Abstract
Comparisons between genomes of closely related bacteria often show large variations in gene content, even between strains of the same species. Such studies have focused mainly on pathogens; here, we examined Thermotoga maritima, a free-living hyperthermophilic bacterium, by using suppressive subtractive hybridization. The genome sequence of T. maritima MSB8 is available, and DNA from this strain served as a reference to obtain strain-specific sequences from Thermotoga sp. strain RQ2, a very close relative (∼96% identity for orthologous protein-coding genes, 99.7% identity in the small-subunit rRNA sequence). Four hundred twenty-six RQ2 subtractive clones were sequenced. One hundred sixty-six had no DNA match in the MSB8 genome. These differential clones comprise, in sum, 48 kb of RQ2-specific DNA and match 72 genes in the GenBank database. From the number of identical clones, we estimated that RQ2 contains 350 to 400 genes not found in MSB8. Assuming a similar genome size, this corresponds to 20% of the RQ2 genome. A large proportion of the RQ2-specific genes were predicted to be involved in sugar transport and polysaccharide degradation, suggesting that polysaccharides are more important as nutrients for this strain than for MSB8. Several clones encode proteins involved in the production of surface polysaccharides. RQ2 encodes multiple subunits of a V-type ATPase, while MSB8 possesses only an F-type ATPase. Moreover, an RQ2-specific MutS homolog was found among the subtractive clones and appears to belong to a third novel archaeal type MutS lineage. Southern blot analyses showed that some of the RQ2 differential sequences are found in some other members of the order Thermotogales, but the distribution of these variable genes is patchy, suggesting frequent lateral gene transfer within the group.
Prokaryotic genomes vary in gene content due, in part, to multiple events of gene acquisition and gene loss (5, 9, 29). Significant variation in gene content can be observed even between strains of the same species: 6 to 25% strain-specific genes have typically been found (Table 1) in studies focused primarily on pathogens (15, 47).
TABLE 1.
Some examples of differences in genome content among closely related strains
| Group and species | % VGa | Comment | Reference(s) |
|---|---|---|---|
| Completely sequenced genomes | |||
| Chlamydia trachomatis | <1 | 2 sequenced genomes; orthologs show ∼90% identity | 46 |
| Escherichia coli | 12-26 | 2 sequenced genomes; orthologs show 98.4% identity | 20, 45 |
| Helicobacter pylori | 6-7 | 2 sequenced genomes; orthologs show 92.6% DNA identity | 2 |
| Salmonella enterica | 10-12 | 5 sequenced strains; orthologs show 99.5-97.6% DNA identity | 11 |
| Group A Streptococcus | 6-9 | 2 sequenced strains; orthologs show 80-100% DNA identity | 51 |
| Microarry hybridization | |||
| Campylobacter jejuni | 0.5-7 | 11 strains; total of 21% variable genes in reference genome | 10 |
| Escherichia coli | 1-10 | 5 strains | 41 |
| Helicobacter pylori | 6-18 | 15 strains; total of 22% variable genes in reference genome | 49 |
| Mycobacterium tuberculosis | 0-0.7 | 16 strains, of which 13 were isolated from patients in San Francisco; total of 1.7% variable genes in reference genome | 28 |
| Salmonella enterica | 8-17 | 3 strains | 37 |
| Staphylococcus aureus | 1-12 | 36 strains; total of 22% variable genes in reference genome | 16 |
| Streptococcus pneumoniae | 3-11 | 20 strains; all strains except one differ by 8-11%; total of 24% variable genes in reference genome | 19 |
| Genomic subtraction | |||
| Escherichia coli | 8 | E. coli K-12 strain used as driver against avian pathogenic strain; subtracted DNA hybridized to a cosmid library | 7 |
| Listeria monocytogenes | 5-6 | Prototype virulent strain genome subtracted from prototype epidemic strain | 21 |
| Salmonella enterica | 2-20 | Genomic DNA from LT2 was subtracted against 4 different strains and hybridized to a cosmid library | 32 |
| Salmonella enterica serovar Typhimurium | 3 | Genomic subtraction using S. enterica serovar Typhi as driver; S. enterica serovar Typhimurium estimated to contain 140 kb not found in S. enterica serovar Typhi | 13 |
% VG refers to the percentage of variable genes (i.e., genes missing from one of the sampled genomes).
It is impractical and expensive to sequence the entire genomes of multiple strains of a species. Comparative sample sequencing (3) provides valuable information on diversity but is no cheaper. Fortunately, there are alternative ways to obtain genome-wide-information once the genome sequence of one strain is available. Micro- and macroarray hybridizations have been used to characterize the genomes of several “unsequenced” bacteria when a “sequenced” close relative exists (Table 1). These methods identify genes with high DNA sequence similarity to genes from the fully sequenced relative but provide no direct sequence information. Genomic subtraction—hybridization of DNA from two genomes and removal of common sequences—yields strain-specific clones that can be sequenced (Table 1). However, in most of the genomic subtraction studies done to date, only a limited number of clones have been sequenced from each strain tested. Here we used a sensitive PCR-based version of this technique, suppressive subtractive hybridization (SSH [1]), and we provide sequence information on many clones, in total, some 48 kbp.
We have chosen to look at bacteria from the genus Thermotoga, hyperthermophiles (Table 2) that live in environments thought to be dominated by Archaea. The complete genome sequence of Thermotoga maritima MSB8 is available, which makes it possible to do SSH experiments with a known reference. The genome sequence revealed that 24% of T. maritima MSB8's open reading frames were most similar to archaeal genes in BLAST analyses, suggesting extensive lateral gene transfer (LGT) between T. maritima and its archaeal neighbors (39). In a previous study (40), we investigated the distribution of two of the archaeal genes in 16 strains of Thermotoga and other related members of the order Thermotogales. These genes, encoding the large subunit of glutamate synthetase (gltB) and myo-inositol 1P synthase (ino-1), appear to have been acquired from different archaeal lineages during the divergence of the Thermotoga lineage. We suggested that most of the archaeal genes in Thermotoga strains might have been obtained during a relatively short evolutionary interval.
TABLE 2.
Strains used in this studya
| Strain | Habitat | Temp (°C) |
|---|---|---|
| Thermotoga | ||
| T. maritima MSB8b | Geothermal heated seafloor, Vulcano, Italy | 55-90 (80) |
| Thermotoga sp. strain RQ2b | Geothermal heated seafloor, Ribeira Quente, the Azores | 76-82 |
| T. naphthophila RKU-10c | Deep subterranean oil reservoir in Niigata, Japan | 48-86 (80) |
| T. petrophila RKU-1c | Deep subterranean oil reservoir in Niigata, Japan | 47-88 (80) |
| T. neapolitana NS-ETd | Shallow submarine hot spring, Naples, Italy | 55-90 (80) |
| T. neapolitana LA4e | Shore of Lac Abbé, Djibouti | 82 |
| T. neapolitana LA10e | Shore of Lac Abbé, Djibouti | 87 |
| Thermotoga sp. strain RQ7b | Geothermal heated seafloor, Ribeira Quente, the Azores | 76-82 |
| Thermotoga sp. strain SG1e | Boiling beach during volcanic eruption, Sangeang Island, Indonesia | 85 |
| Thermotoga sp. strain SL7f | Oil reservoir, Paris basin | 50-75 (70) |
| Thermotoga sp. strain KOL6e | Submarine hydrothermal system, Kolbeinsey ridge, north of Iceland | 90 |
| T. thermarum LA3g | Djibouti | |
| T. subterranea SL1h | Continental oil reservoir, Paris Basin | 50-75 (70) |
| Fervidobacterium | ||
| F. islandicumi | Hot spring, Iceland | 50-80 (65) |
| F. pennavoransj | Hot spring, San Miguel, the Azores | 50-80 (70) |
| F. nodosumk | Hot spring, New Zealand | 65-80 (70) |
| Thermosipho africanusl | Marine hydrothermal system, Obock, Djibouti | 35-77 (75) |
T. maritima MSB8 and Thermotoga sp. strain RQ2 were used in the subtraction study, while the other strains were included in the Southern analysis. For characterized strains, the temperature range and optimal temperature (in parentheses) are given.
Reference 22. Cell mass from RQ2 and RQ7 was a gift from Karl O. Stetter. MSB8 DNA was obtained from The Institute for Genomic Research.
Reference 54. DNA was a gift from Yoh Takahata.
Reference 25. Cell mass was a gift from Karl O. Stetter.
Personal communication from Karl O. Stetter. For this strain the temperature at the isolation site is given. Cell mass was a gift from Karl O. Stetter.
Personal communication from Stéphane L'Haridon. DNA was a gift from Stéphane L'Haridon and Christian Jeanthon.
Reference 59. Cell mass was a gift from Karl O. Stetter.
Reference 26. DNA was a gift from Stéphane L'Haridon and Christian Jeanthon.
Reference 24. Cell mass was a gift from Karl O. Stetter.
Reference 17. DNA was a gift from Fiona Duffner.
Reference 43. DNA was a gift from Stéphane L'Haridon and Christian Jeanthon.
Reference 23. Cell mass was a gift from Karl O. Stetter.
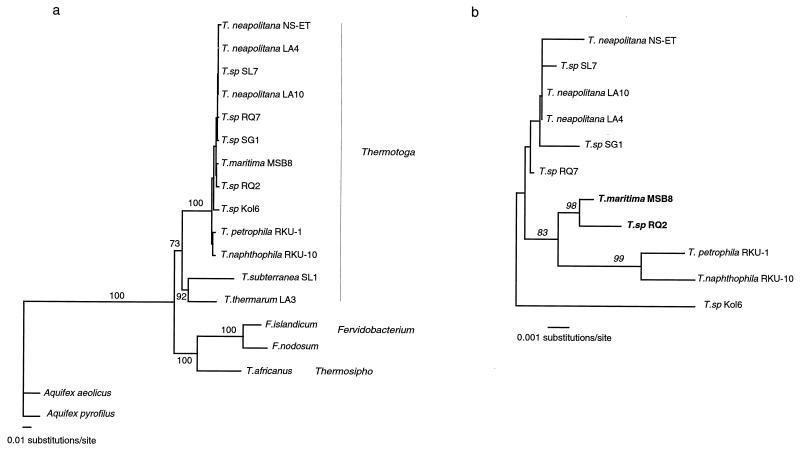
Hence, we expanded comparisons of Thermotoga strains, by using SSH, to identify genes present in different Thermotoga strains that do not have homologs in the sequenced T. maritima genome. Such genes are candidates for recent LGT and loss events in Thermotoga spp. Results from SSH studies of Thermotoga sp. strain RQ2 are presented here. This strain is the most closely related, of those in our collection, to the sequenced T. maritima strain MSB8, differing by only 0.3% in the 16S rRNA gene (Fig. 1 and reference 40). Orthologous protein-coding genes shared between RQ2 and MSB8 are, on average, 96% identical at the DNA level and thus almost completely identical at the amino acid level (Nesbø and Doolittle, unpublished data; for a comparison to the level of divergence of orthologs in other bacterial species, see Table 1).
FIG. 1.
Small-subunit rRNA gene phylogenies of all the Thermotogales strains (with the exception of F. pennivorans, for which there is no sequence available in the GenBank database) included in this study (a) and the strains most closely related to T. maritima MSB8 and Thermotoga sp. strain RQ2 (b). The value on a branch is the number of times the branch was recovered in a bootstrap analysis using 100 bootstrap replicates. Both trees were estimated in PAUP∗ version 4.08ba (53) by using the heuristic search option with 10 stepwise additions. The tree in panel a was estimated by using logdet distances and rooted by the two Aquifex sequences. The tree in panel b was estimated by using Kimura 2P distances and rooted by midpoint rooting.
MATERIALS AND METHODS
Bacterial strains and DNA extraction.
The bacterial strains used are listed in Table 2. DNA was extracted by the protocol of Charbonnier and Forterre (8).
SSH.
Genomic subtraction was carried out by using the PCR-Select Bacterial Genome Subtraction Kit (Clontech), with T. maritima MSB8 as the driver and Thermotoga sp. strain RQ2 as the tester. Briefly, 2 μg of genomic DNA from each strain—the driver (T. maritima MSB8) and the tester (Thermotoga sp. strain RQ2)—is digested with RsaI and two different PCR adaptors are ligated to two different aliquots of the tester (Thermotoga sp. strain RQ2) DNA. Two hybridizations are then performed. In the first hybridization, an excess of driver (T. maritima MSB8) DNA is added to each of the adaptor-ligated tester DNAs (Thermotoga sp. strain RQ2). The samples are then heat denatured and allowed to anneal. After this hybridization, single-stranded DNA will be enriched for tester (Thermotoga sp. strain RQ2)-specific DNA, as DNA fragments that are not tester specific will form hybrid molecules with the driver DNA. In the second hybridization, the two primary hybridization reaction mixtures are mixed together without denaturing. Only subtracted single-stranded tester-specific DNA should reassociate to make hybrids with different adaptors on each end, and only molecules with different adaptors on each end can be amplified exponentially in subsequent steps of the procedure.
Two independent subtractions, differing in hybridization temperature, were performed. In the first experiment, the protocol supplied with the kit was followed, using a hybridization temperature of 63°C in both the first (1.5-h) and second (overnight) hybridizations. In the second experiment, cycling of the temperature was explored by using the following program: an initial 63°C for 20 min and 60°C for 1 min, followed by six cycles of 58°C for 5 min, 57°C for 30 s, 56°C for 30 s, 55°C for 5 min, 56°C for 30 s, and 57°C for 30 s. The second hybridization was performed at 55°C overnight. Cycling was included in the first step in order to facilitate annealing of sequences containing mixtures of highly similar and divergent sequences (for instance, intergenic regions between highly similar genes) and excluded in the second step assuming that most of these sequences would anneal in the first hybridization. However, as there was no significant difference in the proportion of differential clones, results from the two experiments were pooled. PCR products obtained after SSH were cloned into TOPO 2.1 (Invitrogen).
Sequencing reactions were carried out in a 10-μl final volume by using 1 μl of BigDye terminator, 3.2 pmol of primer, 3 μl of template, 2 μl of 5× CSA buffer (PE Applied Biosystems), and enough deionized water to bring the final volume to 10 μl. The cycling conditions used for sequencing reactions were 96°C for 2 min, followed by 40 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Cycle sequencing was done with an MJ Research DNA Engine Tetrad PTC-225 thermal cycler (MJ Research, Watertown, Mass.). After cycle sequencing, chain-terminated products were precipitated with isopropanol, washed with 70% ethanol, dried, rehydrated in deionized water, and analyzed on a 3700 automated DNA sequencer (Applied Biosystems, Foster City, Calif.) with run modules at default settings, with the exception of the cuvette temperature, which was set at 42°C.
Blot hybridization.
Samples were prepared for electrophoresis by digesting 2 μg of genomic DNA with restriction enzymes in a total of 20 μl, and the DNA fragments were separated in a 1% agarose gel. Two samples of each strain were electrophoresed, one cut with RsaI or EcoRI and one cut with HindIII. Five gels were run, and most strains were represented on two gels. Thermotoga sp. strain RQ2 and T. maritima MSB8 samples were run on each gel. The gels were blotted onto GeneScreen membranes (Dupont) in accordance with the manufacturer's protocol. Probes were made from PCR products amplified from the original subtraction clone and labeled with the DIG High Prime DNA labeling kit (Roche). Prehybridization and hybridization were done with DIG Easy Hyb (Roche) at 37 to 39°C in a rotary oven. Washes were preformed in a 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-equivalent buffer at 50 to 55°C. The digoxigenin-labeled probe was detected with CSPD or CPD∗ (Roche), and exposures were usually overnight or shorter, depending on the signal strength.
Data analysis.
The sequences were used as probes to search the T. maritima MSB8 genome at The Institute for Genomic Research Blast (http://tigrblast.tigr.org/cmr-blast/) and GenBank at National Center for Biotechnology Information Blast (http://www.ncbi.nlm.nih.gov/BLAST/). The sequences were categorized by using the following criteria: sequences with no match in the T. maritima MSB8 genome when doing Blast-N searches against the genome sequence were classified as RQ2-specific differential sequences, and sequences that did have a DNA match in the MSB8 genome were classified as false positives if the percent similarity to the DNA sequence of the MSB8 homolog was greater than 85% along the whole sequence and divergent if the percent similarity was less than 85% or if the sequence was rearranged compared to the RQ2 sequence.
The cutoff of 85% was chosen arbitrarily, as this seemed to be the limit of the ability of this hybridization method to distinguish between differential and nondifferential sequences in earlier studies (4, 31).
The ratio of synonymous to nonsynonymous mutations (ds/dn ratio) was determined by using the Nei-Gojobori method (38) in SNAP (synonymous/nonsynonymous analysis program) (30) at http://hiv-web.lanl.gov/SNAP/WEBSNAP/SNAP.html. The size of the library was estimated as described by Bogush et al. (4) and Lev G. Nikolaev (personal communication) by using the formula N = n/P, where N is the estimated library size (in clones), n is the number of clones sequenced, and p is the probability of finding x number of independent clones among the n sequenced clones. P was calculated as follows: P = [(c1 − 1) + (c2 − 1) + (c3 − 1) + Σ +(cm − 1)]/m, where c1 is the number of observations of clone 1 and m is the number of clones sequenced. In our study, this translated into a P value of 0.16 (71 multiple hits) and a total library size of about 2,600 clones. The number of differential clones was estimated to be 39% of the total library. The number of differential genes was estimated by multiplying the number of differential clones by the average sequence length (365 bp) of the clones and dividing the result by the average T. maritima MSB8 gene length (947 bp) (39). Phylogenetic trees were estimated by using PAUP∗ version 4.0.8b (53), TREE PUZZLE 4.0 (52), and PHYLIP 3.6 (14). The proximity test for shared orthologs was done by using a modification of the algorithm in covARES (http://hades.biochem.dal.ca/Rogerlab/Software/software.html).
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the EMBL nucleotide sequence database and assigned accession numbers AJ458574 to AJ458934.
RESULTS
Subtractive hybridization of Thermotoga sp. strain RQ2 versus T. maritima MSB8.
Genomic subtraction was carried out as described in Materials and Methods. A total of 426 clones were sequenced, and among these, 71 sequences were encountered more than once. General features of the clones sequenced are shown in Table 3.
TABLE 3.
Breakdown of the 426 clones sequenced
| Classification of clones | No. of clones | Avg length (bp) | % G+C (Pa) | Total no. of bp | % Identityb |
|---|---|---|---|---|---|
| All differential clones (no DNA match in the MSB8 genome) | 166 | 370 | 43.3 (<0.001) | 48024 | |
| No protein match in databasesc | 47 | 267 | 40.1 (<0.001) | 10443 | |
| Significant protein match in databases | 119 | 406 | 44.4 (<0.001) | 37581 | |
| Highest protein similarity to a T. maritima MSB8 gene or a gene from other Thermotoga strainsd | 35 | 409 | 45.1 (0.238) | 12671 | |
| All clones with a DNA match in the T. maritima MSB8 genome | 260 | 360 | 46.6 (0.015) | 81074 | 87.5 |
| False positives (DNA similarity of >85%) | 130 | 353 | 46.7 (0.013) | 39745 | 94.2 |
| Divergent clones (DNA similarity of ≤85%) | 93 | 372 | 46.5 (0.320) | 28129 | 76.6 |
| Low quality | 37 | 372 | NA | 13200 | 83.6 |
The mean G+C content was compared to the mean G+C content of the T. maritima MSB8 genome (46%) by using a t test. NA, not applicable.
Percent identity indicates the average percent DNA identity to the T. maritima MSB8 homolog.
Expect value, >0.01.
In BLAST searches against protein databases using the translation of the nucleotide sequence of the clone (i.e., BLASTX searches).
On the basis of the number of clones sequenced and the number of clones observed multiple times (see Materials and Methods), we estimated that the pooled RQ2 subtraction library contains roughly 1,000 independent differential clones, or about 350 to 400 differential genes assuming an average gene sequence length similar to that in T. maritima MSB8 (947 bp [39]), and an average clone length of 365 bp.
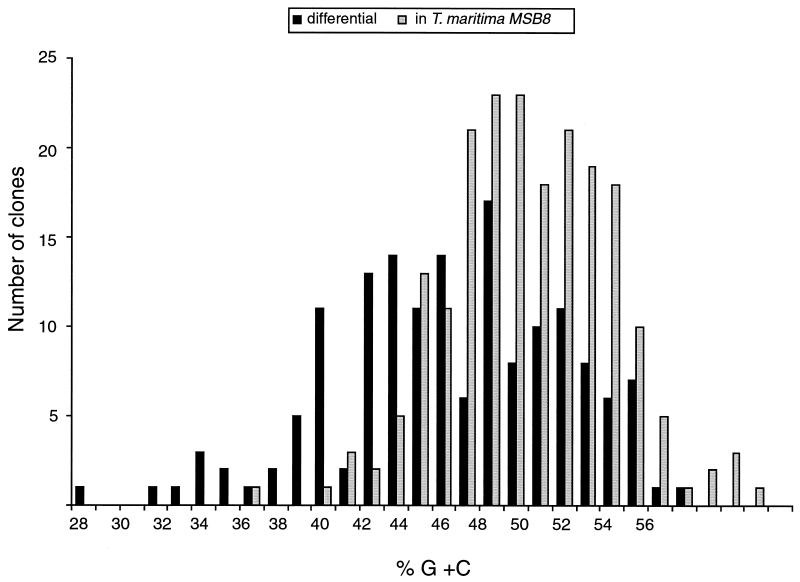
The G+C composition of the false positives and the divergent clones was very similar to that observed for the T. maritima MSB8 genome (46%) (Table 3). The differential clones, however, showed a slightly but significantly lower average G+C ratio (Table 3). The difference in G+C content was more apparent when the G+C content was plotted against the number of clones with and without a DNA match in the MSB8 genome (Fig. 2), suggesting that many of the RQ2-specific sequences have been acquired by LGT from organisms with a lower G+C content than T. maritima.
FIG. 2.
Comparison of G+C contents of clones with (divergent and false-positive clones) and without (differential clones) a DNA match in the T. maritima MSB8 genome.
A large proportion of Thermotoga sp. strain RQ2-specific sequences are involved in sugar transport and synthesis/degradation.
Of the differential clones with a significant match in the GenBank or The Institute for Genomic Research database (Table 4), 36 showed the greatest similarity to 21 proteins from T. maritima MSB8 or other Thermotoga strains in BLASTX searches. Since these genes showed no significant match at the DNA level (whereas shared orthologs, including both false positives and divergent clones show, on average, 88% DNA identity [Table 3]), they are probably the result of either transfer of genes from other members of the Thermotogales, lineage-specific duplications in RQ2, or differential loss of paralogs. Notably, nine of these genes encode ABC transporters, seven of which are predicted to be involved in sugar transport. In addition to this expansion by duplication or transfer from other members of the Thermotogales, RQ2 appears to have acquired at least seven sugar transporters from more distantly related genomes.
TABLE 4.
RQ2-specific sequences with significant protein matches in The Institute for Genomic Research and/or The National Center for Biotechnology Information protein databases
| Group and clonea | bpb | First BLASTX hitc | Organism | Expect value | % Identityd | Note |
|---|---|---|---|---|---|---|
| Transporters | ||||||
| 2D4 | 210 | TM0114 sugar ABC transporter | T. maritima MSB8 | 3.9E−04 | 38 | TM0114 also among divergent clones |
| TAD66 | ||||||
| KJ3D8 | 327 | 7.7E−26 | 48 | |||
| TAB05 | ||||||
| TAF50 | 225 | TM0115 sugar ABC transporter | T. maritima MSB8 | 5.5E−19 | 58 | |
| TAA05 | 584 | a, TM0115 sugar ABC transporter | T. maritima MSB8 | 7.0E−22 | 50 | In T. neapolitana, sequence b is linked to XloA in C6 |
| b, mannosidase | Thermotoga neapolitana | 1.0E-104 (DNA) | 100 (DNA) | |||
| TAB16 | 215 | TM0430 sugar ABC transporter | T. maritima MSB8 | 5.5E−16 | 52 | |
| TXX1 | ||||||
| 2H5 | 197 | TM0432 sugar ABC transporter | T. maritima MSB8 | 2.7E−20 | 80 | |
| TAA71 | ||||||
| TAE94 | 430 | TM0595 sugar ABC transporter | T. maritima MSB8 | 1.0E−4 | 33 | |
| 2A2 | 335 | TM0955 ribose ABC transporter | T. maritima MSB8 | 4.1E−20 | 67 | |
| TAD76 | 263 | TM1066 oligopeptide ABC transporter | T. maritima MSB8 | 3.0E−26 | 79 | Probably two different loci; TM1066 is also represented among the divergent clones |
| TAE32 | 167 | 1.0E−20 | 74 | |||
| TAE41 | 466 | TM1067 oligopeptide ABC transporter | T. maritima MSB8 | 7.6E−10 | 64 | |
| TAE80 | 172 | 8.2E−11 | 44 | |||
| TAD15 | 561 | TM1854 sugar ABC transporter | T. maritima MSB8 | 2.0E−10 | 39 | Start of clone shows no similarity |
| 3A4 | 220 | Hypothetical sugar ABC-transporter gene | Agrobacterium rhizogenes | 1.0E−14 | 52 | |
| 4E9 | ||||||
| TAB60 | 632 | Ribose ABC transporter | Lactococcus lactis | 1.0E−31 | 68 | |
| 2A1 | 354 | Bifunctional carbohydrate binding and transport protein | Streptomyces coelicolor A3 | 2.0E−08 | 50 | |
| TAC78 | 180 | 5.0E−07 | 60 | |||
| TAC69 | 170 | Galactoside ABC transporter | Vibrio cholerae | 2.0E-06 (DNA) | 60 (DNA) | |
| TAE46 | 237 | Lactose transport system permease protein | Synechocystis sp. | 5.0E−04 | 32 | |
| 3F3 | 500 | ABC transporter, periplasmic substrate-binding protein | Deinococcus radiodurans | 3.0E−44 | 47 | RQ2 sequence cluster with Bacillus halodurans homolog in phylogenetic trees; also found in T. neapolitana, linked to clone E6 |
| TAC20 | 567 | 1.0E−44 | 47 | |||
| TAC08 | 461 | Sugar transporter sugar binding protein | Mesorhizobium loti | 1.0E−07 | 29 | |
| TXX3 | 612 | Probable membrane transport protein | Clostridium perfringens | 3.0E−20 | 32 | |
| Degradation of polysaccharides | ||||||
| C6 | 198 | Thermotoga neapolitana xylosidase (XloA) and TM0076 xylosidase | Thermotoga neapolitana, T. maritima MSB8 | 6.0E−18 | 63 | Most likely two copies of this gene, B1 and 2A11, show 93% identity to T. neapolitana gene in DNA searches |
| TAF78 | ||||||
| B1 | 430 | 5.0E-61 (DNA) | 93 (DNA) | |||
| 2A11 | ||||||
| AE28 | 492 | 1.0E−26 | 69 | |||
| E6 TAF85 | 723 | β-Glucosidase (bglA) | T. maritima MSB8 | 0 | 98 | Not reported in genome; see accession no. S34570; linked to clone 3F3 |
| KJ3B6 | 457 | Family 10 xylanase gene xynC | Thermotoga sp. strain FjSS3-B.1 | 1.0E−92 (DNA) | 89 (DNA) | Last 50 bp match noncoding region between TM1270 and TM1271 in T. maritima MSB8 |
| TAA59 | ||||||
| TAA42 | 531 | 0 (DNA) | 95 (DNA) | |||
| A8 | 354 | BH1878—unknown conserved protein (781 aa) | Bacillus halodurans | 2.0E−21 | 55 | PSI BLAST searches suggest that BH1878 is an arabinosidase |
| A12 | 404 | 8.0E−16 | 40 | |||
| TAE65 | ||||||
| TAE79 | There are at least 2 copies of this gene in the RQ2 genome | |||||
| C1 | 185 | 3.0E−7 | 54 | |||
| TAF19 | ||||||
| TAA19 | 114 | 6.0E−04 | 60 | There also appear to be rearrangements compared to BH1878; the clones also show similarity to an arabinan endo-1,5-α-l-arabinosidase homolog (yxiA from Bacillus subtilis | ||
| TAB07 | 251 | 3.0E−27 | 66 | |||
| TAC44 | 626 | 6.0E−26 | 44 | |||
| TAC83 | 533 | 1.0E-09 | 40 | |||
| TAE11 | 240 | 1.0E-08 | 40 | |||
| TAE30 | 241 | 1.0E−04 | 43 | |||
| KJ3D1 | 292 | a, TM0280 hypothetical protein | a, T. maritima MSB8 | 1.0E−14 (DNA) | 94 (DNA) | Sequence a covers the end of TM0280; in T. maritima MSB8, TM0281 is an arabinosidase |
| b, BH1878—unknown conserved protein (781 aa) | b, Bacillus halodurans | 0.94 (DNA) | 67 (DNA) | |||
| KJ3B9 | 248 | Hypothetical 52.5-kDa protein in hutP-bglP intergenic region | Bacillus subtilis | 1.0E−03 | 41 | Second hit, BH1878—unknown conserved protein |
| TAA52 | 565 | Arabinan endo-1,5-α-l-arabinosidase | Bacillus subtilis | 2.0E−66 | 64 | Second hit, BH1878—unknown conserved protein; two different copies of the gene |
| TAF05 | 533 | 3.5E−29 | 68 | |||
| TAF15 | 151 | 2.0E−11 | 65 | |||
| KJB3 | 521 | Arabinosidase | Bacteroides ovatus | 3.0E−23 | 37 | KJB3, TXX2, TAE82, and TAD18 may be from the same gene |
| TXX2 | 483 | 5.0E−48 | 53 | |||
| TAE82 | 297 | α-Arabinofuranosidase I | Streptomyces chartreusis | 3.0E−03 | 30 | |
| TAD18 | 505 | a, sugar ABC transporter | a, Listeria innocua | 6.0E−13 | 43 | |
| b, probable secreted arbinosidase | b, Streptomyces coelicolor | 5.0E−09 | 56 | |||
| TAC01 | 493 | β-Xylosidase | Clostridium acetobutylicum | 1.0E−17 | 34 | Encoded on plasmid pSOL1 in C. acetobutylicum |
| 2A10 | 568 | Pectin methylesterase-like protein | Pectobacterium chrysanthemi | 7.0E−20 | 40 | RQ2 clusters with Pectobacterium chrysanthemi in phylogenetic trees |
| 2A5 | ||||||
| KJ3D6 | ||||||
| TAC87 | 354 | 4.0E−20 | 50 | |||
| Surface polysaccharides | ||||||
| TAF75 | 411 | dTDP-l-rhamnose synthase, rmlD gene | Lactococcus lactis | 1.8E−07 (DNA) | 58 (DNA) | |
| E9 | 444 | dTDP-4-dehydrorhamnose 3,5-epimerase, rmlC gene | Shigella flexneri | 3.0E−38 | 59 | |
| 2A4 | ||||||
| TAA18 | 397 | dTDP-6-deoxy-d-glucose-3,5-epimerase, rmlC gene | Salmonella enterica | 2.0E−10 | 62 | Identical to E9 for bp 1-277 |
| TAA12 | 320 | dTDP-glucose 4,6-dehydratase, rmlB gene | Methanobacterium thermoautotrophicum | 8.0E−27 | 68 | |
| TAE89 | ||||||
| TAC74 | 313 | Glucose-1-phosphate thymidylyltransferase rmlA gene | Salmonella enterica | 2.0E−25 | 59 | |
| TAA67 | 539 | Rhamnosyltransferase, wbaQ gene | Deinococcus radiodurans | 7.0E−12 | 36 | Might be a pseudogene frameshift at aa 73; no similarity after aa 83 |
| KJ3D12 | 757 | Type 2 capsule locus | Streptococcus pneumoniae | 6.0E−03 | 25 | Similar to Wzx, flippase |
| TAC91 | 443 | a, spore coat polysaccharide synthesis, SpsK | Bacillus subtilis | 1.0E−05 | 50 | Second hit of sequence a, RmlD from Listeria monocytogenes |
| b, TM0522 heat shock protein HslU | T. maritima MSB8 | 1.4E−33 (DNA) | 94 (DNA) | TM0522 and TM0523 rearranged, fused | ||
| c, TM0523 hypothetical protein | T. maritima MSB8 | 5.6E−12 (DNA) | 98 (DNA) | |||
| TAF39 | 209 | EpsF, WbnE putative glycosyl transferase | Streptococcus thermophilus | 2.0E−13 | 61 | |
| Other | ||||||
| A11 | 367 | V-ATPase C subunit | Pyrococcus horikoshii | 2.0E−01 | 30 | |
| TAD64 | ||||||
| 2B5 | 765 | a, V-ATPase E subunit | Desulfurococcus sp. strain SY | 3.9 | 27 | |
| b, V-ATPase A subunit | Methanobacterium thermoautotrophicum | 2.0E−09 | 46 | |||
| KJ3D4 | 309 | V-ATPase A subunit | Treponema pallidum | 1.0E−40 | 76 | |
| TAB56 | 491 | V-ATPase A subunit | Archaeoglobus fulgidus | 5.0E−44 | 55 | |
| TAC60 | 106 | V-ATPase A subunit | Desulfurococcus mobilis | 1.0E−10 | 91 | |
| TAB82 | 581 | V-ATPase E subunit | Desulfurococcus sp. strain SY | 3.9 | 24 | |
| 5A12 | 658 | V-ATPase F subunit | Treponema pallidum | 2.0E−03 | 28 | 400 last bp of clone have no similarity |
| KJB7 | 321 | V-ATPase D subunit | Pyrococcus horikoshii | 1.0E−12 | 35 | |
| 3A1 | ||||||
| 4E10 | ||||||
| KJ2B4 | 354 | V-ATPase 1 subunit | Halobacterium sp. strain NRC-1 | 0.62 | 24 | Not significant but included since a complete V-type operon is likely |
| 5A11 | 525 | V-ATPase B subunit | Sulfolobus solfataricus | 5.0E−62 | 62 | |
| D5 | 879 | MutS-like ATPase involved in mismatch repair | Thermoplasma volcanium | 2.0E−46 | 37 | Phylogenetic analyses suggest that this gene belongs to a third group of prokaryote mutS homologs (Fig. 3) |
| TAC19 | 174 | 4.0E−05 | 37 | |||
| TAF09 | 255 | 1.0E−19 | 55 | |||
| TAD43 | 527 | 2.0E−17 | 33 | |||
| TAA79 | 305 | TM0758 flagellin | T. maritima MSB8 | 1.0E−07 | 38 | |
| A5 | 733 | TM0037 conserved hypothetical protein | T. maritima MSB8 | 2.5E−04 | 31 | |
| TAD45 | ||||||
| TAA71 | 195 | TM0432 hypothetical protein | T. maritima MSB8 | 1.0E−22 | 75 | |
| TAE29 | 738 | TM0619 conserved hypothetical protein | T. maritima MSB8 | 1.8E−21 | 64 | |
| TAC72 | 307 | TM0941 hypothetical protein | T. maritima MSB8 | 2.0E−24 | 50 | |
| B11 | 574 | a, TM0945 hypothetical protein | T. maritima MSB8 | 1.6E−07 | 54 | 91-415, no hit; TM0946 deleted? |
| TAC95 | b, TM0947 hypothetical protein | 6.0E−04 | 63 | |||
| TAA68 | 234 | Hypothetical protein | Thermotoga neapolitana | 1.0E−106 (DNA) | 99 | |
| TAF74 | 682 | a, hypothetical protein YtaP | Bacillus subtilis | 1.0E−11 | 29 | |
| b, TM0076 xylosidase | T. maritima MSB8 | 4.0E−04 (DNA) | 81 (DNA) | |||
| 2B8 | 489 | Long-chain primary alcohol dehydrogenase | Thermoanaerobacter ethanolicus | 6.0E−43 | 57 | RQ2 clusters with Thermoanaerobacter ethanolicus, Thermococcus hydrothermalis, and Giardia intestinalis to exclusion of T. maritima MSB8 homologs TM0111, TM0820, and TM0920 |
| TAB19 | 302 | 1.0E−33 | 70 | |||
| TAD49 | 102 | 3.5E−02 | 48 | |||
| KJ3C6 | 191 | |||||
| 2A6 | 489 | 1-Phosphofructokinase | Pseudomonas aeruginosa | 8.0E−21 | 37 | RQ2 sequence and T. maritima MSB8 homolog TM0828 separated in phylogenetic tree |
| KJ3D10 | 749 | Hypothetical protein | Aquifex aeolicus | 4.5E−02 | 28 | |
| 2C2 | 250 | Alkaline phosphatase | Synechocystis sp. | 3.0E−03 | 35 | |
| TAD30 | 726 | Immunoglobulin A-specific serine endopeptidase | Neisseria meningitidis | 3.0E−03 | 25 | |
| E11 | 450 | a, TM1044 transposase, IS605-TnpB family | T. maritima MSB8 | 3.2E−15 (DNA) | 95 (DNA) | BLASTX with sequence b: TM0972 conserved hypothetical protein, GGDEF domain |
| TAD44 | b, conserved hypothetical protein aq_035 | Aquifex aeolicus | 1.0E−06 (DNA) | 59 (DNA) | ||
| TAB30 | 250 | Hypothetical protein PH0104 | Pyrococcus horikoshii | 2.0E−03 | 31 | |
| TAC56 | 410 | Hypothetical protein PH0976 | Pyrococcus horikoshii | 7.0E−21 | 40 | |
| E2 | 651 | a, MTH323 hypothetical protein | Methanobacterium thermoautotrophicum | 1.0E−04 | 45 | |
| 2C12 | b, MTH324 hypothetical protein | 7.0E−05 | 31 | |||
| TAC62 | 464 | Hypothetical protein MTH328 | Methanobacterium thermoautotrophicum | 1.0E−03 | 39 | |
| TAF14 | 363 | Hypothetical protein MTH296 | Methanobacterium thermoautotrophicum | 4.0E−08 | 29 | |
| TAD42 | 708 | Integrin alpha chain, alpha 6 | Homo sapiens | 1.4E−02 | 23 | |
| B5 | 212 | Methyl-accepting chemotaxis protein | Pyrococcus abyssi | 7.0E−7 | 45 | Probably from the same gene |
| TAD52 | 221 | Clostridium acetobutylicum | 3.0E−7 | 40 |
Clones starting with T were sequenced at The Institute for Genomic Research; the remaining clones were sequenced at Dalhousie University.
For identical clones, the length is given for the first clone listed.
If more than one gene was covered by the clone, they are given as follows: a, gene X; b, gene Y; c, gene Z.
Percent identity refers to protein identity if not otherwise noted.
aa, amino acids.
A large number of clones (13 clones) show similarity to a 2,343-bp-long hypothetical gene from Bacillus halodurans (BH1878), which shows a high level of similarity (77% similarity, expect value, E−174) to an arabinan endo-1,5-α-l-arabinosidase (from Bacillus subtilis and Clostridium acetobutylicum), as well as different endo-1,4-β-xylanases. Alignment of the RQ2 sequences with those of BH1878 showed that there are at least three different homologs of (parts of) this gene in the RQ2 genome. Some of the sequences also appear to be rearranged compared to the BH1878 sequence (Table 4; Nesbø and Doolittle, unpublished).
Notably, there are at least four different arabinosidase genes among the RQ2 subtractive clones, while T. maritima MSB8 has only one arabinosidase—TM0281—quite dissimilar in sequence from these. Five additional RQ2-specific genes are predicted to be involved in the degradation of polysaccharides such as cellulose and hemicellulose (Table 4). This large number of poly- and oligosaccharide-degrading enzymes, along with the large number of sugar ABC transporters, suggests that degradation of polysaccharides might be more important to the biology of RQ2 than to that of MSB8.
Large variation in genes involved in production of surface structures.
Nine of the subtractive clones show the highest similarity to the four genes from the rhamnose biosynthetic pathway (Table 4). Rhamnose is widely distributed in O antigens of gram-negative bacteria and in the capsular polysaccharides of gram positives (33, 35). Genes homologous to these RQ2 sequences have earlier been suggested to be frequently transferred among and within bacterial groups such as Streptococcus pneumoniae, Salmonella enterica, and Escherichia coli (27, 33, 55). T. maritima MSB8 contains only one of the rhamnose biosynthetic genes, TM0862, which encodes glucose-1-phosphate thymidylyltransferase (RmlA). Moreover, the sequence from the RQ2 subtractive clone shows only 48% similarity to this MSB8 homolog in BLASTX searches (expect value, 1.1E−06), suggesting that RQ2 has acquired the complete rhamnose pathway by LGT.
There are also several other genes encoding proteins involved in the production of surface structures among the RQ2-specific subtractive clones (Table 4). Large variation in genes encoding surface structures, often due to LGT, has been observed in comparisons of closely related genomes of pathogens (50, 55-57) and has been suggested to be a result of diversifying selection pressure to evade host immune defenses (36, 50, 57). The high proportion of genes involved in surface structures among the RQ2-specific genes, however, suggests that variation in surface structures may be a more general feature of closely related bacterial strains.
V-ATPase in Thermotoga sp. strain RQ2.
Fourteen clones showed significant similarity to seven different archaeal/vacuolar-type ATPase (V-ATPase) subunits, typically found in archaea and eukaryotes. T. maritima MSB8 has only the F-type ATPase typically found in bacteria. V-ATPases are known in a few bacteria, but the distribution is scattered, indicative of lateral transfer (42), and the outgroup closest to RQ2 and MSB8 for which there is information, Fervidobacterium islandicum, has an F-type ATPase (34). Although the simplest scenario would invoke LGT of a V-ATPase gene cluster into RQ2 after its divergence from MSB8, MSB8 does contain a V-ATPase subunit D pseudogene (TM1725). Both the RQ2 V-ATP-A and V-ATP-B subunit sequences clustered strongly (100% bootstrap support) within the archaeal type V-ATPases in all phylogenetic analyses (Nesbø and Doolittle, unpublished).
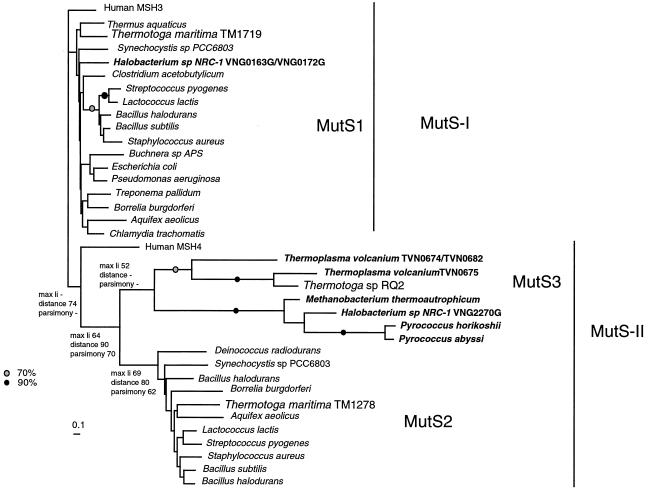
A third archaeal type of prokaryote MutS homologs.
Eisen (12) identified two main prokaryotic MutS lineages, MutS1 and MutS2, where only MutS1 has been shown to be involved in mismatch repair (58). T. maritima MSB8 has homologs of both of these, TM1719 and TM1278. Both genes are also found in the RQ2 genome: one clone matching part of the TM1719 sequence (92% similarity; accession no. AJ458843) was found among the subtractive clones classified as false positives, and a gene with 89% similarity to TM1278 was found in a genomic lambda library made from RQ2 (Nesbø and Doolittle, unpublished). In addition, four RQ2 differential clones showed significant similarity to a MutS homolog from Thermoplasma volcanium. A phylogenetic tree based on the most conserved parts (motifs I to IV in Fig. 1 of reference 8) of the region covered by the RQ2 subtractive clones is shown in Fig. 3 and identifies a new clade of MutS homologs (MutS3) possibly obtained by RQ2 from an archaean. Frequent transfer of MutS1-encoding genes has been described in E. coli and interpreted in the context of a general theory of the role of mismatch repair mutants in permitting LGT (e.g., see reference 6). The function of MutS3-encoding genes is unknown, but we find this evidence of transfer suggestive. In this connection, we note that the Halobacterium genome contains two bacterial MutS1-encoding gene homologs. No other archaeal genomes appear to contain MutS1-encoding genes.
FIG. 3.
Phylogeny of prokaryotic MutS homologs. Two eukaryotic MutS homologs, human MSH3 from the MutS-I group and human MSH4 from the MutS-II group, are included as references. The tree is a Fitch tree made from distances estimated by using a JTT+ Γ +Ι model in PHYLIP version 3.6 (14). The α parameter and the proportion of invariable sites (I) were estimated in PUZZLE version 4.0 (52). The maximum-likelihood tree calculated by using Proml in PHYLIP version 3.6 (with a substitution matrix provided by E. Tillier [personal communication] and the same Γ + Ι model as used in the distance analysis) and parsimony trees estimated by using PAUP∗ (53) gave very similar topologies. The overall topology also agrees with the tree presented by Eisen (12), who used a larger alignment. In all of these analyses, 10 random additions of the sequences and global rearrangements were used. The value at a node is the number of times the node was recovered in 100 bootstrap replicates. Black dots on branches indicate that the bootstrap support was greater than 90% in all of the analyses, while gray dots indicate greater than 70% support. Only the most conserved part of the alignment was used to construct trees (120 amino acids covering motifs I to IV). Some genomes contain multiple copies of the same type of MutS-encoding genes (the Thermoplasma and Halobacterium genomes), and for these, the gene number is indicated. Archaeal sequences are in boldface type. Some possible MutS homologs were not included in the tree, as they formed extremely long branches in a preliminary analysis (H. pylori and Campylobacter jejuni) or did not contain the part of the sequence used in the alignment (Deinococcus radiodurans).
Some of the RQ2-specific sequences are also found in other Thermotoga strains.
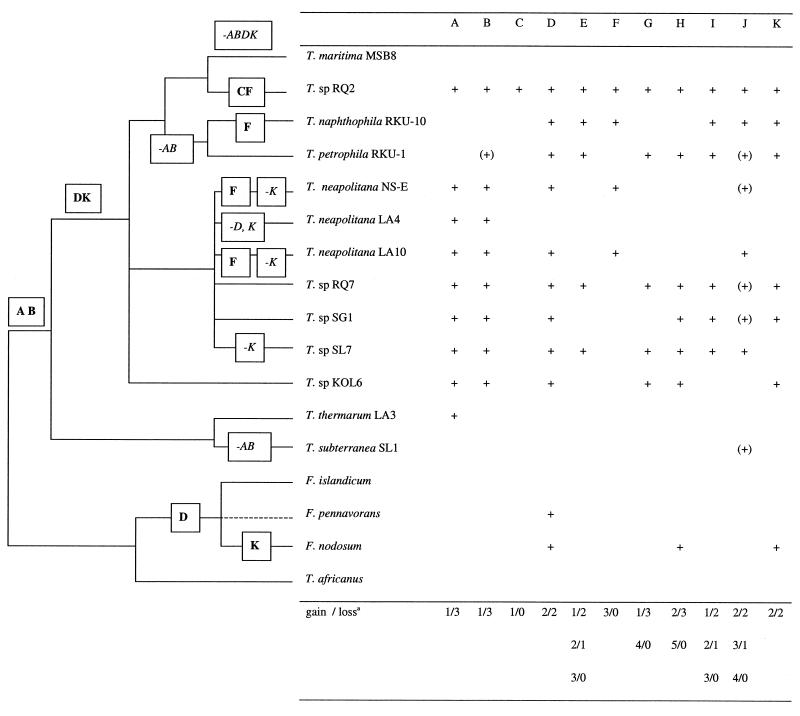
In order to investigate if the RQ2-specific sequences are also found in other Thermotoga strains, we performed Southern blot analyses. Eleven subtractive clones were hybridized to gels with genomic DNAs from 17 different strains of the genus Thermotoga and other members of the order Thermotogales (Fig. 4). All of the genes showed scattered distributions not explainable by simple gene loss from T. maritima MSB8. The minimum number of events required to explain the pattern observed is summarized for each probe, and in total, a minimum of 17 to 27 gains and 10 to 22 losses are needed to explain the hybridization pattern of these 11 probes.
FIG. 4.
Occurrence of RQ2-specific sequences in other members of the order Thermotogales as observed by Southern blot analysis. (A) V-ATP-B clone probe made from T. neapolitana LA4 clone (Nesbø and Doolittle, unpublished). (B) V-ATP-D from Thermotoga sp. strain RQ2 clone 3a1 (the signal from RKU-1 was weak). (C) Pectin methyltransferase-like protein from Thermotoga sp. strain RQ2 clone 2a10. (D) ABC transporter periplasmic substrate-binding protein from Thermotoga sp. strain RQ2 clone 3F3. (E) MutS3 from Thermotoga sp. strain RQ2 clone D5. (F) RmlC from Thermotoga sp. strain RQ2 clones E9 and 2a4. (G) Probable arabinosidase BH1878 from Thermotoga sp. strain RQ2 clone A8. (H) Probable arabinosidase BH1878 from Thermotoga sp. strain RQ2 clone A12 (the signal from F. nodosum was weak). (I) Methyl-accepting chemotaxis protein from Thermotoga sp. strain RQ2 clone B5. (J) Two hypothetical Methanobacterium thermoautotrophicum genes (MTH323 and MTH324) from Thermotoga sp. strain RQ2 clone 2C12 (the signal from most strains was weak). (K) Alcohol dehydrogenase from Thermotoga sp. strain RQ2 clone 2B8. A schematic representation of the small-subunit phylogeny in Fig. 1 is on the left. F. pennivorans is represented by a dotted line because we had no small-subunit sequences from this species. Since the phylogeny of the different T. neapolitana and Fervidobacterium strains is unresolved, loss or gain involving strains from these groups was counted as one event. For instance, for probe D, we assumed one transfer involving a common ancestor of SL7 and RQ7. For the probes where a single most parsimonious pattern could be resolved, the pattern of gain and loss is shown. Genes gained are in boldface type, and genes lost are in italics.
Thermotoga sp. strain RQ2 genes with a DNA match in T. maritima MSB8.
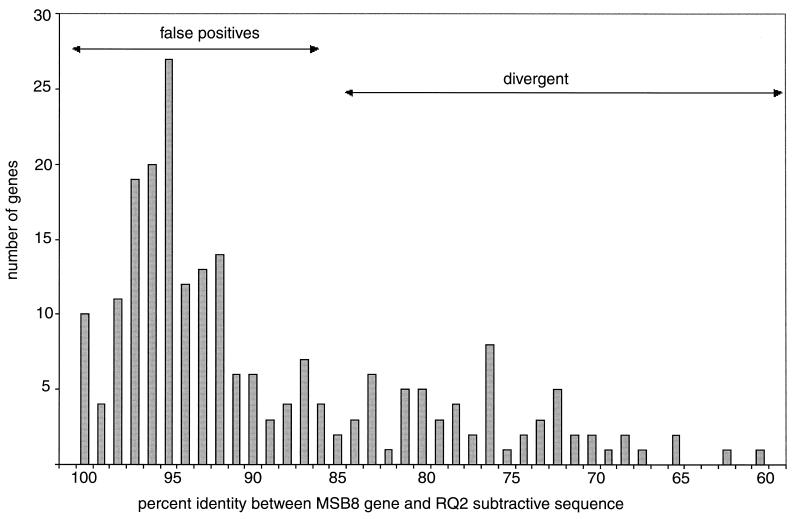
Two hundred sixty clones, corresponding to 243 different genes, did have a DNA match in T. maritima MSB8 (Table 3). On hundred thirty of these shared sequences had greater than 85% DNA identity (average, 94%) and were classified as false positives. Ninety-three clones were classified as divergent, as they showed less than 85% DNA identity (average, 77%) to the MSB8 homolog. As observed for the differential clones, a high number of ABC transporter genes were observed among the divergent clones; 22 of these clones showed significant identity to ABC transporter genes. Thirty-seven clones were judged to be too low in quality to be used in the average-identity calculations (the average identity of these to the MSB8 homolog was 84%).
Figure 5 shows the distribution of the levels of identity of the false positives and the divergent clones to their T. maritima MSB8 homolog. We suspect that some of the divergent genes originated as laterally transferred genes from different members of the order Thermotogales. In support of this, one of the RQ2-specific clones (B1, 2a11; Table 4) shows 82% similarity to TM0076 from T. maritima and 93% similarity to XloA from T. neapolitana. (This clone would have been listed as divergent; however, because it also contains 100 bp with no hit in any databases, it was classified as differential.) Another explanation for an elevated level of divergence would be positive selection. To check for positive selection among the genes, we have estimated the ratio of synonymous to nonsynonymous mutations—the ds/dn ratio. A ds/dn ratio of less than 1 is usually taken as evidence of positive selection (e.g., see reference 18). Most genes show very high ds/dn ratios (average ds/dn ratio, 11.76), a pattern consistent with purifying selection. This was also true for the highly divergent genes (average ds/dn ratio, 14.18), suggesting that they have experienced a normal mutation pattern, most consistent with the notion that they have been acquired by LGT from other members of the order Thermotogales. Only four clones (corresponding to TM0969, TM0134, TM1044, and TM1147) had a ds/dn ratio of less than 1, but there were no clear cases of positive selection. The RQ2 sequence overlapping that of T. maritima MSB8 was either too short (TM1044), or the open reading frame was almost completely deleted in RQ2 and probably corresponded to a pseudogene (TM0969, TM1147, and TM0134).
FIG. 5.
Histogram of the number of RQ2 subtractive sequences with a significant DNA match in the MSB8 genome plotted against percent similarity to the MSB8 homolog. The number of clones is plotted along the y axis, and percent similarity is plotted along the x axis. Low-quality sequences were excluded. Where multiple clones covered the same T. maritima MSB8 gene, an average was calculated. Sequences of clones that covered more than one gene were divided into the respective number of subsequences. This resulted in 222 comparisons between MSB8 and RQ2 coding regions.
Assuming a gene order similar to that seen in the MSB8 genome for the shared genes, the divergent genes tended to occur in clusters in the RQ2 genome (P < 10−16). These data suggest that there are islands of divergent genes that could represent areas with high levels of recombination and/or LGT from other members of the order Thermotogales.
DISCUSSION
SSH proves to be an easy and quick way to derive information on differences in lifestyle and metabolism of closely related prokaryotic strains when one strain's genome has been completely sequenced. About 40% of the sequenced clones did not have a DNA match in the T. maritima MSB8 genome, which translates into twofold enrichment (assuming 20% RQ2-specific genes). This method is clearly most economical in detecting differences in gene content between very similar strains (in terms of identity of shared orthologs). Including the divergent clones, 60% of the clones sequenced are potentially interesting with regard to strain-specific adaptations.
Here we describe RQ2-specific DNA corresponding to a total of 48 kb and identify 72 RQ2-specific genes with a match in the GenBank database (Table 3). We have sequenced only a fraction of the clones in our library, and repeated sampling would uncover further strain-specific genes. A simple calculation (see Materials and Methods) indicates that the RQ2 genome carries 350 to 400 genes not found in the sequenced genome of MSB8. The MSB8 genome bears 1,877 genes: if the RQ2 genome is the same size, this would correspond to about 20% of its total gene complement. Similar values are found in comparisons of other bacterial species; for instance, 6 to 18% for Helicobacter pylori (2, 49), up to 20% for strains of S. enterica (32), and 22% variable genes in the genome of Staphylococcus aureus (16) (Table 1).
The large number of differential genes predicted to be involved in sugar import and polysaccharide degradation strongly suggests that degradation of polysaccharides is more important in the biology of RQ2 than in that of MSB8. There is considerable commercial interest in polysaccharide-degrading enzymes in Thermotoga because of their potential use in high-temperature kraft pulping, and polymorphism in the number and type of genes has been observed earlier in studies focused on individual genes (48). For instance, variation in the number and type of xyn genes has been reported among T. maritima MSB8, T. neapolitana NS-ET, and Thermotoga sp. strain FjSS3B.1 (48). Notably, clone C6, TAF78, and B1 probably encode yet another new Thermotoga xyn gene (Table 4). SSH should provide an economical way to uncover new activities in many systems.
Sugar ABC transporters were particularly abundant among the differential clones, some of which had T. maritima MSB8 proteins as the best match in translated searches against protein databases and some of which showed best matches to proteins from other, more distantly related, prokaryotes. Expansion of ABC transporter gene families was previously observed in the T. maritima MSB8 genome with a notable expansion of oligopeptide transporters (39). Indeed, T. maritima MSB8 had the highest proportion of ABC transporters in a comparison of 18 completely sequenced prokaryote genomes (44). The large number RQ2-specific sugar ABC transporter genes suggests that RQ2 has experienced an independent lineage-specific expansion of sugar transporters; this may be a common way for Thermotoga strains to adapt to their environment.
Many of the differential or highly divergent genes in Thermotoga sp. strain RQ2 are likely to have been acquired through LGT from either Thermotoga strains, other members of the order Thermotogales, or more distantly related bacteria and archaea. This conclusion rests on the patchy distribution seen in the Southern hybridizations (Fig. 4) and on phylogenetic analyses. Although most sequenced clones from RQ2 are not suitable for phylogenetic analyses because they are too short or because the GenBank database holds too few homologs, we have been able to reconstruct phylogenies for some of the genes listed in Table 4 (as noted there). Particularly interesting were the sequences for which homologs were present in the MSB8 genome (Table 4). For instance, phylogenetic analysis of the alcohol dehydrogenase gene (clone 2b8, TAB19, TAD49, and KJ3C6) showed the MSB8 and RQ2 sequences to fall into very distinct sequence clusters, indicating that at least one of the strains has acquired its copy through LGT. A high level of lateral transfer of this gene was also suggested by the Southern analysis. For some other genes, loss in the MSB8 genome appears to be the more likely explanation; this seems to be the case for the β-glucosidase gene encoded by the sequences in clone E6 and TAF85 (Table 4). For still other sequences, a complex pattern of both LGT and differential loss can be inferred. This is, for instance, the case for the ABC transporter detected in nine other Thermotoga strains, as well as two of the Fervidobacterium strains (Table 4), and for the V-ATPase operon. As we get more sequences from other subtraction experiments involving other Thermotoga strains, we will be able to more firmly quantify the relative proportions of gene loss and LGT.
The Southern blot analyses of the differential genes suggest that the two strains sharing the greatest number of sequences defined as RQ2 specific here (that is, not in MSB8) are Thermotoga sp. strains RQ7 and SL7. SL7 was isolated from an oil basin outside Paris, France, while RQ7 was isolated from the same environment as RQ2 (the Azores) (Table 2). Both of these strains cluster with the T. neapolitana strains in small-subunit phylogenies (Fig. 1), as well as for two protein coding genes (40). Overall, the strains from the T. neapolitana clade appear to share more of these variable genes than do T. petrophila RKU-1 and T. naphthophila RKU-10, which nevertheless have a more recent ancestor in common with RQ2 and MSB8 than the T. neapolitana strains do in the small-subunit phylogeny (Fig. 1). A closer phylogenetic relationship among RQ2, T. petrophila RKU-1, and T. naphthophila RKU-10 than among RQ2, RQ7, and SL7 was also observed by analyses of flagellar genes (Boudreau, Nesbø, and Doolittle, unpublished data). Thus, it appears that T. maritima strains frequently exchange genes with their T. neapolitana neighbors. This is also supported by the recombination observed for the ino-1 gene observed previously (40). Hence, for these variable genes, geography and ecology are probably more important than phylogeny.
Acknowledgments
This work was supported by a postdoctoral fellowship to C.L.N. from the Canadian Institute for Health Research and by funds from the Canadian Institutes for Health Research (MOP 4467), Genome Canada, and DOE (DEFC029ER61962).
We are grateful to Christian Blouin for doing the proximity analysis of shared orthologs, Marlena Dlutek for excellent technical assistance, and Lesley Davis for the sequencing done at Dalhousie University. We thank Yan Boucher and Yuji Inagaki for critical reading of the manuscript. We also thank Karl O. Stetter, Yoh Takahata, Stéphane L'Haridon, Christian Jeanthon, and Fiona Duffner for donating Thermotogales DNA or cell mass.
REFERENCES
- 1.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834-846. [DOI] [PubMed] [Google Scholar]
- 3.Beja, O., E. V. Koonin, L. Aravind, L. T. Taylor, H. Seitz, J. L. Stein, D. C. Bensen, R. A. Feldman, R. V. Swanson, and E. F. DeLong. 2002. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 68:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogush, M. L., T. V. Velikodvorskaya, Y. B. Lebedev, L. G. Nikolaev, S. A. Lukyanov, A. F. Fradkov, B. K. Pliyev, M. N. Boichenko, G. N. Usatova, A. A. Vorobiev, G. L. Anders, and E. D. Sverdlov. 1999. Identification and localization of differences between Escherichia coli and Salmonella typhimurium genomes by suppressive subtractive hybridization. Mol. Gen. Genet. 262:721-729. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, Y., C. L. Nesbø, and W. F. Doolittle. 2001. Microbial genomes: dealing with diversity. Curr. Opin. Microbiol. 4:285-289. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. W., J. E. LeClerc, B. Li, W. L. Payne, and T. A. Cebula. 2001. Phylogenetic evidence for horizontal transfer of mutS alleles among naturally occurring Escherichia coli strains. J. Bacteriol. 183:1631-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, P. K., and R. Curtiss III. 1996. Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 93:11149-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charbonnier, F., and P. Forterre. 1995. Protocol 12: Purification of plasmids from thermophilic and hyperthermophilic archaea, p. 87-90. In F. T. Robb and A. R. Place (ed.), Archaea: a laboratory manual—thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124-2129. [DOI] [PubMed] [Google Scholar]
- 10.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, J. A. 1998. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 26:4291-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 2001. PHYLIP phylogeny inference package, version 3.6 ed. Department of Genetics, University of Washington, Seattle.
- 15.Fitzgerald, J. R., and J. M. Musser. 2001. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 9:547-553. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich, A. B., and G. Antranikian. 1996. Keratin degradation by Fervidobacterium pennivorans, a novel thermophilic anaerobic species of the order Thermotogales. Appl. Environ. Microbiol. 62:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fudyk, T. C., I. W. Maclean, J. N. Simonsen, E. N. Njagi, J. Kimani, R. C. Brunham, and F. A. Plummer. 1999. Genetic diversity and mosaicism at the por locus of Neisseria gonorrhoeae. J. Bacteriol. 181:5591-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain, K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 21.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, R., T. A. Langworthy, H. Konig, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 23.Huber, R., C. R. Woese, T. A. Langworthy, H. Fricke, and K. O. Stetter. 1989. Thermosipho africanus gen. nov. represents a new genus of thermophilic eubacteria within the Thermotogales. Syst. Appl. Microbiol. 12:32-37. [Google Scholar]
- 24.Huber, R., C. R. Woese, T. A. Langworthy, J. K. Kristjansson, and K. O. Stetter. 1990. Fervidobacterium islandicum sp. nov., a new extremely thermophilic eubacterium belonging to the Thermotogales. Arch. Microbiol. 154:105-111. [Google Scholar]
- 25.Jannasch, H. W., R. Huber, S. Belkin, and K. O. Stetter. 1988. Thermotoga neapolitana sp. nov. of the extremely thermophilic, eubacterial genus Thermotoga. Arch. Microbiol. 150:103-104. [Google Scholar]
- 26.Jeanthon, C., A. L. Reysenbach, S. L'Haridon, A. Gambacorta, N. R. Pace, P. Glenat, and D. Prieur. 1995. Thermotoga subterranea sp. nov., a new thermophilic bacterium isolated from a continental oil reservoir. Arch. Microbiol. 164:91-97. [PubMed] [Google Scholar]
- 27.Jiang, S. M., L. Wang, and P. R. Reeves. 2001. Molecular characterization of Streptococcus pneumoniae type 4, 6B, 8, and 18C capsular polysaccharide gene clusters. Infect. Immun. 69:1244-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koonin, E. V., L. Aravind, and A. S. Kondrashov. 2000. The impact of comparative genomics on our understanding of evolution. Cell 101:573-576. [DOI] [PubMed] [Google Scholar]
- 30.Korber, B. 2001. HIV signature and sequence variation analysis, p. 55-72. In A. G. Rodrigo and G. H. Learn (ed.), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Lai, Y. C., S. L. Yang, H. L. Peng, and H. Y. Chang. 2000. Identification of genes present specifically in a virulent strain of Klebsiella pneumoniae. Infect. Immun. 68:7149-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan, R., and P. R. Reeves. 1996. Gene transfer is a major factor in bacterial evolution. Mol. Biol. Evol. 13:47-55. [DOI] [PubMed] [Google Scholar]
- 33.Li, Q., and P. R. Reeves. 2000. Genetic variation of dTDP-l-rhamnose pathway genes in Salmonella enterica. Microbiology 146:2291-2307. [DOI] [PubMed] [Google Scholar]
- 34.Ludwig, W., O. Strunk, S. Klungbauer, N. Klugbauer, M. Weiznegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 35.Ma, Y., R. J. Stern, M. S. Scherman, V. D. Vissa, W. Yan, V. C. Jones, F. Zhang, S. G. Franzblau, W. H. Lewis, and M. R. McNeil. 2001. Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob. Agents Chemother. 45:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiden, M. C. J., J. Suker, and I. M. Feavers. 1997. Horizontal genetical exchange in the evolution of Neisseria meningitidis outer membrane proteins, p. 15-34. In B. A. M. van der Zeijst, W. P. M. Hoekstra, and J. D. A. van Embden (ed.), Ecology of pathogenic bacteria: molecular and evolutionary aspects. Royal Netherlands Academy of Arts and Sciences, Amsterdam.
- 37.McClelland, M., K. E. Sanderson, J. Spieth, S. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, S. Tamberlyn, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Science 413:852-856. [DOI] [PubMed] [Google Scholar]
- 38.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, C. M. Fraser, et al. 1999. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 40.Nesbø, C. L., S. L'Haridon, K. O. Stetter, and W. F. Doolittle. 2001. Phylogenetic analyses of two “archaeal” genes in Thermotoga maritima reveal multiple transfers between archaea and bacteria. Mol. Biol. Evol. 18:362-375. [DOI] [PubMed] [Google Scholar]
- 41.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olendzenski, L., L. Liu, O. Zhaxybayeva, R. Murphey, D.-G. Shin, and J. P. Gogarten. 2000. Horizontal transfer of archaeal genes into the Deinococcaceae: detection by molecular and computer-based approaches. J. Mol. Evol. 51:587-599. [DOI] [PubMed] [Google Scholar]
- 43.Patel, B. K. C., H. W. Morgan, and R. M. Daniel. 1985. Fervidobacterium nodosum gen. nov. and spec. nov., a new chemoorganotrophic, caldoactive, anaerobic bacterium. Arch. Microbiol. 141:63-69. [Google Scholar]
- 44.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 45.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glansner, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 46.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Read, T. D., S. R. Gill, H. Tettelin, and B. A. Dougherty. 2001. Finding drug targets in microbial genomes. Drug Discov. Today 6:887-892. [DOI] [PubMed] [Google Scholar]
- 48.Reeves, R. A., M. D. Gibbs, D. D. Morris, K. R. Griffiths, D. J. Saul, and P. L. Bergquist. 2000. Sequencing and expression of additional xylanase genes from the hyperthermophile Thermotoga maritima FjSS3B.1. Appl. Environ. Microbiol. 66:1532-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selander, R. K. 1997. DNA Sequence analysis of the genetic structure and evolution of Salmonella enterica, p. 191-213. In B. A. M. van der Zeijst, W. P. M. Hoekstra, and J. D. A. van Embden (ed.), Ecology of pathogenic bacteria: molecular and evolutionary aspects. Royal Netherlands Academy of Arts and Sciences, Amsterdam.
- 51.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 53.Swofford, D. L. 2001. PAUP∗ phylogenetic analysis using parsimony (∗and other methods), version 4 ed. Sinauer Associates, Sunderland, Mass.
- 54.Takahata, Y., M. Nishijima, T. Hoaki, and T. Maruyama. 2001. Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. Int. J. Syst. E vol. Microbiol. 51:1901-1909. [DOI] [PubMed] [Google Scholar]
- 55.Tarr, P. I., L. M. Schoening, Y. L. Yea, T. R. Ward, S. Jelacic, and T. S. Whittam. 2000. Acquisition of the rfb-gnd cluster in evolution of Escherichia coli O55 and O157. J. Bacteriol. 182:6183-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 57.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 58.Vijayvargia, R., and I. Biswas. 2002. MutS2 family protein from Pyrococcus furiosus. Curr. Microbiol. 44:224-228. [DOI] [PubMed] [Google Scholar]
- 59.Windberger, E., R. Huber, A. Trincone, H. Fricke, and K. O. Stetter. 1989. Thermotoga thermarum sp. nov. and Thermotoga neapolitana occurring in African continental solfataric springs. Arch. Microbiol. 151:506-512. [Google Scholar]