Abstract
The trans-acting antigenomic delta ribozyme, isolated from the human hepatitis delta virus, was shown to be highly stable and active in vitro, as well as in mammalian cell lines. However, the stability and gene-targeting competence of this small ribozyme have not been studied previously in bacterial cells. In this paper we describe the use of two variants of the trans-acting antigenomic delta ribozyme targeting the abundant EF-Tu mRNA in the industrially important gram-positive bacterium Lactococcus lactis. These two delta ribozyme variants were expressed at significant levels and were shown to be highly stable in vivo. The half-life of the EF-Tu mRNA was slightly but consistently reduced in the presence of the classical delta ribozymes (7 to 13%). In contrast, delta ribozymes harboring a specific on/off riboswitch (SOFA-delta ribozymes) targeting the same sites on the EF-Tu mRNA considerably reduced the half-life of this mRNA (22 to 47%). The rates of catalysis of the SOFA-delta ribozymes in L. lactis were similar to the rates determined in vitro, showing that this new generation of delta ribozymes was highly efficient in these bacterial cells. Clearly, SOFA-delta ribozymes appear to be an ideal means for development of gene inactivation systems in bacteria.
Ribozymes are RNA molecules that possess catalytic activity. Thus, they are considered to be RNA enzymes that have great potential for in vivo applications (2, 9). The ability of ribozymes to recognize and catalyze the cleavage of specific RNA molecules (Fig. 1A) makes them attractive tools for both gene therapy and functional genomics (30, 45). Even if they are less stable than oligodeoxyribonucleotides (ODNs), ribozymes have several advantages for engineering versatile gene inactivation systems: (i) because most ribozymes recognize their substrates through a series of Watson-Crick base pairs, their binding sequences can be modified to target almost any RNA molecule; (ii) they cleave and induce degradation of their substrates; (iii) they exhibit turnover (i.e., one ribozyme may successively cleave several substrate molecules); (iv) their effect is at the mRNA level and is thus reversible; (v) their effect can be triggered at any time by overexpression, thus allowing inactivation of essential genes; and (vi) their expression levels can be regulated to control the level of expression of the target gene (partial knockout).
FIG. 1.
Ribozyme-based gene inactivation strategy and δRz secondary structure. (A) The ribozyme binds specifically to an accessible single-stranded region of the target mRNA through a series of Watson-Crick base pairs. Some mRNA target sites are less accessible to the ribozyme depending on their secondary and tertiary structures. Moreover, the association of proteins (solid ovals) with the mRNA in vivo may also prevent ribozyme binding to potential target sites. After binding, the second step consists of specific cleavage of the target mRNA by the ribozyme. Following cleavage, the mRNA is rapidly degraded by the host nucleases (“PacMan” symbols). (B) Secondary structure of the trans-acting antigenomic δRz bound to its substrate or target. The stems are designated P1 to P4. The P1.1 pseudoknot is indicated by dotted lines. The homopurine base pair (G • • G) at the top of the P4 stem and the wobble base pair (G • U) in the substrate recognition stem (P1) are also indicated. The arrow indicates the substrate cleavage site. H at position −1, in reference to the cleavage site, indicates nucleotide A, C or U, while N indicates any nucleotide (A, C, U, or G).
In a ribozyme-based gene targeting system, the ribozyme must interact with nucleotides in an accessible region of its target and make a series of complementary base pairs (Fig. 1A). The most accessible ribozyme binding sites should be single-stranded regions of the target that are not involved in either secondary or tertiary structure interactions and that are not occluded by bound proteins (Fig. 1A). Upon recognition, the target mRNA is cleaved by the ribozyme and then is rapidly degraded by cellular nucleases. In bacteria, in which posttranscriptional gene silencing (RNA silencing) is not known to happen, ribozyme-based gene inactivation systems appear to be a suitable approach.
Hepatitis delta virus is the only animal virus known to contain a single-stranded circular RNA genome (length, 1.7 kb) (39). This genome replicates through a DNA-independent rolling circle mechanism, which involves production of multimeric genomic and antigenomic RNA strands that self-cleave and circularize using ribozyme motifs to generate new progeny. Like other small self-cleaving (cis-acting) motifs, delta motifs catalyze cleavage of one of their own phosphodiester bonds, yielding reaction products harboring 5′-hydroxyl and 2′,3′-cyclic phosphate termini. Both the genomic and antigenomic self-cleaving motifs have been separated into two molecules to develop trans-acting systems in which one molecule possesses the catalytic properties required to cleave several molecules of substrate. The trans-acting antigenomic delta ribozyme (δRz) folds into a secondary structure called the pseudoknot (Fig. 1B) (47). The substrate specificity of δRz depends primarily on the formation of the P1 stem, which consists of one G • U wobble base pair adjacent to the cleavage site, followed by six Watson-Crick base pairs (Fig. 1B) (4). The presence of a single nucleotide located 5′ of the cleaved phosphate (position −1) was shown to be sufficient to allow cleavage. Only C, U, or A is tolerated at this position, although the level of cleavage varies depending on the nucleotide (18). The level of cleavage also depends on the identities of the nucleotides at positions −2 to −4 adjacent to the cleavage site; however, the effects of these nucleotides are less important (18). Following substrate recognition and formation of the P1 stem, the J1/4 junction and the P3 loop (L3) that were single-stranded in the initial stages of folding are subsequently involved in the formation of a 2-bp pseudoknot (P1.1 stem) (Fig. 1B) (19, 21, 50). This conformational change is necessary to promote substrate cleavage (1, 19, 50).
trans-acting antigenomic δRzs have been reported to specifically cleave in vitro both the mRNA encoding the delta antigen (43) and the pregenome RNA strand of hepatitis B virus (5). These results suggested that δRz has potential for further development of a system to inhibit gene expression. The potential to cleave substrates in trans was subsequently demonstrated in vivo with various mRNA targets (16, 29, 46). For example, δRzs expressed in a stable mouse cell line completely knocked out expression of the subtilisin proprotein convertase 2 gene (SPC2) (16). It was therefore proposed that δRz is active and very stable in mammalian cell lines because it resides within an animal virus and evolved in the eukaryotic cell environment (36). Thus, this ribozyme has potential for development of gene inactivation approaches in eukaryotic systems. In bacteria, in which the silencing RNA approach cannot be used to perform functional genomic studies, δRz-based gene inactivation systems appear to be a suitable alternative. However, this has not been investigated previously in prokaryotes.
The goal of this work was to explore the abilities of various engineered trans-acting antigenomic δRzs to specifically cleave mRNAs in the industrially important gram-positive bacterium Lactococcus lactis. Because the mRNA encoding the EF-Tu protein is one of the most abundant RNAs in bacteria, it appeared to be a suitable target for such an evaluation. The advantage of targeting an abundant transcript is that even if the ribozymes are extremely active, they should not be able to cleave all the substrates available, allowing a real evaluation of their maximum activity. Microarray analysis showed that EF-Tu mRNAs of the tufA and tufB genes account for almost 10% of the total RNA in Escherichia coli (E. Massé, personal communication). In contrast, the EF-Tu protein is encoded by only one gene (tuf) in L. lactis IL1403 (8). This translation factor functions as a molecular switch (GTPase activity) during the elongation step (31). Here we show that different engineered trans-acting δRzs can be expressed at significant levels and are highly stable in L. lactis. The expression of classical δRzs targeting the EF-Tu mRNA reduces slightly but consistently the half-life of this mRNA. However, a new generation of δRzs harboring a specific on/off adaptor module (SOFA-δRzs) and targeting the same sites is more efficient and reduces the half-life of the EF-Tu mRNA by as much as 47%. This study reveals the great advantages of using modified SOFA-δRzs for development of bacterial gene inactivation systems.
MATERIALS AND METHODS
Strains and plasmids.
L. lactis strains NZ9800, NZ9000, NZ3900, and IL1403 were grown without shaking in M17 supplemented with 0.5% glucose at 30°C or 37°C. E. coli strains DH5α and DH10β, which were used for cloning, were grown with shaking at 37°C in LB broth. The different plasmids were introduced into L. lactis by electroporation using a Bio-Rad MicroPulser electroporator (51).
The tuf gene was amplified by PCR from IL1403 genomic DNA using sense (5′-GGGGTACCATGGCTAAAGAAGTATACG-3′) and antisense (5′-CGGGTACCTTAAGCTTTGATTTCAGC-3′) primers. The resulting PCR product was cloned downstream of the T7 promoter in the pBluescript II SK vector (Amp) (Stratagene) between the KpnI and BamHI restriction sites. The resulting plasmid was designated pBS-EFTu, and its identity was confirmed by DNA sequencing (T7 sequencing kit; United States Biochemicals).
The ribozyme expression cassettes (HH1-δRz-HH2) (Fig. 2A) were first cloned into the pBluescript II SK vector under control of the T7 promoter. Briefly, both self-cleaving hammerhead motifs (HH1 and HH2) were cloned using two oligodeoxyribonucleotides corresponding to the hammerhead sequences and harboring NcoI and BamHI restriction sites to allow subsequent cloning of the different δRzs (Fig. 2B). The δRzs were then inserted between HH1 and HH2 (NcoI and BamHI) using two complementary oligodeoxyribonucleotides, creating the pBS-HH1-δRz-HH2 plasmids. For in vivo assays, the nisin promoter (PnisA) was amplified from the pLEIItd+KR′′ plasmid (15) by PCR using sense (5′-GTCCCCCGGGCTAGTCTTATAACTATACTGACAATAG-3′) and antisense (5′-GTCCCCCGGGGGAATTCCTCTCGAGGTAGTTCCTTCGAACGAAATCATTG-3′) primers. The resulting PCR product was cloned (SmaI) in the pDL278 shuttle plasmid (Spc) (35), creating pDL-PnisA. The expression cassettes (HH1-δRz-HH2) were excised with XhoI and EcoRI from the pBS-HH1-δRz-HH2 plasmids and subcloned into pDL-PnisA. Plasmids harboring δRzs were designated pDL-PnisA-HH1-δRz-EX-HH2, where X is the position of the target site within the EF-Tu mRNA. δRz-E232 not embedded between the hammerhead motifs was excised with BamHI and NcoI from the pBS-HH1-δRz-E232-HH2 plasmid, blunted, and subcloned into pDL-PnisA directly downstream from the nisin promoter (EcoRI).
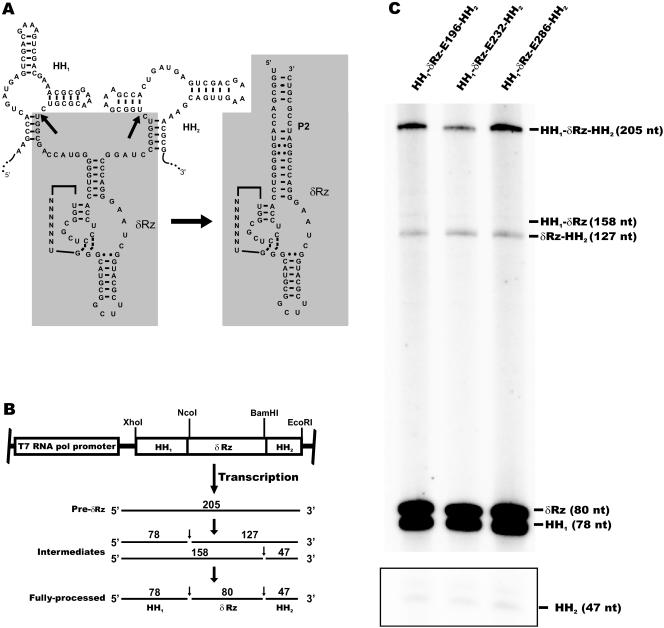
FIG. 2.
Ribozyme expression cassettes. (A) Schematic representation of the δRz expression strategy. The secondary structures of the cis (hammerhead) and trans (delta) (shaded areas) ribozymes are shown. Transcription of the cis-trans-cis cassette produces a chimeric RNA that self-cleaves at two specific sites (HH1 and HH2), releasing the trans-acting δRz (δRz). The arrows indicate the hammerhead (HH1 and HH2) cleavage sites. (B) In vitro transcription of the cis-trans-cis δRz constructs. Expected RNA products generated by self-cleavage of the hammerhead motifs during transcription are shown. The positions of the full-length precursor (Pre-δRz, 205 nt), intermediates (127 and 158 nt), and fully processed transcript (HH1, 78 nt; HH2, 47 nt; δRz, 80 nt) are shown. The positions of restriction enzyme sites used for cloning are indicated. (C) Autoradiogram of a denaturing 8% polyacrylamide gel containing in vitro transcripts of the three cis-trans-cis δRz expression cassettes. The area enclosed in a box was taken from the lower part of the same autoradiogram to show the relative intensity of the small band (47 nt) corresponding to the released HH2.
The specific on/off adapter HH1-SOFA-δRz-E232-HH2 construct was created using the strategy described above for the classical δRzs. SOFA-δRzs that were not embedded between the hammerhead motifs and cloned directly downstream from the nisin promoter (EcoRI) (SOFA-δRz-E196, -E232, -E286, and -SD) were obtained using two partially overlapping oligodeoxyribonucleotides (EcoRI), sense oligodeoxyribonucleotide 5′-CGGAATTCCCAGCTAGTTTNNNNNNNNNNNNNNNNCAGGGTCCACCTCCTCGCGGTNNNNNNNGGGC AT-3′ and antisense oligodeoxyribonucleotide 5′-CGGAATTCCCAGCTAGAAAGGGTCCCTTAGCCATCCGCGAACGGATGCCCNNNNNNNACCGCGAGGAG-3′; the complementary nucleotides are indicated by boldface type, the biosensor (12 nucleotides [nt]) is underlined, the P1 stem (7 nt) is double underlined, and the blocker sequence (4 nt) is indicted by italics. The biosensor sequences correspond to TTCAGTTTCGTA, GTGACCTGGAGC, GATTGCACCGTC, and TTCTTTAGCCAT for SOFA-E196, -E232, -E286, and -SD, respectively. In the sense oligodeoxyribonucleotides, the target binding sequence of δRz corresponds to TGTGAGT, TGTGGGT, GGGCAGT, and AAATGTT for SOFA-E196, -E232, -E286, and -SD, respectively. The inactive SOFA-δRz-E232 control was also constructed using two complementary oligodeoxyribonucleotides. In this case, nucleotide C76 was replaced by A (C76A), while the two cytosines of the L3 loop, which are involved in the formation of the P1.1 pseudoknot, were replaced by two guanosines (C40G and C41G) (see Fig. 7A). Each pair of oligodeoxyribonucleotides was annealed and subsequently extended using T4 DNA polymerase. The pMSP3535 plasmid (Erm), harboring the nisK and nisR genes, was used to allow overexpression of the δRz expression cassettes upon nisin induction in IL1403 (10). Selective medium contained antibiotics at the following concentrations: ampicillin (Amp), 100 μg/ml; spectinomycin (Spc), 300 μg/ml; and erythromycin (Erm), 150 μg/ml.
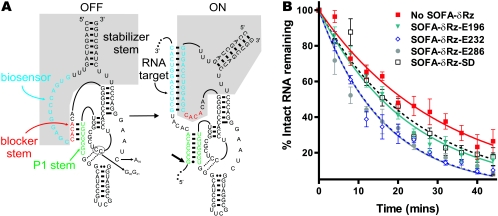
FIG. 7.
Structure and effect of SOFA-δRz. (A) Secondary structures of the off and on conformations of SOFA-δRz-E232. The shaded area indicates the SOFA module. The P1 stem, the biosensor, and the blocker sequence of the ribozyme are indicated by green, blue, and red, respectively. Upon recognition of its specific target, the biosensor (12 nt) of the SOFA module base pairs 12 nt downstream from the cleavage site. The interaction between the target and the biosensor of the ribozyme disrupts the blocker stem and therefore liberates the substrate recognition sequence of the ribozyme, allowing formation of the P1 stem. The stabilizer stem was designed to mimic the P2 stem in classic δRz, allowing base pairing of both 5′ and 3′ extremities. The small arrow indicates the ribozyme cleavage site. The C76-to-A, C40-to-G, and C41-to-G mutations present in the inactive SOFA-δRz-E232 control are indicated. (B) Graphic representation of EF-Tu mRNA half-lives in the presence of four different SOFA-δRzs.
Bioinformatic analyses of the EF-Tu mRNA from L. lactis.
Potential target sites were selected as previously described (5). Briefly, the RNAfold algorithm of the RNA Structure 3.7 program (38) was used to predict the most stable secondary structures of the full-length EF-Tu mRNA sequence from L. lactis subsp. lactis IL1403 (8) (GenBank accession number NC_002662.1). The predicted structures were then analyzed for the probability of binding to complementary 7-mer oligodeoxyribonucleotides (single-stranded regions) using the OligoWalk software (37).
In vitro transcription.
The EF-Tu mRNA and HH1-δRz-HH2 cassettes were transcribed in vitro using linearized pBS-EFTu (BamHI) and pBS-HH1-δRz-HH2 (EcoRI) as templates. Runoff transcription was performed using the Riboprobe in vitro transcription system (Promega) according to the manufacturer's protocols in the presence of 50 μCi of [α-32P]GTP (3,000 Ci/mmol; Amersham Biosciences).
RNase H assays.
RNase H reactions were performed using full-length 32P-labeled EF-Tu mRNA. Briefly, the EF-Tu transcripts (∼0.1 μM, ∼50,000 cpm) and a series of oligodeoxyribonucleotides (seven nucleotides, 5 μM) were preincubated for 10 min at 25°C in an 8-μl (final volume) mixture containing 20 mM Tris-HCl (pH 7.5), 20 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, and 0.1 mM dithiothreitol. RNase H (2 μl, 0.5 U; United States Biochemicals) was then added, and the samples were incubated at 37°C for 30 min. The reactions were quenched by adding loading buffer (97% formamide, 1 mM EDTA [pH 8.0], 0.025% xylene cyanol, 0.025% bromophenol blue), fractionated by denaturing 5% polyacrylamide gel electrophoresis (7 M urea), and analyzed with a PhosphorImaging system (Molecular Dynamics).
Ribozyme expression in vivo.
Different L. lactis strains (NZ9800, NZ9000, NZ3900, and IL1403) containing δRz expression plasmids were grown without shaking at either 30°C or 37°C. Overnight saturated cultures were diluted (1/25) and grown to an optical density at 600 nm (OD600) of 0.5. Ribozyme expression was then induced by addition of nisin to the cultures to a final concentration of 10 ng/ml, and aliquots were removed at different times. Cells were recovered by centrifugation, and total RNA was extracted. For mRNA decay experiments, diluted cultures (1/25) were grown to an OD600 of 0.5, and ribozyme expression was induced with nisin (10 ng/ml). Rifampin (150 μg/ml) was added to the culture 1 h after nisin induction. Aliquots were then removed at different times for extraction of total RNA.
RNA isolation and Northern blot hybridization.
Total RNA was isolated from L. lactis using TRIzol (Invitrogen Life Technologies). The cell pellets were mixed with 500 μl of TRIzol and 250 μg of glass beads (Sigma). The mixtures were incubated at 55°C for 5 min, and this was followed by 3 min of vortexing. This treatment was repeated three times. For Northern blot hybridization, samples containing 10 μg of total RNA were run on 1% agarose or denaturing (7 M urea) 8% polyacrylamide gels and transferred to nylon membranes (Hybond-N; Amersham Biosciences). The membranes were hybridized with either the 5′-end-labeled (32P) EF-Tu probe (5′-CGTGTGGTTTGCTACGGTCG-3′) or one of the δRz ribozyme probes (5′-GGCTCCCTTAGCCATGCGAAGCC-3′ [δRz-short] and 5′-GGGTCCCTTAKCCATGCGAAGCCGCATGCCCATVYSSVACCGGGAGGAGGTGGACCC-3′ [δRz-long], where K is T or G, S is C or G, V is A, C, or G, and Y is C or T). These oligodeoxyribonucleotides (10 pmol) were labeled in a 10-μl (final volume) mixture containing 6.4 pmol of [γ-32P]ATP (6,000 Ci/mmol; Amersham Biosciences) and 5 U of T4 polynucleotide kinase (New England Biolabs) at 37°C for 1 h and then purified on Sephadex G-50 columns. The membranes were rehybridized with either the 23S rRNA probe (5′-ACCCGACAAGGAATTTCGC-3′) (26) or the 5S rRNA probe (5′-GGTGTATCTCCATCGCAATGATGACC-3′) as loading controls. To estimate the relative concentrations of EF-Tu mRNA (EF-Tu and 23S rRNA), δRz-E196 (δRz-short and 5S rRNA), and the EF-Tu mRNA/δRz-E196 ratio (EF-Tu and δRz-short), two probes were used in the same hybridization. In each case, equimolar amounts of the two probes were added, and the signal intensities were adjusted based on the relative labeling level of the corresponding probe. The membranes were exposed on phosphor screens, and the contents were revealed with Molecular Imager Fx (Bio-Rad). The bands were quantified with the Quantity One software (Bio-Rad). RNA half-lives were determined using the one-phase exponential decay equation of GraphPad Prism, version 3.2 (http://www.graphpad.com/prism/Prism.htm).
RESULTS
Identification of potential ribozyme target sites in the EF-Tu mRNA from L. lactis.
Single-stranded regions of target mRNAs should be more accessible to δRz base pairing and cleavage than double-stranded regions (Fig. 1A) (43). Therefore, selection of prospective target sites within mRNAs is the initial step in designing efficient trans-acting δRzs. To identify these accessible sites, we used a procedure that included both bioinformatic tools and biochemical assays (5).
First, using the RNA folding algorithm from the RNA Structure 3.7 software (38), a series of potential secondary structures for the EF-Tu mRNA from the IL1403 strain of L. lactis were obtained. The five most stable structures were then analyzed using the OligoWalk software, which provided the degree of accessibility (single strand) for all 7-nt windows along the different structures (37). A 7-nt window was used because this length corresponds to the length of the binding domain of δRz (P1 stem) (Fig. 1B). When only the 5′ half of the EF-Tu mRNA (positions 1 to 600 of 1,188 nt) was considered, 79 sites appeared to be mostly single stranded. These target sites were analyzed further to identify which ones fulfilled the essential criteria for efficient cleavage by a trans-acting antigenomic δRz. The first base pair of the P1 stem (target recognition stem) had to be G • U, and a G adjacent to the cleavage site (position −1) was avoided since it would hinder substrate cleavage (Fig. 1B) (18). These bioinformatic analyses identified nine potential target sites in the first half of the EF-Tu mRNA from L. lactis (Table 1).
TABLE 1.
Summary of the RNase H cleavage assays
| Site | Position in EF-Tu mRNA | mRNA sequencea | RNase H cleavage |
|---|---|---|---|
| 1 | 55-61 | 5′-cGGACACG-3′ | + |
| 2 | 130-136 | 5′-aGCGACUG-3′ | + |
| 3 | 154-160 | 5′-uGCUGCUC-3′ | NSb |
| 4 | 196-202 | 5′-uGCUCACA-3′ | + |
| 5 | 232-238 | 5′-uGCCCACA-3′ | ++ |
| 6 | 244-250 | 5′-cGCUCCAG-3′ | NS |
| 7 | 250-256 | 5′-aGGUCACG-3′ | + |
| 8 | 256-262 | 5′-cGCGGACU-3′ | + |
| 9 | 286-292 | 5′-uGGCUGCC-3′ | ++ |
The lowercase nucleotides are at position-1 in reference to the cleavage site. The uppercase nucleotides base pair with δRz to form the P1 stem (Fig. 1B).
NS, nonspecific cleavage was observed.
In order to validate these predicted sites, biochemical assays were carried out in vitro with the full-length EF-Tu mRNA produced by runoff transcription. We performed RNase H assays using 7-mer oligodeoxyribonucleotides corresponding to the nine potential recognition domains of δRz. RNase H specifically cleaves the RNA of RNA-DNA heteroduplexes (27). Hence, this enzyme can be used to verify whether specific ODNs bind to the target mRNA, suggesting what regions of the target might indeed be single stranded and could be more accessible to base pair with the ribozyme (5). Internally labeled EF-Tu mRNA was preincubated with unlabeled ODNs, digested by RNase H, and analyzed on denaturing polyacrylamide gels. Cleavage was observed with all ODNs, although at different levels (Table 1). However, some ODNs generated additional bands corresponding to nonspecific cleavage (sites 3 and 6) (Table 1), indicating that they base paired with the EF-Tu mRNA at more than one position even though there was only one perfect complementary sequence for each ODN along the mRNA. The lack of specificity observed in some of our assays may be explained by the fact that E. coli RNase H is active on RNA-DNA heteroduplexes as short as 4 bp (27). Among the ODNs directing specific RNase H cleavage of the EF-Tu mRNA, the two most accessible sites were the sites at positions 232 to 238 and 286 to 292 (sites 5 and 9) (Table 1). These two sites were retained for subsequent in vivo analyses along with a less accessible site, site 4 (positions 196 to 202) (Table 1), for comparison.
Design of δRz expression cassettes.
Additional nucleotides at either the 5′ or 3′ termini of the catalytic core of trans-acting hammerhead ribozymes were shown to impair the catalytic activity, both in vitro and in vivo (7, 44). These extra bases could potentially reduce the specificity and binding efficiency of ribozymes with their target mRNAs by allowing nonspecific hybridization or promote misfolding of the ribozymes, leading to inactive three-dimensional conformations and/or degradation. Common transcription strategies produce transcripts with variable 5′ and 3′ termini because RNA polymerases do not always start and stop at the same position. Moreover, multiple cloning sites between promoters and transcriptional terminators append additional nucleotides at both ends of the desired transcript. In order to circumvent these potential problems when δRzs were expressed in vivo, we designed a cis-trans-cis expression cassette (11, 24). Each δRz was embedded between two self-cleaving hammerhead motifs, forming HH1-δRz-HH2 expression cassettes (Fig. 2A). After transcription, the hammerhead sequences should self-cleave to release δRzs that are exactly 80 nt long with the desired 5′ and 3′ ends (Fig. 2A and B). The hammerhead motifs were designed in such a way that sequences at both ends of the liberated δRzs base pair together, extending the P2 stem (Fig. 2A). Previous studies have demonstrated that δRz is extremely stable in cell culture mainly because its 5′ and 3′ termini base pair and form the P2 stem (36). Furthermore, the catalytic activity of δRz has also been shown to be unaffected by an extended P2 stem (D. Lévesque and J. P. Perreault, unpublished data).
To test this HH1-δRz-HH2 expression strategy, in vitro runoff transcripts were obtained using the different pBS-HH1-δRz-HH2 plasmids linearized by EcoRI as templates (Fig. 2B). Most of the full-length transcripts (205-nt bands) were processed efficiently during transcription, producing the following three fragments: 80-nt fragments corresponding to the released trans-acting δRzs and 78- and 47-nt fragments corresponding to HH1 and HH2, respectively (Fig. 2C). Only trace amounts of intermediates, in which one of the two hammerhead sequences did not self-cleave, were detected (Fig. 2C, cf. 127- and 158-nt bands). Accumulation of the 127-nt intermediate, combined with detection of a relatively faint 47-nt band, suggested that the HH2 self-cleaving motif is less efficient than the HH1 motif (Fig. 2C). More importantly, these results demonstrated that in vitro, the three δRzs (δRz-E196, -E232, and -E286) were precisely and efficiently released after self-cleavage of their flanking hammerhead motifs.
Optimization of δRz expression in L. lactis.
Few inducible promoters in L. lactis have been well characterized (20). The nisin promoter (PnisA) is the best-characterized and most commonly used L. lactis inducible promoter. Nisin is a small antimicrobial peptide (lantibiotic) that is produced naturally by strains of L. lactis harboring the nisin biosynthetic gene cluster (32). Expression of genes under control of the nisin promoter is regulated by signal transduction through a two-component regulatory system, which is composed of a transmembrane sensor kinase (NisK) and a response regulator (NisR) (32). Based on this activation scheme, different strains of L. lactis were engineered to control gene expression using the PnisA inducible promoter (e.g., NZ9800, NZ9000 and NZ3900) (17, 33). Other strains of L. lactis (for example, IL1403) do not harbor a chromosomal copy of the nisR or nisK gene. Nevertheless, these strains can be transformed with the pMSP3535 shuttle plasmid carrying these two genes, allowing nisin-dependent promoter activation (10).
The levels of expression of δRz-E196 in four L. lactis strains (NZ9800, NZ9000, NZ3900, and IL1403/pMSP3535) were determined by Northern blot hybridization (Fig. 3). All strains contained the ribozyme expression cassette under control of the nisin promoter (pDL-PnisA-HH1-δRz-E196-HH2). Cells were diluted from fresh overnight cultures and grown to an OD600 of 0.5, and ribozyme expression was induced with nisin for 1 h. The expression efficiencies in these different backgrounds were compared to that of the IL1403 control strain, in which the promoter cannot be activated upon nisin induction. The fully processed δRz and a precursor (Pre-δRz) were observed in all strains except the IL1403 control strain, in which only a faint δRz band was observed (Fig. 3). The presence of relatively large amounts of precursor suggested that one of the two hammerhead ribozymes was less active in L. lactis than in vitro (Fig. 2C). Use of probes that specifically recognize either the HH1-δRz junction or the δRz-HH2 junction revealed that the accumulated precursor band (Fig. 3, Pre-δRz) contained almost exclusively the HH1-δRz junction (data not shown). This showed that in contrast to what was observed in vitro (Fig. 2C), HH1 self-cleavage is less efficient than HH2 self-cleavage in L. lactis.
FIG. 3.
Ribozyme expression from the nisin promoter: autoradiogram of Northern hybridization blot showing levels of δRz-E196 expression in different L. lactis strains. Cells were diluted from fresh overnight cultures and grown to an OD600 of 0.5, and ribozyme expression was induced with nisin for 1 h. Strains NZ9800, NZ9000, and NZ3900 contain a unique chromosomal copy of both the nisK and nisR genes and trigger expression of δRz-E196 from the nisin promoter (PnisA). Control strain IL1403 lacks the nisK and nisR genes. The pMSP3535 plasmid carries nisK and nisR, which allows gene expression from the nisin promoter in IL1403. Levels of ribozyme expression were normalized against the amount of 5S rRNA, and then the levels in different strains were compared using the amount of fully processed δRz-E196 produced in IL1403 as the basal level (1×).
Using 5S rRNA to normalize the signals, we determined that the amount of fully processed δRz was largest in the IL1403 strain containing the pMSP3535 plasmid (Fig. 3, compare the δRz bands). The level of δRz produced in this strain was estimated to be 25-fold higher than the level produced in the same strain lacking the two-component system (IL1403) and 5- to 12-fold higher than the levels produced in the NZ3900, NZ9000, and NZ9800 strains, which contained a single chromosomal copy of nisK and nisR (Fig. 3). Using equimolar amounts of 5S rRNA and δRz probes in combined Northern blot hybridizations and taking into consideration the fact that each rRNA (5S, 16S, and 23S) accounts for approximately 20% of the total RNA in bacteria (14, 23), we estimated that the fully processed δRz and Pre-δRz accounted for 0.3% and 0.6% of the total RNA, respectively (data not shown). These results demonstrated that high levels of fully processed δRz-E196 could be detected in IL1403/pMSP3535 upon nisin induction. Moreover, it is possible that partially processed δRz-E196 could also recognize and cleave its target. Interestingly, the bands corresponding to the fully processed δRz-E196 and its precursor were unique and sharp in all strains, suggesting that these RNAs are stable in L. lactis (Fig. 3).
We then tested different conditions to optimize the expression of δRz in L. lactis. First, we investigated the possibility of stimulating HH1 self-cleavage to produce more of the fully processed δRz and to prevent the accumulation of the HH1-δRz intermediate (Pre-δRz) (Fig. 3). The ribozyme expression assay shown in Fig. 3 was performed at 30°C, the optimal growth temperature for L. lactis (49); however, the optimal temperature for hammerhead ribozyme self-cleavage in vitro is more than 40°C (40). Since L. lactis grows relatively well at 37°C, we compared the levels of fully processed δRz produced in IL1403/pMSP3535 grown at 30°C and at 37°C. The overall levels of Pre-δRz and δRz were reduced 2.5-fold at 37°C compared to the levels at 30°C, and the level of the fully processed δRz was reduced 4.5-fold (Fig. 4A). These results suggested that either the HH1 ribozyme is less efficient (twofold reduction) or δRz is less stable at 37°C than at 30°C. This experiment demonstrated that the optimal temperature for expression of δRz in L. lactis is 30°C. We thus limited our studies of the effect of δRzs on their targets to experiments performed at 30°C, the optimal growth temperature of L. lactis.
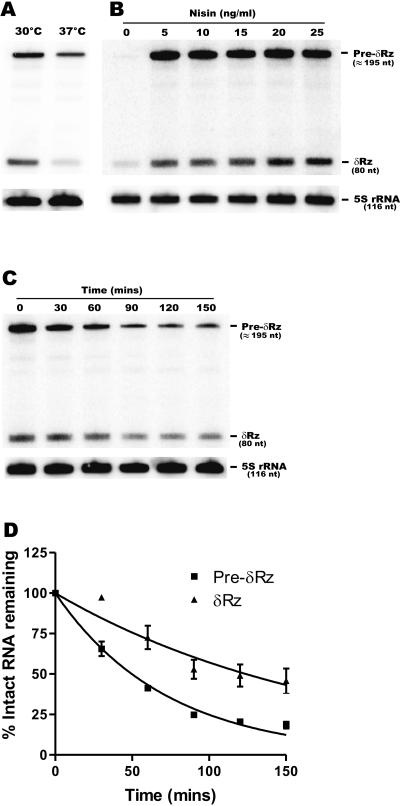
FIG. 4.
Levels of δRz-E196 expression in different conditions. Levels of ribozyme expression were assessed by Northern blot hybridization with total RNA from L. lactis (IL1403/pMSP3535) grown at different temperatures (30°C and 37°C) (A) and with different nisin concentrations (B). (C) Stability of δRz in strain IL1403/pMSP3535. Ribozyme expression was triggered by addition of nisin for 1 h. Then transcription was stopped using rifampin, and aliquots were removed at different times and subsequently analyzed by Northern blot hybridization. (D) Graphic representation of the stability (panel C) of δRz and its precursor (Pre-δRz) as a function of time.
It has been demonstrated previously that the levels of expression from the PnisA promoter are directly related to the nisin concentration (17). We therefore analyzed the levels of δRz expression at nisin concentrations ranging from 5 and 25 ng/ml (Fig. 4B). Nisin induction at a concentration greater than 5 ng/ml did not significantly increase δRz production in IL1403/pMSP3535. Since IL1403 does not harbor the nisin gene cluster, it lacks the nisin immunity genes (nisI and nisFEG) (33). This strain is thus nisin sensitive, and a reduction in growth was observed following addition of nisin at concentrations of 15 ng/ml or greater. Although there was no significant difference between ribozyme expression in the presence of 5 ng/ml of nisin and ribozyme expression in the presence of 10 ng/ml of nisin, 10 ng/ml was chosen for our studies because this concentration was shown to be sublethal for L. lactis (34).
Finally, we investigated the stability of δRz in L. lactis. One of the main drawbacks in the development of several other ribozymes as molecular tools has been their instability in vivo (42). Conversely, δRz was recently shown to be highly stable in human-derived cell lines (36). Ribozyme stability was monitored for 150 min after addition of rifampin, an efficient and widely used transcriptional inhibitor that is active in both gram-positive and some gram-negative bacteria (48). The estimated half-lives of Pre-δRz and fully processed δRz were 50 and 125 min, respectively (Fig. 4C and D). The unusual stability of these RNA molecules may result from the high degree of secondary structure and the compact folding of the δRz catalytic core. The stability of both RNAs is astonishingly high compared to that of most bacterial mRNAs. In Bacillus subtilis, mRNAs of about 1,500 genes studied simultaneously by microarray analysis had an average half-life of 5 min; most mRNAs (80%) had half-lives of less than 7 min, while more than 30 mRNAs had half-lives of 15 min or more (25). Similarly, in E. coli, the mRNA half-lives range from a few seconds to 0.5 h, and the average is about 3 min (3).
Effect of δRz on the half-life of EF-Tu mRNA.
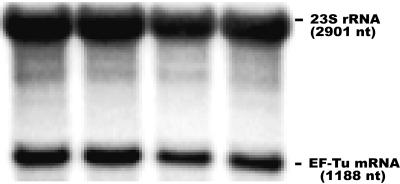
We first confirmed by combined Northern blot hybridization, using equimolar amounts of the 23S rRNA and EF-Tu probes, that EF-Tu mRNA accounts for approximately 10% (11.4% ± 1.3%) of the total RNA in L. lactis (Fig. 5). This is similar to the level of EF-Tu mRNA in E. coli (E. Massé, personal communication), even though L. lactis has only one EF-Tu gene and E. coli has two EF-Tu genes (tufA and tufB).
FIG. 5.
Level of EF-Tu mRNA in L. lactis: combined Northern blot using equimolar amounts of specific probes for 23S rRNA and EF-Tu mRNA. The intensity of the bands was quantified with the Quantity One software (Bio-Rad) and was adjusted to the labeling of the corresponding probe.
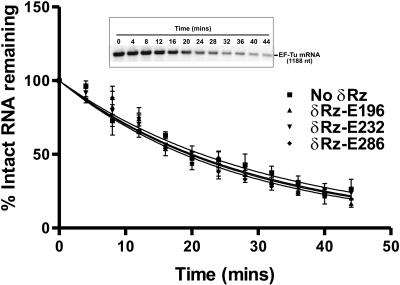
We then determined the half-life of EF-Tu mRNA in L. lactis. Cells containing an empty control plasmid (pDL-PnisA) were grown to an OD600 of 0.5, nisin was added to the medium, and 1 h later, transcription was stopped by addition of rifampin. The stability of EF-Tu mRNA was assessed at different times by Northern blot hybridization (three independent experiments). Using the 23S rRNA as a loading control, we found that in the absence of δRz, EF-Tu mRNA was very stable in L. lactis, with an estimated half-life of 22 min (21.5 ± 2.0 min) (Fig. 6 and Table 2).
FIG. 6.
Effects of three δRzs on the EF-Tu mRNA half-life in L. lactis: graphic representation showing the decay of EF-Tu mRNA as a function of time. The half-lives were determined from three independent experiments using the one-phase exponential decay equation of GraphPad Prism, version 3.2. The inset shows the autoradiogram of a representative Northern hybridization blot. Each signal was normalized with its corresponding 23S rRNA signal.
TABLE 2.
EF-Tu mRNA half-lives
| Targeted site | EF-Tu mRNA half-lives (min)a
|
|
|---|---|---|
| δRz | SOFA-δRz | |
| None | 21.5 ± 2.0 | 21.5 ± 2.0 |
| E196 | 19.8 ± 2.3 | 15.8 ± 1.1 |
| E232 | 20.0 ± 1.8 | 11.4 ± 0.9 |
| E286 | 18.8 ± 1.4 | 11.3 ± 1.0 |
| SDb | NDc | 16.8 ± 1.6 |
Average of three independent experiments.
SD, Shine-Dalgarno.
ND, not determined.
We expressed δRz-E232 from the cis-trans-cis expression cassette shown in Fig. 2 or directly cloned downstream from the nisin promoter. Overexpression of δRz-E232 from pDL-PnisA-HH1-δRz-E232-HH2 led to a slight decrease (about 7.0%) in the half-life of EF-Tu mRNA (20.0 ± 1.8 min) (Fig. 6 and Table 2). In contrast, expression of the same ribozyme not flanked by the HH1 and HH2 motifs led to a significant increase (55%) in the EF-Tu mRNA half-life (33.4 ± 2.3 min), suggesting that this version of δRz-E232 was inactive but could bind the EF-Tu mRNA and stabilize it (data not shown). These results validated the use of the hammerhead expression cassette to express δRzs in L. lactis. Overexpression of δRzs targeting three different sites within EF-Tu (δRz-E196, δRz-E232, and δRz-E286) led to minor but consistent decreases in the EF-Tu mRNA half-life (7.9% [19.8 ± 1.8 min], 7.0% [20.0 ± 1.8 min], and 12.6% [18.8 ± 1.4 min], respectively) (Fig. 6 and Table 2). The small reductions in the half-life of EF-Tu mRNA (around 10%) (Table 2) in the presence of the different ribozymes were not statistically significant and corresponded to reductions of 2% (δRz-E232), 3% (δRz-E196), and 5% (δRz-E286) in the amount of the EF-Tu mRNA 20 min after the addition of rifampin (Fig. 6). These results suggest either that the target sites within EF-Tu mRNA are not as accessible in vivo as they are in vitro (Table 1) or that the δRzs are not highly active in bacterial cells. The potential target sites chosen may be buried within the tertiary structure of the mRNA or occluded by associated proteins (Fig. 1A) (41).
Activity of modified δRzs in bacterial cells.
In attempts to improve the degradation of EF-Tu mRNA, a new and improved δRz variant designated SOFA-δRz (6) was tested in L. lactis. SOFA-δRz (Fig. 7A) was shown to be highly specific and is activated by the presence of its target in vitro (6). In SOFA-δRz, the classic δRz is linked to a target-dependent module that acts as a biosensor. In the absence of the target, the blocker forms an intramolecular stem with the P1 strand, preventing δRz binding and cleavage (“off” conformation). Upon addition of the substrate, the biosensor recognizes and binds the target, releasing the P1 strand, which can subsequently hybridize with its substrate and catalyze cleavage (“on” conformation). The biosensor thus acts as a riboswitch regulating the catalytic activity of δRz (Fig. 7A). The cleavage efficiency of a specific SOFA-δRz was shown to be fivefold higher than the cleavage efficiency of the original δRz in vitro (6). Furthermore, kinetic analyses revealed a second-order rate constant (kcat/Km) that was 25-fold higher than that of the original δRz.
Specific SOFA-δRzs targeting the E196, E232, and E286 sites of the L. lactis EF-Tu mRNA were constructed, which allowed direct comparison of the activities of these new SOFA-δRzs and the original δRzs studied. The half-lives of EF-Tu mRNA were determined in the presence of the different SOFA- δRzs as described above. Both SOFA-δRz-E232 expressed from the cis-trans-cis expression cassette and SOFA-δRz-E232 cloned directly downstream from the nisin promoter were used in order to reassess the importance of using the cis-trans-cis expression strategy. Surprisingly, the effects on the half-life of EF-Tu mRNA were completely different (as determined in three independent experiments). SOFA-δRz-E232 expressed from the cis-trans-cis expression cassette had a limited effect on the half-life of EF-Tu mRNA (19.8 ± 1.2 min) similar to the effect observed with the classical δRz-E232 (20.0 ± 1.8 min) also expressed from the cis-trans-cis expression cassette (Table 2). On the other hand, SOFA-δRz-E232 embedded in a longer transcript was considerably more efficient, reducing the half-life of EF-Tu mRNA by almost 50% (11.4 ± 0.9 min). These results showed that this SOFA-δRz is significantly more active when it is present in a longer transcript and do not justify the use of the cis-trans-cis expression cassette.
The other two SOFA-δRzs (SOFA-δRz-E286 and -E196) were thus cloned directly downstream of the nisA promoter. Ribozyme expression assays showed that the SOFA-δRzs were expressed as long transcripts (∼275 nt) (data not shown). In contrast to the original δRzs, all three SOFA-δRzs induced substantial decreases in the half-life of EF-Tu mRNA, although at different levels (Fig. 7B and Table 2). The SOFA-δRzs targeting the sites found to be the most accessible were indeed the most efficient SOFA-δRzs (SOFA-δRz-E232 and -E286) (Table 1). They reduced the half-life of EF-Tu mRNA by approximately 50%, from 21.5 ± 2.0 min to 11.4 ± 0.9 min and 11.3 ± 1.0 min, respectively. SOFA-δRz-E196, targeting a less accessible site, was correspondingly less efficient, and it reduced the EF-Tu mRNA half-life by only 27% (15.8 ± 1.1 min). These results showed that there was a good correlation between site accessibility, as determined in vitro by RNase H assays (Table 1), and the in vivo cleavage efficiencies of the SOFA-δRzs in L. lactis (Table 2). This validated the in vitro approach used to identify accessible sites in mRNA targets.
Only a few cases of efficient targeting and cleavage using the hammerhead ribozyme have been reported previously in bacteria (22, 28). The lack of success in development of a general method of inhibition of bacterial gene expression using ribozymes could result from lower accessibility of mRNA to binding by ribozymes because of the tight coupling between transcription and translation (7, 12, 13). To test this hypothesis, we designed a SOFA-δRz targeting the initiator codon (AUG) of the EF-Tu mRNA with a biosensor that recognized a portion of the Shine-Dalgarno sequence (SOFA-δRz-SD). This SOFA-δRz interacts mostly in the 5′ untranslated region of the EF-Tu mRNA. Overexpression of this SOFA-δRz decreased the half-life of the EF-Tu mRNA by approximately 20% (16.8 ± 1.6 min) (Table 2). This value is comparable to the decrease in the half-life induced by SOFA-δRz-E196, suggesting that the presence of ribosomes in the coding regions of mRNA substrates does not significantly reduce the target accessibility to SOFA-δRzs. Through their biosensor domain, SOFA-δRzs bind to an additional segment (12 nt) of the target RNA, which may actually help unwind tertiary and secondary structures in the vicinity of the cleavage site and favor P1 stem formation (7 nt). This longer pairing interaction between the ribozyme and its substrate (19 nt) may also help in removing ribosomes that may still be associated with the mRNAs.
As a control, an inactive SOFA-δRz (δRzC76A/-P1.1-E232) (Fig. 7A) was constructed by replacing the cytosine at position 76 with an adenine (C76A) and by replacing the cytosines at positions 40 and 41 with guanines (C40G and C41G), which disrupted the formation of the P1.1 stem (Fig. 7A) (19). Both of these features are essential for cleavage activity (4, 47). Thus, the resulting δRz should exhibit the same substrate binding properties as the active equivalent but should have had no cleavage activity. This control was used to verify that the decreases in the half-lives of EF-Tu mRNA observed upon SOFA-δRz overexpression were caused by the catalytic activity of the SOFA-δRzs. No effect on the half-life of EF-Tu mRNA was detected in the presence of the SOFA-δRzC76A/-P1.1-E232 control (21.2 ± 1.8 min). This confirmed that the effects previously observed (Table 2) are specific to the catalytic activities of the SOFA-δRzs on their substrates. However, one could have expected stabilization of the target, as seen with δRz-E232 not flanked by hammerhead motifs. The presence of a biosensor in SOFA-δRz-E232 significantly changes the interaction of this ribozyme with the target mRNA compared to that of the classic δRz-E232. The additional 12 bp between SOFA-δRz-E232 and its substrate seems to influence the stability of the EF-Tu mRNA differently.
We also monitored bacterial growth upon overexpression of SOFA-δRz-E232 and its catalytically inactive control. No significant effect was observed in L. lactis growth curves when these two ribozymes were expressed compared to the same strain carrying the empty plasmid. Moreover, similar growth was observed whether the production of the ribozymes was induced or not induced with nisin. These results suggest either that the reduction in the EF-Tu mRNA levels induced by SOFA-δRz-E232 is not sufficient to affect the L. lactis growth rate or that there is a feedback effect that stimulates the expression of this essential gene to compensate for the reduction in the EF-Tu mRNA levels.
DISCUSSION
This study showed that different trans-acting antigenomic δRzs and SOFA-δRzs expressed in the industrially important gram-positive bacterium L. lactis were extremely stable and active. Furthermore, several ribozymes significantly reduced the half-life of the unusually abundant and stable EF-Tu mRNA, which was estimated to account for 11.4% ± 1.3% of the total RNA in L. lactis.
SOFA-δRz appears to be suitable for development of gene inactivation systems in L. lactis.
All SOFA-δRzs studied were significantly more active than their classic δRz counterparts targeting the same sites on the L. lactis EF-Tu mRNA. The SOFA-δRzs were 3.4-fold (SOFA-δRz-E196), 6.7-fold (SOFA-δRz-E232), and 3.8-fold (SOFA-δRz-E286) more efficient in reducing the half-life of EF-Tu mRNA (Table 2). It is noteworthy that the most accessible target sites in EF-Tu mRNA (E232 and E286), both predicted in vitro (Table 1) and confirmed in vivo (SOFA-δRzs) (Table 2), gave the best targeting improvement between the classic δRzs and SOFA-δRzs (Table 2). This validated the in vitro approach to site accessibility prediction and confirmed that even if the SOFA-δRzs are considerably more active than the classical δRzs, it is still worthwhile to identify the most accessible sites in potential mRNA targets. These results also suggest that the formation of both biosensor (12-bp) and P1 (7-bp) stems between the SOFA-δRzs and their substrates is more efficient in opening structures within the EF-Tu mRNA than the formation of the P1 stem alone is. Substrate secondary structure and accessibility analyses for the SOFA-δRz should take into account the formation of both a biosensor and a P1 stem between the SOFA-δRz and its potential substrate and thus assess substrate accessibility using 20- to 25-nt windows instead of 7-nt windows, as previously used for the classic δRzs. However, it appears that predicting potential secondary structures of the biosensor sequences is unnecessary. The biosensor sequences of the different SOFA-δRzs are most likely single stranded because these short unpaired regions (10 to 12 nt) are flanked by stems that limit the formation of stable secondary structures. Oligonucleotide hybridization and RNase H probing assays confirmed that the biosensor regions of the different SOFA-δRzs studied are mostly single stranded (6a).
The use of SOFA-δRzs to target mRNAs in bacteria should be more specific than the use of classical δRzs. Based on computer analyses, we determined that the 7-nt recognition sequence of each classical δRz studied (E196, E232, and E286) was found in two or three other locations in the IL-1403 L. lactis genome (2.4 × 106 bp) (8). These analyses showed that recognition sequences that are only 7 nt long are not stringent enough to guarantee absolute target specificity in L. lactis and likely in bacteria in general. Moreover, these analyses probably overestimate the specificity of the classic δRzs since other nonspecific binding sites could be recognized using a subset of the 7-nt recognition sequence (e.g., 6 nt). Analyses using the biosensor (12-nt) and P1 recognition (7-nt) sequences of three SOFA-δRzs (SOFA-δRz-E196, SOFA-δRz-E232, and SOFA-δRz-E286) demonstrated that the SOFA-δRzs were all highly specific, without any other potential cleavage sites present in the IL-1403 genome. However, this does not prevent the biosensor from partially base pairing with other RNA molecules. In such a situation, the P1 region, if liberated, would be in the wrong environment and would most probably not bind to and cleave the nonspecific target. The SOFA-δRzs should thus provide significantly improved substrate specificity in vivo, as demonstrated previously in vitro (6).
Taking into consideration the total amount of stable ribozymes produced in L. lactis upon nisin induction (Pre-δRz and δRz accounted for 0.9% of the total RNA) (Fig. 3 and 4) and the abundance of the EF-Tu mRNA (11.4% of the total RNA), the ribozyme/target ratio corresponds to approximately 1:12.7. In order to verify the calculated ribozyme/EF-Tu mRNA ratio (1:12.7), we used a different approach. Using equimolar amounts of δRz- and EF-Tu-specific probes in combined Northern blot hybridizations, we directly calculated a ribozyme/target ratio of 1:13.5 (data not shown). The observed reductions in the amount of EF-Tu mRNA upon induction of these various δRzs (3% for δRz-E196, 2% for δRz-E232, and 5% for δRz-E286) (Fig. 6), in conjunction with the difference in the relative amounts of these molecules (δRz/EF-Tu mRNA ratio, ∼1:13 [7.7%]), suggest that each ribozyme (Pre-δRz plus δRz) cut less than one molecule of substrate after 20 min. The situation appears to be different for the SOFA-δRzs. The amount of EF-Tu mRNA was reduced by 23% in the presence of either SOFA-δRz-E286 or SOFA-δRz-E232 at 20 min (Fig. 7B). This suggests that during this 20-min period one molecule of SOFA-δRz-E286 or -E232 cut approximately 3.0 molecules of substrate, corresponding to a catalysis rate of 0.15 target/min. Using the same approach, we calculated that SOFA-δRz-E196 and -SD, which reduced EF-Tu mRNA levels by 12% and 9%, respectively, cut approximately 1.6 and 1.2 molecules of substrates in 20 min, corresponding to catalysis rates of 0.08 and 0.06 target/min, respectively. We have to keep in mind that as the experiment proceeded, the concentration of substrate decreased faster than the concentration of the highly stable SOFA-δRzs. The calculated catalysis rates are thus conservative rough estimates since the rates were probably higher in the first minutes of the assays (Fig. 7B). Interestingly, rate constants (kobs) of several SOFA-δRzs were determined to be in the same range in vitro under single-turnover conditions (0.10 to 0.12 min−1) (L. J. Bergeron and J.-P. Perreault; unpublished data). This suggests that these ribozymes are as active in vivo as they are in vitro.
Taken together, these results show that in addition to being significantly more active than the classic δRz, the SOFA-δRz is obviously more specific than its predecessor in L. lactis, making it a good molecular tool for specific gene inactivation in bacterial cells. Since usual mRNAs in bacterial cells are at least 2,000-fold less abundant than the EF-Tu mRNA, since the different SOFA-δRzs studied significantly reduced the half-life of an unusually abundant mRNA, and since these ribozymes seem to be as active in vivo as they are in vitro, it is conceivable that this approach could be used to perform complete specific gene knockdown in the industrially important gram-positive bacterium L. lactis. Moreover, the findings obtained with L. lactis suggest that SOFA-δRzs may also be active in other gram-positive bacteria, thus broadening the significance of this gene knockdown approach.
Acknowledgments
We thank G. Dunny for kindly providing the pMSP3535 plasmid and L. J. Bergeron for access to unpublished data. We also thank P. Landry, J. F. Lussier, R. Strasser, O. Didur, D. Lévesque, and K. Yam for technical assistance and N. Acheson, K. Belhocine, C. Quiroga, K. Yam, and E. Massé for providing comments on the manuscript.
The Sherbrooke RNA group is supported by the Université de Sherbrooke and Génome Québec. This work was supported by grants from the CIHR to B.C. and J.P.P. K.F. was a recipient of a doctoral fellowship from FCAR FRSQ (Québec). J.P.P. is a CIHR Scholar, and B.C. is a CIHR New Investigator Scholar and a McGill William Dawson Scholar.
REFERENCES
- 1.Ananvoranich, S., and J.-P. Perreault. 2000. The kinetic and magnesium requirements for the folding of the antigenomic δ ribozymes. Biochem. Biophys. Res. Commun. 270:600-607. [DOI] [PubMed] [Google Scholar]
- 2.Bagheri, S., and M. Kashani-Sabet. 2004. Ribozymes in the age of molecular therapeutics. Curr. Mol. Med. 4:489-506. [DOI] [PubMed] [Google Scholar]
- 3.Belasco, J. G. 1993. mRNA degradation in prokaryotic cells: an overview of control of messenger RNA stability, p. 3-12. In J. G. Belasco and G. Brawerman (ed.), Control of mRNA stability. Academic Press, New York, N.Y.
- 4.Bergeron, L. J., J. Ouellet, and J.-P. Perreault. 2003. Ribozyme-based gene-inactivation system required a fine comprehension of their substrate specificities; the case of delta ribozyme. Curr. Med. Chem. 10:1241-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergeron, L. J., and J.-P. Perreault. 2002. Development and comparison of procedures for selection of delta ribozyme cleavage sites within the hepatitis B virus. Nucleic Acids Res. 30:4682-4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergeron, L. J., and J.-P. Perreault. 2005. Target-dependent on/off switch increases ribozyme fidelity. Nucleic Acids Res. 33:1240-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bergeron, L. J., C. Reymond, and J. P. Perreault. 2005. Functional characterization of the SOFA delta ribozyme RNA 11:1858-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand, E., R. Pictet, and T. Grange. 1994. Can hammerhead ribozymes be efficient tools to inactivate gene function? Nucleic Acids Res. 22:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breaker, R. R. 2004. Natural and engineered nucleic acids as tools to explore biology. Nature 432:838-845. [DOI] [PubMed] [Google Scholar]
- 10.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:83-90. [DOI] [PubMed] [Google Scholar]
- 11.Bussière, F., S. Ledû, M. Girard, M. Héroux, J.-P. Perreault, and D. P. Matton. 2003. Development of an efficient cis-acting ribozyme cassette in plants. Plants Biotechnol. 1:423-435. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., G. Ferbeyre, and R. Cedergren. 1997. Efficient hammerhead ribozyme and antisense RNA targeting in a slow ribosome Escherichia coli mutant. Nature 15:432-435. [DOI] [PubMed] [Google Scholar]
- 13.Chuat, J. C., and F. Galibert. 1989. Can ribozymes be used to regulate prokaryote gene expression. Biochem. Biophys. Res. Commun. 162:1025-1029. [DOI] [PubMed] [Google Scholar]
- 14.Coudon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cousineau, B., S. Lawrence, D. Smith, and M. Belfort. 2000. Retrotransposition of a bacterial group II intron. Nature 404:1018-1021. [DOI] [PubMed] [Google Scholar]
- 16.D'Anjou, F., L. J. Bergeron, N. B. Larbi, I. Fournier, M. Salzet, J.-P. Perreault, and R. Day. 2004. Silencing of SPC2 expression using an engineered δ ribozyme in the mouse βTC-3 endocrine cell line. J. Biol. Chem. 279:14232-14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Ruyter, P. G. G. A., O. P. Kuipers, M. M. Beerthuyzen, I. Van Alen-Boerrigter, and de W. M. Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deschênes, P., D. A. Lafontaine, S. Charland, and J.-P. Perreault. 2000. Nucleotides −1 to −4 of the hepatitis delta ribozyme substrate increases the specificity of the ribozyme cleavage. Antisense Nucleic Acid Drug Dev. 10:53-61. [DOI] [PubMed] [Google Scholar]
- 19.Deschênes, P., J. Ouellet, J. Perreault, and J.-P. Perreault. 2003. Formation of the P1.1 pseudoknot is critical for both the cleavage activity and the substrate specificity of an antigenomic trans-acting delta ribozyme. Nucleic Acids Res. 31:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djordjevic, G. M., and T. R. Klaenhammer. 1998. Inducible gene expression systems in Lactococcus lactis. Mol. Biotechnol. 9:127-139. [DOI] [PubMed] [Google Scholar]
- 21.Ferré d'Amaré, A. R., K. Zhou, and J. A. Doudna. 1998. Crystal structure of a hepatitis δ virus ribozyme. Nature 395:567-574. [DOI] [PubMed] [Google Scholar]
- 22.Fujita, S., T. Koguma, J. Ohkawa, K. Mori, T. Kohda, H. Kise, S. Nishikawa, M. Iwakura, and K. Taira. 1997. Discrimination of a single base change in a ribozyme using the gene for dihydrofolate reductase as a selective marker in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gourse, R. L., T. Gaal, M. S. Bartlette, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 24.Guerrier-Takada, C., and S. Altman. 2000. Inactivation of gene expression using ribonuclease P and external guide sequences. Methods Enzymol. 313:442-456. [DOI] [PubMed] [Google Scholar]
- 25.Hambraeus, G., C. von Wachefeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Gen. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, M. C., A. K. Nielsen, S. Molin, K. Hammer, and M. Kilstrup. 2001. Changes in rRNA levels during stress invalidates results from mRNA blotting: fluorescence in situ rRNA hybridization permits renormalization for estimation of cellular mRNA levels. J. Bacteriol. 183:4747-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogrefe, H. H., R. I. Hogrefe, R. I. Walder, and J. A. Walder. 1990. Kinetic analysis of Escherichia coli RNase H using DNA/RNA-DNA/DNA substrate. J. Biol. Chem. 265:5561-5566. [PubMed] [Google Scholar]
- 28.Inokuchi, Y., N. Yuyama, A. Hirashima, S. Nishikawa, J. Ohkawa, and K. Taira. 1994. A hammerhead ribozyme inhibits the proliferation of an RNA coliphage SP in Escherichia coli. J. Biol. Chem. 269:11361-11366. [PubMed] [Google Scholar]
- 29.Kato, Y., T. Kuwabara, M. Warashina, H. Toda, and K. Taira. 2001. Relationships between the activities in vitro and in vivo of various kinds of ribozyme and their intracellular localization in mammalian cells. J. Biol. Chem. 276:15378-15385. [DOI] [PubMed] [Google Scholar]
- 30.Khan, A. U., and S. K. LaI. 2003. Ribozymes: a modern tool in medicine. J. Biomed. Sci. 10:457-467. [DOI] [PubMed] [Google Scholar]
- 31.Krab, I. M., and A. Parmeggiani. 1998. EF-Tu, GTPase odyssey. Biochim. Biophys. Acta 1443:1-22. [DOI] [PubMed] [Google Scholar]
- 32.Kuipers, O. P., M. M. Beerthuyzen, P. G. G. A. de Ruyter, E. J. Luesnik, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 33.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 34.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing controlling gene expression in the lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 35.Leblanc, D., L. N. Lee, and A. Abu al Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon pVa380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 36.Lévesque, D., S. Choufani, and J.-P. Perreault. 2002. Delta ribozyme benefits from a good stability in vitro that becomes outstanding in vivo. RNA 8:464-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews, D. H., M. E. Burkard, S. M. Freier, J. R. Wyatt, and D. H. Turner. 1999. Predicting oligonucleotide affinity to nucleic acid targets. RNA 5:1458-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependency of thermodynamic parameters improved prediction of RNA structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 39.Mercure, S., D. Lafontaine, G. Roy, and J.-P. Perreault. 1997. Le motif autocatalytique delta de l'hépatite delta humaine. Med. Sci. 13:662-669. [Google Scholar]
- 40.Peracchi, A. 1999. Origins of the temperature dependence of hammerhead ribozyme catalysis. Nucleic Acids Res. 27:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce, M. L., and D. E. Ruffner. 1998. Construction of a directed hammerhead ribozyme library: towards the identification of optimal target sites for antisense-mediated gene inhibition. Nucleic Acids Res. 26:5093-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu, L., A. Moreira, G. Kaplan, R. Levizt, J. Y. Wang, and K. Drlica. 1998. Degradation of hammerhead ribozymes by human ribonucleases. Mol. Gen. Genet. 4:352-362. [DOI] [PubMed] [Google Scholar]
- 43.Roy, G., S. Ananvoranich, and J.-P. Perreault. 1999. Delta ribozyme has the ability to cleave in trans an mRNA. Nucleic Acids Res. 27:942-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz, J., C. H. Wu, Y. Ito, and G. Y. Wu. 1997. Design and preparation of a multimeric self-cleaving hammerhead ribozyme. BioTechniques 22:338-345. [DOI] [PubMed] [Google Scholar]
- 45.Schubert, S., and J. Kurreck. 2004. Ribozyme- and deoxyribozyme-strategies for medical applications. Curr. Drug Targets 8:667-681. [DOI] [PubMed] [Google Scholar]
- 46.Sheng, J., S. Al-Anouti, and S. Ananvoranich. 2004. Engineered delta ribozymes can simultaneously knock down the expression of the genes encoding uracil phosphoribosyltranferase and hypoxanthine-xanthine-guanine phosphoribosyltransferase in Toxoplasma gondii. Int. J. Parasitol. 34:253-263. [DOI] [PubMed] [Google Scholar]
- 47.Shih, I. H., and M. D. Been. 2002. Catalytic strategies of hepatitis delta virus ribozymes. Annu. Rev. Biochem. 71:887-917. [DOI] [PubMed] [Google Scholar]
- 48.Tavormina, P. L., W. S. Reznikoff, and C. A. Gross. 1996. Identifying interacting regions in the beta subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 258:213-223. [DOI] [PubMed] [Google Scholar]
- 49.Teuber, M. 1995. The genus Lactococcus. The genera of lactic acid bacteria, p. 173-234. In B. J. B. Wood and W. H. Holzapfel (ed.), The lactic acid bacteria. Blackie Academic, Glasgow, Scotland.
- 50.Wadkins, T. S., A. T. Perrotta, A. R. Ferré d'Amaré, J. A. Doudna, and M. D. Been. 1999. A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA 6:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells, J. M., P. W. Wilson, and R. W. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]