Abstract
The sale of small turtles is banned by the Food and Drug Administration from the U.S. market due to concerns about their excretion of Salmonella spp. To produce a safe pet for the export market, the Louisiana pet turtle industry uses gentamicin sulfate baths (1,000 μg/ml) to eradicate Salmonella spp. from turtle eggs. In 1999, we analyzed bacterial samples recovered from turtle farms and found that strains of Salmonella enterica subsp. arizonae and other bacteria, such as Enterobacter cloacae, Citrobacter freundii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia, were resistant to high concentrations of gentamicin (>2,000 μg/ml) and to other aminoglycosides. The goal of this study was to identify the gene(s) which contributes to the high-level gentamicin resistance phenotype observed in bacteria from environmental samples with turtle farming activity, particularly the salmonellae, and to estimate the incidence of such genes in these bacteria. R plasmids from gentamicin-resistant strains were transferred by conjugation and transformation to naive Escherichia coli cells. Cloning and sequencing of the gentamicin resistance determinants on these plasmids revealed the presence of the aminoglycoside acetyltransferase genes aac(3)-IIa and aac(3)-VIa; the latter was present as a gene cassette of a class 1 integron. Multiplex PCR assays showed that every gentamicin-resistant isolate carried one of these acetyltransferase genes. Pulsed-field gel electrophoresis and restriction enzyme digestion analysis of R plasmids carrying these genes revealed different restriction profiles and sizes, indicating a dissemination of the gentamicin resistance genes through mobile molecular elements. The data presented highlight the need to develop an alternate method for the eradication of Salmonella spp. from turtle eggs.
Turtle farming has been an important economic activity in the state of Louisiana since the 1930s (39). The sale of small carapace turtles, Pseudemys (Trachemys) scripta elegans, was banned in 1975 in the United States' domestic pet market by the Food and Drug Administration (FDA) due to concerns about their excretion of the human pathogen Salmonella spp. (9, 35, 38). Estimates in the 1970s indicated that approximately 3 × 105 cases of human salmonellosis could be traced to pet turtles (9, 38), reportedly accounting for 14% of Salmonella species infections per year (9). In spite of the domestic ban on the sale of baby turtles, the export market has continued to thrive. Around 12 million hatchlings enter the export market each year for Western Europe, South America, and Asia (22, 26, 37). Prior to export, hatchlings must be certified Salmonella free by independent FDA-approved laboratories (37). This test involves enrichment and isolation of Salmonella spp. by classic bacteriological assays performed on representative turtle samples obtained at the farms (37-39). In order to eradicate Salmonella spp. from turtle eggs, turtle farms follow a modified version of the protocol used in the turkey industry to treat eggs, known as the pressure-differential gentamicin (GEN) egg-dip procedure (15, 19, 21, 32). This FDA-approved treatment involves cleaning freshly picked eggs by immersing them in a 380-ppm Clorox bath and next treating them with a gentamicin sulfate (Garasol; Shering Corp., Bloomfield, N.J.) solution at 1,000 ppm (μg/ml) (26, 37, 39). The use of gentamicin for the treatment of turtle eggs presents various advantages at a commercial turtle farm level, since it does not lose its antimicrobial activity when in solution or when exposed to freezing or autoclaving temperatures (26, 37).
Gentamicin belongs to the aminoglycoside family of antibiotics. The two major classes of 2-deoxystreptamine aminoglycosides are the 4,5-disubstituted, which includes neomycin (NEO), paramomycin, and ribostamycin, and the 4,6-disubstituted, which includes gentamicin, kanamycin (KAN), and amikacin (AMK) (36, 42, 45). Aminoglycosides are highly potent, broad-spectrum, bactericidal antibiotics that are often used in combination therapy with cell wall active agents to treat infections caused by gram-positive organisms, such as Staphylococcus aureus and streptococci (27, 42, 45), as well as intracellular bacteria (24, 42). Gram-negative aerobic bacilli are also sensitive to these antibiotics, which makes them a treatment of choice for these infections (42, 45). Aminoglycoside antibiotics act by inhibiting prokaryotic protein synthesis (12, 25). They induce misreading of the genetic code by binding to the 16S rRNA A site in bacteria (6, 28). The most important mechanism of aminoglycoside resistance is the enzymatic modification of the antibiotic by aminoglycoside-modifying enzymes, which are divided according to their enzymatic activity into the aminoglycoside phosphoryltransferases, the aminoglycoside nucleotidyltransferases, and the aminoglycoside acetyltransferases (AACs) (12, 36, 42, 45). In spite of the resistance problems that have arisen since the early 1970s and their relatively high toxicity for the host, they are still clinically antibiotics of choice, especially in combined therapeutic use with β-lactams (27, 42, 45).
Antibiotic use constitutes a selective pressure which can ultimately result in the emergence of resistant strains; thus, bacteria resistant to a particular antibiotic are often detected following its implementation or use (11, 23, 25, 27). Gentamicin resistance in Salmonella spp. isolated from pet turtles was originally reported in Canada, 1990, in a study stating that turtles imported from Louisiana carried gentamicin-resistant Salmonella spp. (10). Concerns on the emergence of gentamicin-resistant Salmonella enterica subsp. arizonae which could survive gentamicin treatment had been previously reported in the turkey industry (15, 21). In July 1999, several Louisiana pet turtle export shipment lots failed to gain certification. We received bacterial isolates from failed-lot turtle samples from the certification laboratory and detected strains that were resistant to concentrations of gentamicin of >2,000 μg/ml. The goal of the present study was to identify the gene or genes which contribute to the high-level gentamicin resistance phenotype observed in enteric bacteria, particularly Salmonella spp., from environmental samples with turtle farming activity in Louisiana and to determine the incidence of such genes in the turtle farm environment.
MATERIALS AND METHODS
Sample recovery and bacterial isolation and identification.
The main geographical areas in Louisiana where turtle farming is done are Pierre Part, Jonesville, and Ponchatoula (22, 26, 37). Samples were taken from various commercial pet turtle farms in Louisiana during the egg-laying season (April through September) from 1999 to 2002. Samples were individually taken at different steps of the egg sanitation process, since each step involved different exposures to GEN: (i) turtle hatchlings which had failed the certification assay (i.e., they were excreting Salmonella spp.) and which were provided by Central Analytical Laboratories (Belle Chase, LA) (26, 38); (ii) eggs, which were collected directly from dirt nests and which had not yet been treated with GEN; (iii) the 1,000-μg/ml GEN solution used to treat eggs; and (iv) water from pet turtle farm ponds. Each sample was taken separately, and only one isolate of each genus and species was recovered per sample, to avoid double inclusion of the same isolate. Samples were enriched for Salmonella spp. using selenite cystine broth (Difco, Detroit, MI) and isolated on bismuth sulfite (Difco) plates as described previously (37, 38). Gram-negative bacteria were routinely grown at 37°C using McConkey (Difco) plates. Isolates were identified using API-20E or API-NE (bioMérieux SA, Marcy l'Etoile, France). Conjugation assays were done using heart infusion media (Difco). Laboratory strains were grown using Luria-Bertani (LB) medium (33). All GEN-resistant strains were propagated in media supplemented with 100 μg/ml GEN. A summary of all laboratory bacterial strains and plasmids used in this study is found in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsd | Source or reference |
|---|---|---|
| Laboratory strains | ||
| E. coli TOP10 | Electrocompetent or chemically competent; F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 (ara-leu)7697 galUgalK rpsL (Strr) endA1 nupG; Gens (MIC, <3.9 μg/ml) | Invitrogen |
| E. coli strain BM14 | Azir; Gens (MIC, <3.9 μg/ml) | 8 |
| Environmental strains | ||
| S. enterica subsp. enterica, strain S5 | Genr Kanr Tobr; GEN MIC, >2,000 μg/ml | Turtle nest eggsa |
| S. enterica subsp. arizonae, strain A40 | Genr Kanr Tobr; GEN MIC, >2,000 μg/ml | Infected turtlesb |
| S. enterica subsp. arizonae, strain PH5 | Genr Kanr Tobr; GEN MIC, >2,000 μg/ml | Turtle pond waterc |
| E. cloacae, strain E19 | Genr Kanr Tobr; GEN MIC, >2,000 μg/ml | Infected turtlesb |
| C. freundii, strain C14 | Genr Kanr Tobr; GEN MIC, >2,000 μg/ml | Turtle nest eggsa |
| Plasmids | ||
| pGEM-7Zf | Cloning and sequencing vector; Ampr | Promega |
| pCR2.1-TOPO | Cloning and sequencing vector; Kanr | Invitrogen |
| pSal5 | R plasmid isolated from S. enterica subsp. enterica, strain S5 | This study |
| pAri40 | R plasmid isolated from S. enterica arizonae, strain A40 | This study |
| pAriPH5 | R plasmid isolated from S. enterica subsp. arizonae, strain PH5 | This study |
| pEnt19 | R plasmid isolated from E. cloacae, strain E19 | This study |
| pCit14 | R plasmid isolated from C. freundii, strain C14 | This study |
| pGSal5h | pGEM-7Zf derivative containing a 5.5-kb HindIII fragment from pSal5 | This study |
| pGEnt19b | pGEM-7Zf derivative containing a 5.0-kb BamHI fragment from pEnt19 | This study |
Supplied by Central Analytical Laboratory, Belle Chase, LA, July 1999.
Isolated from turtle breeding pond water in Louisiana, June 2000.
Abbreviations: Ampr, ampicillin resistant; Azir, sodium azide resistant; Genr, gentamicin resistant; Gens, gentamicin sensitive; Kanr, kanamycin resistant; Tobr, tobramycin resistant.
Antibiotic susceptibility assays.
The MIC assays were performed with microdilution plates, using Mueller-Hinton (MH) broth (Difco), following the procedure recommended by the CLSI (formerly NCCLS) (29), using the following aminoglycoside antibiotics: GEN, KAN, streptomycin, NEO, tobramycin (TOB), amikacin (AMK), netilmicin, and sisomicin (Sigma, St. Louis, MO). In all cases, the starting antibiotic concentration tested was 2,000 μg/ml, serially diluted twofold to 3.9 μg/ml.
The Kirby-Bauer disk diffusion assay was done using MH agar plates according to CLSI standards (29), using the following antibiotics: GEN (10 μg), KAN (30 μg), TOB (10 μg), AMK (30 μg), ampicillin (AMP) (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), polymixin B (300 units), cephalothin (30 μg), cefotaxime (30 μg), trimethoprim (5 μg), and nitrofurantoin (300 μg) (Difco). Inhibition zone breakpoints were interpreted according to information supplied by the manufacturer and to CLSI standards (29).
R-plasmid isolation and characterization.
Conjugation was done to detect plasmids carrying GEN resistance determinants. In each bacterial mating experiment, a GEN-resistant organism was used as the donor. Representative isolates of each genus and species (chosen based on similar resistance phenotypes) were used as donors and included S. enterica subsp. enterica serovar Enteritidis, S. enterica subsp. arizonae, Enterobacter cloacae, Citrobacter freundii, Pseudomonas aeruginosa, Acinetobacter baumanii, Stenotrophomonas maltophilia, Proteus vulgaris, and Providencia stuartii. Escherichia coli strain BM14, resistant to sodium azide (AZI), was used as the recipient (8). Bacterial mating was done by mixing 1 ml of a log-phase culture of GEN-resistant donor cells with 4 ml of log-phase recipient AZI-resistant E. coli BM14 cells and incubating at 37°C for 16 h, with no shaking. Transconjugant AZI-resistant, GEN-resistant E. coli colonies were selected on heart infusion plates supplemented with 100 μg/ml GEN and 500 μg/ml AZI.
R plasmids from GEN-resistant bacteria were purified using QIAfilter plasmid kits (QIAGEN, Valencia, CA), following instructions provided by the manufacturer for low-copy-number plasmids. Plasmids were eluted in molecular-grade water at 65°C and stored at −20°C. To minimize the possibility of working with strains carrying more than one plasmid (mobilizable plasmids that might cotransfer during conjugation) and to better assess the resistance phenotypes (AZI-resistant transconjugant E. coli has a slower growth rate), plasmids isolated from transconjugants were used to transform naive OneShot TOP10 E. coli cells (Invitrogen, Carlsbad, CA), using a Bio-Rad Gene Pulser II electroporator (Bio-Rad, Hercules, CA), following recommendations from the manufacturer. Briefly, 50 μl electrocompetent cells were transferred to a chilled 0.1-cm cuvette with 1 to 2 μl of diluted plasmid DNA. Pulses were given at 2.0 kV, 25 μF, and 100 to 200 Ω for 5 ms. Transformant GEN-resistant E. coli cells were selected on LB plates supplemented with 100 μg/ml GEN.
To compare their restriction fragment profiles, R plasmids were isolated as described above from environmental GEN-resistant isolates, transconjugant E. coli, and transformant E. coli cells. The DNA concentration of uncut plasmids was estimated by comparison to a Supercoiled Plasmid DNA ladder (Bayou Biolabs, Harahan, LA) after 0.5% Agarose I (Sigma) in 0.5× Tris-borate-EDTA (TBE) (Bio-Rad) gel electrophoresis migration. R plasmids were digested separately with EcoRI, HindIII, or BamHI (New England Biolabs, Beverly, MA), following recommendations from the manufacturer for each restriction enzyme. Restriction enzyme digestion profiles were assessed using 0.5% agarose III (Amresco, Solon, OH) gel electrophoresis in TAE buffer (33). The 1-kb DNA ladder and λ DNA-HindIII Digest (New England BioLabs) were used as molecular size standards. Image analysis was done by UV transillumination of ethidium bromide-stained gels using Quantity One imaging software (Bio-Rad).
The sizes of the large R plasmids were estimated using pulsed-field gel electrophoresis (PFGE). Briefly, 0.8% Pulse Field Electrophoresis agarose plugs (Sigma) were made with R plasmids isolated as described above. Agarose plugs were loaded onto a 90-ml, 1% agarose (Sigma) gel in 0.5× TBE buffer and run in 0.5× TBE buffer using a CHEF-DR II system (Bio-Rad). Electrophoresis was carried out for 22 h at 6 V/cm, 120° and 14°C, with switch times ramping 1 to 25 s. The Midrange I PFG marker (New England BioLabs) and the BAC-Tracker Supercoiled DNA ladder (Epicentre Biotechnologies, Madison, WI) were used as molecular size standards. Image analysis was done after staining with ethidium bromide, using Quantity One imaging software.
Cloning and identification of GEN resistance gene(s).
A vast number of genes are known to confer GEN resistance in gram-negative bacteria (36, 42, 45). To determine which gene(s) confer GEN resistance in the isolates of this study, these were cloned and sequenced as follows. R plasmids isolated as described above were digested with EcoRI, HindIII, or BamHI, according to the manufacturer's recommendations. Restriction fragments were purified using QIAquick spin columns (QIAGEN) and ligated into previously digested pGEM-7Zf (Promega, Piscataway, NJ). Ligation reactions were used to transform E. coli TOP10 chemically competent cells (Invitrogen), following the protocol provided by the manufacturer. Recombinant clones carrying GEN resistance determinants were selected on LB plates supplemented with 100 μg/ml GEN and 50 μg/ml AMP. Recombinant plasmids were purified with QIAprep spin columns (QIAGEN), and inserts were sequenced using the vector's internal sequencing primers.
Sequencing and computer analysis.
Sequencing was done with fluorescent dye-labeled dideoxynucleotides with an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, CA). Sequence analysis was done using VectorNTI (Invitrogen), BLAST (2), and GenBank (National Center for Biotechnology Information, available through http://www.ncbi.nlm.nih.gov/BLAST/).
Screening for GEN resistance genes.
Multiplex PCR assays were designed to screen environmental isolates for the GEN resistance genes that were identified through cloning and sequencing of GEN resistance determinants. Multiplex PCRs included primers to detect resistance genes aac(3)-IIa and aac(3)-VIa, as well as a fragment of the 16S rRNA gene as an internal control for each reaction (43). Primers were designed using Vector NTI (Invitrogen). PCR screening of genes intI and sul1, commonly associated with class 1 integrons (40, 44), and of armA, rmtA, and rmtB, the only genes reported to confer high-level aminoglycoside resistance in gram-negative bacteria (47), was done in separate reactions using published primers (13, 18, 34, 41, 47, 48). All primers used are summarized in Table 2. To prepare DNA for PCR, pelleted cells from 1 ml of overnight cultures were resuspended in 100 μl distilled H2O and lysed through two 5-min freeze-boil cycles. Two microliters of the supernatants were used as templates in 50-μl PCRs. PCR was done using the FailSafe PCR system, buffer “G” (Epicentre Biotechnologies), according to instructions provided by the manufacturer. Thermocycler conditions were set as follows: (1 cycle at 96°C for 5 min; 35 cycles at 96°C for 30 s, 58°C for 30 s, and 1 final cycle at 70°C for 60 s; and 70°C for 7 min). To amplify the armA gene, PCR was done using the same conditions stated above, but various annealing temperatures (40°C, 42.2°C, 44.1°C, 46.9°C, 48.9°C, 50.2°C, and 51°C) were assayed in different reactions. PCR products were visualized by agarose gel electrophoresis using 2% Agarose I in 0.5× TBE buffer and staining with ethidium bromide. Some PCR products were sequenced to confirm amplification of the appropriate genes and to compare with published sequences in the databases. For this, PCR products were purified using QIAquick spin columns and ligated into pCR2.1-TOPO using the TOPO-TA cloning kit (Invitrogen), as suggested by the manufacturer. Sequencing of appropriate clones was done using the internal vector sequencing primers, as described above.
TABLE 2.
Primers used for PCR and sequencing
| PCR target | Primer | Amplicon size (bp) | Sequence (5′ → 3′) | Accession no. (reference or source)a |
|---|---|---|---|---|
| aac(3)-IIa | aacIIaF | 900 | GGG AAT TCA GAG GAG ATA TCG CGA TGC ATA CG | X13543 (*) |
| aacIIaR | CAT TGT CGA CGG CCT CTA ACC | |||
| aac(3)-VIa | aacVIaF | 465 | CGC TCA GGC GAT ATG GTG AT | L22613 (*) |
| aacVIaR | CAT AAT GGA GCG CGG TGA CT | |||
| armA | metF | 777 | CAA ATG GAT AAG AAT GAT GAT GAT | AY220558 (18) |
| metR | TTA TTT CTG AAA TCC ACT | |||
| rmtA | RMTA-F | 634 | CTA GCG TCC ATC CTT TCC TC | AB083212 (46) |
| RMTA-R | TTT GCT TCC ATG CCC TTG CC | |||
| rmtB | MBH-F | 771 | GGA ATT CCA TAT GAA CAT CAA CGA TGC CCT | AB103506 (13) |
| MBH-R | CCG CTC GAG TCC ATT CTT TTT TAT CAA GTA | |||
| intI1 | intIF | 845 | CTA CCT CTC ACT AGT GAG GGG CGG | X15852 (34, 40) |
| intIR | GGG CAG CAG CGA AGT CGA GGC | |||
| sulI | sulFI | 840 | ATG GTG ACG GTG TTC GGC AT | U12441 (18, 40) |
| sulR | CTA GGC ATG ATC TAA CCC TC | |||
| rrn | rrn1A | 330 | TAA CAC ATG CAA GTC GAA CG | (43) |
| rrn2B | CCC ATT GTG CAA TAT TCC CC | |||
| M13F (−20) | variable | GTA AAA CGA CGG CCA G | (Invitrogen) | |
| M13R | CAG GAA ACA GCT ATG AC |
*, E. C. Achberger, personal communication.
RESULTS
Gentamicin resistance in turtle farms.
Bacterial genera and species recovered from the turtle farm setting are summarized in Table 3 and include S. enterica subsp. enterica, S. enterica subsp. arizonae, E. cloacae, C. freundii, P. aeruginosa, A. baumanii, S. maltophilia, Alcaligenes faecalis, P. vulgaris, and P. stuartii. The antibiotic resistance phenotypes of the organisms tested show that in addition to resistance to GEN, these organisms exhibited resistance to other aminoglycosides, such as KAN and TOB, but not to AMK. Some organisms were also resistant to AMP (Table 3). Organisms such as P. aeruginosa and S. maltophilia were resistant to the majority of the antibiotics assayed.
TABLE 3.
Prevalence of gentamicin resistance genes among bacterial isolates from turtle farmse
| Samplea | Identified organism | No. of isolates collected | No. (%) of Genr isolates | Resistance phenotypeb | GEN MIC (μg/ml)c | % Incidenced
|
|
|---|---|---|---|---|---|---|---|
| aac(3)-IIa | aac(3)-VIa | ||||||
| Pond water | Citrobacter freundii | 43 | 7 (16) | Genr Kanr Tobr | >2,000 | 43 | 57 |
| Enterobacter cloacae | 20 | 3 (15) | Genr Kanr Tobr | >2,000 | 100 | ||
| Pseudomonas aeruginosa | 4 | 2 (50) | Genr Kanr Tobr Ampr (Chlr) (Ctxr) Nitr (Tetr) Tmpr | >2,000 | 100 | ||
| S. enterica subsp. arizonae | 38 | 4 (11) | Genr Kanr Tobr | >2,000 | 100 | ||
| Stenotrophomonas maltophilia | 7 | 3 (43) | Genr Kanr Tobr Ampr (Chlr) (Ctxr) Nitr (Tetr) Tmpr | >2,000 | 100 | ||
| Nest eggs | Citrobacter freundii | 49 | 6 (12) | Genr Kanr Tobr | >2,000 | 33 | 67 |
| Enterobacter cloacae | 44 | 2 (5) | Genr Kanr Tobr | >2,000 | 100 | ||
| Escherichia coli | 3 | 1 (33) | Genr Kanr Tobr | >2,000 | 100 | ||
| Proteus vulgaris | 1 | 1 (100) | Genr Ampr Nitr Pmbr (Tmpr) | 500 | 100 | ||
| Pseudomonas aeruginosa | 12 | 5 (42) | Genr Kanr Tobr Ampr (Chlr) (Ctxr) Nitr (Tetr) Tmpr | >2,000 | 100 | ||
| S. enterica subsp. enterica | 4 | 2 (50) | Genr Kanr Tobr | >2,000 | 100 | ||
| S. enterica subsp. arizonae | 53 | 11 (21) | Genr Kanr Tobr | >2,000 | 100 | ||
| GEN soln. | Acinetobacter baumannii | 1 | 1 (100) | Genr Kanr Tobr Ampr Tmpr | >2,000 | 100 | |
| Alcaligenes xylosoxidans | 2 | 2 (100) | Genr Kanr Tobr Ampr (Chlr) Ctxr Nitr Tetr Tmpr | >2,000 | 100 | ||
| Pseudomonas aeruginosa | 3 | 3 (100) | Genr Kanr Tobr Ampr (Chlr) Ctxr Nitr Tetr Tmpr | >2,000 | 100 | ||
| Pseudomonas fluorescens | 2 | 2 (100) | Genr Kanr Tobr Ampr (Chlr) (Ctxr) Nitr (Tetr) Tmpr | >2,000 | 100 | ||
| Turtles | Alcaligenes faecalis | 1 | 1 (100) | Genr Kanr Tobr Ampr Tmpr | >2,000 | 100 | |
| Citrobacter freundii | 4 | 4 (100) | Genr Kanr Tobr | >2,000 | 100 | ||
| Enterobacter cloacae | 3 | 3 (100) | Genr Kanr Tobr | >2,000 | 100 | ||
| Escherichia coli | 1 | 1 (100) | Genr Kanr Tobr | >2,000 | 100 | ||
| Proteus vulgaris | 2 | 2 (100) | Genr Ampr Nitr Pmbr (Tmpr) | 500 | 100 | ||
| Providencia stuartii | 3 | 3 (100) | Genr Ampr Nitr Pmbr (Tmpr) | 500 | 100 | ||
| Pseudomonas aeruginosa | 8 | 5 (63) | Genr Kanr Tobr Ampr (Chlr) (Ctxr) Nitr (Tetr) Tmpr | >2,000 | 100 | ||
| S. enterica subsp. arizonae | 28 | 28 (100) | Genr Kanr Tobr | >2,000 | 100 | ||
Samples were taken from nest egg homogenate, turtle breeding pond water, and GEN solution, as described previously (26, 37, 38), at turtle farms in Louisiana, 1999 to 2002. Infected turtles' isolates were supplied by Central Analytical Laboratory, Belle Chase, LA, July 1999.
Resistance phenotype displayed by >90% of GEN-resistant isolates, for each genus and species.
MIC value displayed by >90% of GEN-resistant isolates, for each genus and species. GEN-sensitive isolates presented GEN MIC of <3.9 to 7.8 μg/ml.
Percent GEN-resistant strains carrying aac(3)-IIa or aac(3)-VIa.
Abbreviations: Amp, ampicillin; Chl, chloramphenicol; Ctx, cefotaxime; Gen, gentamicin; Genr; GEN resistance; GEN soln., GEN egg dip solution (1,000 μg/ml); Kan, kanamycin; Nit, nitrofurantoin; Pmb, polymixin B; Tet, tetracycline; Tmp, trimethoprim; Tob, tobramycin. Parentheses around antibiotics denote intermediate resistance.
As summarized in Table 3, the majority of GEN-resistant bacteria displayed high-level resistance to GEN (MIC of >2,000 μg/ml). GEN-sensitive bacteria were primarily recovered from untreated eggs and breeding ponds and presented GEN MICs from <3.9 to 7.8 μg/ml (Table 3), whereas the majority of isolates recovered from contaminated turtles and the GEN dip solutions showed elevated resistance to GEN. Nevertheless, GEN-resistant isolates recovered from untreated eggs and breeding ponds also expressed a high level of resistance. E. coli strain BM14, E. coli OneShot TOP10, and E. coli ATCC 25922, used as GEN-sensitive controls, had GEN MICs of <3.9 to 7.8 μg/ml (Table 1).
R-plasmid isolation.
When checked for plasmids, environmental GEN-resistant isolates showed one or more plasmids. Nevertheless, transconjugant E. coli resistant to GEN and AZI were obtained only from the strains of S. enterica subsp. enterica, S. enterica subsp. arizonae, E. cloacae, and C. freundii. Transconjugants' GEN MICs were all >2,000 μg/ml. Likewise, the antibiograms were similar between donor organisms and their E. coli transconjugants. Furthermore, OneShot TOP10 E. coli cells transformed with R plasmids isolated from these transconjugants acquired the same resistance phenotypes as the transconjugants that the R plasmids had been isolated from. The resulting phenotypes and genotypes of these transformant E. coli cells are summarized in Table 4.
TABLE 4.
R plasmids isolated from bacteria recovered from turtle farms
| Plasmid | Size (kb) | RFLPa type | Resistance phenotypeb | GEN MIC (μg/ml)b | Presence of GEN resistance and integron-associated genec
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aac(3)-IIa | aac(3)-VIa | armA | rmtA | rmtB | intI1 | sulI | |||||
| pSal5 | ca. 45 | A | Genr Kanr Tobr | >2,000 | + | − | − | − | − | − | + |
| pAri40 | ca. 145 | B | Genr Kanr Tobr | >2,000 | − | + | − | − | − | + | + |
| pAriPH5 | ca. 145 | B | Genr Kanr Tobr | >2,000 | − | + | − | − | − | + | + |
| pEnt19 | ca. 115 | C | Genr Kanr Tobr | >2,000 | − | + | − | − | − | + | + |
| pCit14 | ca. 45 | D | Genr Kanr Tobr | >2,000 | + | − | − | − | − | − | − |
RFLP, restriction fragment length polymorphism.
Determined using transformant TOP10 E. coli cells.
As determined by PCR and/or sequencing.
Identification of GEN resistance gene(s).
R plasmid pSal5 was isolated from GEN-resistant E. coli obtained after transformation with plasmids from S. enterica (strain S5) (Table 4). The recombinant plasmid pGSal5h was obtained from a 5.5-kb HindIII restriction fragment of pSal5 that was ligated into pGEM-7Zf. Sequence analysis of this GEN-resistant clone revealed the presence of the aminoglycoside resistance gene aac(3)-IIa, which was 98% identical to that in the GenBank database, accession no. X13543 (1, 4, 42).
Preliminary screening done to detect gene aac(3)-IIa in other environmental isolates revealed that some GEN-resistant organisms did not carry aac(3)-IIa. Thus, R plasmid pEnt19, originally from E. cloacae (strain E19) (Table 4), was used for cloning and sequencing, since this plasmid conferred GEN resistance but did not carry aac(3)-IIa. The recombinant plasmid pGEnt19b was obtained from a 5-kb BamHI fragment of the R plasmid pEnt19 ligated into pGEM-7Zf. Sequence analysis of pGEnt19b revealed the presence of the aminoglycoside acetyltranferase gene aac(3)-VIa, which was 99% identical to that in the GenBank database, accession no. L22613 (31). Sequencing and PCR additionally revealed the integron class 1-associated genes intI1 and sul1 (Table 4), indicating the presence of aac(3)-VIa as a gene cassette of a class 1 integron.
The genes aac(3)-IIa and aac(3)-VIa have not been reported to confer high-level GEN resistance (1, 4, 31, 42). We thus attempted to amplify by PCR the recently reported 16S rRNA methylase genes which do confer a high-level aminoglycoside resistance phenotype in gram-negative bacteria, namely, armA (18), rmtA (46, 48), and rmtB (13). Nevertheless, these genes were not detected in the isolates tested (Table 4).
Distribution of GEN resistance genes in bacterial isolates from turtle farms.
Multiplex PCR assays designed to screen for the presence of the resistance genes aac(3)-IIa and aac(3)-VIa on bacterial isolates from turtle farms (n = 336) revealed that every GEN-resistant organism tested carried one of these acetyltransferase genes (Table 3). The gene aac(3)-IIa was present in 37% of the GEN-resistant isolates tested (n = 101); it was detected mainly in nonfermentors but was also found in S. enterica subsp. enterica and C. freundii. Gene aac(3)-VIa accounted for 94% of GEN resistance in Salmonella spp. (n = 35). Neither gene was detected in GEN-sensitive organisms, including GEN-sensitive controls previously described. IntI1 was detected by PCR in all GEN-resistant bacteria carrying aac(3)-VIa but not in organisms carrying aac(3)-IIa.
Plasmid heterogeneity.
The R plasmids pSal5, pAri40, and pEnt19 showed different HindIII and BamHI restriction fragment length polymorphism profiles (Fig. 1). Multiplex PCR of the resistance genes aac(3)-IIa and aac(3)-VIa (Table 4) plus hybridization of these genes with digested R plasmids (Fig. 1) showed that pSal5 carried aac(3)-IIa and that pAri40 and pEnt19 carried aac(3)-VIa, confirming that these genes are plasmid borne. The mentioned R plasmids differed in size as well, as visualized after PFGE and ethidium bromide staining. These results are summarized in Table 4.
FIG. 1.
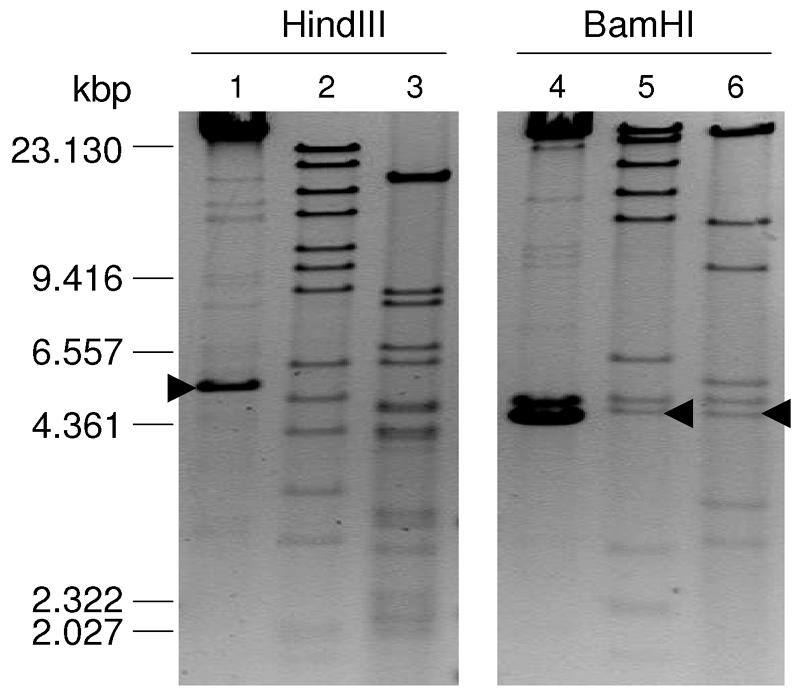
HindIII and BamHI digestion profiles of R plasmids pSal5 (lanes 1 and 4), pAri40 (lanes 2 and 5), and pEnt19 (lanes 3 and 6). A λ-DNA-HindIII digest, used as a molecular standard, is indicated on the left. Arrowheads indicate bands carrying the genes aac(3)-IIa (▸) and aac(3)-VIa (◂).
DISCUSSION
Turtles and other reptiles are known to excrete S. enterica subsp. arizonae and other salmonellae (9, 10, 14). The GEN resistance that has emerged in the last decade presents a threat to the efficacy of the GEN sulfate treatment used by the turtle industry to eradicate Salmonella spp. In this study, we found two genes which are carried by bacteria recovered from environmental samples associated with turtle farming activity which exhibit high-level GEN resistance. These genes are aac(3)-IIa and aac(3)-VIa, which code for aminoglycoside acetyltransferase enzymes (1, 4, 31). The gene aac(3)-IIa presents the AAC(3)-II aminoglycoside resistance pattern phenotype, which involves resistance to GEN, TOB, sisomicin, netilmicin, and dibekacin (1, 4, 36, 42). These genes are reported to be detected in gram-negative bacteria in the clinical setting with a relatively higher frequency (18 to 60%) than genes for other aminoglycoside resistance patterns (27, 36, 42, 45). The gene aac(3)-IIa, originally identified in R plasmids (1), reportedly accounts for 85% of the AAC(3)-II phenotype (27, 42, 45). The AAC(3)-VI resistance pattern includes resistance to GEN (31, 36, 42, 45). The gene aac(3)-VI was originally cloned from a conjugative plasmid of E. cloacae and rarely occurs among clinical isolates (31, 42). Its deduced amino acid sequence shares 50% amino acid similarity with the aac(3)-IIa gene (31, 36, 42, 45).
The majority of aminoglycoside-modifying enzymes can provide effective resistance to aminoglycosides, but only certain phosphotransferase enzymes detected in gram-positive bacteria have been reported to produce high levels of resistance to these antibiotics (7, 16, 36, 42, 45). When we started this study, there had been no reports, to the best of our knowledge, of the acquisition of genes conferring high-level aminoglycoside resistance in environmental gram-negative bacteria (M. A. Diaz, E. C. Achberger, V. R. Srinivasan, and R. J. Siebeling, Abstr. 100th Am. Soc. Microbiol. Gen. Meet., abstr. Z-4, 2000). Recently, however, Galimand et al. (18) found that the high-level aminoglycoside resistance phenotype in a clinical strain of Klebsiella pneumoniae was due to the presence of the aminoglycoside resistance gene armA. This strain of K. pneumoniae also presented the genes aac(3)-II, blaTEM1, blaCTX-M, dfrA12, sul1, and ant(3″)-9, conferring resistance to aminoglycosides, β-lactams, trimethroprim, sulfonamides, and streptomycin-spectinomycin, respectively (18). In this study, we did not detect the gene armA by PCR in any of the isolates screened, in spite of detecting certain shared elements in some of the strains, such as the genes aac(3)-IIa and sul1 (Table 4). We also screened for two other high-level aminoglycoside resistance genes: rmtA, cloned and identified from a clinical isolate of P. aeruginosa AR-2 in Japan (48), and plasmid-borne 16S rRNA methylase gene rmtB, isolated from Serratia marcescens (13). Nevertheless, we did not detect by PCR any other resistance gene that could act in synergy with the already-identified aac(3)-IIa or aac(3)-VIa gene products or by itself to give the high-level GEN resistance phenotype observed in the bacteria we isolated in the turtle farms.
The current study is the first to report the genes which confer GEN resistance to bacteria in the turtle farm environment, namely aac(3)-IIa and aac(3)-VIa. PCR and sequencing experiments confirmed that these resistance genes are present on mobile genetic elements that can facilitate their horizontal transfer among bacteria in the environment (1, 5, 17, 20, 31, 44). Both genes are found on R plasmids of the Enterobacteriacea (1, 4, 31), and yet until recently (18), only the gene aac(3)-VIa had been reported as a gene cassette within a class 1 integron (31, 44, 45, 47). Recent studies, however, have also detected the aac(3)-II gene surrounded by integron elements (18). In fact, the molecular environment of the armA gene has been depicted as a composite or complex integron environment and is found downstream of orf513 coding for a putative recombinase or transposase (30). We are currently conducting research to examine the molecular environment of the genes found.
GEN-resistant isolates were more prevalent in samples where exposure to GEN was greater but were also detected in turtle pond water and nest eggs, consonant with the notion that antibiotic use can favor the selection of resistant strains (23, 27). The presence of these genes on mobile molecular elements can facilitate their transfer and spread among different bacteria in the turtle farm environment (5, 17, 20). However, additional studies are required to determine if the interspecies dissemination of these genes occurred at the turtle farms. This study illustrates the potential for transfer of antibiotic resistance genes, particularly GEN resistance, between different genera in the turtle farms, a matter of particular public health concern as it involves the human pathogen Salmonella spp. The epidemiological data presented stress the need to develop an alternative method for the eradication of Salmonella spp. from turtle eggs. This is the first study to describe the genes involved in GEN resistance in S. enterica subsp. arizonae. It is important to monitor the presence of these resistance genes and molecular transfer elements in bacteria like the salmonellae, since these cause zoonosis and represent possible resistance reservoirs and can thus be important in the development of multiresistance in bacteria (3, 5, 14), as well as in its transfer between animals and humans.
Acknowledgments
This research was conducted in partial fulfillment of requirements for the doctorate of M. A. Díaz.
Funding was provided by the Louisiana Agricultural Experiment Station under project number LAB03329. This publication was approved as manuscript number 05-44-0285.
We thank E. Jolivet for assistance with PFGE and E. C. Achberger for valuable advice and PCR help. M. A. Díaz thanks the LSU Agricultural Center, the Dept. of Biological Sciences, and the Dept. of Veterinary Science for support in the completion of this work, particularly given the unexpected death of her late mentor and friend, R. J. Siebeling.
REFERENCES
- 1.Allmansberger, R., B. Brau, and W. Piepersberg. 1985. Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. Mol. Gen. Genet. 198:514-520. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brau, B., U. Pilz, and W. Piepersberg. 1984. Genes for gentamicin-(3)-N-acetyltransferases III and IV. I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol. Gen. Genet. 193:179-187. [DOI] [PubMed] [Google Scholar]
- 5.Carattoli, A. 2003. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 5:113-122. [PubMed] [Google Scholar]
- 6.Carter, A. P., W. M. Clemons, Jr., D. E. Brodersen, R. J. Morgan-Warren, T. Hartsch, B. T. Wimberly, and V. Ramakrishnan. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291:498-501. [DOI] [PubMed] [Google Scholar]
- 7.Chow, J. W. 2000. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31:586-589. [DOI] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., S. Baucheron, G. Flaujac, S. Schwarz, C. Kehrenberg, J. L. Martel, and E. Chaslus-Dancla. 2000. Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 44:2858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, M. L., M. Potter, R. Pollard, and R. A. Feldman. 1980. Turtle-associated salmonellosis in the United States. Effect of public health action, 1970 to 1976. JAMA 243:1247-1249. [PubMed] [Google Scholar]
- 10.D'Aoust, J. Y., E. Daley, M. Crozier, and A. M. Sewell. 1990. Pet turtles: a continuing international threat to public health. Am. J. Epidemiol. 132:233-238. [DOI] [PubMed] [Google Scholar]
- 11.Davies, J., and D. I. Smith. 1978. Plasmid-determined resistance to antimicrobial agents. Annu. Rev. Microbiol. 32:469-518. [DOI] [PubMed] [Google Scholar]
- 12.Davis, B. D. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebani, V. V., D. Cerri, F. Fratini, N. Meille, P. Valentini, and E. Andreani. 2005. Salmonella enterica isolates from faeces of domestic reptiles and a study of their antimicrobial in vitro sensitivity. Res. Vet. Sci. 78:117-121. [DOI] [PubMed] [Google Scholar]
- 15.Ekperigin, H. E., S. Jang, and R. H. McCapes. 1983. Effective control of a gentamicin-resistant Salmonella arizonae infection in turkey poults. Avian Dis. 27:822-829. [PubMed] [Google Scholar]
- 16.Ferretti, J. J., K. S. Gilmore, and P. Courvalin. 1986. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 167:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 18.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenfield, J., D. J. Sands, C. W. Wood, and K. R. Macdonald. 1975. Comparison of pressure differential procedures for dipping turkey hatching-eggs in gentamicin solution. Poult. Sci. 54:1254-1257. [PubMed] [Google Scholar]
- 20.Heuer, H., E. Krogerrecklenfort, E. M. H. Wellington, S. Egan, J. D. V. Elsas, L. V. Overbeek, J.-M. Collard, G. Guillaume, A. D. Karagoun, T. L. Nikolakopoulou, and K. Smalla. 2002. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol. Ecol. 42:289-302. [DOI] [PubMed] [Google Scholar]
- 21.Hirsh, D. C., J. S. Ikeda, L. D. Martin, B. J. Kelly, and G. Y. Ghazikhanian. 1983. R plasmid-mediated gentamicin resistance in salmonellae isolated from turkeys and their environment. Avian Dis. 27:766-772. [PubMed] [Google Scholar]
- 22.Izadjoo, M. J., C. O. Pantoja, and R. J. Siebeling. 1987. Acquisition of Salmonella flora by turtle hatchlings on commercial turtle farms. Can. J. Microbiol. 33:718-724. [DOI] [PubMed] [Google Scholar]
- 23.Levy, S. B. 1998. The challenge of antibiotic resistance. Sci. Am. 278:46-53. [DOI] [PubMed] [Google Scholar]
- 24.Maurin, M., and D. Raoult. 2001. Use of aminoglycosides in treatment of infections due to intracellular bacteria. Antimicrob. Agents Chemother. 45:2977-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michael-Marler, S., M. L. Brown, and R. J. Siebeling. 1983. Eradication of Arizona hinshawii from artificially infected turtle eggs. Appl. Environ. Microbiol. 45:748-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, G. H., F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu, K. J. Shaw, et al. 1997. The most frequent aminoglycoside resistance mechanisms—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clin. Infect. Dis. 24(Suppl. 1):S46-S62. [DOI] [PubMed] [Google Scholar]
- 28.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 29.NCCLS. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animal. Approved standard (M31-A). NCCLS, Wayne, Pa.
- 30.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rather, P. N., P. A. Mann, R. Mierzwa, R. S. Hare, G. H. Miller, and K. J. Shaw. 1993. Analysis of the aac(3)-VIa gene encoding a novel 3-N-acetyltransferase. Antimicrob. Agents Chemother. 37:2074-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saif, Y. M., and S. M. Shelly. 1973. Effect of gentamicin sulfate dip on Salmonella organisms in experimentally infected turkey eggs. Avian Dis. 17:574-581. [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schwocho, L. R., C. P. Schaffner, G. H. Miller, R. S. Hare, and K. J. Shaw. 1995. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shane, S. M., R. Gilbert, and K. S. Harrington. 1990. Salmonella colonization in commercial pet turtles (Pseudemys scripta elegans). Epidemiol. Infect. 105:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siebeling, R. J., D. Caruso, and S. Neuman. 1984. Eradication of Salmonella and Arizona species from turtle hatchlings produced from eggs treated on commercial turtle farms. Appl. Environ. Microbiol. 47:658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siebeling, R. J., P. M. Neal, and W. D. Granberry. 1975. Evaluation of methods for the isolation of Salmonella and Arizona organisms from pet turtles treated with antimicrobial agents. Appl. Microbiol. 29:240-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siebeling, R. J., P. M. Neal, and W. D. Granberry. 1975. Treatment of Salmonella-Arizona-infected turtle eggs with terramycin and chloromycetin by the temperature-differential egg dip method. Appl. Microbiol. 30:791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 41.Sunde, M., and H. Sorum. 1999. Characterization of integrons in Escherichia coli of the normal intestinal flora of swine. Microb. Drug Resist. 5:279-287. [DOI] [PubMed] [Google Scholar]
- 42.Vakulenko, S. B., and S. Mobashery. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vliegenthart, J. S., P. A. Ketelaar-van Gaalen, and J. A. van de Klundert. 1990. Identification of three genes coding for aminoglycoside-modifying enzymes by means of the polymerase chain reaction. J. Antimicrob. Chemother. 25:759-765. [DOI] [PubMed] [Google Scholar]
- 44.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright, G. D., A. M. Berghuis, and S. Mobashery. 1998. Aminoglycoside antibiotics. Structures, functions, and resistance. Adv. Exp. Med. Biol. 456:27-69. [PubMed] [Google Scholar]
- 46.Yamane, K., Y. Doi, K. Yokoyama, T. Yagi, H. Kurokawa, N. Shibata, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Genetic environments of the rmtA gene in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 48:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamane, K., J. Wachino, Y. Doi, H. Kurokawa, and Y. Arakawa. 2005. Global spread of multiple-aminoglycoside-resistance genes. Emerg. Infect. Dis. 11:951-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]


