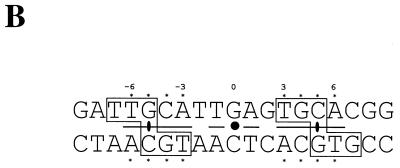During the last decade, the number of plasmids found to replicate by a rolling-circle mechanism has experienced an enormous increase. This group of plasmids now includes more than 200 replicons isolated from a variety of gram-positive and gram-negative bacteria as well as from archaea. All rolling circle-replicating plasmids isolated so far are small (less than 12 kb), indicating that economy of genetic information could be an important aspect of these replicons. Among these plasmids a family has been defined, with pMV158 as a prototype, in which control of replication involves two plasmid-encoded elements: an antisense RNA and a small transcriptional repressor Cop protein (6, 7). The pMV158-encoded CopG protein has been analyzed in some detail. The hydrodynamic behavior of the native homodimeric protein was studied, and the three-dimensional structures of CopG that are free and bound to the central part of its target DNA have been determined (1, 10). In spite of the smallness of CopG (45 residues for each subunit), there is a relatively large extension of DNA (about 50 bp) contacted by the bound protein, which results from cooperative binding of several CopG dimers to this 50-bp operator. Generation of this nucleoprotein complex does not only rely on the cooperative CopG dimer-dimer interactions but also on the specific recognition of the sequence and/or the structure of the entire operator. In that way, a small protein, representing minimal genetic information, could exhibit a high specificity of binding to its target DNA.
The aim of this review is to present a summary of the most relevant molecular aspects of CopG, which is the prototype of the Cop proteins encoded by plasmids of the pMV158 family. As shown for CopG, all these small Cop proteins could exhibit a ribbon-helix-helix (RHH) arrangement, with the two antiparallel β-strands of the ribbon, each coming from one of the protein monomers, involved both in dimer formation and in specific interactions with DNA bases in the operator. Modeling of these Cop proteins on the determined structure of CopG shows that dimer-dimer interfaces similar to that of CopG could arise from specific oligomerization upon binding of the Cop dimers to their target DNAs. Thus, these Cop proteins would represent the minimal functional plasmid-encoded element able to inhibit RNA polymerase activity by binding to the operator DNA in a specific and highly cooperative way.
FEATURES OF CopG AND COMPARISON WITH OTHER RHH TRANSCRIPTIONAL REPRESSORS
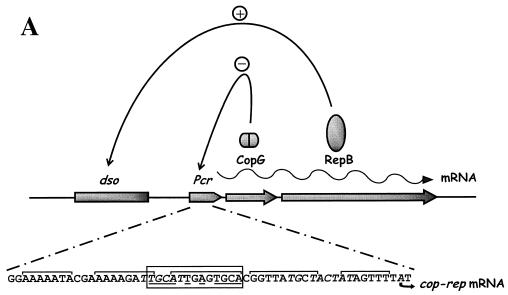
The copG gene of plasmid pMV158 encodes the transcriptional repressor CopG, a homodimeric 45-residue protein that constitutes the smallest natural transcriptional repressor characterized so far. The existence of such a small gene was proposed early (13), even though, in general, orfs that encode polypeptides smaller than 50 residues were not taken into account when researchers were looking for genes involved in replication and its control. CopG is the prototype of a series of repressor proteins encoded by plasmids that exhibit a similar genetic structure at their leading strand initiation and control regions (8). Protein CopG binds to and represses the single Pcr promoter that directs the synthesis of a bicistronic mRNA for CopG and the RepB initiator of replication (Fig. 1A). Thus, by binding to the copG-repB promoter region, CopG regulates its own synthesis and that of RepB (9). The specific target of CopG spans about 50 bp and includes a central 13-bp pseudosymmetric element (inverted repeat) that encompasses most of the −35 box of the promoter (Fig. 1A). Each half-site of the symmetric element contains the palindromic sequence 5′-TGCA-3′ (Fig. 1B), so that two self-symmetrical subsites are present within the element (9, 10). The hydrodynamic behavior of the native CopG shows that the protein is a dimer in solution at concentrations ranging from 10 to 800 μM, with no detectable monomers or association states with a higher number of molecules than dimers. CopG dimers are nearly spherical, with a deduced Stokes radius of around 16 Å (1). With these features, a CopG dimer bound to its target DNA occupies approximately one helical turn (Fig. 1C). The three-dimensional structures of CopG free and bound to either a 19-bp (10) or a 22-bp (5) double-stranded DNA containing the pseudosymmetric element have been determined. These constituted the first examples of a solved plasmid-encoded transcriptional repressor crystal structure. In the cocrystals, one CopG dimer is bound to each half-site of the pseudosymmetric element. Although the protein dimer and the two half-sites have twofold symmetry, the contacts made by each CopG dimer on the DNA are essentially asymmetric, both in the sequence of the contacted bases (Fig. 1B) and in the amino acids involved in these interactions (5, 10).
FIG. 1.
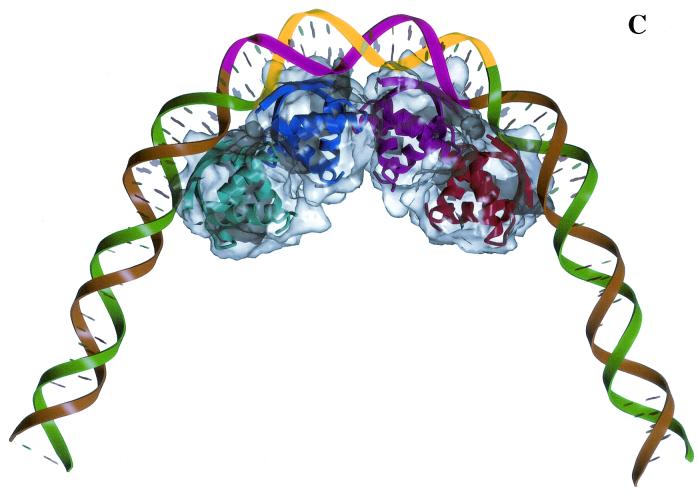
Protein-mediated regulatory circuits controlling replication of plasmid pMV158 and the role of CopG in regulation. (A) The positive elements are constituted by protein RepB (the initiator of replication) and its target DNA (the origin of replication, dso). In the same mRNA, the negative element protein CopG is codified. CopG (shown as a dimer) binds to and represses transcription from the single Pcr promoter. The DNA sequence of this region is shown below: the −35 and extended −10 boxes of the promoter (italics), the regions protected by CopG from DNase I cleavage (brackets), and the initiation of transcription point (arrow). (B) Bases contacted by CopG (boxes) in the 19-bp oligonucleotide used to obtain the CopG-DNA cocrystals. The inverted repeat (underlined) in the 13-bp pseudosymmetric element has its center at the G:C base pair (filled circle), which is numbered 0. The symmetry of this element coincides with that of the bound CopG tetramer. Within the 13-bp element, there are two subsites with perfect symmetry (asterisks). The CopG dimer dyad axes are also indicated (filled ellipses). (C) Model of CopG binding to its target DNA. The DNA strands are displayed in green and brown, except the region corresponding to the 13-bp symmetric element, which is labeled in yellow and magenta. The structure of the CopG dimers is indicated by ribbon plots within a transparent Connolly surface. Four CopG dimers bind to four successive DNA helix turns, and, as a result, the target DNA is bent at a total angle of about 120°.
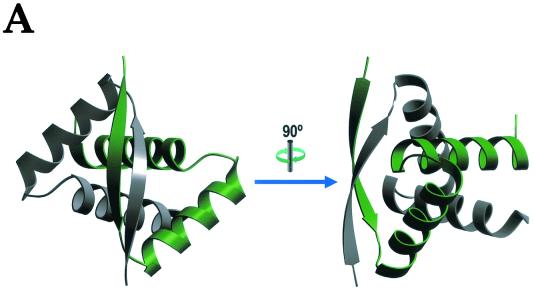
CopG dimers show an RHH arrangement (Fig. 2A) similar to those previously determined for the transcriptional repressors Arc (53 residues) and Mnt (82 residues) of Salmonella enterica serovar Typhimurium bacteriophage P22 and 104-residue MetJ of Escherichia coli (3, 4, 20). All these proteins share a dimerizing 40- to 45-residue RHH motif (Fig. 2B) in which the two-stranded antiparallel β-sheet fits snugly into the major groove of the target DNA to establish specific interactions (Fig. 3). Within the RHH motif, the peptide backbone of the CopG dimer fits almost exactly that of Arc (Fig. 2B), while fewer similarities are found between the topologically equivalent Cα atoms of CopG and Mnt dimers. Although MetJ lacks a glycine-mediated turn (which in Arc, Mnt, and CopG connects the two α-helices included in the RHH signature) and has a longer loop instead, there is a good match between the Cα atoms of CopG and those equivalent in MetJ (Fig. 2B), once the insertion in this latter protein is considered (10). In spite of CopG exhibiting a high structural similarity with the RHH motif of Arc, Mnt, and MetJ, the plasmid-encoded protein shows major differences with the three other repressors in fundamental aspects, such as the regions involved in contacts to the operator DNA or in cooperative dimer-dimer interactions. Direct contacts of CopG to the target DNA are necessarily restricted to the RHH motif, because it comprises the entire protein (Fig. 3A). Essential interactions with their operators, including direct contacts with the bases, originate mainly from the RHH motifs of Arc, Mnt, and MetJ. However, unlike CopG, these three proteins contain additional regions also involved in contacts with their target DNAs. In Arc (Fig. 3B), the six N-terminal residues, which are disordered in the unbound dimer, form tandem reverse turns (a 310 helix) that interact with the DNA in the Arc-operator complex (21). A flexible loop immediately preceding each β-strand of the MetJ dimeric holorepressor changes its conformation upon binding to the target DNA (Fig. 3C) and wraps around and contacts the phosphate backbone (23). Although the structure of Mnt bound to its target DNA has not been determined, biochemical experiments also show that Mnt uses an N-terminal arm to wrap around and to contact the center of its operator (11, 19). Thus, CopG, containing exclusively the RHH motif, represents the minimal DNA-binding structure within this superfamily of repressor proteins. While this review was being written, the structures of two plasmid-encoded repressor proteins, both of them included in modules for better-than-random segregation of the plasmid copies, were determined (18). Protein ω (71 residues), from the streptococcal plasmid pSM19035, has been crystallized alone, whereas ParD (82 amino acids), from plasmid RK2, has been determined by nuclear magnetic resonance. Both proteins have the RHH motif that would be involved in binding to DNA (17, 18). Protein ω has a 27-residue-long, flexible, N-terminal region before the RHH that could interact with DNA, although most of this part is not seen in the crystal structure (17). In the case of ParD, the dimeric protein exhibited a well-structured N-terminal domain and its first residues could be aligned with the entire length of CopG; the C-terminal moiety of ParD turned out to be relatively unstructured.
FIG. 2.
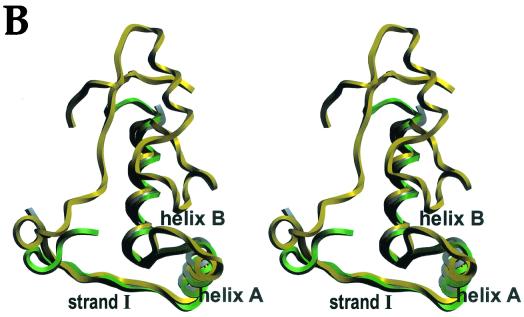
The three-dimensional structure of CopG. (A) Ribbon plot picture of a CopG dimer made up by a green monomer and a grey monomer. Arrows denote the N-terminal β-strands, and the α-helices are depicted as ribbons. Two views are shown: top of the surface constituting the primary interaction region with DNA (left) and after a vertical clockwise 90-degree rotation (right). (B) Stereo ribbon plot picture of the superimposition of the monomeric structures of CopG (grey), Arc (green), and MetJ (yellow), with the regular secondary structure elements of the RHH motif labeled. CopG constitutes the smallest structure, Arc further displays an N-terminal helical region, and MetJ displays both N- and C-terminal elongations.
FIG. 3.
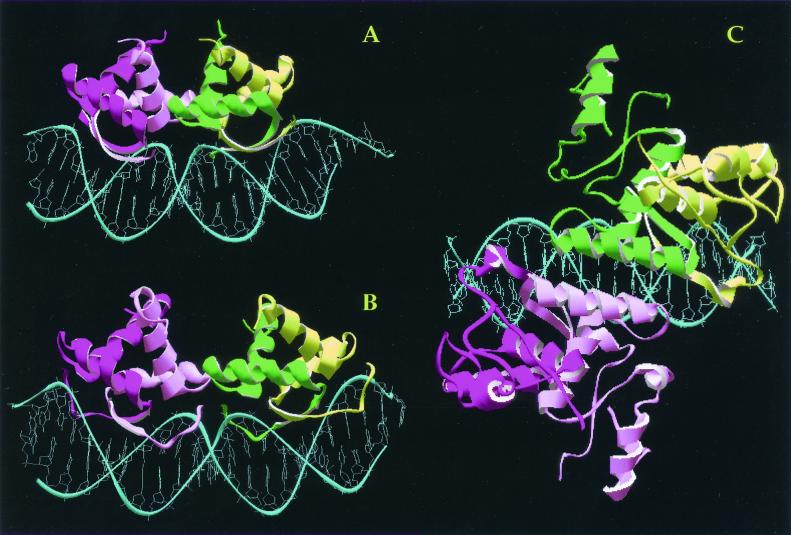
The structures of CopG (A), Arc (B), and MetJ (C) in association with their DNA targets. The DNAs are shown in blue, and the proteins are depicted as ribbon plot pictures, with each subunit of the same dimer displaying a different intensity of green or magenta.
Although association states with a higher number of molecules than dimers are only found in solutions of free Mnt (which is a tetramer), some oligomerization occurs upon cooperative binding of dimeric Arc, MetJ, and CopG to their cognate target DNAs. Arc and Mnt are tetramers when bound to the operator DNA, whereas, in MetJ-operator complexes, each repressor dimer binds to one of the two to five tandem 8-bp Met boxes that constitute the operator (23). Comparative analysis of different natural MetJ operators indicates that the number of Met boxes tends to increase as their similarity to the consensus Met box decreases (20). Dimer-dimer contacts that account for the observed cooperative DNA binding involve protein regions that are different in the repressor/operator complexes of Arc and MetJ (Fig. 3B and C): Arc dimers make contacts through the loop between helices A and B of the RHH motif, whereas MetJ dimers interact along the entire length of helix A (21, 23). These differences would accommodate the distinct spacing between consecutive binding sites of Arc or MetJ dimers, which, in turn, results in an Arc tetramer bound to one face of the DNA, while arrays of MetJ dimers wrap around the DNA helix (Fig. 3B and C). As in Arc and MetJ, CopG oligomerization states higher than dimers seem to depend on the presence of the target DNA in the solution. In spite of this, two similar (although not identical) kinds of dimer-dimer interfaces (related either by a crystallographic or a local dyad) are observed in the crystal structure of the unbound CopG (Fig. 4), resulting in the generation of a right-handed helical superstructure that encompasses six CopG dimers per turn (10). In contrast, the unliganded crystal structures of Arc and MetJ do not show these protein superstructures, indicating looser interdimer contacts in these repressors. Cocrystals of CopG bound to the operator central region, which encompasses the 13-bp pseudosymmetric inverted repeat (Fig. 3A), show a protein tetramer interacting through the β-ribbons with the major groove at two consecutive DNA helix turns on the same face of the DNA (5, 10). Dimers in this bound tetramer interact in the same way as the dimers related by a crystallographic dyad in the unbound crystal structure of CopG (Fig. 4). Despite the general resemblance between the crystal structures of CopG bound to DNA and those of Arc-operator complexes, CopG regions involved in dimer-dimer interactions span farther than those in Arc (compare Fig. 3A and B) and include residues located in the loops between the three regular secondary structure elements, in helix A, and in almost the entire length of helix B (10). This results in a dimer-dimer interaction surface that is almost twice as much as that observed in the Arc-operator complex. As a consequence of the strong dimer interpenetration, a 50 to 60° DNA bending is induced upon binding of the CopG tetramer, involving compression not only of the DNA minor groove in the center of the target (which is the case in the Arc-operator complex) but also of the major groove at two successive helical turns interacting with the protein (Fig. 3A). In the Arc-bound DNA, compression of the major groove facing the protein is prevented by interaction of the N-terminal 310 helix of Arc, so that the major grooves are even widened with respect to the idealized B-DNA (Fig. 3B). This different architecture of the Arc-operator complex with the looser dimer-dimer interaction results in an induced DNA bend of only 22°.
FIG. 4.
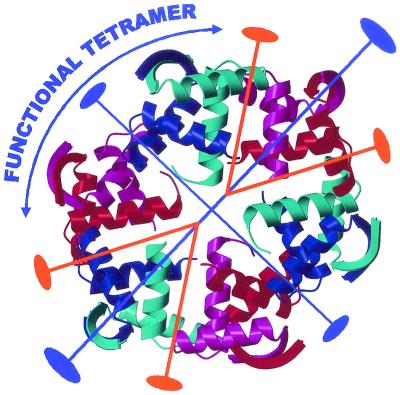
Oligomeric structure of CopG protein within the unbound crystals rendering a right-handed protein superhelix. The DNA-binding β-ribbons are exposed to the superhelical outer surface and are almost equidistant to each other (about 30 Å). Dimers, representing the minimal structural unit, are made up by red/magenta and cyan/blue monomers. The dimer of dimers making up the functional tetramer (as seen in the DNA complexes) is labeled. The protein oligomer is made up by crystallographic (blue) and local (orange) dyad axes.
The solved crystal structure of the CopG-DNA complex reveals only, unfortunately, the central region of the entire operator bound by CopG, and attempts at obtaining cocrystals of CopG complexed to a 55-bp oligonucleotide comprising the whole operator remain unsuccessful. Information on the interactions of CopG with the DNA regions adjacent to the inverted repeat arises from footprinting and preliminary stoichiometry analyses. The present model implies cooperative binding of four CopG dimers on the same face of the operator double helix, inducing a 120° global DNA bend towards the protein (Fig. 1C). The two central dimers correspond to the tetramer bound to the inverted repeat of the operator (as seen in the solved cocrystals), while one more dimer is bound at each side of this central tetramer interacting, by the same face of the DNA, with the major groove adjacent to the inverted repeat (10). Binding of the two outer CopG dimers seems to depend, directly or indirectly, on the nucleotide sequence, as it could be prevented by changing the DNA adjacent to the inverted repeat. Recombinant DNAs containing only the central region of the wild-type CopG operator are bound very inefficiently by the protein, suggesting that binding of the outer CopG dimers might stabilize the nucleoprotein complex by dimer-dimer cooperative interactions (G. del Solar, unpublished results). In this sense, cooperative interactions seem to have an even greater influence on the affinity of CopG binding than the strict conservation of the inverted repeat DNA sequence, as deduced from the rather slight effect arising from point mutations in this element (including some changes that affect CopG-contacted bases). In contrast, systematic mutation of each base in a double met box operator showed that the largest reduction in MetJ binding affinity results from mutations at bases that make direct contacts to the protein (23). Also, mutations in the TAGA boxes located at the Arc operator half-sites and involved in contacts with Arc have been found to be extremely deleterious to binding of this repressor (21).
Cop PROTEINS ENCODED BY PLASMIDS OF THE pMV158 FAMILY
A sequence alignment of CopG with the other Cop proteins of the pMV158 family of plasmids (Fig. 5) shows that they all display features that are compatible with a dimeric RHH structure. Up to 22 Cop proteins (out of the 24 members of the family) were found, although some of them were identical: those of pFX2, pWV01, and pSH71 (once a single mistake in the nucleotide sequence of the latter is assumed) and also those of pLF1311 and pLF14. A putative Cop protein can be proposed to be encoded by pLH2, assuming that a single mistake in the nucleotide sequence would result in an initiation codon for a cop gene. Only plasmids pHPK255 and pHP489 (both from Helicobacter pylori) seem to lack a cop gene, while the other plasmids of the pMV158 family would encode Cop proteins ranging in size between 44 and 56 residues. CopG from pMV158 and its closely related pSSU1-Cop, pLH2-Cop, and pSMQ172-Cop are the shortest proteins, perhaps constituting the minimal structure unit for these repressors. In the other Cop proteins, extensions at their N and/or C termini are observed (Fig. 5), so that involvement of these regions in additional contacts with the DNA (as is the case with N-terminal segments of Arc, MetJ, and Mnt) or in oligomerization cannot be discarded. All of these Cop sequences (with the exception of CopE of pE194) have the glycine-mediated turn connecting the two α-helices, and residues of similar nature are located at those positions involved in the maintenance of the hydrophobic core structure of the dimer (Fig. 5).
FIG. 5.
Plasmid-encoded CopG-like proteins. Cop proteins encoded by rolling circle-replicating plasmids of the pMV158 family (upper part) or encoded by other replicons (lower part) were aligned, taking as a fixed position the turn (boxed and shaded) in CopG, and not allowing any gap. Numbers in the upper part indicate the amino acid positions relative to those of CopG. The indicated secondary-structure regions correspond to those determined for CopG. Conserved hydrophobic positions (heavily shaded) and CopG residues that interact specifically with the DNA bases (•) are indicated. The indicated amino acid residues in plasmids pLH2 and pSH71 (boxed) would be present only if a mistake in the sequence were assumed. Plasmid-encoded proteins whose structure has been solved are indicated with asterisks. In the case of the ω protein of plasmid pSM19035, the first 21 residues (not visible in the crystal structure) are omitted. The accession numbers of the different plasmids can be found in reference 20.
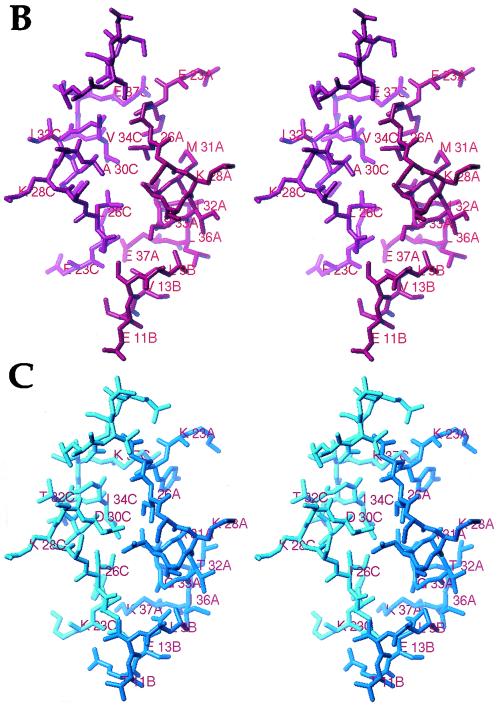
An interesting and still unanswered question arises on whether the dimer-dimer interface of these proteins is similar to that observed in the CopG cocrystals. If so, either conservation of residues at the relative positions constituting the dimer-dimer interaction surface or compensatory changes affecting both sides of this interface should be expected. The residues involved in interdimer interactions to render a functional tetramer mainly comprise positions 23 to 37 (molecule A) and 9 to 13 (molecule B) of one CopG dimer. They constitute, together with the same segments of molecules C and D, respectively, the contact surface (Fig. 6A). On inspecting these positions in the sequences of the Cop family members (Fig. 5), it can be seen that all cases are compatible with our experimental pMV158 CopG/cognate double-stranded DNA structure, so that they should be capable of building up structurally similar tetramers (Fig. 3A). Wherever nonconservative substitutions occur, these changes are compensated for by appropriate substitutions at other positions among those that make up the dimer-dimer interface. To address this issue in more detail, we have made a model of the most divergent Cop family member, CopE from pE194 (12). Among the residues engaged in interface shaping, only four residues are conserved relative to CopG, five are replaced by similar amino acids, and 11 substitutions are nonconservative (Fig. 5 and 6B and C). CopE is the only member of the family not displaying the characteristic Gly at the relative position 25 in the turn connecting the two helices of the RHH motif. If one looks at the modeled interface (Fig. 6), only one position appears that could produce a slight sterical hindrance, precisely around relative position 25, which is an Asn residue in CopE. All other amino acid substitutions are compatible with our structure. This substitution is, in principle, possible, as the main chain angles flanking this residue in the experimental structure (φ and Ψ = 54 and 40°) lie within a most-favored region of a Ramachandran plot, corresponding to a left-handed α-helix. In the model, the Cβ atoms of positions 10 and 25 would be just 2.5 Å from each other. But a minimum rearrangement of the main chain at either position could easily compensate for this. According to our model, the side chain of Asn in position 25 (in CopE) would be perfectly accommodated by the main chain of segment between relative positions 9 and 12 (Leu-Glu), with its Oδ1 hydrogen atom bonded by the main chain amide nitrogen atoms and with its Nδ2 atom interacting with Glu12 Oδ1, also different from the experimental CopG structure (Ser12). A further noteworthy change, Ala (CopG) to Asp (CopE), occurs at position 30, and the locations of these positions in both chains A and C are rather close to each other (the Cβ-to-Cβ distance is 4.5 Å). To minimize electrostatic repulsion of the side chains, a rotamer could be thought of where both carboxylate groups lie in parallel planes. These positions would be further stabilized by an interaction of Oδ1 of Asp in position 30 with Oγ of Ser in position 27, practically in the same rotamer position as that of CopG.
FIG.6.
Dimer-dimer interface of Cop proteins. (A) Interaction surface between dimers as observed in the functional tetramer of CopG. Each dimer is represented as a Cα plot and superimposed with its Connolly dot surface. The N-terminal methionine residues of each chain (termed A to D) are labeled. (B) Detail of the structure of functional CopG tetramers around the interface between the two homodimers (molecules A and B in red and C and D in magenta) displaying the chain segments mainly involved. Some of the residues are labeled. (C) The same region corresponding to the model made for CopE from pE194 (molecules A and B in blue and molecules C and D in cyan). After substitution of the side chains, only allowed rotamers were tested to find optimal interdigitation and favorable interactions.
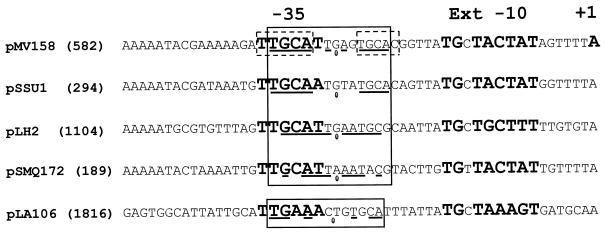
With regard to the interactions of Cop proteins with their cognate DNA targets, a subgroup of Cop proteins consisting of those from pMV158, pSSU1, pLH2, pSMQ172, and pLA106 can be clearly distinguished, based on the conservation of residues at relative positions that, in CopG, make contacts with the target DNA (Fig. 5). These residues are identical in the Cop proteins encoded by pMV158, pSSU1, pLH2, and pSMQ172, while a Ser is found instead of the equivalent conserved Thr at the relative position 8 in pLA106-Cop (Fig. 5). Conservation of amino acids involved in contacts with the target DNA should correlate with conservation of the Cop operator sequences in these five plasmids. This prompted us to try to locate these operators under the following constraints: (i) they must be placed in a region overlapping the corresponding putative cop-rep promoters, and (ii) they should have the 5′-TGCA-3′ half-sites within a pseudosymmetric 13-bp element. Boxes −35 and −10 of putative cop-rep promoters had been proposed for plasmids pSSU1, pSMQ172, and pLA106 (GenBank accession numbers AB019522, AF295100, and D88438, respectively). However, and with the above criteria, we could locate a promoter different from that proposed for pSMQ172 and could find the putative −10 and −35 boxes of the pLH2 cop-rep promoter (see below). All these promoters would contain an extended −10 box, as is the case with the pMV158 Pcr promoter (22). As expected, each of these plasmids was found to possess a 12- or 13-bp pseudosymmetric element that overlaps the proposed −35 box with the same relative location as that of the inverted repeat of pMV158 (Fig. 7). These elements also contain, at either one or both half-sites, the 5′-TGCA-3′ sequence, which, in pMV158, is involved in specific interactions with CopG. The dissimilarities of the Cop operators of pSSU1, pLH2, pSMQ172, and pLA106 with respect to the pMV158-CopG operator are not so unexpected if we take into account that some of these changes (even those affecting bases contacted by the protein) have been found not to impair significantly the binding of CopG to its target DNA (del Solar, unpublished). A search for copG-like genes in the sequenced bacterial genomes did not show any significant homology. In addition, we did not find any sequence similar to the 13-bp pseudosymmetric element, the primary target of CopG.
FIG. 7.
Proposed cop-rep promoters and Cop operators for plasmids closely related to pMV158. The positions of the −35 and −10 regions of the Pcr promoter and the transcription initiation site of the cop-rep mRNA of pMV158 are in bold. Also in bold are the putative cop-rep promoters for the other plasmids. The pseudosymmetric elements (boxed), the bases participating in the symmetry (underlined), and the center of symmetry (ellipse) are indicated. The broken-lined boxes on the pMV158 sequence indicate positions where the displayed base and/or its complement is involved in direct contacts with residues of CopG as seen in the cocrystal of the protein-DNA complex.
OTHER PLASMID-ENCODED CopG-LIKE PROTEINS
Although CopG is the best known plasmid-encoded RHH protein, the structures of the plasmid-encoded repressor proteins ω and ParD have recently been determined and shown to belong to the same RHH superfamily (17, 18). A search for proteins with structure and function similar to that of CopG in plasmids unrelated to pMV158 revealed the existence of four more proteins encoded by the rolling circle-replicating plasmids pHD2, pRN1, pRN2, and pDL10, the last three of which have been isolated from archaeal hosts (references 14 and 15 and references therein). Sequence similarities between CopG and these proteins are much lower than those observed among the Cop proteins of the pMV158 plasmid family, and the involvement of these proteins in the control of plasmid replication remains to be proved. However, the Cop protein of plasmid pRN1 binds to a DNA region containing the putative cop-rep promoter (14). Based on sequence homology, the TraY and TrwA proteins of conjugative plasmids have been proposed to contain the RHH motif (3, 16), thus increasing the number of plasmid-encoded proteins belonging to the Arc/MetJ superfamily from the small number reported earlier (19) to the 30 proteins reported here (Fig. 5). We conclude that this class of proteins is likely to be far more widespread. Because they are encoded by very small replicons, like the 1,717-bp mycoplasma plasmid pADB201 (2), these proteins may represent an efficient solution found to problems of gene regulation, such as the delicate function of controlling the synthesis of an essential initiator.
Acknowledgments
Thanks are due to Jose A. Ruiz-Masó for his help and to members of our labs for discussions.
Research was funded by European Union-Ministerio de Ciencia y Tecnología (MCyt) grant 2FD97-0518 (to M.C. and M.E.) and by grants from MCyT (BMC2000-0550 to M.E., PB98-1631 to M.C., and BIO2000-1659 to F.X.G.-R.), the Generalitat de Catalunya (SGR188 to M.C.), and the Comunidad de Madrid (CAM 07/B/49/99 to G.D.S.).
REFERENCES
- 1.Acebo, P., M. Garcia de Lacoba, G. Rivas, J. M. Andreu, M. Espinosa, and G. del Solar. 1998. Structural features of the plasmid pMV158-encoded transcriptional repressor CopG, a protein sharing similarities with both helix-turn-helix and β-sheet DNA binding proteins. Proteins Struct. Funct. Genet. 32:248-261. [DOI] [PubMed] [Google Scholar]
- 2.Bergemann, A. D., J. C. Whitley, and L. R. Finch. 1989. Homology of mycoplasma plasmid pADB201 and staphylococcal plasmid pE194. J. Bacteriol. 171:593-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breg, J. N., J. H. J. van Ophesden, M. J. Burgering, R. Boelens, and R. Kaptein. 1990. Structure of Arc repressor in solution: evidence for a family of β-sheet DNA-binding proteins. Nature 346:586-589. [DOI] [PubMed] [Google Scholar]
- 4.Burgering, M. J. M., R. Boelens, D. E. Gilbert, J. N. Breg, K. L. Knight, R. T. Sauer, and R. Kaptein. 1994. Solution structure of dimeric Mnt repressor (1-76). Biochemistry 33:15036-15045. [DOI] [PubMed] [Google Scholar]
- 5.Costa, M., M. Solá, G. del Solar, R. Eritja, A. M. Hernández-Arriaga, M. Espinosa, F. X. Gomis-Rüth, and M. Coll. 2001. Plasmid transcriptional repressor CopG oligomerises to render helical superstructures unbound and in complexes with oligonucleotides. J. Mol. Biol. 310:403-417. [DOI] [PubMed] [Google Scholar]
- 6.del Solar, G., P. Acebo, and M. Espinosa. 1995. Replication control of plasmid pLS1: efficient regulation of plasmid copy number is exerted by the combined action of two plasmid components, CopG and RNA II. Mol. Microbiol. 18:913-924. [DOI] [PubMed] [Google Scholar]
- 7.del Solar, G., and M. Espinosa. 1992. The copy number of plasmid pLS1 is regulated by two trans-acting plasmid products: the antisense RNA II and the repressor protein, RepA. Mol. Microbiol. 6:83-94. [DOI] [PubMed] [Google Scholar]
- 8.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarría, M. Espinosa, and R. Díaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Solar, G., J. Pérez-Martín, and M. Espinosa. 1990. Plasmid pLS1-encoded RepA protein regulates transcription from repAB promoter by binding to a DNA sequence containing a 13-base pair symmetric element. J. Biol. Chem. 265:12569-12575. [PubMed] [Google Scholar]
- 10.Gomis-Rüth, F. X., M. Solá, P. Acebo, A. Párraga, A. Guasch, R. Eritja, A. González, M. Espinosa, G. del Solar, and M. Coll. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 17:7404-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight, K. L., and R. T. Sauer. 1989. Identification of functionally important residues in the DNA binding region of the Mnt repressor. J. Biol. Chem. 264:13706-13710. [PubMed] [Google Scholar]
- 12.Kwak, J.-H., and B. Weisblum. 1994. Regulation of plasmid pE194 replication: control of cop-repF operon by Cop and of repF translation by countertranscript RNA. J. Bacteriol. 176:5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacks, S. A., P. López, B. Greenberg, and M. Espinosa. 1986. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J. Mol. Biol. 192:753-765. [DOI] [PubMed] [Google Scholar]
- 14.Lipps, G., M. Stegert, and G. Krauss. 2001. Thermostable and site-specific DNA binding of the gene product ORF56 from the Sulfolobus islandicus plasmid pRN1, a putative archaeal plasmid copy control protein. Nucleic Acids Res. 29:904-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDowell, D. G., and N. H. Mann. 1991. Characterization and sequence analysis of a small plasmid from Bacillus thuringiensis var. kurstaki strain HD1-DIPEL. Plasmid 25:113-120. [DOI] [PubMed] [Google Scholar]
- 16.Moncalián, G., G. Grandoso, M. Llosa, and F. de la Cruz. 1997. OriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J. Mol. Biol. 270:188-200. [DOI] [PubMed] [Google Scholar]
- 17.Murayama, K., P. Orth, A. B. de la Hoz, J. C. Alonso, and W. Saenger. 2001. Crystal structure of ω transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 Å resolution. J. Mol. Biol. 314:789-796. [DOI] [PubMed] [Google Scholar]
- 18.Oberer, M., K. Zangger, S. Prytulla, and W. Keller. 2002. The anti-toxin ParD of plasmid RK2 consists of two structurally distinct moieties and belongs to the ribbon-helix-helix family of DNA binding proteins. Biochem. J. 361:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pabo, C. O., and R. T. Sauer. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61:1053-1095. [DOI] [PubMed] [Google Scholar]
- 20.Rafferty, J. B., W. S. Somers, I. Saint-Girons, and S. E. V. Phillips. 1989. Three dimensional crystal structures of Escherichia coli Met repressor with and without corepressor. Nature 341:705-710. [DOI] [PubMed] [Google Scholar]
- 21.Raumann, B. E., M. A. Rould, C. O. Pabo, and R. T. Sauer. 1994. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature 367:754-757. [DOI] [PubMed] [Google Scholar]
- 22.Sabelnikov, A. G., B. Greenberg, and S. A. Lacks. 1995. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250:144-155. [DOI] [PubMed] [Google Scholar]
- 23.Somers, W. S., and S. E. V. Phillips. 1992. Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature 359:387-393. [DOI] [PubMed] [Google Scholar]