Abstract
The Pseudomonas aeruginosa LasR protein functions in concert with N-3-oxo-dodecanoyl-l-homoserine lactone (3O-C12-HSL) to coordinate the expression of target genes, including many genes that encode virulence factors, with cell density. We used a LexA-based protein interaction assay to demonstrate that LasR forms multimers only when 3O-C12-HSL is present. A series of LasR molecules containing internal deletions or substitutions in single, conserved amino acid residues indicated that the N-terminal portion of LasR is required for multimerization. Studies performed with these mutant versions of LasR demonstrated that the ability of LasR to multimerize correlates with its ability to function as a transcriptional activator of lasI, a gene known to be tightly regulated by the LasR-3O-C12-HSL regulatory system. A LasR molecule that carries a C-terminal deletion can function as a dominant-negative mutant in P. aeruginosa, as shown by its ability to decrease expression of lasB, another LasR-3O-C12-HSL target gene. Taken together, our data strongly support the hypothesis that LasR functions as a multimer in vivo.
An increasing number of bacterial species have been shown to utilize a cell density-dependent mechanism known as quorum sensing (QS) to regulate the expression of various genes (13, 28). The opportunistic pathogen Pseudomonas aeruginosa contains two QS systems, las and rhl, and the former is the best-characterized QS system in this organism (25, 27). The las system is composed of the transcriptional activator LasR and its cognate autoinducer (AI) molecule, N-3-oxo-dodecanoyl-l-homoserine lactone (3O-C12-HSL). Many of the target genes regulated by the LasR-3O-C12-HSL system or by RhlR and its cognate AI molecule, N-butyryl-l-homoserine lactone (C4-HSL), encode virulence factors, thereby implicating QS as an important constituent of P. aeruginosa pathogenesis.
LasR and RhlR belong to the LuxR protein family, which was named after the regulator of bioluminescence (lux) in Vibrio fischeri (36). Members of this family act as transcriptional activators and exhibit homology with LuxR in two distinct regions of the protein. The first region appears to be required for AI binding and is located in the N-terminal portion of the protein. The second region resides within the carboxyl-terminal portion of the protein and appears to characterize a DNA-binding domain (DBD) containing a helix-turn-helix motif (36).
The current paradigm for the activation of gene expression in QS suggests that the protein-AI complex interacts with a specific DNA element and, perhaps through contacts with RNA polymerase, stimulates target gene expression (35, 36). The binding of AI by the activator protein is believed to result in conformational changes that allow formation of a functional form of the protein. Several studies have indicated that proteins belonging to this family interact with DNA (16, 17, 34-36, 42, 44), and the existence of dyad symmetry in the proposed DNA element suggests that members of the LuxR family of proteins might form dimers. In support of this idea are studies of LuxR, of Agrobacterium tumefaciens TraR, and of the Erwinia carotovora and Erwinia stewartii CarR and EsaR proteins, respectively (17, 40, 43, 44).
Here we describe evidence for the multimerization of LasR and show that the multimer form is important for LasR function. Using LasR molecules harboring internal deletions or amino acid substitutions, we have begun to identify regions and specific amino acids involved in LasR-LasR interactions. In addition, we found that a dominant-negative form of LasR can interfere with the production of proteases by P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
Recombinant DNA work was carried out by using Escherichia coli DH5α (41) as the host. The wild-type P. aeruginosa PAO230 and lasI null mutant PAO231 strains were gifts from H. Schwiezer (14). Both of these strains carry a chromosomally integrated lasB::lacZ gene fusion. The E. coli MG4 λI14 lysogen (33) carries a lasI::lacZ fusion as a prophage. Multimerization assays were carried out by using E. coli SU101 (6), which carries a chromosomally integrated sulA::lacZ fusion, as the reporter strain. Cultures were routinely grown at 37°C in Luria-Bertani (LB) medium (30) or PTSB medium (20) containing, when required, 12 μg of tetracycline per ml or 100 μg of ampicillin per ml for E. coli or 200 μg of carbenicillin per ml for P. aeruginosa. Synthetic 3O-C12-HSL and C4-HSL were prepared as previously described (23, 24).
Recombinant DNA techniques.
Standard recombinant DNA techniques were used (30). Restriction endonucleases and DNA-modifying enzymes were obtained from GibcoBRL Life Technologies (Gaithersburg, Md.) and New England Biolabs (Beverly, Mass.). All constructs were verified by appropriate endonuclease mapping and, when required, DNA sequencing.
PCR parameters.
PCR was performed by using Taq (GibcoBRL Life Technologies) or Vent DNA polymerase (New England Biolabs). Each reaction mixture (final volume, 100 μl) contained 0.1 μg of DNA template, 100 pmol of each primer, each deoxynucleoside triphosphate at a concentration of 0.25 mM, 3 mM MgCl2 (Taq) or 3 mM MgSO4 (Vent), 10 μl of manufacturer-supplied PCR buffer, and 1 U of enzyme. Amplification was carried out with a Sprint thermal cycler (Hybaid, Franklin, Mass.) by using 30 cycles of 30 s at 94°C, 45 s at 55°C, and 1 min at 72°C.
Construction of plasmids for use in the LasR multimerization studies.
Intact lasR or lasR harboring deletions was fused to a region encoding the LexA DBD (lexADBD) and placed under control of the lac promoter on plasmid pSR658 (4). Unless otherwise noted, all primers used in the construction of plasmids also encoded XhoI and PvuII sites to facilitate cloning.
To create pLXR, the full-length lasR gene was PCR amplified from pKDT1.7 (38) and ligated into appropriately digested pSR658. To create a series of deletions within the N-terminal portion of LasR, pKDT37 (22) was digested with StyI and treated with S1 nuclease. The deletions were designed to maintain the first three amino acid residues to ensure proper expression of the gene. Religated, S1 nuclease-treated plasmids were recovered, and the DNA fragments encoding LasR were cloned into pKDT34 (22) to construct a derivative in which the lasR gene contained an internal deletion. Constructs were sequenced to determine deletion endpoints and to select the constructs that maintained the proper reading frame. The resultant plasmids, pKDT37(Δ4-29), pKDT37(Δ4-87), pKDT37(Δ4-160), and pKDT37(Δ4-172), served as templates for PCRs performed with appropriate primers to obtain the truncated lasR fragments which were subsequently cloned into pSR658 to construct lexADBD-lasR fusions. Plasmids pLXR2, pLXR3, pLXR4, and pLXR5 carry the lasR structural gene with deletions of amino acid residues 4 to 29, 4 to 87, 4 to 160, and 4 to 172, respectively. Plasmid pLXR was used as a PCR template to generate pLXR6, which carries lasR harboring a deletion of the region encoding amino acids 176 to 237 (lasRΔ176-237) and in which the terminal two LasR amino acids (amino acids 238 and 239) are maintained. Expression of the fusion proteins by the host strain when it was grown in the presence of 3O-C12-HSL was confirmed by Western blot analysis performed with polyclonal anti-LasR antisera (data not shown).
Point mutations in LasR were introduced by using overlap extension (29) or megaprimer (1) PCR-based site-directed mutagenesis. The amino acids to be altered were selected based on their degrees of conservation in previously published alignments of 15 LuxR family proteins (36). Tyr-64, Asp-73, Pro-74, and Gly-113 are identical in the 15 LasR homologues, while Ala-105 is present in 13 LasR protein homologues. Trp-19, Trp-88, Leu-110, and Pro-117 are functionally similar in 15 LasR protein homologues, and Phe-7 and Cys-79 are functionally similar in more than eight LasR protein homologues. The mutations introduced were Phe-7-Thr (pLXRF7T), Trp-19-Ala (pLXRW19A), Tyr-64-His (pLXRY64H), Asp-73-Glu (pLXRD73E), Pro-74-Leu (pLXRR74L), Pro-74-Gln (pLXRP74Q), Cys-79-Ser (pLXRC79S), Trp-88-Tyr (pLXRW88Y), Ala-105-Val (pLXRA105V), Leu-110-Ile (pLXRL110I), Gly-113-Val (pLXRG113V), Gly-113-Asp (pLXRG113D), and Pro-117-Phe (pLXRP117F). In each case the 0.75-kb product obtained from PCRs was digested and ligated into appropriately digested pSR658.
Plasmid pEXR1 carries both lasRΔ176-237 under control of the tac promoter and full-length lasR under control of its own promoter. This plasmid was constructed by PCR amplification of lasRΔ176-237 from pLXR6 by using primers that also encoded EcoRI or HindIII recognition sites. The desired product was digested and ligated into similarly digested pEXRR (5) to place expression of lasRΔ176-237 under control of the tac promoter. A second construct, pEXRΔC, which carries only the lasRΔ176-237 region under control of the tac promoter, was constructed by excising the full-length lasR from pEXR1 as a BamHI fragment and then religating the plasmid.
Protein interaction assay.
The LexA-based bacterial protein interaction system used in this work has been described previously (4, 6). lasR DNA fragments of interest were cloned in frame, distal to the LexA DBD-encoding region on pSR658 to place expression of the resultant hybrid protein under control of the isopropyl-β-d-galactopyranoside (IPTG)-inducible lac promoter. The ability of hybrid proteins to multimerize and function as repressors was monitored by determining the inhibition of expression of the chromosomally encoded, LexA-repressible sulA::lacZ gene fusion in E. coli SU101 (6). As a positive control, we utilized pSR658-neuD, which encodes a fusion of the LexA DBD to NeuD, a protein which is required for synthesis of the sialic acid capsule of E. coli K1 (4) and which has been shown to form dimers (4). The reporter strain carrying pSR658 was used as a vector control to monitor endogenous expression of the sulA::lacZ fusion located on the chromosome. Strains were grown overnight in LB medium containing 12 μg of tetracycline per ml at 37°C with shaking; this was followed by subculturing at a 1:100 ratio into fresh medium containing 12 μg of tetracycline per ml and 1 mM IPTG and growth to the mid-log phase (optical density at 600 nm [OD600], 0.7). Cells were harvested and assayed for β-galactosidase (β-Gal) activity as described previously (18). To study the role of AI in multimerization, 3O-C12-HSL (100 nM) or C4-HSL (500 nM) was added to the subculture.
Functional studies of LasR.
To determine whether the LexA DBD-LasR constructs were functional, the appropriate plasmids were introduced into E. coli MG4 λΙ14, which harbors a λ-borne lasI::lacZ fusion integrated as a prophage (33). Overnight cultures in LB medium containing 12 μg of tetracycline per ml were subcultured (1:100) into fresh medium in the presence or absence of 100 nM 3O-C12-HSL, 12 μg of tetracycline per ml, and 1 mM IPTG and grown at 37°C with shaking to an OD600 of 0.7. β-Gal activity was assayed (18) to determine expression of lasI.
Demonstration of dominant-negative activity.
To study the dominant-negative effect of LasRΔ176-237, plasmids pEX1.8 (25), pEXRΔC, and pEXR1 were introduced into P. aeruginosa PAO230 (14). Cultures were grown in PTSB medium with carbenicillin (200 μg/ml) at 37°C to the late log phase (OD540, 1.2). When needed, 3O-C12-HSL was added to cultures at a concentration of 1 or 10 μM. LasRΔ176-237 expression was induced by adding 1 mM IPTG to the cultures. Cells were harvested, and β-Gal assays (18) performed to determine lasB expression.
Agar plate assay of protease activity.
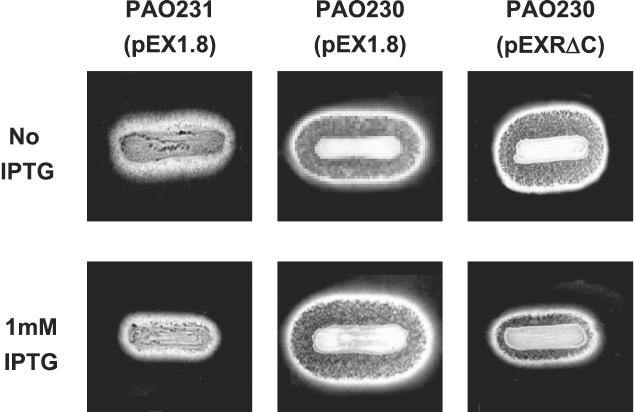
The proteolytic activity of P. aeruginosa PAO230 or PAO231 (14) carrying pEX1.8 or pEXRΔC was visualized on 10% skim milk agar plates (Becton-Dickinson, Sparks, Md.) supplemented with 200 μg of carbenicillin per ml. Strains were grown in PTSB medium containing 200 μg of carbenicillin per ml to an OD540 of 1.2, and a 10-μl aliquot was streaked onto the agar surface of each plate. The plates were incubated at 30°C for at least 36 h. Proteolysis was visible as clear zones around the growing bacteria.
RESULTS AND DISCUSSION
The LasR protein is a major transcriptional activator of P. aeruginosa QS and plays a pivotal role in the activation of many virulence genes. While the complete mechanism by which LasR activates its target genes has not been clearly elucidated, genetic and biochemical studies suggest that members of the LuxR family of proteins may need to multimerize in order to function as activators of transcription.
LasR is capable of forming multimers in the presence of 3O-C12-HSL.
In order to examine the ability of LasR to form multimers, we utilized a LexA-based protein interaction assay that was initially described in studies of Fos and Jun interactions (6). Subsequently, this assay system was used to demonstrate E. coli Neu protein multimerization (4) and E. coli FecA-FecR and FecR-FecI interactions (8). The LexA protein consists of two domains, an N-terminal DBD and a C-terminal domain responsible for dimerization. LexA functions as a repressor when it is in a dimeric form (32). In the LexA interaction system, lasR DNA fragments of interest were cloned in frame distal to the LexA DBD-encoding region, placing expression of the resultant hybrid protein under control of the IPTG-inducible lac promoter. Hybrid proteins that are capable of forming multimers result in the formation of a functional LexA DBD dimer, whose ability to inhibit expression of a sulA::lacZ gene fusion (6) can be monitored in the E. coli SU101 reporter strain.
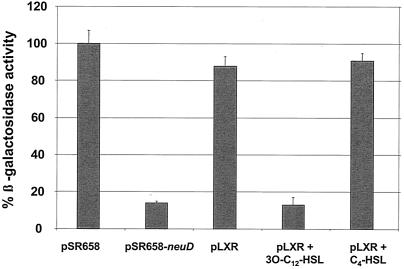
Interestingly, LasR formed multimers only in the presence of 3O-C12-HSL (Fig. 1). LasR multimerization was not seen in the presence of C4-HSL, in agreement with previous studies that indicated that C4-HSL cannot interact with LasR (11, 22). LasR multimerization was detectable with as little as 1 nM 3O-C12-HSL, and the assay was saturable at 10 nM (data not shown), indicating that the concentration used in the present study was sufficient to ensure complete multimerization. Furthermore, 100 nM 3O-C12-HSL has been shown to be sufficient to stimulate expression of several LasR-regulated genes in E. coli (33). It was previously reported that in E. coli LasR-regulated, full expression of lasI, which encodes the 3O-C12-HSL synthase, requires 5 to 10 nM 3O-C12-HSL (33). These concentrations correlate well with the levels which we have observed for LasR multimerization in our studies.
FIG. 1.
LasR forms multimers in the presence of 3O-C12-HSL. The multimerization of LasR was determined by the ability of LexA-LasR protein fusions to inhibit lacZ expression in the reporter strain E. coli SU101. β-Gal activity from the strains was normalized to the activity of E. coli SU101 containing the vector control (pSR658). A strain carrying the LexA DBD-NeuD protein encoded on pSR658-neuD was used as a positive control. When required, 100 nM 3O-C12-HSL or 500 nM C4-HSL was added to cultures. The results are averages for three independent assays. pSR658, LexA DBD; pSR658-neuD, LexA DBD-NeuD; pLXR, LexA DBD-LasR.
Elucidation of specific regions required for LasR multimerization.
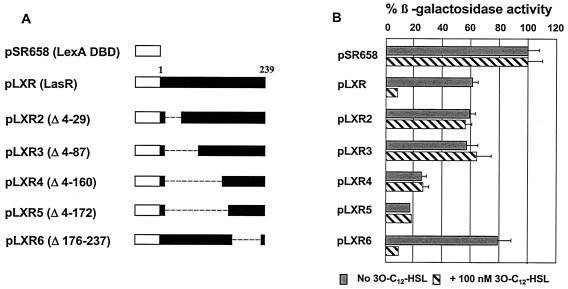
A series of LexA DBD-LasR fusions (Fig. 2A) were examined for the ability to form multimers. The data obtained (Fig. 2B) indicate that deletion of amino acids 4 to 29 or 4 to 87 eliminated the ability of LasR to multimerize even when 3O-C12-HSL was present. Further deletion of the N-terminal region (deletions Δ4-160 and Δ4-172) resulted in proteins which consisted of only the C-terminal region (amino acids 173 to 239) but retained partial multimerization activity even in the absence of 3O-C12-HSL. These observations suggest that the C-terminal portion of LasR may promote or strengthen multimerization of the intact LasR protein subunits. Taken together, these data indicate that LasR has two regions, one in the N-terminal portion of the protein and the other in the C-terminal portion, which act as determinants of multimerization. In the absence of 3O-C12-HSL, neither full-length LasR nor LasR containing deletion Δ4-29 or Δ4-87 exhibited multimerization even though the C-terminal domain, which can partially multimerize, was present. In fact, multimerization of the C-terminal region was observed only when a significant portion of the LasR N-terminal region was removed, suggesting that the region between amino acids 88 and 160 may contain elements that hinder multimerization of the C-terminal domain. This finding is in agreement with studies of LuxR and TraR (17, 36), in which the authors proposed that in the absence of the appropriate AI, the N-terminal portion of the protein may mask the C-terminal DBD and inhibit binding to DNA. Based on our data, LasR may function in an analogous manner. Removal of amino acids 176 to 237 (LasRΔ176-237) did not interfere with the 3O-C12-HSL-dependent multimerization, thus supporting the proposed role of the N-terminal domain in 3O-C12-HSL-dependent multimerization. These observations also support the role of this region in the interaction with AI, consistent with hypotheses put forward for other LuxR homologues (36).
FIG. 2.
Multimerization of LasR deletion constructs. (A) Plasmids pLXR to pLXR6 encoding the lasR deletion constructs. (B) Abilities of the LexA DBD-LasR constructs to express protein capable of forming multimers in the presence or absence of 100 nM 3O-C12-HSL. The results are the averages for three independent determinations.
Multimerization is required for function of LasR.
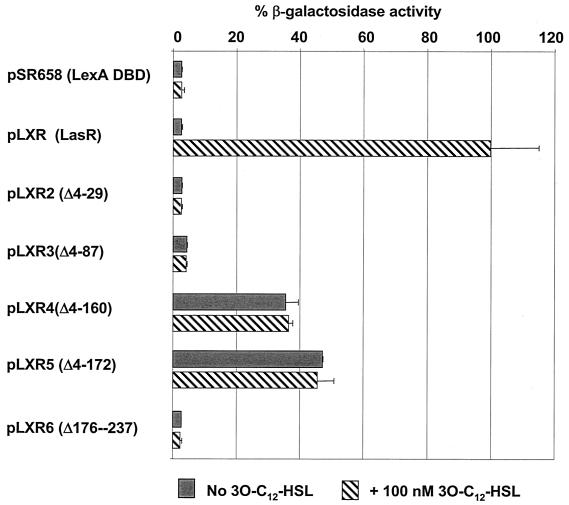
To examine whether multimerization was required for LasR to function as a transcriptional activator, the LexA DBD-LasR constructs shown in Fig. 2A were monitored for their ability to activate expression of a lasI::lacZ transcriptional gene fusion (Fig. 3).
FIG. 3.
Abilities of LasR deletions to function as activators of lasI. The truncated LasR mutants (pLXR2 to pLXR6), as well as full-length LasR (pLXR), were examined for the ability to activate lasI expression from λI14 in E. coli strain MG4 carrying a lasI::lacZ fusion. Expression of lasI was determined by assaying β-Gal activity in the presence or absence of 100 nM 3O-C12-HSL, and the data are expressed relative to the value for the positive control (pLXR), which was normalized to 100%. The results are averages for three independent assays.
As expected, full-length LasR (pLXR) was able to stimulate lasI expression in the presence of 3O-C12-HSL (Fig. 3). Both LasRΔ4-29 (pLXR2) and LasRΔ4-87 (pLXR3) cannot multimerize (Fig. 2B) and did not function. Interestingly, deletions Δ4-160 (pLXR4) and Δ4-172 (pLXR5) exhibited partial activation of lasI expression, which correlated well with their apparent intermediate levels of multimerization (Fig. 2B). pLXR6, the construct in which amino acids 176 to 237 were deleted, exhibited 3O-C12-HSL-dependent multimerization (Fig. 2B) but did not activate lasI expression (Fig. 3). This was most likely due to the absence of the LasR DBD in this construct. The data illustrate that there is a strong correlation between the ability of LasR to form a multimer and its ability to function, indicating that the multimer is the active form of LasR.
Identification of residues important for multimerization and function.
Our observations indicate that the N-terminal portion of LasR is involved in 3O-C12-HSL binding and multimerization of LasR. Alignment of 15 LuxR family proteins (36) identified a number of highly conserved amino acid residues within the N-terminal region. To more finely examine the role of these residues, a series of LasR mutants, each containing an alteration at a single amino acid, were examined for the ability to multimerize and function as activators (Table 1). Proteins which contained alterations of Trp-19 (pLXRW19A), Tyr-64 (pLXRY64H), Asp-73 (pLXRD73E), Pro-74 (pLXRP74L and pLXRP74Q), Trp-88 (pLXRW88Y), Ala-105 (pLXRA105V), Gly-113 (pLXRG113V and pLXRG113D), or Pro-117 (pLXRP117F) could not multimerize even in the presence of 3O-C12-HSL. All of the mutations except Ala-105-Val also severely affected the function of LasR; the protein with the Ala-105-Val mutation exhibited partial activity. It should be noted that the E. coli lysogen reporter system is extremely sensitive to the presence of 3O-C12-HSL (33). As a result, there may be some transient multimerization of the Ala-105-Val mutant that is reflected in the functional assay results but not detected in the protein interaction assay. From the data for these mutants, it is not possible to specifically correlate loss of multimerization ability or function to either a structural alteration or the inability of the protein to bind AI. It may be a combination of both. Three mutations, Phe-7-Thr (pLXRF7T), Cys-79-Ser (pLXRC79S), and Leu-110-Ile (pLXRL100I), had no effect on the ability of LasR to multimerize or function. Implicit in this finding is that Phe-7, Cys-79, and Leu-110 most likely do not play a role in the interaction of LasR with AI. Taken together, the data suggest that many of the N-terminal conserved residues are important for LasR multimerization and, ultimately, the function of the protein. These findings also support our hypothesis that LasR is functional only when it is in a multimeric form.
TABLE 1.
Effects of mutations in conserved amino acids on LasR multimerization and function
| Plasmid | % sulA expressiona
|
% Changeb | % lasI expressionc
|
% Changeb | ||
|---|---|---|---|---|---|---|
| No 3O-C12-HSL | With 3O-C12-HSL | No 3O-C12-HSL | With 3O-C12-HSL | |||
| pSR658 | 100 ± 7 | 98 ± 3 | −2 ± 7.8 | 5 ± 1 | 6 ± 1 | 1 ± 1.4 |
| pLXR | 87 ± 3 | 17 ± 3 | −70 ± 4.2d | 6 ± 2 | 100 ± 12 | 94 ± 12.2d |
| pLXRF7T | 90 ± 5 | 15 ± 2 | −75 ± 5.4d | 6 ± 3 | 90 ± 5 | 84 ± 5d |
| pLXRW19A | 79 ± 1 | 79 ± 4 | 0 ± 4.1 | 6 ± 2 | 8 ± 2 | 2 ± 2.8 |
| pLXRY64H | 87 ± 4 | 92 ± 6 | 5 ± 7.2 | 6 ± 2 | 10 ± 1 | 4 ± 2.2 |
| pLXRD73E | 79 ± 8 | 77 ± 4 | −2 ± 8.9 | 7 ± 1 | 9 ± 1 | 2 ± 1.4 |
| pLXRP74L | 91 ± 8 | 105 ± 12 | 14 ± 14.4 | 9 ± 1 | 10 ± 2 | 1 ± 1.7 |
| pLXRP74Q | 94 ± 6 | 96 ± 2 | 2 ± 6.3 | 8 ± 1 | 10 ± 1 | 2 ± 1.4 |
| pLXRC79S | 95 ± 5 | 15 ± 1 | −75 ± 5.1d | 9 ± 1 | 90 ± 17 | 81 ± 17d |
| pLXRW88Y | 77 ± 4 | 79 ± 1 | 5 ± 4.1 | 7 ± 2 | 12 ± 1 | 5 ± 2.2 |
| pLXRA105V | 86 ± 3 | 81 ± 10 | −5 ± 10.4 | 8 ± 2 | 32 ± 4 | 24 ± 4.5d |
| pLXRL110I | 90 ± 3 | 20 ± 2 | −70 ± 3.6d | 9 ± 2 | 97 ± 20 | 89 ± 1.4d |
| pLXRG113V | 86 ± 5 | 82 ± 4 | −4 ± 6.4 | 11 ± 1 | 11 ± 2 | 0 ± 1.7 |
| pLXRG113D | 105 ± 5 | 110 ± 10 | 5 ± 11.2 | 10 ± 1 | 10 ± 2 | 0 ± 1.7 |
| pLXRP117F | 88 ± 4 | 90 ± 3 | 2 ± 5 | 9 ± 1 | 10 ± 2 | 1 ± 1.7 |
Expression of sulA::lacZ from E. coli SU101 in the absence or presence of 100 nM 3O-C12-HSL when a plasmid was present. Repression of sulA expression indicates multimer formation by mutant LasR expressed from a plasmid. The values are averages ± standard deviations for three individual experiments and were normalized to the value for parent plasmid pSR658, which expressed only the LexA DBD. The value for the parent plasmid was defined as 100%.
Net percent change in sulA::lacZ or lasI::lacZ expression in the presence of 100 nM 3O-C12-HSL. Negative numbers indicate a decrease. The values are averages ± standard deviations for three individual experiments.
Expression of lasI::lacZ in E. coli MG4(λI14), expressed as a percentage relative to the expression with parent plasmid pLXR, which expresses full-length wild-type LasR. An increase in expression indicates that LasR can function as a transcriptional activator in the absence or presence of 100 nM 3O-C12-HSL. The values are averages ± standard deviations for three individual experiments.
The change in expression was significant.
A C-terminal truncation can function as a dominant-negative form of LasR.
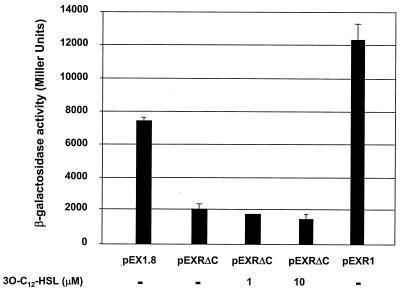
The data (Fig. 2B and 3) demonstrate that LasRΔ176-237 (pLXR6) can form 3O-C12-HSL-dependent, nonfunctional multimers. Given that the lack of function of this construct is most likely due to the absence of the LasR DBD, we postulated that this molecule could exert a dominant-negative effect by complexing with full-length LasR present in the cell to form a nonfunctional heteromultimer. The ability of LasRΔ176-237 to function as a dominant-negative form was assayed by monitoring its ability to inhibit lasB expression in P. aeruginosa PAO230, which is a wild-type strain but carries a chromosomally integrated lasB::lacZ fusion. Expression of lasB, which encodes a P. aeruginosa elastase, is highly sensitive to the LasR-3O-C12-HSL system (26). As expected, PAO230(pEX1.8) exhibited a significant level of β-Gal activity (Fig. 4). Expression of excess LasRΔ176-237, from pEXRΔC, resulted in decreased lasB expression from PAO230. LasRΔ176-237 was able to reduce lasB expression (Fig. 4) even when 3O-C12-HSL was present at a concentration (1 or 10 μM) greater than that required to activate expression of lasB in P. aeruginosa (25, 27). Overexpression of full-length LasR concomitant with expression of LasRΔ176-237 (pEXR1) resulted in an increase in lasB expression compared to the expression when only truncated LasR is expressed (pEXRΔC). This finding indicates that the decrease in lasB expression is not due to a titration of available 3O-C12-HSL by LasRΔ176-237; rather, LasRΔ176-237 titrates out full-length LasR via the formation of nonfunctional heteromultimers. The ability of LasRΔ176-237 to function as a dominant-negative mutant implies that a strain in which it is overexpressed should exhibit a decrease in LasR function and thus should exhibit reduced or no proteolysis (2, 9, 10, 19, 21, 25, 27, 37, 38). Indeed, when plated on skim milk agar medium, strain PAO230(pEXRΔC) exhibited reduced proteolysis only when IPTG was present to induce expression of the lasRΔ176-237 allele (Fig. 5). The data indicate that the decreased proteolysis was directly due to the presence of LasRΔ176-237. As expected, the strain carrying the parent vector (pEX1.8) exhibited clearing regardless of the presence of IPTG. P. aeruginosa PAO231, a lasI null mutant of strain PAO230 harboring the vector control pEX1.8, exhibited little proteolytic activity regardless of whether IPTG was present. This was expected since this strain should not produce 3O-C12-HSL and thus could not significantly activate lasB expression. Some proteolysis was still detectable since not all proteases are dependent upon the las system to the same degree (28). In fact, there may be as-yet-unidentified proteases that may not be controlled by the las QS system. From our results obtained with PAO230(pEXR1), which expresses both LasR and LasRΔ176-237, it appears that the formation of functional homomultimers is favored, given that expression of lasB is increased even though it can be assumed that some nonfunctional heteromultimers or LasRΔ176-237 homomultimers can still form. This could be explained by the functional LasR homomultimer being a more stable structure than the nonfunctional multimers. Alternatively, functional homomultimers may exhibit tighter binding to the lasB regulatory element due to the presence of two LasR DBD domains. This notion is supported by the ability of LasR C-terminal constructs (Δ4-160 and Δ4-172) to form functional multimers (Fig. 2 and 3). These data support our hypothesis that the C-terminal region of LasR also plays a role in the multimerization process and may be important for formation of a stable multimer.
FIG. 4.
LasR carrying a C-terminal deletion can function as a dominant-negative mutant to inhibit multimerization of LasR. The ability of a LasR mutant (LasRΔ176-237) to interfere with the formation of functional LasR multimers was determined by measuring the expression of lasB::lacZ in P. aeruginosa PAO230. Expression of lasB was determined by measuring β-Gal activity in strains harboring pEX1.8 (vector control), pEXRΔC (ptac-lasRΔ176-237), or pEXR1 (plasR-lasR ptac-lasRΔ176-237). 3O-C12-HSL was not added or was added to a final concentration of 1 or 10 μM as indicated. The results are averages for three independent assays.
FIG. 5.
LasRΔ176-237 inhibits proteolysis in P. aeruginosa PAO230. P. aeruginosa PAO230 carrying either pEX1.8 (vector control) or pEXRΔC (ptac-lasRΔ176-237) was assayed for proteolytic activity by plating on skim milk agar in the presence or absence of 1 mM IPTG, which was used to induce expression of LasRΔ176-237. Proteolysis was visualized as a clear zone surrounding the bacterial streak. P. aeruginosa PAO231(pEX1.8), a lasI null mutant, was utilized as a negative control.
The results presented in this paper implicate 3O-C12-HSL in LasR multimerization and demonstrate that the ability to form multimers correlates with the ability of LasR to function as a transcriptional activator. Based on our findings, we propose that the multimerization of LasR is controlled via the N-terminal portion of the protein in the presence of 3O-C12-HSL but that the C-terminal portion of LasR also contains elements important for the formation of a functional multimer. Whether LasR must actually bind 3O-C12-HSL to initiate multimer formation or whether 3O-C12-HSL acts to stabilize the multimer or both is not clear. Our results are in agreement with results (17) indicating that the interactions of TraR and LuxR with their respective cognate AIs result in the formation of stable homodimers. Furthermore, TraR binding and LuxR binding to their DNA target elements have been shown to require the cognate AIs (7, 16, 17). While there is no published data, the requirement for 3O-C12-HSL for LasR function suggests that binding of the AI also determines the ability of LasR to bind to DNA. Recent studies of the TraR protein demonstrate that a region spanning residues 49 to 156 is required for dimerization (17), and genetic analysis of LuxR suggests that an N-terminal portion of the protein is a portion that is important for multimerization (3). The N-terminal region has also been implicated in the binding of AIs by LuxR (12, 31) and TraR (17). Our data for LasR agrees with the previously published data.
Interestingly, the E. carotovora and E. stewartii LuxR homologues (CarR and EsaR, respectively) appear to form dimers in the absence of their cognate AI molecules (17, 40). However, in contrast to LasR, TraR, and LuxR, both CarR and EsaR function as repressors when binding of the AI appears to result in derepression (39, 40). Furthermore, the ability of CarR and EsaR to act as repressors in the absence of the AI suggests that AI binding is not required for DNA binding.
Our findings are consistent with a model in which the LasR protein is inactive until 3O-C12-HSL interaction with the N-terminal domain occurs, which results in structural changes that lead to a protein conformation capable of binding DNA and stimulating transcription of the target gene. Studies of TraR dimerization (17) suggest that the inactive, monomeric form of the protein is associated with the cytoplasmic membrane, with the N-terminal region of the protein inserted in the membrane. A similar suggestion has been put forward for LuxR (15). Such an arrangement would render the protein unable to interact with AI within the cytoplasmic compartment and thereby prevent premature activation of target genes. Subsequent activation would occur when the AI signal entering the cell from the environment interacts with the N-terminal domain, resulting in structural changes which allow the protein to leave its membrane location and dimerize to form a functional complex. While there is no direct evidence that LasR uses a similar mechanism, it is intriguing to speculate that this may be the case, based on the structural and functional similarities among the members of the LuxR family of proteins. In addition, while we have been careful to describe our findings with LasR as a multimerization process, it appears that given the findings obtained with both TraR and CarR, a dimer form is most likely the active form.
To our knowledge, we describe here the first study of a QS system in which formation of a multimer is directly correlated with the ability of a LuxR protein family member to function as a transcriptional activator. Our data provide further insight to another stage of the mechanism that P. aeruginosa utilizes to regulate virulence gene expression. Given that the number of LuxR homologues is steadily increasing, our results and their implications are not necessarily restricted to LasR and P. aeruginosa but may extend to many, if not all, of these homologues and their related QS systems. The approaches used here should be applicable to any member of the LuxR protein family, especially when purified protein is not available.
Acknowledgments
We thank B. Iglewski, R. Silver, C. Benyajati, M. Granger-Schnarr, D. Daines, and H. Schwiezer for strains, plasmids, comments, and suggestions.
This study was supported in part by a grant from the Cystic Fibrosis Foundation (to L.P.). K.D.T. was the recipient of a Cystic Fibrosis Foundation postdoctoral fellowship, and P.K. was the recipient of a Royal Thai Government scholarship and a grant from the Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand.
REFERENCES
- 1.Barik, S. 1996. Site-directed mutagenesis in vitro by megaprimer PCR, p. 203-216. In M. K. Trower (ed.), In vitro mutagenesis protocols, vol. 57. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 2.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, S. H., and E. P. Greenberg. 1992. Genetic evidence for multimerization of LuxR, the transcriptional activator of Vibrio fischeri luminescence. Mol. Mar. Biol. Biotechnol. 1:408-413. [Google Scholar]
- 4.Daines, D., and R. P. Silver. 2000. Evidence for multimerization of Neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J. Bacteriol. 182:5267-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dmitrova, M., G. Younes-Cauet, P. Oertel-Buchheit, D. Porte, M. Schnarr, and M. Granger-Schnarr. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205-212. [DOI] [PubMed] [Google Scholar]
- 7.Egland, K. A., and E. P. Greenberg. 2000. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J. Bacteriol. 182:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambello, M. J., S. Kaye, and B. H. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray, K. M., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1994. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J. Bacteriol. 176:3076-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanzelka, B. L., and E. P. Greenberg. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177:815-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardman, A. M., G. S. Stewart, and P. Williams. 1998. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and non-pathogenic bacteria. Antonie Leeuwenhoek 74:199-210. [DOI] [PubMed] [Google Scholar]
- 14.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa, site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 15.Kolibachuk, D., and E. P. Greenberg. 1993. The Vibrio fischeri luminescence gene activator LuxR is a membrane-associated protein. J. Bacteriol. 175:7307-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, Z. Q., and S. K. Farrand. 1999. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc. Natl. Acad. Sci. USA 96:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, Z. Q., Y. Qin, and S. K. Farrand. 2000. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275:7713-7722. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Ochsner, U. A., A. K. Koch, A. Fiechter, and J. Reiser. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell to cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 22.Passador, L., K. D. Tucker, K. Guertin, M. Journet, A. S. Kende, and B. H. Iglewski. 1996. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J. Bacteriol. 178:5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pesci, E. C., and B. H. Iglewski. 1999. Signaling in Pseudomonas aeruginosa, p. 105-115. In R. England, G. Hobbs, N. Bainton, and D. M. Roberts (ed.), Microbial signalling and communication. Cambridge University Press, Cambridge, United Kingdom.
- 27.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pesci, E. H., and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa, p. 147-155. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 29.Poguliz, R. J., A. N. Vallejo, and L. R. Pease. 1996. In vitro recombination and mutagenesis by overlap extension PCR, p. 167-176. In M. K. Trower (ed.), In vitro mutagenesis protocols, vol. 57. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schaefer, A. L., B. L. Hanzelka, A. Eberhard, and E. P. Greenberg. 1996. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J. Bacteriol. 178:2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnarr, M., and M. Granger-Schnarr. 1993. LexA, the self-cleaving transcriptional repressor of the SOS system, p. 170-189. In E. Eixkstein and D. M. J. Lilley (ed.), Nucleic acids and molecular biology, vol. 7. Springer-Verlag, Berlin, Germany.
- 33.Seed, P. C., L. Passador, and B. H. Iglewski. 1995. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shadel, G. S., and T. O. Baldwin. 1992. Identification of a distantly located regulatory element in the luxD gene required for negative autoregulation of the Vibrio fischeri luxR gene. J. Biol. Chem. 267:7690-7695. [PubMed] [Google Scholar]
- 35.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 91:12619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens, A. M., and E. P. Greenberg. 1999. Transcriptional activation by LuxR, p. 231-242. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 37.Toder, D. S., S. J. Ferrell, J. L. Nezezon, L. Rust, and B. H. Iglewski. 1994. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect. Immun. 62:1320-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toder, D. S., M. J. Gambello, and B. H. Iglewski. 1991. Pseudomonas aeruginosa LasA; a second elastase gene under transcriptional control of lasR. Mol. Microbiol. 5:2003-2010. [DOI] [PubMed] [Google Scholar]
- 39.von Bodman, S. B., D. R. Majerczak, and D. L. Coplin. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. USA 95:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welch, M., D. E. Todd, N. A. Whitehead, S. J. McGowan, B. W. Bycroft, and G. P. Salmond. 2000. N-Acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strain for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.You, Z., J. Fukushima, T. Ishiwata, B. Chang, M. Kurata, S. Kawamoto, P. Williams, and K. Okuda. 1996. Purification and characterization of LasR as a DNA-binding protein. FEMS Microbiol. Lett. 142:301-307. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, J., and S. C. Winans. 1998. Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol. Microbiol. 27:289-297. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]