When Apirion first proposed that mRNA decay in Escherichia coli involves a series of endo- and exonucleolytic events (2), the general working assumption was that the turnover of transcripts is a simple salvage pathway that is necessary for recycling of ribonucleotides. Although experimental data at that time indicated that mRNAs are rapidly degraded (11, 40) and that decay of individual transcripts is independent of length (9), the number and specificities of the enzymes that actually carry out transcript degradation were still open questions. Twenty-nine years and many experiments later, a much different picture has emerged. Not only is the pathway of mRNA decay far more complex than originally envisioned, but it apparently also plays an integral role in regulating the expression of many genes. While many important features of this system remain to be elucidated, this prospective attempts to convey the current state of knowledge. In addition, it focuses primarily on those areas where there are disagreements regarding important features of the mRNA decay process.
INITIATION OF mRNA DECAY
Of all of the aspects associated with mRNA decay in E. coli, the question of how the process is initiated has sparked the most controversy. Not only have there been conflicting views regarding which enzyme(s) is responsible for the initial endonucleolytic cleavage events, but there have also been disagreements regarding the importance of certain multiprotein complexes. E. coli is known to contain at least five endoribonucleases: RNases III, E, G, and I/M.
RNase III was first discovered as a protein that cleaves double-stranded RNA (94). In vivo, RNase III specifically degrades stem-loop structures, particularly those in intercistronic regions (34, 88). It plays a major role in processing of the 30S rRNA precursor (34). The enzyme has been shown to indirectly affect the half-lives of a limited number of transcripts (7, 36, 88), primarily by eliminating a stem-loop structure, usually upstream of the translation start site. However, deletion of the RNase III structural gene (rnc) does not lead to any significant change in the degradation of total pulse-labeled RNA or specific transcripts (5). Thus, it is probably not a major player in mRNA decay.
In 1979, Ono and Kuwano (84) isolated a temperature-sensitive mutation, called ams-1 (for altered mRNA stability), that led to a slowing in the decay of total pulse-labeled RNA. Independently, Ghora and Apirion (37) identified RNase E on the basis of its role in the processing of 9S rRNA into a 5S form. More than a decade later, several laboratories showed that the ams-1 and rne-3071 mutations are alleles of the same gene, now called rne (6, 68, 77, 105).
In addition to being required for the maturation of 9S rRNA, RNase E has also been shown to be involved in the processing of the 5′ end of 16S rRNA (54, 111), the maturation of the RNA subunit of RNase P (63), the degradation of the antisense inhibitor (RNA I) of plasmid colE1 DNA replication (107), and the processing of tRNAs (53, 85, 92). As such, it is intimately involved in the processing of a large number of nontranslated E. coli transcripts.
Besides all of these functions, RNase E has also been strongly implicated in the decay of total pulse-labeled RNA (3, 84), as well as a number of specific transcripts (3, 42, 65, 75, 78). In fact, analysis of mRNA decay in mutations carrying extensive deletions in the carboxy-terminal region of the RNase E protein has shown that mRNA decay is seriously deficient, even under conditions in which the cell is still viable (62, 86).
The RNase E protein contains 1,061 amino acids (21, 22) comprising at least three distinct domains (28). The amino-terminal 500 amino acids encode the catalytic activity of the protein (67). Farther downstream is the so-called arginine-rich RNA binding site (amino acids [aa] ∼597 to 684) that has been shown to bind RNA in vitro (67, 104). The C-terminal third of RNase E (aa ∼734 to 1061) functions as a scaffold for the assembly of a multiprotein complex called the degradosome (109). Originally identified by Carpousis et al. (20), the E. coli degradosome has been shown by immunoprecipitation experiments to contain RNase E, PNPase, the RhlB RNA helicase, and the glycolytic enzyme enolase (69, 89, 90).
While it was originally suggested that an RNase E cleavage site encompasses a 10-nucleotide (nt) region (ACAGA/UAUUUG) (107), subsequent analysis of many more sites now indicates that RNase E prefers single-stranded regions that are typically, but not always, A-U rich (25). Additionally, it is now known that RNase E is a 5′-end-dependent endoribonuclease (64, 66) that prefers substrates with monophosphorylated 5′ ends to triphosphorylated termini (57, 64, 66, 101, 106).
On the basis of the catalytic properties of RNase E and its association with PNPase and an RNA helicase whose activity is stimulated by its association with RNase E (27), it became a working hypothesis that the RNase E-based degradosome is the primary mechanism for mRNA degradation in E. coli (8, 91). However, recent results have brought this view into question. In the first place, although the N-terminal region of RNase E is highly conserved in prokaryotes, the C-terminal degradosome scaffolding region is not (49). More importantly, RNase E mutants lacking the degradosome scaffolding region exhibit normal mRNA decay properties (86). Thus, questions remain about whether the degradosome actually participates in mRNA decay.
If the actual role of RNase E in mRNA decay is still not fully understood, the identification of its homologue, called RNase G, has only complicated the story. Originally discovered as the product of cafA (for cytoplasmic axial filament), the protein has extensive sequence similarity (49.5% sequence similarity and 34.1% sequence identity) to the first 498 aa of RNase E (54, 83, 111). Furthermore, in vivo experiments have suggested some functional overlap between RNases E and G (110). In fact, it has recently been shown that RNase G is involved in decay of the adhE transcript (108). Also, several groups have shown that RNase G is also a 5′-end-dependent endoribonuclease (48, 106). Thus, the bacterium has two 5′-end-dependent enzymes that seem to play a role in mRNA decay. In addition, both enzymes have been highly conserved in a large number of prokaryotes (51).
RNase I, encoded by the rna gene, is a relatively nonspecific endoribonuclease that is found primarily in the periplasmic space of the bacterium (80). As such, it has not been viewed as a prime candidate for playing a significant role in mRNA decay. However, it has been subsequently found that a modified form of the enzyme, called RNase I*, is present in the spheroplast fraction (15). Although these observations indicated that the enzyme could participate in mRNA decay, analysis of mutants carrying the rna-19 allele showed that there were no significant changes in the decay of either total pulse-labeled RNA or specific transcripts (C. Arraiano, S. D. Yancey, and S. R. Kushner, unpublished results). In addition, Zhu et al. (115) did not observe any change in the phenotype of an rna deletion mutant.
The final endoribonuclease that has been hypothesized to participate in mRNA decay is RNase M. On the basis of an extensive series of experiments, Kennell and coworkers concluded that the decay of each transcript is initiated by endonucleolytic attack near the 5′ terminus (1, 10, 55, 56). When subsequent work indicated that most of the cleavages occur between pyrimidine and adenosine residues (16), Cannistraro and Kennell (14) undertook the identification and purification of RNase M, an enzyme whose properties are similar to those of pancreatic RNase A. When combined with subsequent data that indicated that the majority of 5′ termini in E. coli have hydroxyl groups and not phosphomonoesters (13), Cannistraro and Kennell argued that RNase M is the major degradative endoribonuclease for mRNA (13).
Although the data to support this hypothesis appear compelling, the failure to identify the structural gene for RNase M has been of considerable concern. In fact, it now appears that RNase M is a mutationally altered form of RNase I, which is found specifically in one strain of E. coli, MRE600. This happens to be the strain that Cannistraro and Kennell (14) used for their purification work. The rna gene in MRE600, which was chosen because it is naturally deficient in RNase I, has been shown by Subbarayan and Deutscher (102) to contain a UGA nonsense mutation at codon 5, as well as three additional changes that result in amino acid substitutions. Their data indicate that deletion of the rna locus leads to complete loss of both RNase I and M activities. Thus, it seems likely that strains such as MG1693 and its derivatives, which have been used for many mRNA decay experiments (3), will contain RNase I but not RNase M.
TERMINAL STEPS IN mRNA DECAY
By the early 1970s, several 3′ → 5′ exonucleases, including RNase II (99) and polynucleotide phosphorylase (PNPase) (41), had already been well characterized as proteins that degrade oligoribonucleotides one at a time. In addition, both PNPase and RNase II were shown to be significantly inhibited by secondary structures (60, 81). Subsequently, many additional 3′ → 5′ exonucleases, including RNases R, BN, PH, D, and T, have been identified (29). Interestingly, many of these enzymes, including RNases BN, D, PH, and T, seem to be exclusively involved in the final maturation of the 3′ ends of tRNAs (52). In addition, unlike Saccharomyces cerevisiae, where the 5′ → 3′ Xrn1 exonuclease plays a major role in mRNA decay (19), E. coli does not appear to contain any exonucleases of this type (30). Thus, the oligoribonucleotides that are generated by endonucleolytic cleavages are exclusively degraded in the 3′ → 5′ direction.
Initial genetic support for this conclusion came from the analysis of a double mutant containing a temperature-sensitive allele in the structural genes for RNase II (rnb-500) and a null mutation in PNPase (pnp-7). At the nonpermissive temperature, large amounts of partially degraded mRNAs accumulated (33). Since a single mutant with either PNPase or RNase II inactivated is viable while a double mutant is not, it has been suggested that the two enzymes can partially substitute for each other (33). A similar situation has also been observed with PNPase and RNase R (24).
One of the interesting questions regarding the exonucleolytic degradation of full-length mRNAs and/or their initial degradation products concerns which enzyme(s) is primarily responsible for this activity. Thirty years ago, Chaney and Boyer (23) demonstrated that RNA degradation in E. coli proceeds primarily via a hydrolytic mechanism. Deutscher and Reuven (31) confirmed that work, showing that at least 90% of the degradative capacity in E. coli is hydrolytic in nature. Since RNase II uses a hydrolytic mechanism (99), in contrast to the phosphorolytic mechanism of PNPase (39), it has been assumed that RNase II, and perhaps RNase R, another hydrolytic enzyme, would be primarily responsible for the bulk of mRNA decay.
However, it has to be kept in mind that mRNAs represent only a small fraction (<10%) of the total RNA population that exists inside the cell (79). In fact, most RNA synthesis involves making rRNAs and tRNAs. These structural RNAs undergo considerable processing to generate their mature forms. Thus, if there were some type of compartmentalization of functions, for example, in which PNPase were primarily involved in mRNA decay and RNase II and RNase R were the major exonucleases used in processing reactions, the net effect would be that the bulk of RNA degradation occurred hydrolytically. Such a division of function has been observed in the case of the degradation of poly(A) tails, where it appears that PNPase prefers tails associated with mRNAs while RNase II works better on tails found on rRNAs (73). In fact, a variety of experiments suggest that PNPase is the primary 3′ → 5′ exoribonuclease involved in the degradation of mRNAs (B. K. Mohanty and S. R. Kushner, unpublished data).
An unanswered question is whether a significant number of E. coli transcripts, particularly those that are monocistronic, are degraded primarily by an exonucleolytic mechanism. Clearly, this is the primary way that S. cerevisiae degrades its mRNAs (19). Since there are many small mRNA transcripts (for example, the lpp mRNA is less than 340 nt in length), one would think that an enzyme like PNPase could easily degrade the transcript without the need for prior endonucleolytic cleavages. However, some short transcripts, such as rpsT, are clearly cut first by RNase E (65). Thus, it may be that only a small fraction of the E. coli transcripts are degraded solely via an exonucleolytic mechanism.
Finally, it is worth noting that there is one additional exoribonuclease involved in mRNA decay. This enzyme, called oligoribonuclease (encoded by orn), is essential for cell viability (38). It turns out that neither PNPase nor RNase II can completely degrade an oligoribonucleotide, leaving 4- to 7-mers as terminal digestion products. These short oligonucleotides accumulate in an orn mutant (38). Whether the accumulation of these small oligomers or some other, yet-to-be-characterized, function of the enzyme is the cause of cell inviability is an open question.
ROLE OF POLYADENYLATION IN mRNA DECAY
Even though the first poly(A) polymerase that was purified came from E. coli (4, 70), few efforts were made to examine in vivo polyadenylation until the late 1970s (96). Furthermore, while Sarkar and her colleagues continued to publish evidence that supported the existence of poly(A) tails on E. coli transcripts (103), it was not until Cao and Sarkar (18) identified pcnB (61) as the structural gene for poly(A) polymerase I that workers began to take seriously the idea that E. coli transcripts are polyadenylated. Subsequent experiments showed that polyadenylation affects the degradation of nontranslated RNAs, such as the DNA replication regulator RNA I of plasmid colE1 (113, 114), as well as a variety of specific mRNAs (26, 43, 82). It was hypothesized that polyadenylation serves as a targeting mechanism for the degradation of individual transcripts (50, 71). More recent experiments have indicated that polyadenylation has some effect on the degradation of most transcripts (Mohanty and Kushner, unpublished).
While these results suggested that polyadenylation plays a role in the degradation of mRNAs, one troubling aspect of the work has been the fact that only a small percentage of the mRNA population (1 to 2%) appears to be polyadenylated at any time (18). Mohanty and Kushner (71) addressed this issue by employing a specially constructed pcnB gene to demonstrate that, for most of the mRNAs tested, decay rates increase with higher levels of polyadenylation. In addition, they showed that increasing the level of poly(A) polymerase can lead to the quantitative polyadenylation of a particular transcript, such as lpp (71). They also showed that poly(A) levels are regulated by a combination of translational control of poly(A) polymerase (71) and the activities of the 3′ → 5′ exonucleases RNase II and PNPase (73). RNase E also participates in the regulation of poly(A) levels by generating new 3′ termini that become targets for polyadenylation (73).
Exactly how polyadenylation participates in mRNA decay is still not clear. On the one hand, it is well known that both RNase II and PNPase are inhibited by secondary structures (60, 81, 100). Furthermore, many E. coli transcripts are terminated in a Rho-independent fashion, which generates a stem-loop structure with a very short 3′ single-stranded extension. Addition of a poly(A) tail would make such molecules far better substrates for these enzymes. In fact, Coburn and Mackie (27) have shown that polyadenylation is necessary to degrade a 174-nt fragment of the rpsT transcript that is generated by an RNase E cleavage event. With an mRNA such as lpp, polyadenylation appears to be required for breakdown of the full-length transcript (71). Thus, it is not certain whether polyadenylation is employed primarily to help degrade full-length transcripts or only degradation products that contain stable secondary structures because they are no longer being translated.
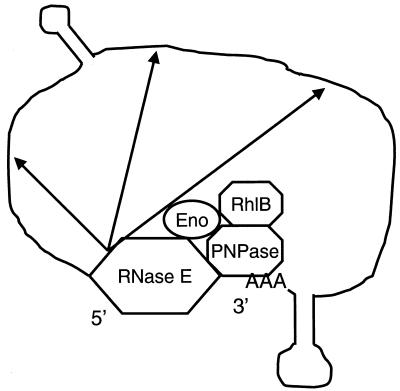
Another unresolved question about polyadenylation involves the in vitro results of Huang et al. (44) and Walsh et al. (112), which indicate that RNase E can act as a deadenylating enzyme. In vivo experiments have not provided any evidence that this reaction actually takes place in the bacterium (73). Even if RNase E is not involved in deadenylation in vivo, since the enzyme is associated with PNPase in the degradosome, binding of PNPase to a poly(A) tail would also bring RNase E into close proximity to the transcript and might facilitate the concerted degradation from both the 5′ and 3′ ends (Fig. 1). While such a mechanism would inherently improve the efficiency of mRNA decay, the fact that degradosome assembly-deficient RNase E mutants have normal mRNA decay rates (86) demonstrates that this model may only describe a special case of a more general pathway of mRNA degradation.
FIG. 1.
Initiation of mRNA degradation by the RNase E-based degradosome. In this model, the degradosome, composed of RNase E, PNPase, the RhlB RNA helicase, and enolase (Eno), is associated with the transcript through the binding of PNPase to the poly(A) tail that has been added following the Rho-independent transcription terminator and the attachment of RNase E to the 5′ end. After the initial endonucleolytic cleavages (arrows) occur, the subsequent steps are similar to those shown in Fig. 2 and 3.
The other area of some controversy relates to the existence and nature of a second poly(A) polymerase. Although the pcnB-encoded poly(A) polymerase has been shown to account for at least 90% of the poly(A) tails in E. coli (71, 82), the question remained as to which enzyme is responsible for the residual polyadenylation. Cao et al. (17) claimed that the f310 gene encodes the backup poly(A) polymerase. However, a careful in vivo analysis of this gene has suggested that while the F310 protein may bind ATP, it does not polyadenylate RNA transcripts (74). Rather, it turns out that PNPase is the responsible party (72). It is not clear what controls the switching of PNPase from a degradative to a biosynthetic enzyme.
Interestingly, PNPase is also responsible for the low level of non-A residues that are found in poly(A) tails at low frequency in wild-type cells, as well as the highly heteropolymeric tails that are isolated from pcnB mutant strains of E. coli (72). Since the two enzymes add poly(A) tails at different locations within the same transcript, Mohanty and Kushner (72) proposed that polyadenylation alters the efficiency of mRNA decay. Finally, it should be noted that while the total lack of polyadenylation only leads to reduced growth rates (72), increased levels of poly(A) polymerase are highly toxic (71).
REGULATION OF mRNA DECAY
The idea that mRNA decay is a constitutively expressed salvage pathway has long been put to rest. In the first place, the synthesis of many of the proteins involved in mRNA decay is under some form of posttranscriptional control. For example, RNase III regulates its own synthesis by cleaving a stem-loop structure that is upstream of the ribosome binding site (7). In addition, it also regulates PNPase synthesis by cleaving a stem-loop structure in the intercistronic region of the rpsO pnp polycistronic operon (88). Additionally, PNPase autoregulates is own synthesis by binding to the 5′ end of its transcript (93) and apparently degrading a secondary structure in the 5′ leader region (47).
Another enzyme that autoregulates its own synthesis is RNase E (46, 76). Although the control mechanism is still not completely understood, RNase E interacts with its 361-nt 5′ untranslated region to help keep intracellular levels of the enzyme at a relatively fixed level (32, 46, 98). However, recent experiments have suggested that autoregulation is far more complicated, involving multiple promoters (87) and portions of the carboxy-terminal region of the RNase E protein (86).
Beyond these forms of direct regulation of individual ribonucleases, there also appear to be additional levels of control. Thus, in the absence of RNase II, PNPase levels increase significantly. Conversely, in the absence of PNPase, RNase II levels increase (116). In addition, there appears to be a specific protein, called Gmr, that regulates RNase II levels (12).
Polyadenylation also plays a role in controlling the degradative capacity of the cell. Specifically, poly(A) levels are important because they alter the ability of RNase E and PNPase to autoregulate their own synthesis. Thus, in the absence of polyadenylation, the half-lives of the rne and pnp transcripts are significantly shorter, resulting in reduced levels of both enzymes (71a). When poly(A) levels increase, the rne and pnp transcripts are stabilized (71), leading to increased levels of both proteins (73).
It appears that the cell has the capacity to alter its degradative capacity, depending on specific intracellular circumstances. Such flexibility would be particularly useful in helping the cell adapt to dramatic changes in its growth environment.
mRNA DECAY MODELS
Although no one has argued that the original ideas of Apirion (2) are incorrect, what has not been clear is exactly which enzymes are involved in the early, and presumably rate-limiting, steps of mRNA decay. There are two schools of thought. The first, represented by the proponents of broad-specificity endoribonucleases, has stated that enzymes like RNases M and I* initiate mRNA decay, cleaving phosphodiester bonds to yield the 5′ OH termini that were identified by Cannistraro and Kennell (13) (Fig. 2). In this model, enzymes like RNase E and its homologue RNase G have specificities that are too stringent to accommodate a broad role in mRNA decay (13).
FIG. 2.
Decay of mRNAs employing nonspecific endoribonucleases. In this model, an enzyme such as RNase M would initiate mRNA decay near the 5′ terminus of the transcript. Following additional cleavages by it and perhaps another endoribonuclease, such as RNase I*, the breakdown products would be exonucleolytically degraded by a combination of PNPase, RNase II, and RNase R. Secondary structures could be removed by the action of an RNA helicase and/or the addition of poly(A) tails, which has been shown to promote the movement of PNPase through a stem-loop structure (27). RBS, ribosome binding site; PAP I, poly(A) polymerase I.
The major problem with this hypothesis relates to the actual existence of RNase M and the importance of RNase I*. It now appears that RNase M is, in fact, a mutant form of RNase I that is probably only found in one strain of E. coli (102). The likelihood of finding a similarly modified rna gene (four independent mutations) in other strains is not high. However, in order to conclusively rule out this enzyme and any other form of RNase I, it is necessary to carefully examine mRNA decay in an rna deletion mutant. The experiments cited earlier were done with an rna-19 mutant strain that still contains 1 to 2% of the wild-type RNase I level (M. Deutscher, personal communication). However, even if RNase M is not a factor, there is the unresolved issue of how to account for the high percentage of 5′ OH termini that were found on RNA oligonucleotides isolated from E. coli (13).
The other school of thought regarding mRNA decay maintains that RNase E, and possibly RNase G, serves as the enzyme that initiates mRNA decay (Fig. 3 [and Fig. 1 if the cell contains full-length RNase E]). In this model, RNase E or G initiates transcript decay by binding to the 5′ terminus and then cleaving at a distance. Since the enzyme prefers a 5′ PO4 to a 5′ triphosphate, which would be found at the primary 5′ terminus of a transcript, the initial cleavage event would be rate limiting. This model would explain why processing of individual transcripts by RNase III would lead to more rapid decay of the downstream transcripts since the 5′ triphosphate would be removed. A well-documented case of this type of behavior is the rps pnp operon, where cleavage by RNase III in the intercistronic region leads to a significant reduction of the half-life of the pnp transcript (88). In addition, it could explain why some 5′ untranslated regions, such as the highly structured ompA leader, impart increased stability to a variety of transcripts (35). Presumably, this type of structure inhibits RNase E or G binding. In contrast, it would not be expected to have any significant impact on a broad-specificity endoribonuclease such as RNase I*.
FIG. 3.
Degradation of mRNAs employing RNases E and G for the initial endonucleolytic cleavages. In this model, RNase E and PNPase are not assumed to be associated in the degradosome. Either RNase E or G can move processively (arrows) in the 5′ → 3′ direction, generating a series of smaller breakdown products that are subsequently degraded as described in Fig. 2. The final step in mRNA decay involves oligoribonuclease hydrolyzing the 4- to 7-mers that do not function as effective substrates for PNPase, RNase II, and RNase R. RBS, ribosome binding site; PAP I, poly(A) polymerase I.
Coburn and Mackie (25) have argued that once RNase E binds to the 5′ terminus of a transcript, it can work its way processively in the 5′ → 3 direction, degrading the mRNA into a series of smaller oligoribonucleotides that would be susceptible to degradation by either PNPase, RNase II, or RNase R (Fig. 3). The final step in the pathway would be the degradation by oligoribonuclease of the short oligoribonucleotides (4 to 7 nt in length) that are resistant to the activity of PNPase, RNase II, and RNase R (38).
A potential problem with this model is that RNase E cleavage generates 5′ PO4 termini (95), a species that is not in high abundance according to Cannistraro and Kennell (13). Furthermore, since RNase E is also involved in tRNA processing (53, 85, 92), it could also be argued that the alterations in mRNA decay observed in rne-1 mutants could be an indirect result of a buildup of ribosomes on transcripts as the cell becomes deprived of mature tRNAs at the nonpermissive temperature. Previous work has shown increased mRNA stability in the presence of drugs, like chloramphenicol, that inhibit translation (58, 97). However, although it might be expected that ribosome stalling could have some effect on mRNA stability, there are several lines of evidence that suggest that this is not the case.
In the first place, some of the arguments against RNase E have centered on the fact that mRNA decay still continues, although at a slower rate, in rne-1 strains at the nonpermissive temperature. However, the isolation of different rne mutations, particularly rneΔ610, has demonstrated that at 37°C, when the cells are still viable, decay rates for individual transcripts can decrease 10- to 20-fold compared to that of a wild-type control (85, 86). These changes cannot be explained by ribosome stalling, since the cell is still actively growing under these conditions. In addition, Ingle and Kushner (45) demonstrated that mRNAs contained on polysomes decayed in vitro at rates comparable to those observed in vivo in the presence of active polyadenylation. Under the conditions of their experiments, translation was blocked (45).
However, even if one subscribes to the RNase E-based model of mRNA decay, there still is the issue of the importance of the degradosome in mRNA decay. The proponents of this model argue that the degradosome contains all of the proteins necessary to degrade an mRNA into small oligoribonucleotides. RNase E initiates decay and produces shorter oligoribonucleotides that can be degraded by PNPase. Since PNPase is inhibited by secondary structures, the presence of the RhlB RNA helicase can remove potential inhibitory structures. In addition, the poly(A) tails present at the 3′ termini of Rho-independent transcription terminators would provide a nice binding site for PNPase. Thus, taken together, the findings indicate that the degradosome could simultaneously bind to both ends of a transcript, leading to its rapid decay (Fig. 1). While there is electron micrographic evidence that the degradosome forms in vivo (59), the experiments of Ow et al. (86) have shown that, in mutants where degradosome formation is blocked, mRNA decay proceeds normally. However, even if degradosome assembly is not critical for mRNA decay, RNase E truncation mutants consistently grow more slowly than wild-type controls (86). It seems likely that a functional degradosome is required for some other pathway in RNA metabolism.
Thus, a number of interesting questions remain to be answered. If RNase E does initiate mRNA decay, why do the 5′ ends of so many oligoribonucleotides contain hydroxy termini? On the other hand, if RNase E is not the enzyme that initiates mRNA decay, what other enzyme accounts for this activity? It does not seem to be RNase M. Subbarayan and Deutscher (102) have reported that, on the basis of computer analysis, there is no RNase I homologue in E. coli. Similarly, other than RNases E and G, there are no homologues of this class of enzyme (Kushner, unpublished). In addition, the role of polyadenylation in mRNA decay appears to be far more complicated than originally thought. A great deal has been learned since 1973, but many important features of mRNA decay remain to be elucidated.
Acknowledgments
This work was supported in part by grant GM57220 from the National Institutes of Health to S.R.K.
REFERENCES
- 1.Achord, D., and D. Kennell. 1974. Metabolism of messenger RNA from the gal operon of Escherichia coli. J. Mol. Biol. 90:581-599. [DOI] [PubMed] [Google Scholar]
- 2.Apirion, D. 1973. Degradation of RNA in Escherichia coli: a hypothesis. Mol. Gen. Genet. 122:313-322. [DOI] [PubMed] [Google Scholar]
- 3.Arraiano, C. M., S. D. Yancey, and S. R. Kushner. 1988. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 170:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.August, J., P. J. Ortiz, and J. Hurwitz. 1962. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J. Biol. Chem. 237:3786-3793. [PubMed] [Google Scholar]
- 5.Babitzke, P., L. Granger, and S. R. Kushner. 1993. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J. Bacteriol. 175:229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babitzke, P., and S. R. Kushner. 1991. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 88:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardwell, J. C. A., P. Regnier, S.-M. Chen, Y. Nakamura, M. Grunberg-Manago, and D. L. Court. 1989. Autoregulation of RNase III operon by mRNA processing. EMBO J. 8:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barlow, T., M. Berkmen, D. Georgellis, L. Bayr, S. Arvidson, and A. Von Gabain. 1998. RNase E, the major player in mRNA degradation, is down-regulated in Escherichia coli during a transient growth retardation (diauxic lag). Biol. Chem. 379:33-38. [DOI] [PubMed] [Google Scholar]
- 9.Blundell, M., E. Craig, and D. Kennell. 1972. Decay rates of different mRNAs in Escherichia coli and models of decay. Nature (London) New Biol. 238:46-49. [DOI] [PubMed] [Google Scholar]
- 10.Blundell, M., and D. Kennell. 1974. Evidence for endonucleolytic attack in decay of lac messenger RNA in Escherichia coli. J. Mol. Biol. 83:143-161. [DOI] [PubMed] [Google Scholar]
- 11.Brenner, S., F. Jacob, and M. Meselson. 1961. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190:576-581. [DOI] [PubMed] [Google Scholar]
- 12.Cairrao, F., A. Chora, R. Zilhao, A. J. Carpousis, and C. M. Arraiano. 2001. RNase II levels change according to the growth conditions: characterization of gmr, a new Escherichia coli gene involved in the modulation of RNase II. Mol. Microbiol. 39:1550-1561. [DOI] [PubMed] [Google Scholar]
- 13.Cannistraro, V. J., and D. Kennell. 1993. The 5′ ends of RNA oligonucleotides in Escherichia coli and mRNA degradation. Eur. J. Biochem. 213:285-293. [DOI] [PubMed] [Google Scholar]
- 14.Cannistraro, V. J., and D. Kennell. 1989. Purification and characterization of ribonuclease M and mRNA degradation in Escherichia coli. Eur. J. Biochem. 181:363-370. [DOI] [PubMed] [Google Scholar]
- 15.Cannistraro, V. J., and D. Kennell. 1991. RNase I*, a form of RNase I, and mRNA degradation in Escherichia coli. J. Bacteriol. 173:4653-4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannistraro, V. J., M. N. Subbarao, and D. Kennell. 1986. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J. Mol. Biol. 192:257-274. [DOI] [PubMed] [Google Scholar]
- 17.Cao, G.-J., J. Pogliano, and N. Sarkar. 1996. Identification of the coding region for a second poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:11580-11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao, G.-J., and N. Sarkar. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. USA 89:10380-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caponigro, G., and R. Parker. 1996. Mechanism and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 60:233-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carpousis, A. J., G. Van Houwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889-900. [DOI] [PubMed] [Google Scholar]
- 21.Casarégola, S., A. Jacq, D. Laoudj, G. McGurk, S. Margarson, M. Tempête, V. Norris, and I. B. Holland. 1994. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp-1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol. 238:867. [DOI] [PubMed] [Google Scholar]
- 22.Casarégola, S., A. Jacq, D. Laoudj, G. McGurk, S. Margarson, M. Tempête, V. Norris, and I. B. Holland. 1992. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol. 228:30-40. [DOI] [PubMed] [Google Scholar]
- 23.Chaney, S. G., and P. D. Boyer. 1972. Incorporation of water oxygens into intracellular nucleotides and RNA. II. Predominantly hydrolytic RNA turnover in Escherichia coli. J. Mol. Biol. 64:581-591. [DOI] [PubMed] [Google Scholar]
- 24.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 273:14077-14080. [DOI] [PubMed] [Google Scholar]
- 25.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 26.Coburn, G. A., and G. A. Mackie. 1996. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem. 271:15776-15781. [DOI] [PubMed] [Google Scholar]
- 27.Coburn, G. A., and G. A. Mackie. 1998. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 279:1061-1074. [DOI] [PubMed] [Google Scholar]
- 28.Cohen, S. N., and K. J. McDowall. 1997. RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 23:1099-1106. [DOI] [PubMed] [Google Scholar]
- 29.Deutscher, M. P. 1993. Promiscuous exoribonucleases of Escherichia coli. J. Bacteriol. 175:4577-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deutscher, M. P., and Z. Li. 2000. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. 66:67-105. [DOI] [PubMed] [Google Scholar]
- 31.Deutscher, M. P., and N. B. Reuven. 1991. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 88:3277-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diwa, A., A. L. Bricker, C. Jain, and J. G. Belasco. 2000. An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev 14:1249-1260. [PMC free article] [PubMed] [Google Scholar]
- 33.Donovan, W. P., and S. R. Kushner. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn, J. J., and F. W. Studier. 1973. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease III. Proc. Natl. Acad. Sci. USA 70:3296-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emory, S. A., P. Bouvet, and J. G. Belasco. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6:135-148. [DOI] [PubMed] [Google Scholar]
- 36.Faubladier, M., K. Cam, and J. P. Bouche. 1990. Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E dependent processing. J. Mol. Biol. 32:461-471. [DOI] [PubMed] [Google Scholar]
- 37.Ghora, B. K., and D. Apirion. 1978. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell 15:1055-1066. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh, S., and M. P. Deutscher. 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. USA 96:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godefroy-Colburn, T., and M. Grunberg-Manago. 1972. Polynucleotide phosphorylase. Enzymes 7:533-574. [Google Scholar]
- 40.Gros, F., H. Hiatt, W. Gilbert, C. B. Kurland, R. W. Risebrough, and J. D. Watson. 1961. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature 190:581-585. [DOI] [PubMed] [Google Scholar]
- 41.Grunberg-Manago, M. 1963. Polynucleotide phosphorylase. Prog. Nucleic Acid Res. 1:93-133. [Google Scholar]
- 42.Hajnsdorf, E., F. Braun, J. Haugel-Nielsen, J. Le Derout, and P. Régnier. 1996. Multiple degradation pathways of the rpsO mRNA of Escherichia coli. RNase E interacts with the 5′ and 3′ extremities of the primary transcript. Biochimie 78:416-424. [DOI] [PubMed] [Google Scholar]
- 43.Hajnsdorf, E., F. Braun, J. Haugel-Nielsen, and P. Régnier. 1995. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang, H., J. Liao, and S. N. Cohen. 1998. Poly(A)- and poly(U)-specific RNA 3′ tail shortening by E. coli ribonuclease E. Nature 391:99-102. [DOI] [PubMed] [Google Scholar]
- 45.Ingle, C. A., and S. R. Kushner. 1996. Development of an in vitro mRNA decay system for Escherichia coli: poly(A) polymerase I is necessary to trigger degradation. Proc. Natl. Acad. Sci. USA 93:12926-12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain, C., and J. G. Belasco. 1995. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 9:84-96. [DOI] [PubMed] [Google Scholar]
- 47.Jarrige, A.-C., N. Mathy, and C. Portier. 2001. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 20:6845-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang, X., A. Diwa, and J. G. Belasco. 2000. Regions of RNase E important for 5′-end-dependent RNA cleavage and autoregulated synthesis. J. Bacteriol. 182:2468-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaberdin, V. R., A. Miczak, J. S. Jakobsen, S. Lin-Chao, K. J. McDowall, and A. von Gabain. 1998. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl. Acad. Sci. USA 95:11637-11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kushner, S. R. 1996. mRNA decay, p. 849-860. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 51.Kushner, S. R. mRNA decay and processing. In P. Higgins (ed.), The bacterial chromosome, in press. ASM Press, Washington, D.C.
- 52.Li, Z., and M. P. Deutscher. 1996. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 86:503-512. [DOI] [PubMed] [Google Scholar]
- 53.Li, Z., and M. P. Deutscher. 2002. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA 8:97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, Z., S. Pandit, and M. P. Deutscher. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim, L. W., and D. Kennell. 1980. Evidence for random endonucleolytic cleavages between messages in decay of Escherichia coli trp mRNA. J. Mol. Biol. 141:227-233. [DOI] [PubMed] [Google Scholar]
- 56.Lim, L. W., and D. Kennell. 1979. Models for decay of Escherichia coli lac messenger RNA and evidence for inactivating cleavages between its messages. J. Mol. Biol. 135:369-390. [DOI] [PubMed] [Google Scholar]
- 57.Lin-Chao, S., and S. Cohen. 1991. The rate of processing and degradation of antisense RNA I regulates the replication of ColE1-type plasmids in vivo. Cell 65:1233-1242. [DOI] [PubMed] [Google Scholar]
- 58.Lindahl, L., and J. Forchhammer. 1969. Evidence for reduced breakdown of messenger RNA during blocked transcription or translation in Escherichia coli. J. Mol. Biol. 43:593-606. [DOI] [PubMed] [Google Scholar]
- 59.Liou, G.-G., W.-N. Jane, S. N. Cohen, N.-S. Lin, and S. Lin-Chao. 2001. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. USA 98:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Littauer, U. Z., and H. Soreq. 1982. Polynucleotide phosphorylase. Enzymes 15:517-553. [Google Scholar]
- 61.Liu, J., and J. S. Parkinson. 1989. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J. Bacteriol. 171:1254-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez, P. J., I. Marchand, S. A. Joyce, and M. Dreyfus. 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33:188-199. [DOI] [PubMed] [Google Scholar]
- 63.Lundberg, U., and S. Altman. 1995. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA 1:327-334. [PMC free article] [PubMed] [Google Scholar]
- 64.Mackie, G. A. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395:720-723. [DOI] [PubMed] [Google Scholar]
- 65.Mackie, G. A. 1991. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the products of the ams gene in vivo and in vitro. J. Bacteriol. 173:2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mackie, G. A. 2000. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 275:25069-25072. [DOI] [PubMed] [Google Scholar]
- 67.McDowall, K. J., and S. N. Cohen. 1996. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding motif. J. Mol. Biol. 255:349-355. [DOI] [PubMed] [Google Scholar]
- 68.Melefors, Ö., and A. von Gabain. 1991. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol. Microbiol. 5:857-864. [DOI] [PubMed] [Google Scholar]
- 69.Miczak, A., V. R. Kaberdin, C.-L. Wei, and S. Lin-Chao. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. USA 93:3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Modak, M. J., and P. R. Srinivasan. 1973. Purification and properties of a ribonucleic acid primer independent polyriboadenylate polymerase from Escherichia coli. J. Biol. Chem. 248:6904-6910. [PubMed] [Google Scholar]
- 71.Mohanty, B. K., and S. R. Kushner. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34:1094-1108. [DOI] [PubMed] [Google Scholar]
- 71a.Mohanty, B. K., and S. R. Kushner. Polyadenylation of Escherichia coli transcripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase. E. Mol Microbiol., in press. [DOI] [PubMed]
- 72.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:11966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol. Microbiol. 36:982-994. [DOI] [PubMed] [Google Scholar]
- 74.Mohanty, B. K., and S. R. Kushner. 1999. Residual polyadenylation in poly(A) polymerase I (pcnB) mutants of Escherichia coli does not result from the activity encoded by the f310 gene. Mol. Microbiol. 34:1109-1119. [DOI] [PubMed] [Google Scholar]
- 75.Mudd, E. A., A. J. Carpousis, and H. M. Krisch. 1990. Escherichia coli RNase E has a role in the decay of bacteriophage T4 mRNA. Genes Dev. 4:873-881. [DOI] [PubMed] [Google Scholar]
- 76.Mudd, E. A., and C. F. Higgins. 1993. Escherichia coli endoribonuclease RNase E: autoregulation of expression and site-specific cleavage of mRNA. Mol. Microbiol. 3:557-568. [DOI] [PubMed] [Google Scholar]
- 77.Mudd, E. A., H. M. Krisch, and C. F. Higgins. 1990. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol. Microbiol. 4:2127-2135. [DOI] [PubMed] [Google Scholar]
- 78.Mudd, E. A., P. Prentki, D. Belin, and H. M. Krisch. 1988. Processing of unstable bacteriophage T4 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 7:3601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neidhardt, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 80.Neu, H. C., and L. A. Heppel. 1964. Some observations on the “latent” ribonuclease of Escherichia coli. Biochem. 51:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nossal, N. G., and M. F. Singer. 1968. The processive degradation of individual polynucleotide chains. J. Biol. Chem. 243:913-922. [PubMed] [Google Scholar]
- 82.O'Hara, E. B., J. A. Chekanova, C. A. Ingle, Z. R. Kushner, E. Peters, and S. R. Kushner. 1995. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:1807-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Okada, Y., M. Wachi, A. Hirata, K. Suzuki, K. Nagai, and M. Matsuhashi. 1994. Cytoplasmic axial filaments in Escherichia coli cells: possible function in the mechanism of chromosome segregation and cell division. J. Bacteriol. 176:917-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ono, M., and M. Kuwano. 1979. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 129:343-357. [DOI] [PubMed] [Google Scholar]
- 85.Ow, M. C., and S. R. Kushner. 2002.. Initiation of tRNA maturation by RNase E is essential for cell viability in Escherichia coli. Genes Dev. 16:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ow, M. C., Q. Liu, and S. R. Kushner. 2000. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 38:854-866. [DOI] [PubMed] [Google Scholar]
- 87.Ow, M. C., Q. Liu, B. K. Mohanty, M. E. Andrew, V. F. Maples, and S. R. Kushner. 2002. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol. Microbiol. 43:159-171. [DOI] [PubMed] [Google Scholar]
- 88.Portier, C., L. Dondon, M. Grunberg-Manago, and P. Regnier. 1987. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is ribonuclease III processing at the 5′ end. EMBO J. 6:2165-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Py, B., H. Causton, E. A. Mudd, and C. F. Higgins. 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14:717-729. [DOI] [PubMed] [Google Scholar]
- 90.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 91.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 92.Ray, B. K., and D. Apirion. 1981. Transfer RNA precursors are accumulated in Escherichia coli in the absence of RNase E. Eur. J. Biochem. 114:517-524. [DOI] [PubMed] [Google Scholar]
- 93.Robert-Le Meur, M., and C. Portier. 1994. Polynucleotide phosphorylase of Escherichia coli induces the degradation of its RNase III processed messenger by preventing its translation. Nucleic Acids Res. 22:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robertson, H. D., R. E. Webster, and N. D. Zinder. 1967. A nuclease specific for double-stranded RNA. Virology 12:718-719. [DOI] [PubMed] [Google Scholar]
- 95.Roy, M. K., B. Singh, B. K. Ray, and D. Apirion. 1983. Maturation of 5S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur. J. Biochem. 131:119-127. [DOI] [PubMed] [Google Scholar]
- 96.Sarkar, N., D. Langley, and H. Paulus. 1978. Isolation and characterization of polyadenylate-containing RNA. Biochemistry 17:3468-3474. [DOI] [PubMed] [Google Scholar]
- 97.Schneider, E., M. Blundell, and D. Kennell. 1978. Translation and mRNA decay. Mol. Gen. Genet. 160:121-129. [DOI] [PubMed] [Google Scholar]
- 98.Sousa, S., I. Marchand, and M. Dreyfus. 2001. Autoregulation allows Escherichia coli RNase E to adjust continuously its synthesis to that of its substrates. Mol. Microbiol. 42:867-878. [DOI] [PubMed] [Google Scholar]
- 99.Spahr, P. F. 1964. Purification and properties of ribonuclease II from Escherichia coli. J. Biol. Chem. 239:3716-3726. [PubMed] [Google Scholar]
- 100.Spickler, C., and G. A. Mackie. 2000. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 182:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spickler, C., V. Stronge, and G. A. Mackie. 2001. Preferential cleavage of degradative intermediates of rpsT mRNA by the Escherichia coli degradosome. J. Bacteriol. 183:1106-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Subbarayan, P. R., and M. P. Deutscher. 2002. Escherichia coli RNase M is a multiply altered form of RNase I. RNA 7:1702-1707. [PMC free article] [PubMed] [Google Scholar]
- 103.Taljanidisz, J., P. Karnik, and N. Sarkar. 1987. Messenger ribonucleic acid for the lipoprotein of the Escherichia coli outer membrane is polyadenylated. J. Mol. Biol. 193:507-515. [DOI] [PubMed] [Google Scholar]
- 104.Taraseviciene, L., G. R. Bjork, and B. E. Uhlin. 1995. Evidence for a RNA binding region in the Escherichia coli processing endoribonuclease RNase E. J. Biol. Chem. 270:26391-26398. [DOI] [PubMed] [Google Scholar]
- 105.Taraseviciene, L., A. Miczak, and D. Apirion. 1991. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol. Microbiol. 5:851-855. [DOI] [PubMed] [Google Scholar]
- 106.Tock, M. R., A. P. Walsh, G. Carroll, and K. J. McDowall. 2000. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 275:8726-8732. [DOI] [PubMed] [Google Scholar]
- 107.Tomcsanyi, T., and D. Apirion. 1985. Processing enzyme ribonuclease E specifically cleaves RNA I, an inhibitor of primer formation in plasmid DNA synthesis. J. Mol. Biol. 185:713-720. [DOI] [PubMed] [Google Scholar]
- 108.Umitsuki, G., M. Wachi, A. Takada, T. Hikichi, and K. Nagia. 2001. Involvement of RNase G in in vivo mRNA metabolism in Escherichia coli. Genes Cells 6:403-410. [DOI] [PubMed] [Google Scholar]
- 109.Vanzo, N. F., Y. S. Li, B. Py, Blum, E., C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wachi, M., G. Umitsuki, and K. Nagai. 1997. Functional relationship between Escherichia coli RNase E and the CafA protein. Mol. Gen. Genet. 253:515-519. [DOI] [PubMed] [Google Scholar]
- 111.Wachi, M., G. Umitsuki, M. Shimizu, A. Takada, and K. Nagai. 1999. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 259:483-488. [DOI] [PubMed] [Google Scholar]
- 112.Walsh, A. P., M. R. Tock, M. H. Mallen, V. R. Kaberdin, A. von Gabain, and K. J. McDowall. 2001. Cleavage of poly(A) tails on the 3′-end of RNA by ribonuclease E of Escherichia coli. Nucleic Acids Res. 29:1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu, F., and S. N. Cohen. 1995. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature 374:180-183. [DOI] [PubMed] [Google Scholar]
- 114.Xu, F., S. Lin-Chao, and S. N. Cohen. 1993. The Escherichia coli pcnB gene promotes adenylylation of antisense RNA I of ColE1-type plasmids in vivo and degradation of RNA I decay intermediates. Proc. Natl. Acad. Sci. USA 90:6756-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu, L., T. Gangopadhyay, K. P. Padmandabha, and M. P. Deutscher. 1990. Escherichia coli rna gene encoding RNase I: cloning, overexpression, subcellular distribution of the enzyme, and use of an rna deletion to identify additional RNases. J. Bacteriol. 172:3146-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zilhao, R., R. Cairrao, P. Régnier, and C. M. Arraiano. 1996. PNPase modulates RNase II expression in Escherichia coli: implications for mRNA decay and cell metabolism. Mol. Microbiol. 20:1033-1042. [DOI] [PubMed] [Google Scholar]