Abstract
We have investigated whether UV-induced mutations are created with equal efficiency on the leading and lagging strands of DNA replication. We employed an assay system that permits measurement of mutagenesis in the lacZ gene in pairs of near-identical strains. Within each pair, the strains differ only in the orientation of the lacZ gene with respect to the origin of DNA replication. Depending on this orientation, any lacZ target sequence will be replicated in one orientation as a leading strand and as a lagging strand in the other orientation. In contrast to previous results obtained for mutations resulting from spontaneous replication errors or mutations resulting from the spontaneous SOS mutator effect, measurements of UV-induced mutagenesis in uvrA strains fail to show significant differences between the two target orientations. These data suggest that SOS-mediated mutagenic translesion synthesis on the Escherichia coli chromosome may occur with equal or similar probability on leading and lagging strands.
Exposure of cells to UV light results in the formation of mutagenic DNA lesions, among which the two most frequent lesions are cyclobutane pyrimidine dimers and 6-4 pyrimidine-pyrimidone photoproducts at adjacent pyrimidines (13). In the bacterium Escherichia coli, mutagenesis by UV light and by a variety of chemical mutagens is mediated by the inducible SOS response (13, 29). The SOS response is a global response involving the coordinated expression of some 30 genes (9). Its general function is to promote cellular survival upon induction of DNA damage, albeit at the cost of increased mutagenesis. The response is controlled by the interplay of the RecA and LexA proteins, the latter constituting the transcriptional repressor of the SOS regulon (13). After blockage of ongoing DNA replication by DNA damage, RecA forms nucleoprotein filaments with the damage-induced single-stranded DNA. This activated form of RecA then mediates the cleavage of LexA, thereby enabling the expression of the SOS genes.
SOS mutagenesis is dependent on the lexA-controlled umuDC operon (17, 18, 32), which upon induction of the system yields the UmuD′2C complex (UmuD′ being the RecA-mediated cleavage product of UmuD). This complex was recently shown to be a DNA polymerase, termed Pol V (26, 28, 34-36). Pol V is thought to promote mutagenesis by mediating mutagenic bypass of the DNA lesions, such as pyrimidine dimers, after DNA Pol III holoenzyme (HE) is blocked at such lesions. Pol V is a member of a recently described class of DNA polymerases (the UmuC/DinB/Rad30/Rev1 superfamily), now named Y-family DNA polymerases, that is found in a broad range of organisms (25). These enzymes are best characterized by their generally low-fidelity synthesis and/or ability to bypass DNA lesions in vitro (for a review, see reference 38). In addition to Pol V, E. coli also possesses another SOS-inducible Y-family DNA polymerase, Pol IV (37). The precise functions of Pol IV are not yet clear, but its role in DNA-damage induced mutagenesis, including UV mutagenesis, appears limited (4, 19, 20, 35).
In addition to mutagenesis at DNA lesions by bypass synthesis (targeted mutagenesis), SOS induction is also characterized by increased mutagenesis on nondamaged DNA. This increased mutagenesis has been called the “SOS mutator activity” or “untargeted mutagenesis” (39, 40). Like targeted mutagenesis, the SOS mutator activity depends on the umuDC genes (5). The effect may result from Pol V-produced replication errors on undamaged DNA (23, 35) or, as we have proposed, from Pol V-mediated extension of Pol III-created terminal mismatches (10, 22).
The precise mechanisms by which the SOS system promotes lesion bypass are not known. Pol V likely performs one or more critical steps in the translesion synthesis in conjunction with RecA, SSB, and the Pol III β,γ complex, an assembly referred to as mutasome (27, 34, 36) or Pol V HE (28). How Pol V might be targeted to the site of template damage or what the relevant interactions are among the various DNA polymerases at the replication fork is unclear. Sutton et al. (33) have proposed that Pol V and Pol III act in coordinated fashion in translesion synthesis, based on physical interaction observed between UmuD, UmuC, and certain Pol III HE subunits. On the other hand, the presence of the Pol III core is not required for Pol V-catalyzed translesion synthesis in vitro (34, 36), and a simple competition between the various polymerases for the lesion-containing primer terminus has also been proposed (1, 24).
In this work, we have attempted to learn more about the interactions at the replication fork by investigating whether SOS-mediated lesion bypass might occur with differential efficiency in the leading and lagging strands of DNA replication. At the replication fork, the leading and lagging strands are the product of distinctly different enzymologies (15, 21), and the SOS system may well affect them differentially. To address this question, we have performed lac reversion assays in pairs of strains differing only in the orientation of the lac operon with respect to the origin of chromosomal replication (Fig. 1). Within each pair, a given lac sequence is replicated by the leading strand machinery in one orientation and by the lagging strand machinery in the other. Differences in UV-induced mutant frequencies within such pairs, if found, will be indicative of differential mutability between the two strands.
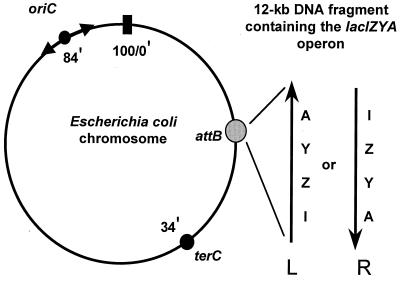
FIG. 1.
Insertion of lac operon into phage lambda attB site on E. coli chromosome in two orientations with regard to chromosomal replication oriC. The orientation in which the lac operon is transcribed in the same direction as the replication fork movement through the target is designated R, while the L orientation indicates lac transcription in the opposite direction (see arrows at attB). The arrows at oriC represent the two forks initiated at this site.
Previously, we applied this system to the question of normal replication fidelity by analyzing the reversion frequencies of several defined lacZ alleles in mismatch-repair-defective strains. Moderate but consistent differences (two- to sixfold, depending on the mutational marker) were observed between the two lac orientations, indicating that normal replication fidelity is different in the two strands (11). It was deduced that on the E. coli chromosome, for the base-substitution errors investigated, lagging strand replication is more accurate than leading strand replication (11). We also investigated the orientation bias for the SOS mutator effect, as mediated by the constitutive recA730 allele. In recA730 strains, the SOS system is induced constitutively, leading to increased mutagenesis in the absence of DNA-damaging treatments (untargeted mutagenesis) (10, 39, 40). We found, interestingly, that the orientation bias is inversed under these conditions and that, apparently, lagging strand replication is the major source of the mutations (22). In the present study, we have investigated the frequency of UV-induced mutations in excision-repair-deficient (uvrA) strains by measuring the UV-induced reversion of six different lacZ alleles. Each of the six alleles proved readily mutable by UV light, but no significant differences in reversion frequency were observed between the two gene orientations. These data suggest that on the E. coli chromosome translesion synthesis of mutagenic UV photoproducts may proceed with equal efficiency in the two DNA strands.
MATERIALS AND METHODS
Media.
Solid and liquid media were as previously described (12). Minimal plates were supplemented with 0.4% glucose or 0.4% lactose as a carbon source, 5 μg of thiamine/ml, and 50 μg of amino acids/ml if required. Antibiotics were added as follows: tetracycline, 12.5 μg/ml; ampicillin, 25 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml.
Strain constructions.
A series of strains containing the lac operon inserted in the chromosomal phage λ attachment site (attL) in the two possible orientations was created by the method described by Diederich et al. (8) as described previously (11, 14). All are derivatives of strain MC4100, which has the lac operon deleted from its normal location near 8 min on the E. coli map (2). Each of the inserted lac operons carries a defined lacZ missense or frameshift mutation that allows scoring of mutagenesis via reversion to lac+. Specifically, we used strains containing the lacZ gene from strains CC102 and CC106 (6), which permitted scoring of G · C→A · T and A · T→G · C base substitutions, respectively, and the lacZ gene from strains CC107, CC108, CC110, and CC111 (7), which permitted scoring of (+1)G, (−1)G, (+1)A, and (−1)A frameshifts, respectively.
The base pair substitution strains EC3114 and EC3120 (carrying the R- and L-oriented lacZ gene from CC102, respectively) and EC3156 and EC3150 (carrying the R- and L-oriented lacZ gene from CC106) have been described (11). For the purpose of the present study, they were made excision repair deficient (uvrA) by P1 transduction using N3055 (uvrA277::Tn10) (E. coli Genetic Stock Center, Yale University) as a donor, using selection for tetracycline resistance.
The frameshift reversion strains are derivatives of the following described strains (14): EC3301 and EC3307 (carrying the R- and L- oriented lacZ gene from strain CC107), EC3313 and EC3319 (carrying the R- and L-oriented lacZ gene from CC108), EC3325 and EC3331 (carrying the R- and L-oriented lacZ gene from CC110), and EC3337 and EC3343 (carrying the R- and L-oriented lacZ gene from CC111). Each of these strains was made excision repair deficient (uvrA) as described above.
Several of the strains were made umuDC or dinB defective by introduction, via P1 transduction, of the ΔumuDC595::cat or ΔdinB::cat allele, respectively. Donor strains were RW82 (41) and YG2247 (20), respectively.
The mutL strains listed in Table 1 were derived from EC3114, EC3120, EC3150, EC3156, EC3313 and EC3319 (see above). To each of those, the mutL::Tn5 allele from strain NR9559 (12) was introduced by P1 transduction.
TABLE 1.
Mutant frequencies in pairs of strains containing lac operon in opposite (L and R) orientations on E. coli chromosomea
| Test group | lac allele (mutation) | lac orientation | UV dose (J/m2) | % Survival | No. of Lac+/108 cells |
|---|---|---|---|---|---|
| UV experiments | |||||
| CC102 (G · C→A · T) | L | 0 | 100 | 0.9 | |
| R | 0 | 100 | 0.6 | ||
| L | 3 | 36 | 124 | ||
| R | 3 | 36 | 139 | ||
| L | 6 | 8 | 200 | ||
| R | 6 | 8 | 220 | ||
| CC102(umuDC::cat) | |||||
| L | 0 | 100 | 0.6 | ||
| R | 0 | 100 | 0.5 | ||
| L | 3 | 5 | 0.6 | ||
| R | 3 | 4.5 | 0.5 | ||
| CC102(dinB::cat) | |||||
| L | 0 | 100 | 0.8 | ||
| R | 0 | 100 | 0.7 | ||
| L | 3 | 39 | 97 | ||
| R | 3 | 38 | 96 | ||
| CC106 (A · T→G · C) | L | 0 | 100 | 0.1 | |
| R | 0 | 100 | 0.1 | ||
| L | 4 | 24 | 110 | ||
| R | 4 | 23 | 127 | ||
| L | 6 | 8 | 220 | ||
| R | 6 | 8 | 260 | ||
| CC107 (+1G) | L | 0 | 100 | 6.0 | |
| R | 0 | 100 | 7.0 | ||
| L | 6 | 11 | 185 | ||
| R | 6 | 11 | 174 | ||
| CC108 (−1G) | L | 0 | 100 | 5.5 | |
| R | 0 | 100 | 7.5 | ||
| L | 3 | 32 | 289 | ||
| R | 3 | 28 | 295 | ||
| CC108(umuDC::cat) | |||||
| L | 0 | 100 | 7.5 | ||
| R | 0 | 100 | 9.5 | ||
| L | 3 | 2 | 3.5 | ||
| R | 3 | 3 | 9 | ||
| CC108(dinB::cat) | |||||
| L | 0 | 100 | 4.5 | ||
| R | 0 | 100 | 8.0 | ||
| L | 3 | 21 | 257 | ||
| R | 3 | 16 | 266 | ||
| CC110 (+1A) | L | 0 | 100 | 1.25 | |
| R | 0 | 100 | 1.2 | ||
| L | 3 | 31 | 212 | ||
| R | 3 | 30 | 185 | ||
| CC110(umuDC::cat) | |||||
| L | 0 | 100 | 1.2 | ||
| R | 0 | 100 | 1.4 | ||
| L | 3 | 4.5 | 0.8 | ||
| R | 3 | 5.0 | 0.6 | ||
| CC110(dinB::cat) | |||||
| L | 0 | 100 | 1.45 | ||
| R | 0 | 100 | 1.3 | ||
| L | 3 | 30 | 281 | ||
| R | 3 | 31 | 244 | ||
| CC111 (−1A) | L | 0 | 100 | 15 | |
| R | 0 | 100 | 14 | ||
| L | 4 | 14 | 86 | ||
| R | 4 | 14 | 80 | ||
| mutL strains | |||||
| CC102 (G · C→A · T) | L | 0 | 100 | 195 (P < 0.001) | |
| R | 0 | 100 | 37 | ||
| CC106 (A · T→G · C) | L | 0 | 100 | 47 (P < 0.001) | |
| R | 0 | 100 | 21 | ||
| CC108 (−1G) | L | 0 | 100 | 2,700 (P < 0.001) | |
| R | 0 | 100 | 11,500 | ||
| recA730 strains | |||||
| CC106 (A · T→G · C) | L | 0 | 100 | 0.11 (P = 0.02) | |
| R | 0 | 100 | 0.06 |
The strains used in these experiments are described in Materials and Methods. All are also uvrA. The presence of the umuDC or dinB deficiency is shown in parentheses following the indicated lac allele. For the UV experiments, each entry is based on the median value of six independent cultures comprising three independent lacZ integrants for each orientation. For any of these experiments, the difference in lac reversion frequency between L and R strains is not statistically significant (P > 0.5). For the mutL and recA730 experiments, each based on 20 independent cultures, the P value (in parentheses) indicates the statistical significance of the frequency difference between the two orientations (see Materials and Methods). See reference 22 for additional, large differences between L and R orientations in recA730 strains.
The recA730 strains listed in Table 1 were derivatives of EC3150 and EC3156. These strains made sulA366 by the two-step procedure described by Fijalkowska et al. (10). These sulA derivatives were then made recA730 in another two-step procedure (10) by first making the strains srl::Tn10 and then srl+ recA730 by using NR11239 (10) as recA730 donor, selecting on minimal sorbitol medium.
UV light irradiation and measurement of lacZ revertant frequencies.
Overnight cultures for each of the strains were diluted in fresh Luria-Bertani (LB) broth, grown to an optical density of 600 nm (OD600) of 0.2 to 0.3 (≈2 × 108 cells/ml), centrifuged, and resuspended in half the volume of ice-cold 0.9% NaCl. These suspensions were UV irradiated (at 254 nm) in 10-ml aliquots in a petri dish, using the Cross-linker UV irradiation system (UV Products). All procedures were performed in subdued light. Duplicate samples were plated before and after irradiation on LB plates to determine the bacterial survival. For Lac+ frequency determination, 1 ml of the irradiated cells was added to 5 ml of fresh LB broth, grown overnight, and plated on minimal-lactose plates to measure lac+ mutants and on minimal-glucose plates to measure total cells. P values for statistical significance of differences between mutant frequencies for strains with L- or R-oriented lac operons were calculated using the nonparametric Mann-Whitney criterion (30) applied to the mutant yield distributions of the independent cultures for each strain using the program STATISTICA (StatSoft).
RESULTS AND DISCUSSION
To obtain further insight into interactions at the replication fork that might affect SOS-mediated translesion synthesis, we have addressed the question whether the bypass of UV lesions occurs with similar efficiency in the leading and lagging strands of chromosomal DNA replication. Previous studies have indicated that the intrinsic replication fidelity in the two strands is not identical (11, 14, 22), which likely finds its origin in the differential replication enzymology in the two strands (15, 21). Interestingly, a profound, opposite strand bias was observed for mutagenesis by the recA730-mediated SOS mutator activity (22). In view of the latter observation, the question arises whether this particular strand bias is a general feature of SOS mutagenesis or unique to the SOS mutator activity.
Our assay system (Fig. 1 and 2) uses pairs of strains containing defined chromosomal lacZ mutations in the two orientations (L and R; Fig. 1) relative to the replication origin. The reversion of these alleles to lac+ proceeds by defined base pair substitution or frameshift mutation (6, 7). Six different lacZ alleles were chosen, two transition and four frameshift alleles, for which the target sequences contain adjacent pyrimidines (Fig. 2) and which have been shown to be responsive to UV irradiation (7). Within each strain pair, depending on the gene orientation, a UV lesion targeting the reversion will be present in the leading or the lagging strand (Fig. 2), and any strand preference for UV mutagenesis should be observable in the form of differential mutability of the two strains. All strains used are also excision repair deficient (uvrA), facilitating interpretation of the results in terms of replication fidelity without complications arising from DNA repair.
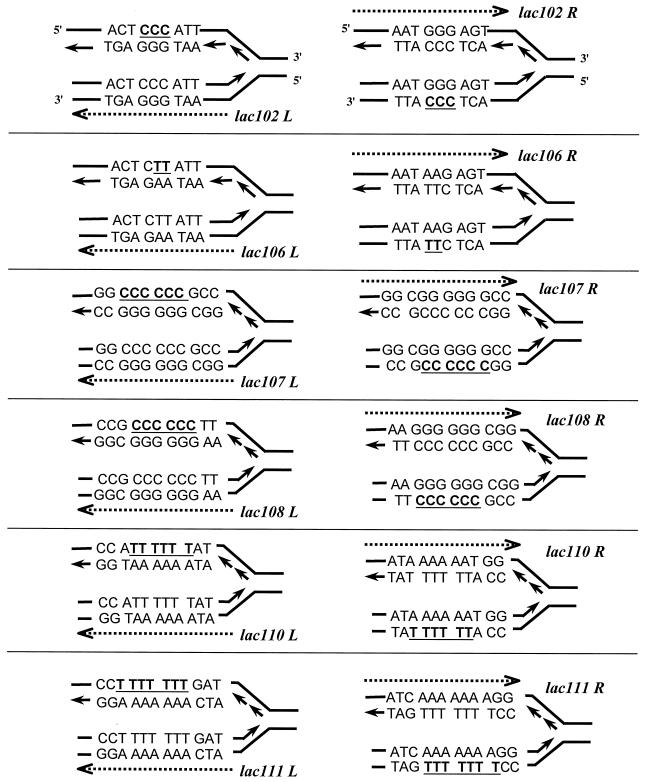
FIG. 2.
Shown are more detailed drawings of the replication fork configurations for R and L orientations for each of the six lacZ alleles used in this paper. lacZ102, lacZ106, lacZ107, lacZ108, lacZ110, and lacZ111 denote the lac alleles as originally present in strains CC102, CC106, CC107, CC108, CC110, and CC111 (6, 7), respectively. The dashed arrows indicate the lacZ coding strand (5′ → 3′). The nucleotides in the target sequences which may create pyrimidine dimers are indicated in bold and underlined.
The results of the experiments are presented in Table 1. All six lac alleles proved readily mutable at moderate UV doses that yielded approximately 10 to 30% survival. The two base substitution alleles that permit measurement of G · C→A · T and A · T→G · C transitions were most strongly enhanced: 200- to 2,000-fold, respectively. However, no significant differences could be observed between the L- and R-oriented strains. The four frameshift alleles were more modestly mutable, the mutant frequency being enhanced between 5- and 100-fold. However, again, no significant differences could be detected between the two orientations. These results contrast clearly to the orientation biases observed when measuring mutagenesis by normal DNA replication errors in mutL strains (Table 1 and references 11 and 14) or mutagenesis resulting from the recA730-mediated SOS mutator (Table 1 and reference 22).
Table 1 also contains results obtained with the umuDC- or dinB-deficient derivatives of several of the strains. The data indicate that UV mutagenesis as measured in these experiments is umuDC dependent and dinB independent, as expected (4, 17-19). These results permit interpretation of the current data in terms of the error-prone action of Pol V (see below).
Based on our results, obtained for six different lac alleles in a variety of DNA sequence contexts, we suggest that UV-induced mutagenesis on the E. coli chromosome occurs with similar likelihood on the leading and lagging strands of replication. In other words, it appears that a given UV lesion has the same probability of (i) blocking ongoing replication and (ii) subsequently being copied by Pol V, regardless of whether it is initially encountered by the leading or the lagging strand replication machinery.
Leading/lagging strand differences for normal replication errors have been assumed to be a consequence of the greater dissociative character of the lagging strand polymerase. This dissociative character provides additional opportunities for removal of Pol III-mediated misinsertion errors (11). In contrast, the greater dissociability of the lagging strand polymerase, as well as the presence of single-stranded DNA regions allowing greater access for the RecA nucleofilament, make the lagging strand a preferred target for Pol V, providing an explanation for the reversed, lagging strand bias of the SOS mutator effect (22). Apparently, in the case of UV (and, likely, other mutagens causing bulky DNA lesions), these (subtle) dissociative differences between the strands cease to be a significant factor for the fate of the stalled complex. Thus, it appears that there is a distinction to be drawn between the transient blockage of DNA polymerases at terminal mismatches, which can be alleviated simply by mismatch removal, and the more persistent blockage at sites of DNA template lesions. At the latter, removal of misinserted bases opposite the lesions is not particularly helpful, as it only leads to futile polymerase cycling. Our data suggest that the access of Pol V to the primer terminus (the presumed rate-limiting step for translesion synthesis) is ultimately the same for either strand, despite the enzymological differences between the two replication modes. This may be consistent with the reported timing of UV mutagenesis, occurring some 45 to 60 min after the initial blockage of replication by the UV irradiation, as deduced from delayed photoreactivation experiments (3) and time course of UmuDC induction/activation (16, 31). At these later times, memory of leading versus lagging strand replication is not likely to be retained. This is in contrast to the transient replication blockages that are thought to underlie the recA730-mediated SOS mutator effect (untargeted mutagenesis), for which a strand bias is observed (22).
Acknowledgments
We thank Malgorzata Bialoskorska for her excellent technical assistance. This research was supported by KBN 6-PO4A-026-16 (to D.G., M.M.T., P.J., and I.J.F.).
REFERENCES
- 1.Becherel, O. J., and R. P. P. Fuchs. 2001. Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc. Natl. Acad. Sci. USA 98:8566-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlyn, M. K. 1998. Linkage map of Escherichia coli, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridges, B. A., and R. Woodgate. 1985. The two-step model of bacterial UV mutagenesis. Mutat. Res. 150:133-139. [DOI] [PubMed] [Google Scholar]
- 4.Brotcorne-Lannoye, A., and G. Maenhaut-Michel. 1986. Role of RecA protein in untargeted UV mutagenesis of bacteriophage λ: evidence for he requirement for the dinB gene. Proc. Natl. Acad. Sci. USA 83:3904-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciesla, Z. 1982. Plasmid pKM101-mediated mutagenesis in Escherichia coli is inducible. Mol. Gen. Genet. 186:298-300. [DOI] [PubMed] [Google Scholar]
- 6.Cupples, C. G., M. Cabrera, C. Cruz, and J. H. Miller. 1990. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration in the lambda attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-25. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez De Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the lexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 10.Fijalkowska, I. J., R. L. Dunn, and R. M. Schaaper. 1997. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J. Bacteriol. 179:7435-7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fijalkowska, I. J., P. Jonczyk, M. Maliszewska-Tkaczyk, M. Bialoskorska, and R. M. Schaaper. 1998. Unequal fidelity of leading and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 95:10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fijalkowska, I. J., and R. M. Schaaper. 1995. Effects of Escherichia coli dnaE antimutator alleles in a proofreading-deficient mutD5 strain. J. Bacteriol. 177:5979-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedberg, E. C., G. C. Walker, and W. Seide. 1995. DNA repair and mutagenesis, p. 28-29. ASM Press, Washington, D.C.
- 14.Gawel, D., M. Bialoskorska, P. Jonczyk, R. M. Schaaper, and I. J. Fijalkowska. Asymmetry of frameshift mutagenesis during leading and lagging strand replication in E. coli. Mutat. Res. 501:129-136. [DOI] [PubMed]
- 15.Glover, B. P., and C. S. McHenry. 2001. The DNA polymerase III holoenzyme: an asymmetric dimeric replicative complex with leading and lagging strand polymerases. Cell 105:925-934. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez, M., and R. Woodgate. 2002. The “tale” of UmuD and its role in SOS mutagenesis. BioEssays 24:141-148. [DOI] [PubMed] [Google Scholar]
- 17.Kato, T., and E. Nakano. 1981. Effects of the umuC36 mutation on ultraviolet-radiation-induced base-change and frameshift mutations in Escherichia coli. Mutat. Res. 83:307-319. [DOI] [PubMed] [Google Scholar]
- 18.Kato, T., and Y. Shinoura. 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 156:121-131. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S.-R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpresssion of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S.-R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266:207-215. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. Freeman, New York, N.Y.
- 22.Maliszewska-Tkaczyk, M., P. Jonczyk, M. Bialoskorska, R. M. Schaaper, and I. J. Fijalkowska. 2000. SOS mutator activity: unequal mutagenesis on leading and lagging strands. Proc. Natl. Acad. Sci. USA 97:12678-12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maor-Shoshani, A., N. B. Reuven, G. Tomer, and Z. Livneh. 2000. Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis. Proc. Natl. Acad. Sci. USA 97:565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napolitano, R., R. Janel-Binz, J. Wagner, and R. P. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohmori, H., E. C. Friedberg, R. P. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Cell 8:7-8. [DOI] [PubMed] [Google Scholar]
- 26.Pham, P., J. G. Bertram, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2001. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature 409:366-370. [DOI] [PubMed] [Google Scholar]
- 27.Rajagopalan, M., C. Lu, R. Woodgate, M. O'Donnell, M. F. Goodman, and H. Echols. 1992. Activity of the purified mutagenesis proteins UmuC, UmuD′ and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc. Natl. Acad. Sci. USA 89:10777-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuven, N. B., G. Arad, A. Maor-Shoshani, and Z. Livneh. 1999. The mutagenic protein UmuC is a DNA polymerase activated by UmuD′, RecA and SSB and is specialized for translesion replication. J. Biol. Chem. 274:31763-31766. [DOI] [PubMed] [Google Scholar]
- 29.Smith, B. T., and G. C. Walker. 1998. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics 148:1599-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokahl, R. R., and F. J. Rohlf. 1981. Biometry, 2nd ed., p. 429-445. Freeman, San Francisco, Calif.
- 31.Sommer, S., F. Boudsocq, R. Devoret, and A. Bailone. 1998. Specific RecA amino acid changes affect RecA-UmuDC interaction. Mol. Microbiol. 28:281-291. [DOI] [PubMed] [Google Scholar]
- 32.Steinborn, G. 1978. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypic characterization in DNA repair and mutagenesis. Mol. Gen. Genet. 165:87-93. [DOI] [PubMed] [Google Scholar]
- 33.Sutton, M. D., T. Opperman, and G. C. Walker. 1999. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD′ interact physically with the replicative DNA polymerase. Proc. Natl. Acad. Sci. USA 96:12373-12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, M. J., I. Bruck, R. Eritja, J. Turner, E. G. Frank, R. Woodgate, M. O'Donnell, and M. F. Goodman. 1998. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc. Natl. Acad. Sci. USA 95:9755-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang, M. J., P. Pham, X. Shen, G. S. Taylor, M. O'Donnell, R. Woodgate, and M. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 36.Tang, M. J., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′2C is an error-prone DNA polymerase. Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner, J., P. Gruz, S.-R. Kim, M. Yamada, K. Matsui, R. P. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Z. 2001. Translesion synthesis by the UmuC family of DNA polymerases. Mutat. Res. 486:59-70. [DOI] [PubMed] [Google Scholar]
- 39.Witkin, E. M. 1976. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol. Rev. 40:869-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witkin, E. M., J. O. McCall, M. R. Volkert, and I. E. Wermundsen. 1982. Constitutive expression of SOS functions and modulation of mutagenesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol. Gen. Genet. 185:43-50. [DOI] [PubMed] [Google Scholar]
- 41.Woodgate, R. 1992. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221-225. [DOI] [PubMed] [Google Scholar]