The amino acid sequence deduced from korB showed 41 to 51% identities and up to 68% similarity with the β subunits of 2-oxoacid:ferredoxin oxidoreductases from the organisms listed above. It is of note that almost all (αβ)2-type 2-oxoacid oxidoreductases identified so far are from extremophiles (with exceptions of KGOR from T. aromatica and a putative 2-oxoacid oxidoreductase deduced from ORFs in the S. aureus genome.)
TABLE 4.
Alignment of conserved amino acid motifs in several (αβ)2-type 2-oxoacid:ferredoxin oxidoreductases which may act as cofactor binding sitesa
| Cofactor | Oxidoreductase | Subunit | Amino acid sequence | Accession no. |
|---|---|---|---|---|
| [4Fe-4S] cluster | TaKGOR | β | 23WCPGCGD29...56IGCSS60...206SPCV209 | |
| BhPOR | β | 18WCPGCGN24...51IGCSS55...199SPCV202 | NP 374405 | |
| HsOOR | β | 29WCPGCGD35...62IGCSG66...211TQCP214 | NP 243239 | |
| TmOOR | β | 13WCPGCGD19...46IGQSA50...195QPCV198 | NP 279532 | |
| StOOR | β | 11WCPGCGN17...44IGCSG48...195QPCP198 | NP 228971 | |
| SaOOR | β | 11WCPGCGD17...43IGCSG47...195QPCP198 | BAA10899 | |
| TPP | TaKGOR | β | 98SGGDGD103..135TKGQAS140 | |
| BhPOR | β | 105YGGDGD110..129TTGQAS134 | ||
| HsOOR | β | 94AGGDGD99..130TKGQAS135 | ||
| TmOOR | β | 88ESGDGC93..124TKGQAS129 | ||
| StOOR | β | 97VSGDGC102..134TKGQAS139 | ||
| SaOOR | β | 88SGGDGD93..126TKGQTS131 | ||
| CoA | TaKGOR | α | 11GSG-GAG16 | |
| BhPOR | α | 9GGQQGEG15 | NP 374406 | |
| HsOOR | α | 11GEA-GDG16 | NP 243240 | |
| TmOOR | α | 10GEA-GQG15 | NP 279533 | |
| StOOR | α | 9GGQQGEG15 | NP 228970 | |
| SaOOR | α | 9GGAQGTG15 | BAA10898 |
TaKGOR, 2-oxoglutarate:ferredoxin oxidoreductase from T. aromatica; BhPOR, pyruvate:ferredoxin oxidoreductase from Bacillus halodurans; 2-oxoacid:ferredoxin oxidoreductases of Halobacterium sp. strain NRC-1 (HSOOR), Thermotoga maritima (TmOOR), Sulfolobus tokodaii strain 7 (StOOR), and S. aureus (SaOOR). The conserved cysteines in bold are considered to ligate the [4Fe-4S] cluster; the glycine-rich motif at the N terminus of the α subunit is considered to form a loop binding the adenine moiety of CoA.
Based on both the biochemical data (subunit architecture, iron content) and the gene sequence analysis, KGOR from T. aromatica can clearly be classified into the (αβ)2 group of 2-oxoacid oxidoreductases.
Three highly conserved putative cofactor binding motifs were identified in the amino acid sequences of the two subunits of T. aromatica KGOR. Table 4 shows the corresponding sequences of different 2-oxoacid oxidoreductases of the (αβ)2 type with high amino acid sequence similarities to KGOR from T. aromatica. Four cysteines conserved in the β subunit of KGOR have been assigned to coordinate a [4Fe-4S]1+/2+ cluster (28, 38, 46). This motif differs from that of the typical ferredoxin-type [4Fe-4S]1+/2+ cluster ligation motif (CXXCXXXCP). The β subunit also harbors conserved residues which represent putative TPP binding sites. The corresponding conserved amino acid motifs are G(S)GDG and TK(T)GQA(T)S (24, 31); the former motif is considered to be involved in divalent cation binding of the TPP cofactor. Finally, the N terminus of the α subunit carries a glycine-rich motif which is similar to the typical adenine recognition loop identified in citrate synthases (39). This loop is proposed to be involved in CoA binding of 2-ketoacid oxidoreductases (46).
Acknowledgments
We are grateful to M. Jahn, C. Schühle, and J. Gescher, Freiburg, Germany, for help in the molecular biological work and to H. Schägger, Frankfurt, Germany, for N-terminal amino acid sequencing. We thank W. Haehnel, Freiburg, Germany, for fluorescence spectroscopic analysis and G. Fuchs, Freiburg, Germany, for helpful suggestions.
This work was supported by the Deutsche Forschungsgemeinschaft (BO 1565).
REFERENCES
- 1.Anders, H. J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 2.Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide plus sulfane in usually stable iron-sulfur proteins. Anal. Biochem. 131:373-378. [DOI] [PubMed] [Google Scholar]
- 3.Bock, A. K., J. Kunow, J. Glasemacher, and P. Schönheit. 1996. Catalytic properties, molecular composition and sequence alignments of pyruvate:ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro). Eur. J. Biochem. 237:35-44. [DOI] [PubMed] [Google Scholar]
- 4.Boll, M., and G. Fuchs. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234:921-933. [DOI] [PubMed] [Google Scholar]
- 5.Boll, M., S. J. P. Albracht, and G. Fuchs. 1997. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. A study of adenosinephosphate activity, ATP stoichiometry of the reaction and EPR properties of the enzyme. Eur. J. Biochem. 244:840-851. [DOI] [PubMed] [Google Scholar]
- 6.Boll, M., and G. Fuchs. 1998. Identification and characterization of the natural electron donor ferredoxin and of FAD as a possible prosthetic group of benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. Eur. J. Biochem. 251:946-954. [DOI] [PubMed] [Google Scholar]
- 7.Boll, M., D. Laempe, W. Eisenreich, A. Bacher, T. Mittelberger, J. Heinze, and G. Fuchs. 2000. Non-aromatic products formed from anoxic conversion of benzoyl-CoA with benzoyl-CoA reductase and cyclohexa-1,5-diene-1-carbonyl-CoA hydratase. J. Biol. Chem. 275:21889-21895. [DOI] [PubMed] [Google Scholar]
- 8.Boll, M., G. Fuchs, C. Meier, A. X. Trautwein, and D. J. Lowe. 2000. EPR and Mössbauer studies of benzoyl-CoA reductase. J. Biol. Chem. 275:31857-31868. [DOI] [PubMed] [Google Scholar]
- 9.Boll, M., G. Fuchs, G. Tilley, F. A. Armstrong, and D. J. Lowe. 2000. Unusual spectroscopic and electrochemical properties of the 2[4Fe-4S] ferredoxin of Thauera aromatica. Biochemistry 39:4929-4938. [DOI] [PubMed] [Google Scholar]
- 10.Boll, M., G. Fuchs, and D. J. Lowe. 2001. Single turnover EPR studies of benzoyl-CoA reductase. Biochemistry 40:7612-7620. [DOI] [PubMed] [Google Scholar]
- 11.Brackmann, R., and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-hydroxybenzoyl-CoA reductase (dehydroxylating) from a denitrifying Pseudomonas species. Eur. J. Biochem. 213:563-571. [DOI] [PubMed] [Google Scholar]
- 12.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Breese, K., M. Boll, J. Alt-Mörbe, H. Schägger, and G. Fuchs. 1998. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium T. aromatica. Eur. J. Biochem. 256:148-154. [DOI] [PubMed] [Google Scholar]
- 14.Brostedt, E., and S. Nordlund. 1991. Purification and partial characterization of pyruvate oxidoreductase from the photosynthetic bacterium Rhodospirillum rubrum grown under nitrogen-fixing conditions. Biochem. J. 279:155-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchanan, B. B., and D. I. Arnon. 1990. A reverse Krebs cycle in photosynthesis: consensus at last. Photosynth. Res. 24:47-53. [PubMed] [Google Scholar]
- 16.Buckel, W. und R. Keese. 1995. Einelektronen-Redoxreaktionen von Coenzym-A-Estern in anaeroben Bakterien—ein Vorschlag für einen neuen Mechanismus. Angew. Chem. 107:1595-1598. [Google Scholar]
- 17.Chabriere, E., M. H. Charon, A. Volbeda, L. Pieulle, E. C. Hatchikian, and J. C. Fontecilla-Camps. 1999. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat. Struct. Biol. 6:182-190. [DOI] [PubMed] [Google Scholar]
- 18.Charon, M.-H., A. Volbeda, E. Chabriere, L. Pieulle, and J. C. Fontecilla-Camps. 1999. Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr. Opin. Struct. Biol. 9:663-669. [DOI] [PubMed] [Google Scholar]
- 19.Drake, H. L., S.-I. Hu, and H. G. Wood. 1981. Purification of five components from Clostridium thermoaceticum which catalyze synthesis of acetate from pyruvate and methyltetrahydrofolate. Properties of phosphotransacetylase. J. Biol. Chem. 256:1137-11144. [PubMed] [Google Scholar]
- 20.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraisse, L., and H. Simon. 1988. Observations on the reduction of non-activated carboxylates by Clostridium formicoaceticum with carbon monoxide or formate and the influence of various viologens. Arch. Microbiol. 150:381-386. [Google Scholar]
- 22.Hans, M., J. Sievers, U. Müller, E. Bill, J. A. Vorholt, D. Linder, and W. Buckel. 1999. 2-hydroxyglutaryl-CoA dehydratase from Clostridium symbiosum. Eur. J. Biochem. 265:404-414. [DOI] [PubMed] [Google Scholar]
- 23.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 24.Hawkins, C. F., A. Borges, and R. N. Perham. 1989. A common structural motif of thiamin pyrophosphate-binding enzymes. FEBS Lett. 255:77-82. [DOI] [PubMed] [Google Scholar]
- 25.Heider, J., and G. Fuchs. 1997. Anaerobic metabolism of aromatic compounds. Eur. J. Biochem. 243:577-596. [DOI] [PubMed] [Google Scholar]
- 26.Hirsch, W., and G. Fuchs. 1998. Phenylglyoxylate:NAD+ oxidoreductase (CoA benzoylating), a new enzyme of anaerobic phenylalanine metabolism in the denitrifying bacterium Azoarcus evansii. Eur. J. Biochem. 251:907-915. [DOI] [PubMed] [Google Scholar]
- 27.Howard, J. B., and D. C. Rees. 1996. Structural basis of biological nitrogen fixation. Chem. Rev. 96:2965-2982. [DOI] [PubMed] [Google Scholar]
- 28.Kletzin, A., and M. W. W. Adams. 1996. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 178:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyritsis, P., O. M. Hatzfeld, T. A. Link, and J.-M. Moulis. 1998. The two [4Fe-4S] clusters in Chromatium vinosum have largely differing reduction potentials. J. Biol. Chem. 273:15404-15411. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lindqvist, Y., and G. Schneider. 1993. Thiamine pyrophosphate-dependent enzymes: transketolase, pyruvate oxidase and pyruvate decarboxylase. Curr. Opin. Struct. Biol. 3:896-901. [Google Scholar]
- 32.Lochmeyer, C., and G. Fuchs. 1990. NADP+ specific 2-oxoglutarate dehydrogenase in denitrifying Pseudomonas species. Arch. Microbiol. 153:226-229. [Google Scholar]
- 33.Lovenberg, W., B. B. Buchanan, and J. Rabinowitz. 1963. Studies on the chemical nature of ferredoxin. J. Biol. Chem. 238:3899-3913. [PubMed] [Google Scholar]
- 34.Menon, S., and S. W. Ragsdale. 1996. Unleashing hydrogenase activity in carbon monoxide dehydrogenase/acetyl-CoA synthase and pyruvate:ferredoxin oxidoreductase. Biochemistry 35:15814-15821. [DOI] [PubMed] [Google Scholar]
- 35.Möbitz, H., and M. Boll. 2002. A Birch-like mechanism in enzymatic benzoyl-CoA reduction—a kinetic study of substrate analogues combined with an ab initio model. Biochemistry 41:1752-1758. [DOI] [PubMed] [Google Scholar]
- 36.Muller, Y. A., Y. Lindqvist, W. Furey, G. E. Schulz, F. Jordan, and G. S. Schneider. 1993. A thiamine diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase. Structure 1:95-103. [DOI] [PubMed] [Google Scholar]
- 37.Penttinen, H. K. 1979. Fluorometric determination of thiamine and its mono-, di- and triphosphate esters. Methods Enzymol. 62:58-59. [DOI] [PubMed] [Google Scholar]
- 38.Plaga, W., F. Lottspeich, and D. Oesterhelt. 1992. Improved purification, crystallization and primary structure of pyruvate:ferredoxin oxidoreductase from Halobacterium halobium. Eur. J. Biochem. 205:391-397. [DOI] [PubMed] [Google Scholar]
- 39.Russel, R. J., D. W. Hough, M. J. Danson, and G. L. Taylor. 1994. The crystal structure of citrate synthase from the thermophilic archaeon. Thermoplasma acidophilum. Structure 2:1157-1167. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Schachter, D., and V. J. Taggart. 1976. Benzoyl coenzyme A and hippurate synthesis. J. Biol. Chem. 203:925-933. [PubMed] [Google Scholar]
- 42.Shiba, H. T., Y. Kawasumi, T. Igarashi, T. Kodama, and Y. Minoda. 1985. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligatory autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141:198-203. [Google Scholar]
- 43.Unciuleac, M., and M. Boll. 2001. Mechanism of ATP-driven electron transfer catalyzed by the benzene ring-reducing enzyme benzoyl-CoA reductase. Proc. Natl. Acad. Sci. USA 98:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahl, R. C., and W. H. Orme-Johnson. 1987. Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme in Klebsiella pneumoniae are similar enzymes. J. Biol. Chem. 262:10489-10496. [PubMed] [Google Scholar]
- 45.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182:157-159. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Q., T. Iwasaki, T. Wakagi, and T. Oshima. 1996. 2-oxoacid:ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J. Biochem. 120:587-599. [DOI] [PubMed] [Google Scholar]