Abstract
Helicobacter pylori encodes three two-component systems and two orphan response regulators (RRs) that are predicted to be involved in transcriptional regulation. The HP1043 gene encodes an essential OmpR-like RR, 1043RR, for which no histidine kinase has been identified. Gel filtration and cross-linking experiments on the purified 1043RR protein reveals that this protein is a dimer and in vivo dimerization assays localize the dimerization to the N-terminal regulatory domain. DNA-binding studies have revealed two targets for specific binding of the 1043RR protein and moreover, phosphorylation of the protein was not needed for the activation of binding. Footprinting analysis demonstrated that the 1043RR protein binds to its own promoter, P1043, overlapping the −35 promoter element from positions −17 to −45, suggesting that this protein is autoregulatory. In addition, it binds at a similar location, spanning nucleotides from positions −22 to −51 at the promoter of the methyl-accepting chemotaxis tlpB gene, PtlpB. A possible inverted repeat was identified in the binding sites of both promoters. In an attempt to overexpress 1043RR in H. pylori, the 10-fold induction in transcription of a second copy of HP1043 with use of an inducible promoter failed to increase cellular levels of the RR protein, suggesting that 1043RR is tightly regulated at a posttranscriptional level. The P1043 and PtlpB promoters were demonstrated to be coordinately regulated in response to growth phase in H. pylori. The essential role of HP1043 in encoding a cell cycle regulator is discussed.
Helicobacter pylori chronically infects >50% of the human population causing, in some cases, diseases such as gastritis, peptic ulcers, and stomach cancer (6, 35, 37). Many of the virulence factors associated with pathogenesis of the organism have been identified, including urease (13), flagellins (48), the vacuolating cytotoxin, VacA, and the cag pathogenicity island gene products (11, 12, 18, 49), as well as factors involved in adhesion (21) and iron metabolism (38, 51). The molecular mechanisms by which H. pylori regulates the expression of genes involved in virulence, as well as physiology and metabolism, however, is little understood.
Sequencing of the genome of two independent isolates of H. pylori has revealed a relatively low number of annotated genes with possible regulatory roles (1, 50). Recent studies have been successful in elucidating the functions of some of these (for a review, see reference 43); however, the effective roles of many of the regulatory genes remain to be elucidated. Included in these annotated regulators are 10 genes that are predicted to code for two-component system regulators. Two-component systems are widely used by prokaryotes to regulate responses to environmental signals and are constituted by two proteins, a histidine kinase (HK) and a response regulator (RR) with functional domains (for a review, see reference 47). The HK protein receives an environmental signal by virtue of its N-terminal input domain and communicates this to its cognate RR protein through a phosphotransfer reaction between the autophosphorylated transmitter domain of the HK and the regulatory domain of the RR. This reaction triggers a conformational change in the RR protein activating its C-terminal output domain, which then elicits the response.
The H. pylori genome encodes analogues of the CheA and CheY proteins, whose role in regulating the random tumbling motion due to flagellar rotation in response to signals from chemotaxis receptors has been well documented in other systems (for a review, see references 2 and 20). Although the CheA/CheY system of H. pylori deviates from the archetype, most notably in that CheA contains an additional CheY-like domain, the roles of these proteins in motility and chemotaxis have been demonstrated (4, 24). The genome contains another three HKs and five RRs, and these proteins are predicted to be involved in transcriptional regulation. The genes of two pairs of HK and RR proteins (HP0165-HP0166 and HP1364-HP1365) have a tandem organization within the genome, suggesting that they constitute cognate partners of individual two-component systems. Analysis of phosphotransfer reactions between purified proteins has demonstrated that, indeed, HP0165-HP0166 form a two-component system and, in addition, a third pair, HP0244-HP0703, although encoded from distant loci, constitute another system (3). Two further genes, HP1043 and HP1021, encode orphan RRs for which no HKs have been identified. Interestingly, both of these regulators have substitutions in the conserved signature amino acids in their regulatory domains. Therefore, it has been hypothesized that they may exert their function in the absence of regulatory domain phosphorylation (43).
Mutational analysis of the two-component regulatory genes has revealed that two of the RRs HP0166 and HP1043 could not be inactivated by allelic replacement, and the mutation of a third HP1021 resulted in major growth defects, suggesting that these regulators control essential functions for growth (3). The essential roles of HP1043 and HP0166 were further confirmed when introduction of a second copy of the gene permitted the gene's disruption (33). To date, the target genes that are regulated by the RRs have been defined for only two of the two-component systems. FlgR, the RR encoded by the HP0244-HP0703 system, acts as the activator for σ54-driven promoters and has been shown to play a major role in the regulation of the flagellar structural genes (3, 44). More recently, the target genes regulated by the HP0165-HP0166 system have been identified, although the function of the targets remains unknown (16).
In this study, we attempt to characterize the function and target of the HP1043 essential gene. We show that this HP1043 protein is a dimeric protein that binds specifically to its own promoter and to the promoter of the tlpB chemotaxis gene, suggesting that it is directly involved in regulating both promoters. Furthermore, the expression of both promoters is coordinately regulated in response to growth phase. In addition, we provide evidence that the expression of HP1043 is tightly regulated at a posttranscriptional level.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. H. pylori strains were recovered from −80°C stocks on Columbia agar plates containing 5% horse blood, 0.2% cycloheximide, and Dent's or Skirrow's antibiotic supplement under microaerophilic conditions (Oxoid) and routinely cultured in a 5% CO2-95% air atmosphere at 95% humidity. Liquid cultures were grown in modified brucella broth containing 5% fetal calf serum and Dent's or Skirrow's antibiotic supplement. When required, kanamycin was added to a final concentration of 20 μg/ml. Escherichia coli cultures were cultured in Luria-Bertani medium, and when required, ampicillin was added to a final concentration of 100 μg/ml. For transformation by naturally competent H. pylori, freshly grown overnight cultures were spotted onto plates and grown for a further 5 h at which point 1 to 5 μg of plasmid DNA was spotted in 20 μl of brucella broth onto the growing strain and incubated overnight. Transformants were then selected on plates containing kanamycin (20 μg/ml), and single colonies were selected for further analysis.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| H. pylori | ||
| G27 | Clinical isolate, wild type | 52 |
| G27vac::Kan | Derivative of G27 in which 2,445 bp of the vacA locus have been substituted with a kanamycin cassette | This study |
| G27vac::Ppfr1043 | Derivative of G27 containing a second copy of HP1043 under the control of the iron-inducible Ppfr promoter inserted in the vacA locus | This study |
| E. coli | ||
| DH5-α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 25 |
| R917 | 71/818 glpT::λORPRlacZ | 17 |
| Plasmids | ||
| ptrcHisA | Expression vector for N-terminal His6 tag cloning; Ampr | Invitrogen, Inc. |
| pGEMT | Vector for cloning of PCR fragments; Ampr | Promega |
| pGEM3Z | Cloning vector; Ampr | Promega |
| ptrc::1043 | Derivative of ptrcHisA expressing the HP1043 response regulator; Ampr | This study |
| pcIcat | Derivative of pBR322 in which the 5′ end of λ cI is fused in frame to the cat gene and under the control of the lac promoter; Ampr | 17 |
| pcI1043 | Derivative of pcIcat in which the cat gene and replaced by the HP1043 gene amplified using primers 1043-C1 and 1043BR such that the 5′ end of λ cI is fused in frame to the HP1043 gene; Ampr | This study |
| pcI1043-N | Derivative of pcIcat in which the 5′ end of λ cI is fused in frame to the N terminus of the HP1043 gene amplified using primers 1043-C1 and cI-N; Ampr | This study |
| pcI1043-C | Derivative of pcIcat in which the 5′ end of λ cI is fused in frame to the C terminus of HP1043 gene amplified by using primers cI-N and 1043BR; Ampr | This study |
| pGEM1043p | Derivative of pGEMT containing a 430-bp PCR fragment resulting from amplification of the HP1043 promoter region with the oligos 1043-PF and 1043-PR from the genome of G27; Ampr | This study |
| pGEM103p | Derivative of pGEMT containing a 394-bp PCR fragment resulting from amplification of the HP0103 (tlpB) promoter region with the oligos 103C and 103r from the genome of G27; Ampr | This study |
| pVac::Km | pGEM3 containing a 1.3-kb BamHI fragment containing the kanamycin cassette from pILL600 flanked with a 564-bp EcoRI-BamHI fragment obtained with the oligos tRNA-S5 and ΔtRNA-S3 corresponding to the region upstream of the vacA gene and a 753-bp BamHI-PstI fragment obtained with the oligos ΔVacA-5 and VacA-3 corresponding to the distal end of the vacA gene; Ampr Kmr | This study |
| pVac::Ppfr1043 | Derivative of pVac::Km containing upstream of the kanamycin cassette a 181-bp BamHI-XhoI fragment carrying the pfr promoter and a 699-bp XhoI-BglIII fragment carrying the HP1043 gene and its ribosomal binding site; Ampr Kmr | This study |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; oligos, oligonucleotides.
Expression and purification of 1043RR protein.
The HP1043 gene was amplified from the H. pylori G27 chromosomal DNA with primers 1043-His and 1043-C (Table 2), digested with NheI and BamHI and cloned into ptrcHisA generating ptrc::1043. A 200-ml volume of Luria-Bertani medium was inoculated with 5 ml of overnight culture of E. coli DH5-α containing the ptrc::1043 plasmid and grown at 37°C with vigorous shaking to an optical density at 600 nm (OD600) of 0.5. Expression of the HP1043 gene was then induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM, and incubation was continued for a further 4 h, at which point the cells were harvested by centrifugation at 8,000 rpm for 10 min in a Beckman JA10 rotor. After storage overnight at −20°C, the pellet was resuspended in 4 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10 mM imidazole). Lysozyme was added to a final concentration of 1 μg/ml, and the suspension was incubated for 30 min on ice. Cells were then lysed by sonication and the cell lysate was recovered after 30 min of centrifugation at 10,000 rpm in a JA25-50 rotor (Beckman) and added to 1 ml of nickel-nitrilotriacetic acid agarose resin (Qiagen), which was preequilibrated in lysis buffer and incubated with gentle agitation at 4°C for 1 h. A 1-ml propylene column was then loaded with the resin-lysate mixture, and after the flowthrough was discharged, the resin was washed twice with 4 ml of wash buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 20 mM imidazole). The bound protein was eluted with 1 ml of elution buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 250 mM imidazole) and dialyzed overnight against footprinting buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 50 mM NaCl, 0.01% Nonidet P-40, 1 mM dithiothreitol) containing 10% glycerol and then again against footprinting buffer containing 50% glycerol. The concentration of the protein preparation was obtained by using the Bradford colorimetric method (Bio-Rad), and the protein was aliquoted and stored at −80°C.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Siteb | Positionc |
|---|---|---|---|
| 1043-His | atattagctagccatATGCGCGTTCTACTGATTGAAAAAAATTC | NheI | 1105416-1105388 |
| 1043-C | atattcggatccACGGAATTTACTCTTCACACGCC | BamHI | 1104738-1104760 |
| 1043C | AAAGCCTTTAACATTAAGCC | 1105341-1105362 | |
| 103C | GAAAAATGAGCGGCACAAAAT | 110796-110816 | |
| 103r | aactgcatCATCAAATACATCACGGAAC | PstI | 111129-111110 |
| 1043-PR | attcagggatccCTCTCTGTTACATCAGCC | BamHI | 1105322-1105339 |
| 1043-PF | agcttctgcagAGCTTAATCGTAATCAAGCGG | PstI | 1105735-1105753 |
| 1043-C1 | attattgtcgaccATGCGCGTTCTACTGATTG | SalI | 1105416-1105398 |
| 1043BR | attcagggatccTTACTCTTCACACGCCGG | BamHI | 1104745-1104760 |
| cI-C | attattgtcgaccTTGGGTTCTAATGTGATTGAA | SalI | 1105070-1105050 |
| cI-N | agtcttggatccTCaAAACCTCAAACGAGCCTC | BamHI | 1105068-1105089 |
| 1043-1 | attattctcgagAGGAGTCATACACCATGCGCG | XhoI | 1105430-1105410 |
| 1043-2 | agcttagatctagaTTACTCTTCACACGCCGGTT | BglII | 1104744-1104764 |
| Ppfr-F | attcaggatccCCCTATTGATGCCAACCC | BamHI | 699473-699456 |
| Ppfr-R | ccgatctcgagTTGTCCCATAATTATAGCATA | XhoI | 699299-699319 |
| tRNA-S5 | cgctcagaattcATGATGGCGAAATCTTGCGCAA | EcoRI | 937638-937673 |
| ΔtRNA-S3 | aattggggatccTTAAAAAAACTTCTCCCAAATCGTG | BamHI | 938166-938202 |
| ΔVacA-5 | gcctaaggatccGGTACCAATGGCATTAGTAATG | BamHI | 940647-940668 |
| VacA-3 | gagttctgcagCTTGTAAGCGTTTGGCGAACG | PstI | 941379-941400 |
| cagN | GTCAATGGTTTCGTTAGTC | 579940-579923 |
Capital letters indicate H. pylori-derived sequences, small letters indicate sequences added for cloning purposes, and underlined letters indicate restriction recognition sites.
Restriction recognition sites.
Nucleotide positions refer to the H. pylori 26695 genome sequence published by Tomb et al. (50).
Gel filtration and cross-linking.
The purified 1043RR protein was dialyzed overnight in phosphate-buffered saline (PBS). An aliquot of 60 μg of the protein was loaded onto a Superdex 75 PC 3.2/30 column (Amersham-Pharmacia Biotech) preequilibrated with PBS, and then the protein was eluted at 0.03 ml per min and collected in 100-μl fractions. The protein molecular weight standards (LMW gel filtration calibration kit; Pharmacia Biotech) were applied to the column under the same conditions, and the elution volume and molecular weight of each standard were used to generate a standard curve to determine the molecular weight of the 1043RR protein peak. For in vitro cross-linking, the dialyzed protein was diluted to 1 μg/μl in PBS. Disuccinimidyl suberate (DSS) was added to 10 μl of protein solution to a final concentration of 2.5 mM, followed by incubation at room temperature for 1 h. To stop the cross-linking reaction, 20 μl of protein sample buffer (42) was added, and the protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Construction of H. pylori 1043RR overexpressing strains.
To generate a construct that would allow homologous recombination into the vacA locus, upstream and downstream flanking regions of the locus were amplified with primers tRNA-S5 and ΔRNA-S3 and primers ΔVacA-5 and VacA-3, respectively. The PCR products were cloned into pGEM3Z as a 564-bp EcoRI-BamHI fragment corresponding to the 3′ end of the cysS gene upstream of vacA and a 753-bp BamHI-PstI fragment corresponding to the distal end of the vacA gene, respectively. The HP1043 gene and 17 bp upstream of the coding sequence containing its ribosomal binding site were then amplified with primers 1043-1 and 1043-2 and cloned as a 699-bp XhoI-BglII fragment in a two-way ligation along with the pfr promoter (Ppfr) as a 181-bp XhoI-BamHI fragment into the BamHI site separating the vacA flanking regions. The kanamycin gene was cloned as a 1.3-kb BamHI cassette into the pGEM3Z derivative containing the vacA flanking regions alone or that containing also Ppfr1043 clones into the remaining BamHI site, generating pVac::Km and pVac::Ppfr1043, respectively. These constructs were transformed into DH5-α and analyzed for expression of 1043RR by Western blotting and transformed into G27 and analyzed for overexpression of 1043RR by primer extension and Western blotting.
DNase I footprinting.
The pGEM1043p and pGEM103p plasmids (Table 1) containing the cloned promoter regions were first digested with BamHI and SpeI, respectively, and then subjected to dephosphorylation by using CIP (New England Biolabs, Inc.). Approximately 1 pmol of each plasmid was then 5′ end labeled by using 2 pmol of [γ-32P]ATP (5,000 Ci/mmol; Amersham) with T4 polynucleotide kinase (New England Biolabs, Inc.), digested with PstI, and the promoter fragment, labeled at one extremity, was extracted from a 6% polyacrylamide gel and eluted in 3 ml of elution buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 300 mM sodium acetate [pH 5.2], 0.2% SDS) at 37°C overnight. The eluted probe was extracted once with an equal volume of phenol-chloroform (1:1) and then ethanol precipitated. Approximately 20,000 cpm of the labeled probe was used in each reaction mixture for the footprinting experiment. Protein-DNA complexes were formed in 50 μl of footprinting buffer with 10% glycerol and containing 1 μg of sonicated salmon sperm DNA as nonspecific competitor DNA and 10 mM acetylphosphate, when indicated, for 15 min at room temperature. DNase I digestion was carried out by addition of 1 μl of DNase I (0.01 U/μl) in footprinting buffer containing 5 mM CaCl2 for precisely 1 min at room temperature. The reaction was stopped by the addition of 140 μl of stop buffer (192 mM sodium acetate, 32 mM Na2EDTA, 0.14% SDS, 64 μg of sonicated salmon sperm DNA/ml). Samples were then extracted once with an equal volume of phenol-chloroform (1:1), ethanol precipitated, and resuspended in 6 μl of sequencing loading buffer (98% deionized formamide, 10 mM EDTA [pH 8.0], 0.025% xylene cyanol FF, 0.025% bromophenol blue). After denaturation at 95°C for 2 min, samples were subjected to electrophoresis on 6% urea-polyacrylamide gels at 1,500 V, dried, and autoradiographed.
RNA preparation.
Total H. pylori RNA was extracted as described previously (45). Briefly, 25 ml of H. pylori grown in modified brucella broth at 37°C was harvested at an OD590 between 0.6 and 1.0 (or otherwise where indicated). Cells were lysed in 3.7 ml of 100 mM Tris-HCl (pH 7.5)-2 mM Na2EDTA-1% SDS for 5 min at 95°C. After cooling, 300 μl of 1 M KCl was added, and after a further incubation on ice for 10 min, cellular debris was removed by centrifugation at 8,000 rpm for 10 min in a JA20 rotor (Beckman). To 3.5 ml of supernatant, 4.56 g of CsCl was added, and the RNA was sedimented by centrifugation in an SW65 rotor for 15 to 20 h at 35,000 rpm. The RNA pellet was resuspended in 500 μl of TE (10 mM Tris-HCl [pH 8], 1 mM Na2EDTA), extracted once with an equal volume of phenol-chloroform (1:1), ethanol precipitated, resuspended in 200 μl of TE, quantified, reprecipitated, and stored in ethanol at −20°C.
In vitro transcription.
Single round in vitro transcription was performed as follows. A total of 1 μg of pGEM1043p DNA was incubated with the E. coli RNA polymerase core enzyme, which was reconstituted to incorporate the H. pylori σ80 factor as described by Beier et al. (5) in transcription buffer (40 mM Tris [pH 7.5], 6 mM MgCl2, 2 mM spermidine, 5 mM dithiothreitol, 10 mM NaCl, 10% glycerol) for 20 min at 37°C. Incubation was continued for 5 min after the addition of 0.4 mM ribonucleotides (rNTPs) and 0.2 mg of heparin/ml and stopped by the addition of an equal volume of 0.3 mg of tRNA/ml and 40 mM EDTA. The reaction mixture was then extracted once with an equal volume of phenol-chloroform, precipitated, and used in primer extension experiments.
Primer extension analysis.
The primer (5 pmol) was 5′ end labeled by using 6 pmol of [γ-32P]ATP (5,000 Ci/mmol) with T4 polynucleotide kinase at 37°C for 30 min. Unincorporated [γ-32P]ATP was removed by passing the labeling reaction mixture through a Chroma Spin+TE-10 column (Clontech). Labeled primer (0.1 to 1.0 pmol) was then added to 10 μg of H. pylori total RNA or half of the in vitro transcription reaction, 2 μl of 2 mM deoxynucleoside triphosphates, and 2 μl of 5× avian myeloblastosis virus (AMV) reverse transcriptase buffer (Roche) to make up a final volume of 9 μl. The reaction mixture was incubated at 95°C and allowed to cool to 43°C; 1 μl of AMV reverse transcriptase (10 U/μl; Roche) was then added, and incubation was continued for a further 30 min. The sample was then incubated for 10 min at room temperature with 1 μl of RNase A (10 mg/ml), extracted once with an equal volume of phenol-chloroform (1:1), ethanol precipitated, and resuspended in 6 μl of sequencing loading buffer. After denaturation at 95°C for 2 min, samples were subjected to electrophoresis on 6% urea-polyacrylamide gels at 1,500 V, dried, and autoradiographed. To ensure correct mapping of the promoters, a sequencing reaction was completed with a T7 sequencing kit (USB Corp.) by using the same primer as in the primer extension reactions and the corresponding cloned promoter region. Quantification of the signals from extension products obtained was performed by using a PhosphorImager and ImageQuant software (Molecular Dynamics).
Western blot analysis.
The total protein of H. pylori cultures grown to logarithmic phase were separated by SDS-PAGE and transferred onto nitrocellulose filters by standard methods (42). Sera containing antibodies to the 1043RR protein were raised in mice by immunization with the purified 1043RR protein. Filters were blocked for an hour at room temperature by agitation in blocking solution (3% skim milk and 0.1% Triton X-100 in PBS) and incubated for a further hour with a 1:1,000 dilution of the 1043RR serum in blocking solution. After being washed, the filters were incubated in a 1:2,000 dilution of peroxidase-conjugated anti-mouse immunoglobulin (Dako) in blocking solution for an hour, and the resulting signal was detected by using the Supersignal West Pico chemiluminescent substrate (Pierce).
Assay for β-galactosidase activity.
Overnight cultures were diluted 20-fold into fresh medium containing 1 mM IPTG and were grown at 37°C to an absorbance at 600 nm of 0.4 to 0.6. Assays and units were performed as described previously (34).
RESULTS
The 1043RR protein is a dimer in solution.
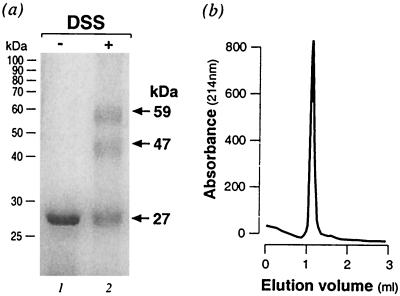
The HP1043 gene from H. pylori is predicted to encode an RR protein (50) that we call 1043RR. To begin characterization of 1043RR, we overexpressed it with a histidine tag in E. coli and purified it under nondenaturing conditions as described in Materials and Methods. To determine the oligomerization state of the purified protein, the protein was treated with the DSS cross-linking agent. From SDS-PAGE analysis, two bands with apparent molecular masses of ca. 49 and 56 kDa (lane 2, Fig. 1a) created by cross-linking were detected after DSS treatment, but the monomer only and no higher order oligomer was observed in the untreated sample (lane 1). The bands observed upon protein cross-link may represent two different products of a dimeric form of the protein, suggesting that 1043RR may form a dimer in solution. To confirm the dimeric form of 1043RR in solution, we performed gel filtration of the purified protein preparation. Figure 1b shows the elution profile in which there is a single well-defined protein peak that is calculated to correspond to a molecular mass of 45 kDa. This indicates that the 1043RR protein is indeed a dimer.
FIG. 1.
In vitro cross-linking and gel filtration of 1043RR. (a) SDS-PAGE of the purified 1043RR protein without (lane 1) or with (lane 2) treatment with the DSS cross-linking reagent. The positions of the molecular mass standards are indicated on the left of the gel. The monomer and cross-linked conformations are indicated with arrows and their apparent molecular mass was reported on the right. (b) Elution profile of the purified 1043RR protein from a Superdex 75 gel filtration column. The molecular mass of the single peak of absorbance at 214 nm was calculated as 45 kDa after calibration of the column with molecular mass standards (13.7, 25, 43, and 67 kDa with elution volumes of 1.39, 1.29, 1.12, and 1.03, respectively).
1043RR dimerizes in vivo by means of its N-terminal regulatory domain.
In order to further analyze the dimerization of 1043RR, we used an E. coli-based in vivo dimerization assay, which exploits the properties of the λ cI repressor to analyze protein-protein interactions (17). Fusion of the cI repressor N-terminal DNA-binding domain to a heterologous protein results in a functional λ repressor able to strongly bind to its operator and repress the lacZ reporter gene only when the heterologous protein dimerizes efficiently. Gene fusions comprising the N-terminal domain of the λ cI repressor gene fused to the complete HP1043 gene, as well as to its N-terminal and C-terminal domains separately, encoding amino acids 1 to 115 or 116 to 223, respectively, were constructed. The respective coding regions were cloned in frame with the λ cI gene encoding the N terminus generating pcI1043, pcI1043-N, and pcI1043-C, respectively (Table 1). The pcIcat plasmid (Table 1) encodes a hybrid of the cI N terminus fused to the cat gene, which is already known to be able to dimerize (17) and was used as a positive control. To test the ability of these protein hybrids to dimerize in vivo, the plasmids were transformed into the E. coli R917 reporter strain and tested for their ability to repress the lacZ gene reporter. The results presented in Table 3 indicate that the complete 1043RR protein both forms a functional λ repressor and represses β-galactosidase synthesis to levels similar to those of the positive control, thus confirming the ability of 1043RR to dimerize in vivo. Furthermore, the N terminus of the protein but not the C terminus of the protein results in repression of the reporter, indicating that the ability of 1043RR to dimerize is encoded within the first 115 amino acids comprising its regulatory domain.
TABLE 3.
In vivo dimerization of 1043RR
| Bacterial strain | Mean β-galactosidase activity (Miller units)a |
|---|---|
| R719 | 4,405 ± 350 |
| R719/pcIcat | 365 ± 74 |
| R719/pcI1043 | 431 ± 116 |
| R719/pcI1043-N | 410 ± 59 |
| R719/pcI1043-C | 2,544 ± 168 |
The values indicated are the means of three independent experiments ± standard deviations.
HP1043 is transcribed from a σ80 promoter.
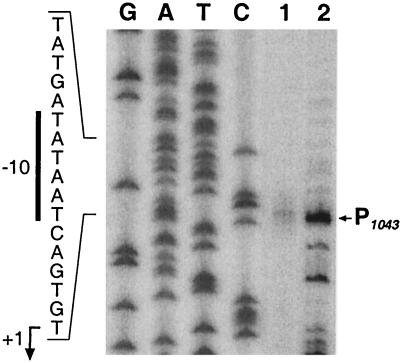
From the genome sequence of H. pylori 26695, it would appear that the HP1043 gene is transcribed monocistronically since there is a 360-bp intergenic region at its 5′ end separating it from the upstream gene and the downstream gene is oppositely oriented so that it converges with the 3′ end of HP1043. In order to map the promoter of HP1043 within the upstream intergenic region, we performed primer extension with RNA extracted from H. pylori G27 strain. Extension of the RNA with a HP1043-specific primer gave rise to a cluster of faint bands, which were centered at 34 nucleotides upstream from the ATG translational start site (Fig. 2, lane 1). Analysis of the sequence upstream of the mapped 5′ ends of RNA reveals a conserved −10 hexamer (TATAAT) with an “extended −10” (TGn) motif (28) and a poorly conserved −35 (CTTAAA) recognized by Eσ70 of E. coli. In order to verify that this is a promoter element, we performed primer extension on RNAs generated in vitro by using pGEM1043p (Table 1) harboring the cloned HP1043 promoter region as a template and E. coli RNA polymerase core enzyme containing the H. pylori σ80 factor. Results from this experiment show that the major transcript in vitro initiates from the same position as the in vivo start point (Fig. 2, lane 2), verifying that HP1043 is transcribed from a σ80-recognized promoter, P1043.
FIG. 2.
Mapping of HP1043 promoter. Total RNA extracted from H. pylori G27 (lane 1), as well as RNA generated in vitro (lane 2), by using E. coli RNA polymerase core enzyme reconstituted with the H. pylori σ80 factor and plasmid pGEM1043p as a template, were hybridized to the radiolabeled oligonucleotide 1043C (Table 2) and elongated with reverse transcriptase. The putative transcriptional start site is marked with an arrow to the right of the panel. To ensure correct mapping of the extension, we sequenced in parallel the cloned promoter in the plasmid pGEM1043p (Table 1) with the same primer (lanes G, A, T, and C). The nucleotide sequence of the sense strand upstream of the transcriptional start site is shown on the left of the panel with the −10 promoter element indicated by a vertical bar; the nucleotide corresponding to the +1 initiation is indicated by a bent arrow.
The 1043RR protein binds to its own promoter.
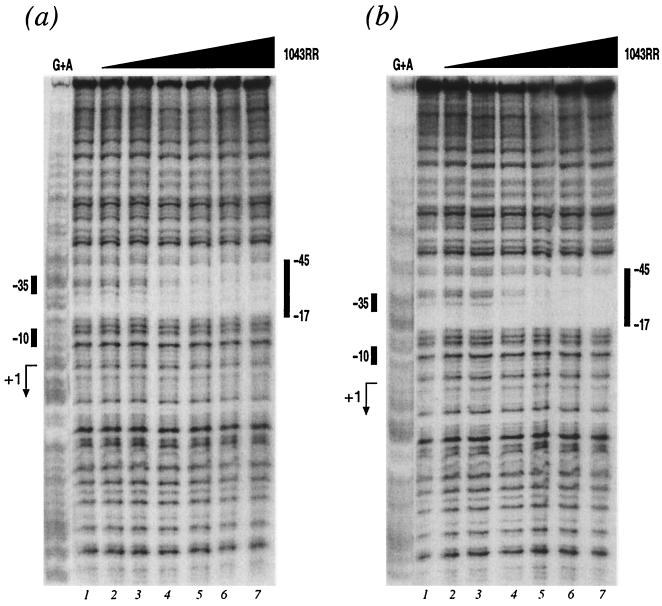
In order to investigate whether the HP1043 regulator has an autoregulatory role, we performed DNA-binding experiments with the purified protein to identify possible direct binding of the 1043RR to its promoter DNA. The HP1043 promoter region, cloned into plasmid pGEM1043p (Table 1), was labeled at one end with [γ-32P]ATP at the BamHI site and DNA gel shift or footprinting experiments were performed with purified 1043RR. In the presence of a 100-fold excess of nonspecific competitor, the mobility of the HP1043 promoter probe was retarded with increasing amounts of 1043RR in gel shift experiments (data not shown), indicating that the 1043RR binds specifically to its own promoter. Footprinting experiments were then performed to identify the binding site of 1043RR protein, and the results are shown in Fig. 3. With increasing 1043RR concentrations we observe the appearance of a very defined protected area. The addition of 0.6 μg of purified 1043RR (Fig. 3a, lane 4) resulted in a partially protected region whose boundaries become clear in reaction mixtures with higher protein concentrations (Fig. 3a, lanes 5 to 7). The protected region spans from positions −17 to −45 with respect to the transcriptional start site, therefore, overlapping the −35 promoter element. Acetyl phosphate has been shown to act as a phosphoryl donor to many RR proteins (30); however, the addition of acetyl phosphate to the footprinting reaction mixtures did not change the affinity of the protein for the DNA and resulted in a similar footprint appearing upon the addition of 0.6 μg of purified 1043RR (Fig. 3b), the major difference being a more complete protection of the nucleotides within the footprint. These data suggest that in the unphosphorylated form the 1043RR protein binds specifically to its own promoter at a site overlapping the −35 promoter element and that acetyl phosphate had the effect of tightening the protein-DNA complex.
FIG. 3.
Binding of 1043RR protein to P1043. DNase I footprint of the 1043RR protein on the P1043 DNA in the absence (a) or presence (b) of acetyl phosphate. The probe used is a 430-bp BamHI-PstI fragment from plasmid pGEM1043p (Table 1) labeled at its BamHI site. The 5′ end-labeled probe was incubated with increasing amounts of 1043RR from lanes 1 to 7 corresponding to 0, 0.06, 0.2, 0.6, 2, 6, and 18 μg of purified protein, respectively. The vertical bars on the right of the panel indicate the areas of DNase I protection. The numbers indicate the distance from the transcriptional start site of the P1043 promoter. The G+A lane is a G+A sequence reaction on the DNA probe used as size marker (32). The bars on the left indicate the positions of the −10 and −35 hexamers, and the bent arrow indicates the transcriptional start site.
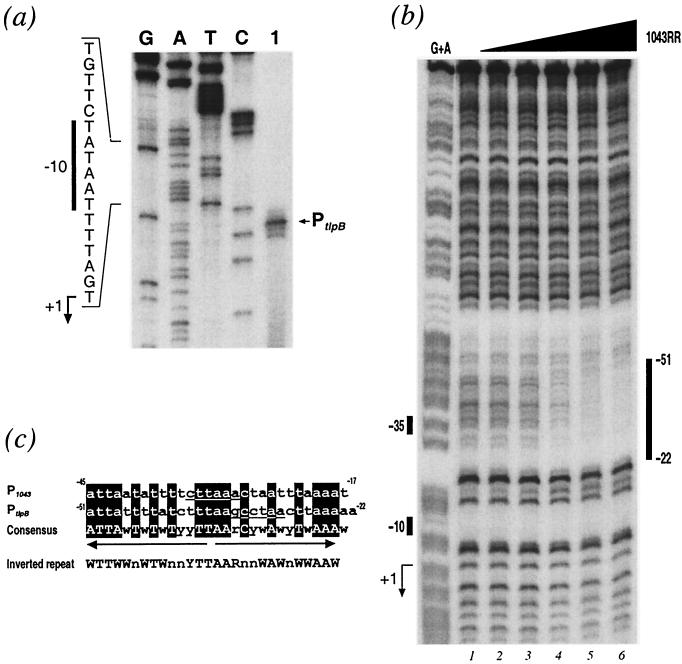
The 1043RR protein binds to the promoter of the methyl-accepting chemotaxis gene tlpB.
In our laboratory, we previously studied the tlbB gene (HP0103), which is predicted to encode a methyl-accepting chemotaxis protein, due to the fact that a high-affinity binding site for the heat shock repressor protein HspR was identified in its 3′ coding sequence (14). The promoter of the tlpB gene was mapped by primer extension and is reported in Fig. 4a. The transcriptional start site of the gene is mapped to 140 nucleotides upstream of the translational ATG start and is preceded by a conserved TATAAT −10 hexamer, suggesting that tlpB is transcribed from a σ80 promoter, PtlpB; however, no conserved −35 region (TTGACA) recognized by σ70 of E. coli could be detected. The nucleotide sequence upstream of the −10 hexamer of this promoter shared apparent similarities to the sequence of the 1043RR-protected nucleotides within the P1043 promoter. We therefore decided to perform DNA-binding experiments to investigate whether 1043RR was capable of binding this promoter. A probe was prepared from the pGEM103p plasmid containing the cloned tlpB promoter (PtlpB) and used in DNase I footprinting experiments with the purified protein in the presence of an excess of nonspecific competitor DNA. The results shown in Fig. 4b indicate that the addition of increasing amounts of 1043RR resulted in protection of nucleotides spanning from positions −22 to −51 with respect to the transcriptional start site of PtlpB. This indicates that 1043RR binds specifically to PtlpB and in a location similar to that of P1043 overlapping the −35 promoter element. Upon aligning the nucleotide sequences that are protected by 1043RR in both promoter regions, we see that 18 of 29 nucleotides are conserved in both sequences, which represent an imperfect inverted repeat, recognized by the 1043RR dimer (Fig. 4c).
FIG. 4.
(a) Mapping of the tlpB promoter, PtlpB. The radiolabeled oligonucleotide 103C (Table 2) was hybridized to total RNA extracted from H. pylori G27 and elongated with reverse transcriptase. The putative transcriptional start site is marked with an arrow to the right of the panel. To ensure correct mapping of the extension, we sequenced in parallel the cloned promoter in the plasmid pGEM103p (Table 1) with the same primer (lanes G, A, T, and C). The nucleotide sequence of the sense strand upstream of the transcriptional start site is shown to the left of the panel with the −10 promoter element indicated by a vertical bar, and the nucleotide corresponding to the +1 initiation is indicated by a bent arrow. (b) Binding of 1043RR protein to PtlpB. DNase I footprint of the 1043RR protein on the PtlpB DNA. The probe used was a 394-bp SpeI-PstI fragment from plasmid pGEM1043p (Table 1) labeled at its SpeI site. The 5′ end-labeled probe was incubated with increasing amounts of 1043RR from lanes 1 to 6 corresponding to 0, 0.2, 0.6, 2, 6, and 18 μg of purified protein, respectively. The vertical bar on the right of the panel indicate the areas of DNase I protection. Numbers indicate the distance from the transcriptional start site of the PtlpB promoter. The G+A lane is a G+A sequence reaction on the DNA probe used as size marker (32). The bars on the left indicate the position of the −10 and −35 hexamers, and the bent arrow indicates the transcriptional start site. (c) Alignment of 1043RR binding sites. The nucleotide sequences corresponding to the regions of DNase I protection from the P1043 and PtlpB from Fig. 3 and 4b, respectively, are aligned: the numbers correspond to distances from the respective transcriptional start sites. Nucleotides corresponding to the respective −35 hexamers are underlined; conserved nucleotides are shaded. The consensus for the alignment is shown underneath. Arrows indicate the position and direction of putative inverted repeat, and the sequence of conserved nucleotides leading to the inverted pattern are shown below in capitals. In silico analysis showed that the conserved inverted repeat TTN4TN4TTAAN4AN4AA can be found 29 times within the genome and that 16 of them are located upstream of genes coding for diverse functions, thus suggesting that this consensus is indicative of a possible binding site.
Construction of putative 1043RR overexpressing strains.
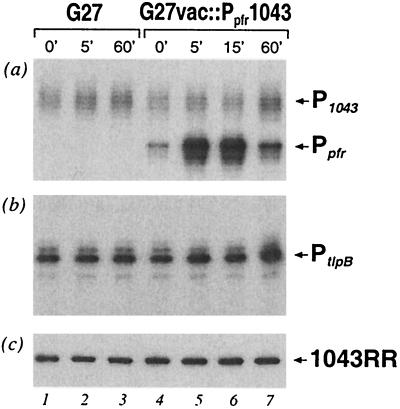
The HP1043 gene exerts an essential role for H. pylori and cannot be deleted (3, 33) which means it is very difficult to assign a function to this gene or to investigate its regulatory role. In an attempt to overcome this problem, we decided on an approach of overexpression of the 1043RR protein in H. pylori that involves the generation of a strain with a second copy of the HP1043 gene under the control of an inducible promoter. We have recently reported the iron-activated indelibility of the Ppfr promoter of the pfr gene (15). This promoter is regulated by the Fur protein in H. pylori and can be induced by the addition of FeSO4 to the growth medium. Since the addition of 1 mM FeSO4 to the growth medium of the wild-type H. pylori G27 culture had no effect on the transcription of either the P1043 or PtlpB promoter (data not shown), the Ppfr promoter can be used as an inducible system to overexpress a second copy of HP1043 for analysis of the effect on the P1043 or PtlpB promoter. The Ppfr promoter was successfully used in our laboratory to complement a fur mutant and overexpression was accomplished on iron induction (data not shown), thus indicating that this system can be used to overexpress any gene integrated in the vacA locus of the H. pylori genome. The HP1043 gene, including its ribosome-binding site, was cloned downstream of the Ppfr promoter and introduced into the vacA locus of the H. pylori G27 chromosome, generating G27vac::Ppfr1043 as described in Materials and Methods. As a control strain, a kanamycin cassette alone was introduced into vacA locus generating G27vac::kan. These two strains were grown to logarithmic phase in liquid culture, and total RNA was extracted from each before (0 min) and at 5, 15, and 60 min after the addition of 1 mM FeSO4. In primer extension experiments the transcription of the second copy of HP1043 from Ppfr, as well as transcription from the innate P1043 and PtlpB promoters, was monitored in the RNA preparations. Figure 5a shows the results of primer extension with an HP1043-specific primer on RNAs from the control and overexpression strains before and after FeSO4 induction. The induction of transcription of the second copy of HP1043 was seen after addition of FeSO4, where transcription from Ppfr was induced 10-fold after 5 min (lane 5) and 11-fold after 15 min (lane 6) but the level of induction had decreased to 3-fold after 1 h (lane 7). However, no significant differences were observed in transcription from the innate P1043 or PtlpB promoter either in the control strain G27vac::kan or in the overexpression strain G27vac::Ppfr1043 (lanes 1 to 7, Fig. 5a and b, respectively). These data suggest that transcription of the second copy of HP1043 gene could be successfully induced by at least 11-fold by using the Ppfr/FeSO4 induction system in the G27vac::Ppfr1043 overexpression strain; however, the induction of transcription of the gene had surprisingly no effect on the regulation of the P1043 or PtlpB promoter.
FIG. 5.
Analysis of the transcription and expression from the putative 1043RR-overexpressing constructs. G27vac::Kan and G27vac::Ppfr1043 strains were grown to logarithmic phase, and 1 mM FeSO4 was added to the growth medium for iron induction of the inducible Ppfr promoter. Total RNA (a and b) and total protein (c) were extracted from strains G27vac::Kan (marked G27 on top of the panels) before iron induction (lane 1) and at 5 and 60 min after induction (lanes 2 and 3, respectively) and from G27vac::Ppfr1043 before (lane 4) and at 5, 15, and 60 min after induction (lanes 5, 6, and 7, respectively). Transcription from the innate P1043 promoter and the Ppfr promoter expressing the second copy of HP1043 was analyzed by primer extension on the RNA preparations with HP1043-specific primer, 1043C (see panel a). Transcription from PtlpB was analyzed by primer extension on the RNA preparations with tlpB-specific primer, 103C (see panel b). Cellular levels of 1043RR protein in each condition were analyzed by Western blotting (see panel c).
In order to assess the effect of the induction of HP1043 transcription on the accumulation of the 1043RR protein in the overexpression strain, we raised antibodies to the purified 1043RR protein in mice for use in Western blot analysis. To assess whether HP1043 under the control of Ppfr was capable of expressing the 1043RR protein, we analyzed whole-cell lysates from E. coli strains carrying the pVac::Ppfr1043 plasmid, which was used for construction of the overexpressing strains in H. pylori, by Western blotting. A strong reacting band of ca. 26 kDa was present in the total protein of DH5-α strains carrying the pVac::Ppfr1043 plasmid, but not in the DH5-α strains without the plasmid (data not shown), indicating that the 1043RR could be expressed from this construct in E. coli. Total protein extracts of the control and overexpression strains of H. pylori from the same induction conditions as reported for the primer extension were analyzed by Western blotting with the polyclonal serum from mice, and the results are reported in Fig. 5c. Antibodies raised against 1043RR recognized a band of ca. 26 kDa in all protein preparations (lanes 1 to 7); however, the amount of recognized protein does not vary significantly between the lanes. This indicates that the cellular levels of the 1043RR protein remain constant even after an 11-fold induction of HP1043 transcript. These data further suggest that in H. pylori the expression of 1043RR is tightly regulated at a posttranscriptional level.
Both HP1043 and tlpB have similar growth-phase-dependent expression profiles.
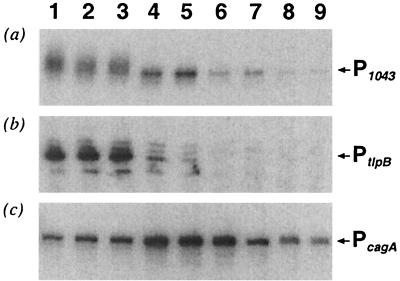
Since it proved impossible to determine the in vivo effect of 1043RR on the P1043 or PtlpB promoter either by reverse genetics (3, 33) or by our strategy of attempted overexpression, we decided to study the regulation of the P1043 and PtlpB themselves in response to various environmental signals. Exponentially growing H. pylori G27 wild-type cells were exposed to various environmental conditions, including heat shock (15 min at 42°C), osmotic shock (15 min in 300 mM NaCl), and acid shock (15 min in pH 4), and treatment with high iron (1 mM FeSO4) or nickel (100 μM and 1 mM NiCl2) and acetyl phosphate (0.2 mM). However, primer extension experiments on RNA extracted before and after each treatment revealed no response of either the P1043 or PtlpB promoter to these signals (data not shown). A time course experiment was then run in which H. pylori G27 culture was grown in liquid over 3 days, and RNA was extracted from G27 cells at the early and late logarithmic phases and the early and late stationary phases of growth. Transcription from the P1043 or PtlpB promoters, as well as the cagA promoter PcagA, as a control was monitored over the time course by primer extension. It was found that both P1043 or PtlpB have similar growth-phase-dependent transcriptional profiles (Fig. 6a and b). The transcription from both P1043 and PtlpB promoters is highest in lanes 1, 2, and 3, which represent experiments performed on RNA preparations from logarithmically growing cells harvested at absorbencies of 0.24, 0.38, and 0.6, respectively. In late log phase and the beginning of stationary phase represented in lanes 4 and 5, respectively, the transcription from both promoters is reduced: that of P1043 is reduced to ca. 50%, whereas PtlpB is reduced to 20 and 10% in lanes 4 and 5, respectively. Progressing through stationary to late stationary phase (represented in lanes 6 to 9), the transcription from both promoters is further coordinately reduced. As a control of this experiment, we also monitored transcription from PcagA, a promoter whose expression has been studied in detail (45). The PcagA promoter is expressed throughout the time course experiment (Fig. 6c) but is induced by fivefold in early stationary phase (lanes 4 to 6), thereby showing a very different growth-phase-dependent expression profile. We conclude that the P1043 and PtlpB promoters are coordinately regulated in response to growth phase.
FIG. 6.
Growth-phase-dependent regulation of the P1043 and PtlpB and PcagA promoters. G27 wild type was grown in liquid medium over a time course, and total RNA was extracted after 7, 10, 13, 23, 29, 35, 47, 55, and 72 h, which corresponded to OD600 absorbencies of 0.26, 0.38, 0.59, 1.1, 1.29, 1.23, 1.2, 1.22, and 1.24, respectively, and in turn correspond to experiments in lanes 1 to 9, respectively. (a) Primer extension of RNA preparations with the HP1043-specific primer, 1043C. (b) Primer extension on the RNA preparations with the tlpB-specific primer, 103C. (c) Primer extension of RNA preparations with the cagA-specific primer, cagN.
DISCUSSION
In this study we show that the 1043RR protein, a member of the OmpR subfamily of RRs (9, 31, 41), is a DNA-binding protein, and we identify two targets for this RR. 1043RR binds specifically to its own promoter (P1043), suggesting an autoregulatory role, and also to the promoter of the tlpB gene (PtlpB). This gene is encoded in the H. pylori 26695 strain by the HP0103 gene and is predicted to encode a methyl-accepting chemotaxis protein (MCP). The essential role of HP1043 is not, however, in the activation of tlpB as this gene can be mutated in the H. pylori chromosome (data not shown). MCPs act as membrane receptors and monitor the chemical composition of the environment, interacting in a supramolecular complex with CheW and CheA, transducing signals to mediate chemotactic responses (for a review, see reference 8). There are three classical MCPs encoded in the H. pylori genome (50), as well as a truncated cytoplasmic MCP encoded by HP0599 (24); however, the chemoattractants or repellents to which each responds are as yet unidentified. Importantly, full motility and chemotaxis play an important role during H. pylori infection. Therefore, regulation of a chemotaxis gene transcription, as well as other still-unidentified coregulated genes, is likely to be required in the variable environment of the stomach for efficient colonization.
The specific targets or binding sites of the 1043RR within these promoters were defined by footprinting analysis (Fig. 3 and 4b). Primer extension studies revealed that both P1043 and PtlpB are σ80 promoters with perfect TATAAT −10 elements (Fig. 2 and Fig. 4a); however, neither the P1043 nor PtlpB promoter has a −35 element that is well conserved with the E. coli consensus, exhibiting three or one conserved nucleotide, respectively. The 1043RR binds specifically and exclusively overlapping the −35 promoter element at both target promoters, protecting nucleotides from −17 to −45 at P1043 and from −22 to −51 at PtlpB. Upon analysis, the nucleotide sequences of the binding sites appear as a conserved inverted repeat with TTWWnWTWnnYTT (TTn4Tn4TT) in each arm of the inverted repeat. This type of binding motif is consistent with the dimeric structure of the protein reported in this study.
Gel filtration and in vitro cross-linking experiments demonstrated that the 1043RR protein is a dimer in solution. Furthermore, in vivo dimerization assays identified the dimerization domain to be within the first 115 amino acids in the N terminus, and deletion of the C-terminal effector domain has no effect on the dimerization ability of 1043RR. NtrC and PhoB have been defined as members of a class of RR wherein the C-terminal domain inhibits the oligomerization functions of the N-terminal domain (22). However, 1043RR appears to belong to a different class in which the two domains function independently, as has been reported for the BvgA and PhoP RRs (7, 29).
Footprinting analysis revealed that the 1043RR protein is capable of specific binding to its targets in the unphosphorylated form (Fig. 3). It is known that other RRs, such as with PhoP or BvgA, may bind DNA in the unphosphorylated form (7, 29, 53); however, the affinity or pattern of binding of RRs usually is dependent on its phosphorylation status. The unusual aspect of this RR is that to date no HK has been identified for 1043RR. Based on previous studies, there was no evidence that any of the three HKs thought to be involved in signaling transcriptional regulators could phosphorylate this protein (5). Furthermore, the CheA protein, although capable of autophosphorylation and phosphorelay to the receiver domain located at its C-terminal region was not able to phosphorylate 1043RR in vitro (data not shown). As such, 1043RR remains an orphan RR. Analysis of its N-terminal regulatory domain reveals deviations in the conserved amino acids that are thought to constitute the acid-pocket active site that in RRs is phosphorylated by a phosphorelay reaction from the cognate HK protein (46). In CheY, these correspond to conserved residues Asp12, Asp13, and Asp57. In 1043RR there is a conserved substitution to Glu at the equivalent of position 12, which is commonly observed in other RR proteins. Nevertheless, the phosphate-accepting aspartic residue Asp57 is shifted by one position, and there is a Lys at the equivalent of position 13. Interestingly, an aspartate-to-lysine mutation at position 13 of CheY resulted in a constitutively active RR (26). All of these observations taken together raise the possibility that 1043RR might be regulated by a mechanism(s) other than phosphorylation. In contrast, the observation that in vitro treatment of the 1043RR protein with acetyl phosphate determined a tighter contact of the protein to its binding site (Fig. 3) might suggest that phosphorylation has a role in activation of transcription and that its cognate sensor kinase remains to be identified.
The location of binding of the 1043RR is typical of regulatory proteins involved in activation of transcription, which bind overlapping or upstream of the promoter elements (10) such as the PhoP RR whose core binding site is between −66 and −17 of the promoters it activates (19). Activation through binding at these proximal sites facilitates contacts between the activator, the RNA polymerase enzyme, and the promoter DNA, thereby allowing the initiation of transcription (40). From the time course experiments we can correlate initiation at the P1043 and PtlpB promoters and the expression of 1043RR gene (Fig. 6). Unfortunately, due to the inability to either knock out or overexpress the HP1043 gene the precise nature of the regulation 1043RR effects at these promoters in vivo remains without evidence. However, our data support a model in which the 1043RR protein activates transcription of the promoters by binding at sites upstream of the −10 promoter element.
Since the in vivo regulatory function of the protein could not be investigated directly, we attempted to identify an environmental signal to which these promoters were coordinately regulated. By analysis of the transcription of these promoters in a time course experiment, we have demonstrated that both promoters are growth phase dependent and, moreover, that P1043 and PtlpB have similar transcriptional profiles. This suggests that these promoters are coregulated, and we propose that 1043RR may be effecting this regulation directly. It is also interesting that the P1043 promoter has an “extended −10” TGn promoter motif. It is known that in E. coli promoters with this motif and a conserved TATAAT allow transcriptional initiation in the absence of a −35 region (28). The relevance of this motif in the transcription of H. pylori promoters has been previously demonstrated (23). This may account for the sustained transcription of P1043 in stationary phase, where the 1043RR protein is likely not to be activating transcription.
Our attempts to overexpress the 1043RR in H. pylori, by adding a second copy of HP1043 under the strong flaA promoter (data not shown) or an inducible promoter (Fig. 6) did not result in increase of cellular levels of 1043RR. Using the second approach, we successfully induced an 11-fold increase in the HP1043 transcript; however, the levels of protein remained constant within the cell, suggesting a very tight posttranscriptional control on expression of the protein. It would appear therefore that not only is the expression of this protein essential for H. pylori (3, 33) but also the cellular level of the protein is critical and highly regulated. We provide evidence that 1043RR is autoregulated at the transcriptional level and, in addition, regulated posttranscriptionally by an unknown mechanism. Interestingly, one of the few other RRs identified with an essential role is CtrA, an OmpR-type regulator of Caulobacter crescentus (39) that activates cell cycle promoters by binding to conserved TTAAn7TTAA motifs upstream of the −10 hexamer (36). Included in its regulon are a major chemotaxis operon and a gene encoding an MCP (27). We have shown in this study that promoters bound by 1043RR are coordinately regulated with respect to growth phase; therefore, it is tempting to suggest that 1043RR may play a similar role to CtrA in the regulation of cell-cycle-related functions in H. pylori. If so, it is possible to predict that 1043RR may regulate functions such as cell division, DNA methylation, membrane and flagellar biosynthesis, and chromosome replication. Further studies exploiting the in vitro DNA-binding ability of this protein may be performed to select and identify other targets and coregulated genes, since the traditional reverse genetics and overexpression approaches are inappropriate for the study of 1043RR.
Acknowledgments
We thank L. Paolozzi for the gift of plasmid pcIcat and strain R917, E. Cartocci for help in the gel filtration experiments, C. Mallia for editing the manuscript, and G. Corsi for artwork.
This work has been supported partially by EU-TMR grant FMRX-CT980164, Chiron, and MURST. I.D. was the recipient of an EU-TMR fellowship (FMRX-CT980164).
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 3.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179:4676-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1998. Functional analysis of the Helicobacter pylori principal sigma subunit of RNA polymerase reveals that the spacer region is important for efficient transcription. Mol. Microbiol. 30:121-134. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1990. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J. Infect. Dis. 161:626-633. [DOI] [PubMed] [Google Scholar]
- 7.Boucher, P. E., K. Murakami, A. Ishihama, and S. Stibitz. 1997. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J. Bacteriol. 179:1755-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bren, A., and M. Eienbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory propogation. J. Bacteriol. 182:6885-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan, R. G. 1993. The winged helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74:773-776. [DOI] [PubMed] [Google Scholar]
- 10.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 11.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cover, T. L., M. K. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 269:10566-10573. [PubMed] [Google Scholar]
- 13.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174:2466-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. In vitro selection of high-affinity HspR-binding sites within the genome of Helicobacter pylori. Gene 283:63-69. [DOI] [PubMed] [Google Scholar]
- 15.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and iron-repressed genes in H. pylori. Mol. Microbiol. 32:1297-1309. [DOI] [PubMed] [Google Scholar]
- 16.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Lallo, G., P. Gherlardini, and L. Paolozzi. 1999. Two-hybrid assay: construction of an Escherichia coli system to quantify homodimerization ability in vivo. Microbiology 145:1485-1490. [DOI] [PubMed] [Google Scholar]
- 18.Dundon, W. G., M. de Bernard, and C. Montecucco. 2001. Virulence factors of Helicobacter pylori. Int. J. Med. 290:647-658. [DOI] [PubMed] [Google Scholar]
- 19.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenbach, M. 1996. Control of bacterial chemotaxis. Mol. Microbiol. 20:903-910. [DOI] [PubMed] [Google Scholar]
- 21.Evans, D. J., Jr., and D. G. Evans. 2000. Helicobacter pylori adhesins: review and perspectives. Helicobacter 5:183-195. [DOI] [PubMed] [Google Scholar]
- 22.Fiedler, U., and V. Weiss. 1995. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of receiver modules. EMBO J. 15:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsyth, M. H., and T. L. Cover. 1999. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. J. Bacteriol. 181:2261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, M., R. B. Bourret, M. I. Simon, and K. Volz. 1997. Uncoupled phospohorylation and activation in bacterial chemotaxis: the 2.3 Å structure of an aspartate to lysine mutant at position 13 of CheY. J. Biol. Chem. 272:11850-11855. [DOI] [PubMed] [Google Scholar]
- 27.Jones, S. E., N. L. Ferguson, and M. R. Alley. 2001. New members of the ctrA regulon: the major chemotaxis operon in Caulobacter is CtrA dependent. Microbiology 147:949-958. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35 recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at “extended minus 10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 29.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 32.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDaniel, T. K., K. C. DeWalt, N. R. Salama, and S. Falkow. 2001. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter 6:15-23. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Nomura, A., G. N. Stemmermann, P. H. Chyou, G. I. Perez-Perez, and M. J. Blaser. 1994. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Intern. Med. 120:977-981. [DOI] [PubMed] [Google Scholar]
- 36.Ouimet, M. C., and G. T. Marczynski. 2000. Analysis of a cell-cycle promoter bound by a response regulator. J. Mol. Biol. 302:761-775. [DOI] [PubMed] [Google Scholar]
- 37.Parsonnet, J., D. Vandersteen, J. Goates, R. K. Sibley, J. Pritikin, and Y. Chang. 1991. Helicobacter pylori infection in intestinal- and diffuse-type gastric adenocarcinomas. J. Natl. Cancer Inst. 83:640-643. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Perez, G. I., and D. A. Israel. 2000. Role of iron in Helicobacter pylori: its influence in outer membrane protein expression and in pathogenicity. Eur. J. Gastroenterol. Hepatol. 12:1263-1265. [DOI] [PubMed] [Google Scholar]
- 39.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 40.Rhodius, V. A., and S. J. Busby. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152-159. [DOI] [PubMed] [Google Scholar]
- 41.Russo, F., and T. J. Silhavy. 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222:567-580. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Scarlato, V., I. Delany, G. Spohn, and D. Beier. 2001. Regulation of transcription in Helicobacter pylori: simple systems or complex circuits? Int. J. Med. Microbiol. 291:107-117. [DOI] [PubMed] [Google Scholar]
- 44.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spohn, G., D. Beier, R. Rappuoli, and V. Scarlato. 1997. Transcriptional analysis of the divergent cagAB genes encoded by the pathogenicity island of Helicobacter pylori. Mol. Microbiol. 26:361-372. [DOI] [PubMed] [Google Scholar]
- 46.Stock, A. M., E. Martinez-Hackert, B. F. Rasmussen, A. H. West, J. B. Stock, D. Ringe, and G. A. Petsko. 1993. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry 32:13375-13380. [DOI] [PubMed] [Google Scholar]
- 47.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 48.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Telford, J. L., P. Ghiara, M. Dell'Orco, M. Comanducci, D. Burroni, M. Bugnoli, M. F. Tecce, S. Censini, A. Covacci, Z. Xiang, E. Papini, C. Montecucco, L. Parente, and R. Rappuoli. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its role in gastric disease. J. Exp. Med. 179:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 51.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 52.Xiang, Z., S. Censisni, P. F. Bayeli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zu, T., R. Manetti, R. Rappuoli, and V. Scarlato. 1996. Differential binding of BvgA to two classes of virulence genes of Bordetella pertussis directs promoter selectivity by RNA polymerase. Mol. Microbiol. 21:557-565. [DOI] [PubMed] [Google Scholar]