Abstract
The DNA region encoding the mature form of a pneumococcal murein hydrolase (LytB) was cloned and expressed in Escherichia coli. LytB was purified by affinity chromatography, and its activity was suggested to be the first identified endo-β-N-acetylglucosaminidase of Streptococcus pneumoniae. LytB can remove a maximum of only 25% of the radioactivity from [3H]choline-labeled pneumococcal cell walls in in vitro assays. Inactivation of the lytB gene of wild-type strain R6 (R6B mutant) led to the formation of long chains but did not affect either total cell wall hydrolytic activity at the stationary phase of growth or development of genetic competence. Longer chains were formed when the lytB mutation was introduced into the M31 strain (M31B mutant), which harbors a complete deletion of lytA, which codes for the major autolysin. Furthermore, the use of this mutant revealed that LytB is the first nonautolytic murein hydrolase of pneumococcus. Purified LytB added to pneumococcal cultures of R6B or M31B was capable of dispersing, in a dose-dependent manner, the long chains characteristic of these mutants into diplococci or short chains, the typical morphology of R6 and M31 strains, respectively. In vitro acetylation of purified pneumococcal cell walls did not affect the activity of LytB, whereas that of the LytA amidase was drastically reduced. On the other hand, the use of a translational fusion between the gene (gfp) coding for the green fluorescent protein (GFP) and lytB supports the notion that LytB accumulates in the cell poles of either the wild-type R6, lytB mutants, or ethanolamine-containing cells (EA cells). The GFP-LytB fusion protein was also able to unchain the lytB mutants but not the EA cells. In contrast, translational fusion protein GFP-LytA preferentially bound to the equatorial regions of choline-containing cells but did not affect their average chain length. These observations suggest the existence of specific receptors for LytB that are positioned at the polar region on the pneumococcal surface, allowing localized peptidoglycan hydrolysis and separation of the daughter cells.
Cell wall hydrolases have been found in practically all known eubacteria. Usually, bacteria have several murein hydrolases that can degrade or remodel the cell wall. These enzymes are classified as muramidases (lysozymes), glucosaminidases, amidases, endopeptidases, and transglycosylases according to the specific bond that is split down in the cell wall. The ubiquitous presence of these enzymes in microorganisms together with their potential function in restructuring the cell wall has suggested many important roles for them (28, 40). Nevertheless, only a limited number of murein hydrolases have been purified, mainly due to their presence in small amounts and/or their high-affinity binding to the cell wall.
Streptococcus pneumoniae is the leading cause of pneumonia and bloodstream infections in the elderly and is one of the main causes of middle ear infections in children. In some parts of the United States resistance to penicillin is as high as 20 to 40%, and, in aggregate, infectious diseases are currently the third-leading cause of death (33). LytA amidase, the major murein hydrolase of S. pneumoniae, is considered an important virulence factor (34). LytA has been well characterized biochemically (22), and the lytA gene is the first example of a gene coding for a bacterial peptidoglycan hydrolase for which molecular characterization has been performed (17). Comparative analysis of the pneumococcal amidase with the lytic enzymes encoded by several pneumococcal phages led us to propose that these cell wall lytic enzymes could be the result of the fusion of two independently active domains (12, 18). Experimental support for this proposal required the construction of active, chimeric phage-bacterium lytic enzymes (7, 8) to demonstrate that the carboxy-terminal domain (choline-binding domain [ChBD]) is responsible for the specific recognition of choline-containing cell walls of pneumococcus whereas the catalytic centers of these enzymes are localized in the amino-terminal parts of the proteins. In spite of the abundance of LytA amidase in S. pneumoniae, from laboratory assays the biological role of this enzyme appears to be quite limited. A mutant lacking lytA (M31) can grow normally, forming small chains (6 to 10 cells in length), and autolysis at the stationary phase of growth is abolished when the mutant is incubated at 37°C (37). Interestingly, M31 displays another autolytic activity, active when incubated at 30°C, which has been recently ascribed to the first lysozyme, LytC, reported in pneumococcus (19). Insights into the strong affinity of these enzymes for the choline cell wall substrate have been recently provided by the elucidation of the crystal structure of the ChBD of LytA, which has revealed a peculiar solenoid-like structure (11).
A critical question remaining to be investigated in this system concerns the kind of separation that might reduce the spreading of strains during colonization and whether some of these strains have an enhanced capacity to disseminate globally (33). In a preliminary account, we reported the construction of a lytB mutant strain of S. pneumoniae characterized by long chains of cells and indicated that the activity of LytB might be responsible for the ultimate physical separation of daughter cells, the final event of the cell division cycle (20). In this study we cloned and expressed the lytB gene, purified LytB, and localized this enzyme at the cell poles. This polar position of the LytB enzyme might be indicative of a critical role in cell separation.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. pneumoniae was grown in C medium (25) supplemented with yeast extract (0.8 mg/ml; Difco Laboratories; C+Y medium) without shaking. Growth was monitored with a Hach 2100N nephelometer. Ethanolamine (EA)-containing pneumococci (EA cells) were grown for at least 10 generations in a chemically defined medium (Cden-EA) (32). Escherichia coli was grown in Luria-Bertani medium with shaking. The transformation procedures for S. pneumoniae (37) and E. coli (36) were as described previously.
TABLE 1.
Bacterial strains, plasmids, and primers
| Material | Descriptiona | Reference or source |
|---|---|---|
| Bacterial strains | ||
| S. pneumoniae | ||
| R6 | Wild type | Rockefeller University |
| M31 | ΔlytA R6 mutant | 37 |
| R6B | R6 (lytB::ermC) | 20 |
| M31C | M31 (lytC::ermC) | 19 |
| M31B | M31 (lytB::ermC) | 20 |
| E. coli | ||
| DH5α | supE44 hsdR17 recA1 endA1 ΔlacU169(Δ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 36 |
| BL21(DE3) | F−bmpT gal [cdm] [lon] hsdSB with DE3 | 43 |
| DH10B | F′mcrA Δ(mrr hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK λ−rpsL endA1 nupG | Life Technologies |
| Plasmids | ||
| pT7-7 | Apr | 45 |
| pGL100 | lytA+, Apr | 15 |
| pRGR5 | lytB+, Apr | This study |
| pUCE191 | Lnr/Err Apr | 1 |
| pGreenTIR | gfp, Apr | 31 |
| pRGR25 | gfp, Apr | This study |
| pRGR25B | gfp lytB+, Apr | This study |
| PRGR25A | gfp lytA+, Apr | This study |
| Primers | ||
| LytB-N1 | GGAAGAAGGCATATGAAGAAAGTAAGATTTATTTTTTTAGC | This study |
| LytB-N2 | GGGTGCACATATGAGTGATGGTACTTGGCAAGGAAAACAG | This study |
| LytB-C | CCGAATTCTTACTAATCTTTGCCACCTAGC | This study |
| GFP51 | GGAATTCGCGGTACCAGTAAAGGAGAAGAACTTTTC | This study |
| GFP52 | AACTGCAGTTTAGGATCCTTTGTATAGTTCATCCATGCC | This study |
| LytB51 | CGGGATCCAGTGATGGTACTTGGCAAGG | This study |
| LytB52 | AACTGCAGTTAATCTTTGCCACCTAGCTTC | This study |
| LytA51 | CGGGATCCGAAATTAATGTGAGTAAATTAAG | This study |
| LytA52 | CAACTGCAGTTATTTTACTGTAATCAAGCCCATCTGGC | This study |
Ap, ampicillin; Ln, lincomycin; Er, erythromycin. Primer sequences are 5′ to 3′. The nucleotides underlined denote the restriction sites.
Preparation of choline-binding proteins (ChBPs) from autolytic pneumococcal walls.
To purify the ChBPs bound to the cell envelope, we followed the procedure described by García et al. (19). In short, an exponential culture of the M31 strain was disrupted in a French pressure cell press (American Instrument Company) and centrifuged, and the pellet was washed twice in 50 mM potassium phosphate buffer (pH 6.5) containing 10 mM MgCl2 and 0.1% Brij 58. After centrifugation, the pellet was resuspended in the same buffer containing 0.1% choline chloride (autolytic walls) and allowed to autolyze by overnight incubation at 30°C. Afterwards, the concentration of choline chloride was raised to 2%, and, after incubation at 4°C for 15 min, the mixture was ultracentrifuged (120,000 × g, 1 h, 4°C). The supernatant was dialyzed against 50 mM potassium phosphate buffer (pH 6.5) applied to a DEAE-cellulose column and washed with 1.5 M NaCl. ChBPs were eluted from the column upon addition of 2% choline chloride (19).
Cloning, nucleotide sequencing, computer analyses, and plasmid constructions.
Routine DNA manipulations were performed essentially as described previously (36). DNA fragments were purified by using the Geneclean II kit (Bio 101). The relevant oligonucleotide primers used are listed in Table 1. DNA sequences were determined by the dideoxy chain termination method (39) with an automated Prism 377 DNA sequencer (Applied Biosystems). All primers for PCR amplification and nucleotide sequencing were synthesized on a Beckman model Oligo 1000 M synthesizer. Amino acid sequences were analyzed with the Protein Analysis Tool at the World Wide Web molecular biology server of the Geneva University Hospital and the University of Geneva. Protein sequence similarity searches were done with the BLASTP program via the National Institute for Biotechnology Information server. Pairwise and multiple protein sequence alignments were done with the ALIGN and CLUSTAL W programs, respectively, at the Baylor College of Medicine Human Genome Center server. To construct pRGR5, we amplified by PCR an M31 DNA fragment by using oligonucleotides LytB-N2 and LytB-C. The resulting fragment was digested with NdeI and EcoRI and ligated to pT7-7, previously treated with the same enzymes. Recombinant plasmid pRGR5, which expressed the mature form of LytB, was selected among the ampicillin-resistant transformants of E. coli DH5α. The accuracy of the construction was checked by restriction analysis, and pRGR5 was retransformed into E. coli BL21(DE3) (Fig. 1A). R6B and M31B mutants were obtained by insertion duplication mutagenesis, as described elsewhere (19). To construct pRGR25B harboring the fusion gfp-lytB gene, we amplified the gfp gene, coding for the green fluorescent protein (GFP), from pGreenTIR using oligonucleotides GFP51 and GFP52. The resulting fragment was digested with EcoRI and PstI and ligated to pT7-7 cut with the same enzymes. The plasmid obtained, pRGR25, was digested with BamHI and PstI and ligated to the PCR-amplified lytB fragment from M31 DNA with oligonucleotides LYTB51 and LYTB52 and digested with the same enzymes. The accuracy of the construction was checked, and pRGR25B was retransformed into E. coli BL21(DE3). Similarly, the lytA gene was amplified by PCR from R6 DNA with oligonucleotides LYTA51 and LYTA52 and digested with BamHI and PstI. Afterwards, this fragment was ligated to pRGR25 digested with the same enzymes, and pRGR25B, coding for fusion GFP-LytA, was obtained.
FIG. 1.
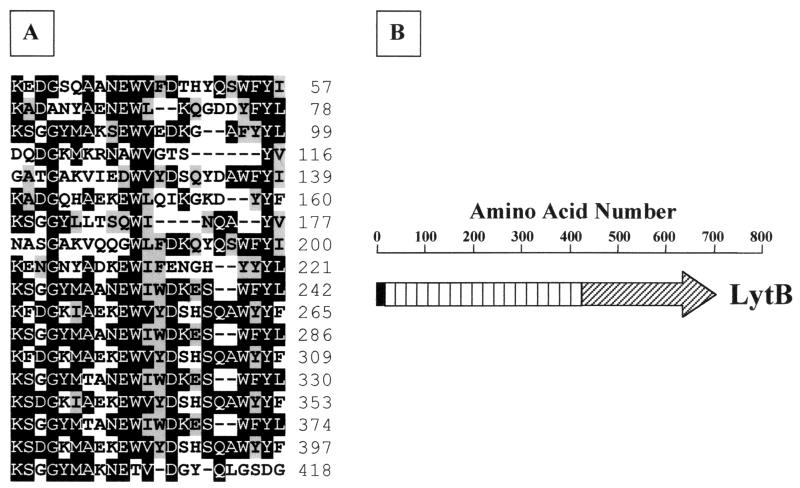
Schematic representation of LytB and self-alignment of the N-terminal part of the deduced sequence of LytB. (A) Residues highlighted in black are identical in at least 7 motifs; grey highlighting indicates conserved substitutions. Numbers at the right correspond to the amino acid positions. (B) Black box, signal peptide; white boxes, repeat motifs; hatched region, catalytic domain.
Purification of the mature LytB.
E. coli BL21(DE3)(pRGR5) was incubated in Luria-Bertani medium containing ampicillin (0.1 mg/ml) up to an A600 of 0.8. At this time, isopropyl-β-d-thiogalactopyranoside (50 μM) was added, and incubation proceeded for 6 h at 25°C to minimize the presence of inclusion bodies. The culture was centrifuged (10,000 × g, 5 min), and the pelleted bacteria were resuspended in 20 mM sodium phosphate buffer (pH 7.0) and disrupted in a French pressure cell press. The insoluble fraction was separated by centrifugation (15,000 × g, 15 min), and the supernatant was loaded onto a DEAE-cellulose column to purify, in a single step, the LytB protein according to the procedure previously described for other ChBPs (38).
Miscellaneous methods.
A polyclonal anti-LytB serum was raised in rabbits by using the purified LytB protein as described previously (13) and diluted 1/1,000 for Western blot analysis (36). Sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis was carried out with the system described by Laemmli (26), and protein bands were visualized by staining with Coomassie brilliant blue R250. DNA probes were labeled with the DIG luminescence detection kit (Boehringer Mannheim). Southern blots and hybridizations were carried out according to the manufacturer's instructions. Pneumococcal cell walls were radioactively labeled with [methyl-3H]choline, [3H]lysine, or [14C]EA, and their cell wall enzymatic degradation products were separated by Sephadex G-75 chromatography (32). The standard assay conditions for the hydrolysis of the labeled pneumococcal cell walls have been described elsewhere (32). Preparation of subcellular fractions was carried out as described previously (19). N-terminal amino acid sequence determination was done according to a published procedure (41). Observation of the cell morphology of S. pneumoniae was carried out, unless otherwise stated, at the mid-log phase of growth, and the cells were photographed. N acetylation of cell walls with acetic anhydride was performed according to a procedure recently published (51). Fluorescence microscopy was directly analyzed by phase-contrast or epifluorescence microscopy with a Zeiss Axioplan Universal microscope with excitation standard fluorescein isothiocyanate set D480/30 and emission TBP 460/530/610 filters.
Transmission electron microscopy.
S. pneumoniae strain M31B cells were grown to the late exponential phase in C+Y medium, centrifuged, washed in 0.1 M potassium phosphate buffer (pH 7.1) (KP buffer), and prefixed with 2.5% glutaraldehyde for 30 min at 0°C. Afterwards, the cells were centrifuged, washed several times in KP buffer, and fixed with 1% OsO4 for 2 h at room temperature. Then, cells were washed by centrifugation, added to agar, and dehydrated in increasing concentrations of ethanol. Cells were embedded in Spurr's resin (42), and ultrathin sections were stained with lead citrate (35) and examined in a Philips EM 300 electron microscope working at 80 kV.
RESULTS
Cloning and expression of lytB.
In a search of the pneumococcal ChBPs, and particularly those tightly bound to the cell envelope, we purified four protein bands with strong choline affinity from strain M31, which lacks the major LytA autolysin (19). One of these bands was excised from the polyacrylamide gel, and the N-terminal sequence was determined. Comparison of this sequence with the translated version of the nucleotide sequence of the S. pneumoniae type 4 genome (strain TIGR4) allowed the localization of a gene, designated lytB (20), between coordinates 804737 and 806713 in the genetic map according to The Institute of Genome Research (46). A 2-kb DNA fragment embracing the lytB gene was amplified by PCR using oligonucleotides LytB-N1 and LytB-C and M31 DNA as a template. The sequence of this fragment revealed that the lytB gene of strain R6 was larger than the lytB gene reported for strain TIGR4. This result agrees with the sequence of the R6 genome recently published (23). This observation could suggest the existence of polymorphism in this gene (23, 46, 52). Nevertheless, the LytB sequences already published need careful reexamination for misread overlaps in view of the large number of highly similar motifs present in this protein.
The amplified lytB gene of strain R6 codes for a putative 81.9-kDa protein (LytB) that, together with the recently characterized LytC, displays a modular organization different from those of all the ChBPs described previously. The ChBD was found in the N-terminal region, whereas the active site of the enzyme was localized in the C-terminal domain. In addition, both enzymes contain cleavable signal peptides, in contrast to the other pneumococcal and phage cell wall lytic enzymes (19). For LytB, the ChBD is built up of 18 imperfect repeat motifs. We have observed that 9 out of these 18 motifs, located more C terminal in the binding domain of the LytB protein (see Fig. 8A), were highly similar to the canonical ChBD motif of C-LytA recently determined by crystallographic analysis (11) whereas the other most N-terminal ones exhibit a higher number of replacements (Fig. 1).
FIG.8.
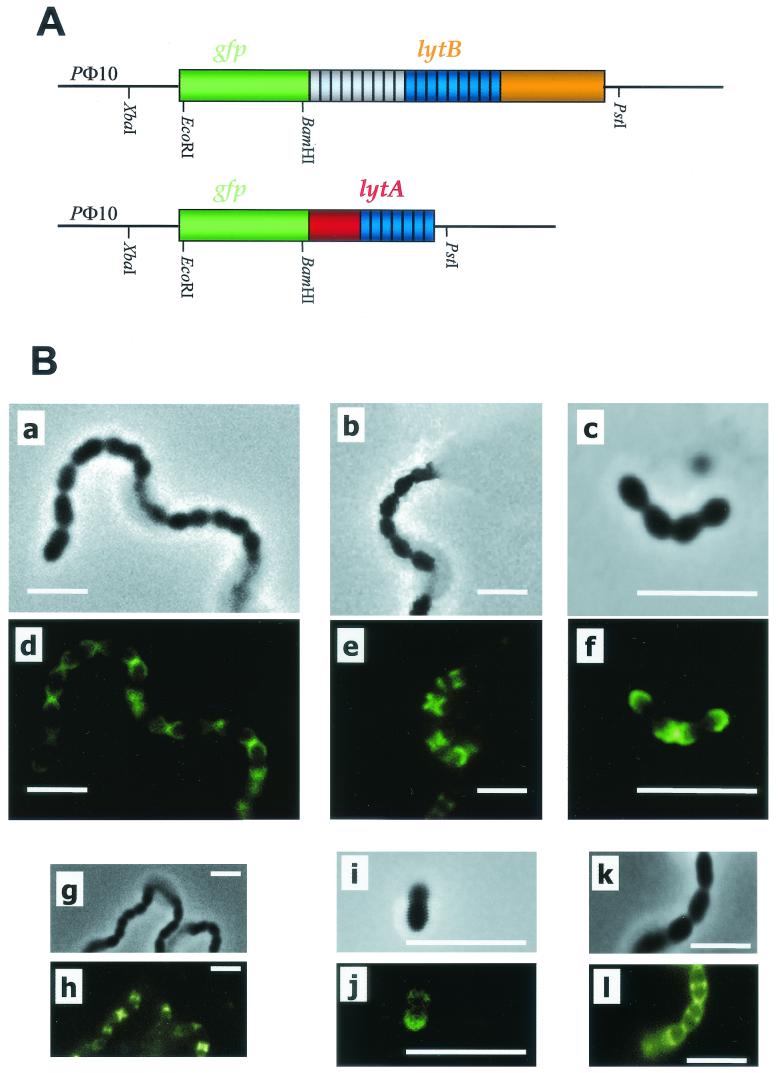
Localization of GFP-LytB and GFP-LytA at the surface of S. pneumoniae. (A) Schematic representation of the translational fusions between gfp and the mature form of lytB or lytA. PΦ10, promoter of gene 10 of the T7 phage. Dark blue shading indicates similar repeat motifs between lytB and lytA. (B) Phase-contrast microscopy examination and the corresponding fluorescence images. Cultures of S. pneumoniae R6B received fusion protein GFP-LytB (9.5 μg/ml) and were incubated at 37°C. Samples were taken at 1, 20, and 30 min, and pictures were taken with a Nikon Eclipse inverted microscope in phase contrast (a, b, and c, respectively) and using fluorescence (d, e, and f, respectively). Pictures g and h show the result of incubation of the same cells grown in EA medium (R6B-EA) with GFP-LytB, and pictures i and j correspond to the wild-type R6 strain incubated with the fusion protein for 30 min at 37°C. Pictures k and l show the result of incubation of M31B cells with GFP-LytA. Bars, 4 μm.
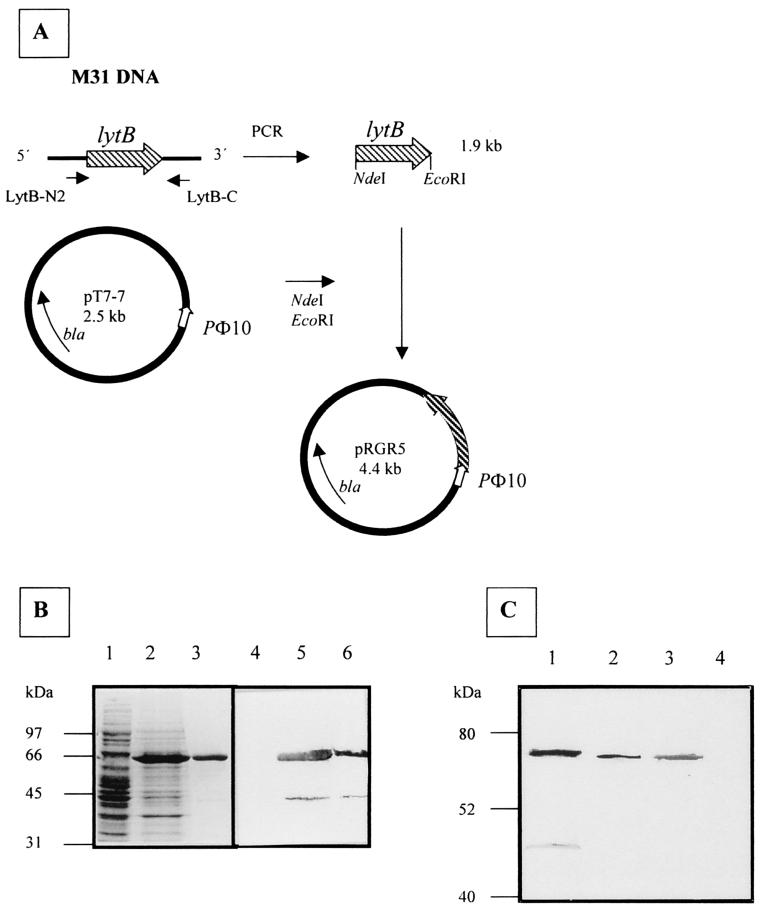
Initially we attempted to clone and express both forms of the lytB gene, encoding either the mature or the unprocessed forms of the LytB protein, using pT7-7 as an inducible expression vector. However, only the mature form could be expressed in E. coli. The strategy used to construct pRGR5 is detailed in Materials and Methods and depicted in Fig. 2A. The mature form of LytB was purified with DEAE-cellulose, and its N-terminal analysis (Ser-Asp-Gly-Thr-Trp-Gln-Gly) confirmed that it corresponded to the expected protein. Interestingly, we observed that in some purification experiments a minor band of about 43 kDa (Fig. 2B, lane 3) eluted together with LytB. When these samples were subjected to a Western blot analysis using anti-LytB serum, both the large (LytB) and the small proteins appeared as clear bands. In fact, the analysis of the N-terminal residues of the 43-kDa protein (Thr-Ala-Asn-Glu-Trp-Ile-Trp-Asp-Lys) strongly suggested that it corresponded to an internal restart of LytB, beginning at position Thr-316 and preceded by an ATG and a ribosome-binding site-like sequence, probably due to overexpression of lytB gene. However, we cannot completely rule out the possibility that the 43-kDa protein resulted from a proteolytic degradation. The comparative Western blot analysis of LytB production in E. coli and the presence of this protein in pneumococcal strains with mutations affecting different murein hydrolases revealed that LytB was absent in the R6B strain. Also, the only species of LytB present in S. pneumoniae corresponded to the high-molecular-weight form found when lytB was overexpressed in E. coli (Fig. 2C).
FIG. 2.
Schematic representation of the construction of pRGR5 and purification of LytB. (A) Plasmids are drawn as circles. PΦ10, promoter of gene 10 of the T7 phage; bla, gene coding the β-lactamase. LytB-N2 and LytB-C are the oligonucleotides used to amplify the lytB gene. (B) Sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gel stained with Coomassie blue (left) and the corresponding Western blot obtained by using a polyclonal antiserum raised against LytB (right). Lanes 1 and 4, crude extracts from BL21(DE3)(pT7-7); lanes 2 and 5, crude extracts from BL21(DE3)(pRGR5); lanes 3 and 6, purified LytB. Standard size markers are indicated on the left. (C) Western blot obtained by using the same LytB antiserum. Lane 1, purified LytB; lane 2, crude extract from M31; lane 3, crude extract from M31C; lane 4, crude extract from M31B. Standard size markers are indicated on the left.
Biochemical properties of LytB.
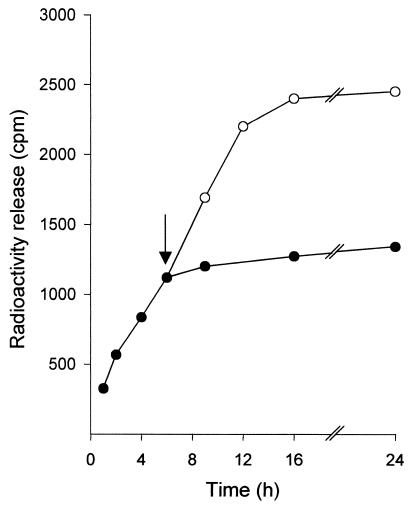
LytB was capable of degrading choline-containing pneumococcal cell walls in a standard in vitro assay. Nevertheless, the degradation reaction of this substrate by LytB was rather atypical because the rate of choline release was slow, reaching a maximum after 6 h incubation, when the liberation of about 25% of the choline residues was achieved (Fig. 3). This is in remarkable contrast with the noticeable activity reported for amidase LytA and for lysozyme LytC, two well-characterized murein hydrolases. Interestingly, LytB was not inactivated at the end of total choline release from cell walls, since addition of new substrate to the reaction mixture resulted in an increase of cell wall degradation. This limited degradation might indicate a restricted recognition and/or degradation of the substrate, probably focused in a selected region of the cell wall (see below). The same behavior was shown by LytB tested on [3H]lysine cell walls; however, there was no detectable degradation of [14C]EA cell walls (data not presented). This enzymatic characteristic leads to an extremely low specific activity compared with the values for the other two murein hydrolases characterized in the pneumococcal system. For comparison, several relevant traits of autolysins LytA and LytC, as well as phage lysozyme Cpl1 (14), are summarized in Table 2. The LytB activity was maximal at 37°C and pH 7.0 in 20 mM phosphate buffer.
FIG. 3.
Hydrolysis of [3H]choline-labeled cell walls with LytB. Hydrolytic activity was assayed at 37°C using incubation mixtures containing 10 μl of purified LytB (30.4 U/mg) with 10 μl of [3H]choline-labeled cell walls (5,500 cpm) as the substrate and 240 μl of 20 mM phosphate buffer, pH 7.0. At the time indicated (arrow) several tubes received fresh substrate (10 μl) (○).
TABLE 2.
Comparison of the biochemical properties of the purified LytB, LytA, LytC, and Cpl1 enzymes
| Catalytic activity | Optimum
|
Choline 50% inhibitory concn (mM) | Autolysis | Signal peptide | Sp act (U/mg) | |
|---|---|---|---|---|---|---|
| Temp (°C) | pH | |||||
| LytB (glucosaminidase) | 37 | 7.0 | 5 | No | Yes | 30.4 |
| LytA (amidase) | 37 | 7.0 | 24 | Yes | No | 2.5 × 105 |
| LytC (lysozyme) | 30 | 6.0 | 30 | Yes | Yes | 6.0 × 103 |
| Cpl1 (lysozyme) | 37 | 6.0 | 2 | Yes | No | 1.2 × 105 |
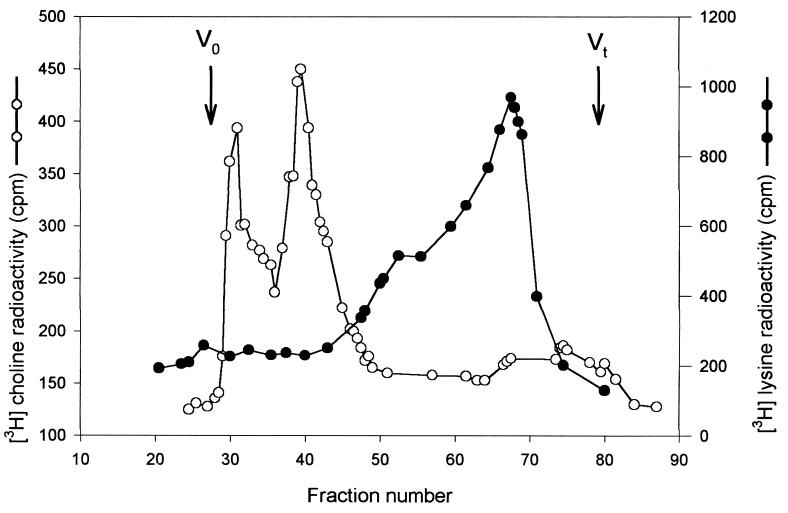
Based on sequence comparison, LytB was proposed to be a glycosidase (20); however this property required confirmation by biochemical analysis. For this purpose we used a well-established method to distinguish pneumococcal murein hydrolases (32). The elution profile of the degradation products obtained from radioactively labeled pneumococcal cell walls digested in vitro with LytB ruled out the possibility that LytB was an amidase or an endopeptidase. However, the elution pattern of [3H]choline-labeled cell walls (Fig. 4) was fully consistent with that of glycosidase, which produces the products retained in the column, in contrast with the excluded products produced by amidases (32).
FIG. 4.
Hydrolysis of [3H]choline- and [3H]lysine-labeled pneumococcal cell walls by LytB. Two hundred microliters of [3H]choline-labeled (○) or [3H] lysine-labeled (•) cell walls and 100 μl of 20 mM phosphate buffer, pH 7.0, were incubated at 37°C for 18 h with 200 μl of the purified LytB (30.4 U/mg). After centrifugation (12,000 × g, 15 min), 450 μl of the supernatant was applied on a Sephadex G-75 column (2 by 45 cm). The elution was carried out with 0.15 M NaCl. Samples (1 ml) of each fraction were measured for radioactivity (500 μl). V0, void volume; Vt, total volume.
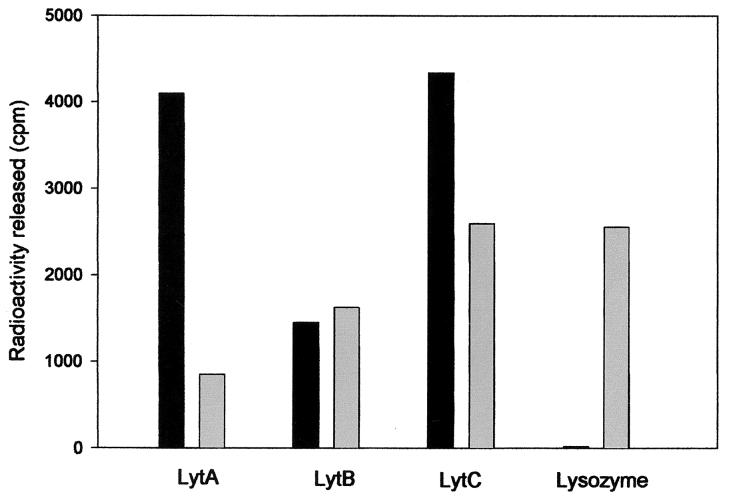
Vollmer and Tomasz (51) have recently characterized a deacetylase gene in pneumococcus which appears to be responsible for the “fine tuning” of partial acetylation of hexosamine residues of the cell wall. Interestingly, the use of N-acetylated cell walls as a substrate revealed that the in vitro activity of LytB was similar to that detected by using cell walls prepared from R6. In contrast, the activity of LytA was practically eliminated and that of LytC was also reduced when acetylated walls were used as the substrate (Fig. 5). As a positive control we also observed that the acetylated cell walls were fully degraded by chicken egg white lysozyme as previously found (51).
FIG. 5.
Effect of chemical acetylation on the susceptibility of pneumococcal peptidoglycan to enzymatic digestion. [3H]choline-labeled cell walls (10 μl), either nonmodified (black bars) or chemically acetylated with acetic anhydride (grey bars), were incubated with 10 μl of LytA (2.5 × 105 U/mg in 240 μl of 20 mM phosphate buffer, pH 7.0) or 10 μl of LytB (30.4 U/mg in 240 μl of 20 mM phosphate buffer, pH 7.0) or 10 μl of LytC (6.0 × 103 U/mg in 240 μl of 20 mM phosphate buffer, pH 6.0) or 10 μl of chicken egg white lysozyme (20 μg/ml in 240 μl of 20 mM phosphate buffer, pH 5.5). Incubations were carried out at 37°C for 20 min (LytA and chicken egg white lysozyme) or at 30°C for 30 min (LytC) or at 37°C for 8 h (LytB). After incubation, samples (200 μl) were taken and assayed for enzymatic activity.
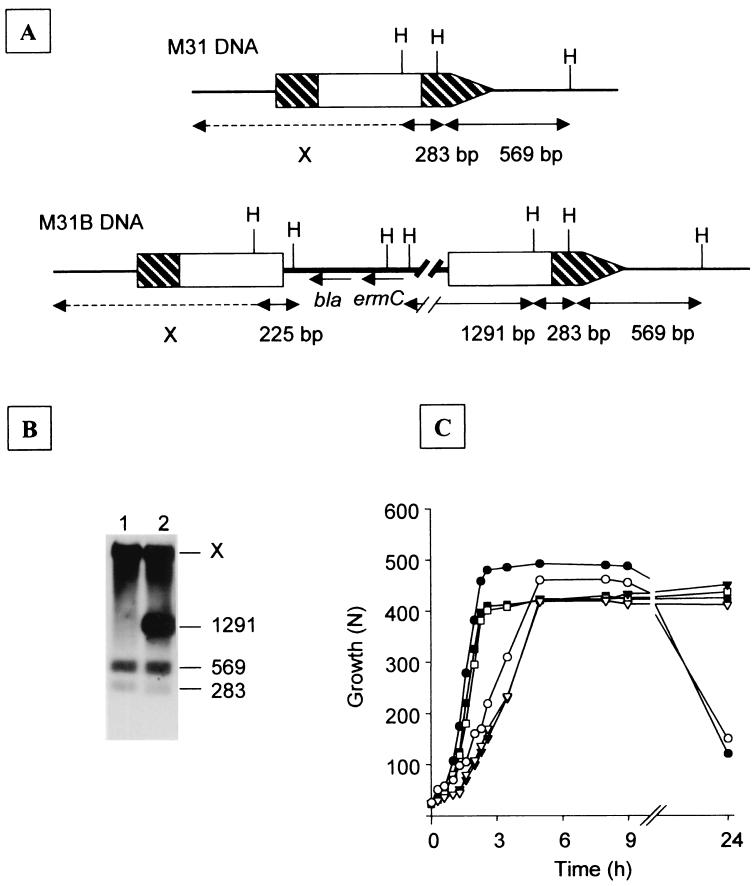
Characterization of chain-dispersing activity. To get more insight into the biological role of the LytB glucosaminidase, we inactivated the lytB gene (Fig. 6A) in strains R6 and M31, which led to the formation of long chains (more than 100 cells in the case of the M31B mutant) (20). The accuracy of these constructions was tested by Southern blot analysis as illustrated in Fig. 6B for the M31B mutant. This chaining effect did not interfere with the autolytic properties of the parental cells, as the R6B mutant autolysed at the end of the stationary phase of growth when incubated at 37°C, mainly due to the activity of LytA whereas the M31B mutant autolysed after several hours of incubation at only 30°C by the action of LytC (Fig. 6C). Moreover, both R6B and M31B mutants divided at approximately the normal frequency and were genetically transformable at rates similar to those for the parental cells (data not shown), which does not suggest a role for LytB in competence development. The addition of 6.5 μg of purified LytB/ml to the cultures of M31 or M31B did not induce lysis at 37°C at the end of the stationary phase of growth, although these cells were able to lyse at 30°C, probably by the action of LytC. The M31C mutant (an M31 derivative that contains an inactivating insertion in the lytC gene) did not lyse at either 30 or 37°C with or without the addition of LytB (Fig. 6C). According to these observations, LytB is the first characterized nonautolytic pneumococcal murein hydrolase.
FIG. 6.
Construction of a lytB pneumococcal mutant, growth curves of mutant strains, and effect of the addition of LytB. (A) Schematic representation of the DNA region surrounding the lytB gene in insertion duplication mutant strain M31B (lytB) and its parental strain, M31. Hatched arrow, lytB gene; white rectangle in lytB, HaeIII fragment used for insertion duplication mutagenesis; thick line, vector plasmid pUCE191. H, HaeII; X, fragment of unknown size; bla and ermC, genes encoding ampicillin and erythromycin-lincomycin resistance, respectively. (B) Southern blot hybridization using lytB as the probe. Lane 1, HaeII-digested M31 DNA; lane 2, HaeII-digested M31B DNA. The sizes (in base pairs) of the DNA fragments are shown on the right. X, fragment of unknown size. (C) Cultures of R6B (•), M31C (▪), or M31C plus 10 μl of LytB (30.4 U/mg) (□) were incubated at 37°C, whereas M31B (○), M31C (▾), or M31C plus 10 μl of LytB (30.4 U/mg) (▿) were incubated at 30°C. LytB was added at N = 100. Growth was monitored by nephelometry. N, nephelometric units.
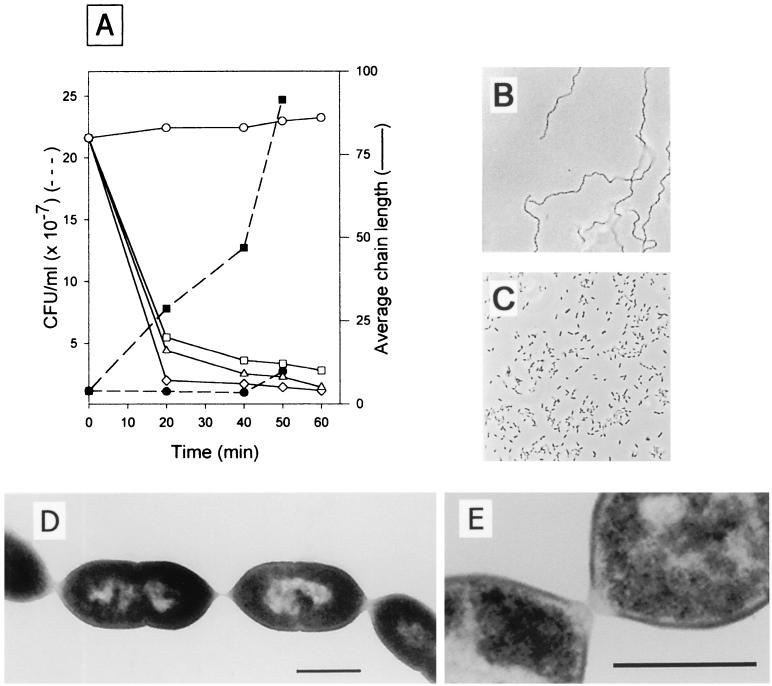
Interestingly, the addition of purified LytB to the R6B and M31B cultures dispersed the chains of these mutants in a dose-dependent manner until the cells largely took on the appearance of typical diplococci or small chains of about 6 to 10 cells corresponding to the parental cell morphologies (Fig. 7A to C). As a consequence of unchaining, the incubation of either R6B and M31B chains in the presence of LytB resulted in a noticeable increase in cell number counts (Fig. 7A). These results clearly demonstrated that the biological role of the LytB glucosaminidase is the separation of daughter cells at the end of cell division. It is noteworthy that the kinetics of dispersion of the chains by the purified LytB is rather quick since about 20 min of incubation was enough to nearly recover the original length of the cells. By contrast, the rate of in vitro degradation of isolated cell walls is such that 6 h of incubation is required for detection of a significant amount of solubilized cell walls (Fig. 3 and 7A). Electron microscopy of the R6B chains showed that the neighboring cells are normally compartmentalized and appear to be held together by thin filamentous material that seems to be an extension of the polar cell walls (Fig. 7D and E), as previously illustrated for pneumococcal cells grown in EA medium (49). Furthermore, differential electron density at the poles with respect to the lateral edges suggests that they are structurally and/or chemically different (Fig. 7E).
FIG. 7.
Kinetics of dispersion of R6B chains by purified LytB, viable counting, and microscopic observations. (A) R6B cultures (n = 80) were treated, at time zero, with 10 (⋄), 1 (▵), or 0.1 μl (□) of LytB (30.4 U/mg). R6B untreated, as a control, was also tested (○). (B and C) Aliquots were removed to determine the average chain length by examination using phase-contrast microscopy and by counting viable cells of cultures treated with 10 μl of LytB (▪) and untreated (•). Purified LytB was added at time zero. The morphology of strain R6B at time zero (B) and 30 min after the addition of 10 μl LytB (C), examined by phase-contrast microscopy, is shown. (D and E) Thin sectioning and electron-microscopic examination of R6B cells. The neighboring cells in the chain are separated by cross walls of normal dimensions and normal depth penetration. Bars, 1 (D) and 0.5 μm (E).
Polar localization of LytB.
To visualize the specific localization of LytB on the cell surface, we constructed a translational fusion between gfp and the region of lytB coding for the mature form of the enzyme (Fig. 8A). The corresponding GFP- LytB fusion protein was purified from a DEAE-cellulose column, as described for the ChBPs (19), and tested for functional activity by its dechaining ability on strain M31B. The chimeric protein was effectively capable of shortening the long chains of this mutant, with kinetics similar to those described above for LytB. The localization of the fluorescence was specifically concentrated on the region close to the polar ends of each cell (Fig. 8B, images a to f). On the other hand, we also confirmed this specific polar localization of the GFP-LytB fusion in wild-type pneumococcal strain R6 (Fig. 8B, images i and j).
The presence of choline residues in the teichoic acid of pneumococci has provided valuable information about the biological role of this aminoalcohol in S. pneumoniae (48). Moreover, it is well documented that choline residues can be biosynthetically replaced in teichoic acid by certain analogs, such as EA, and that pneumococci growing in EA show striking physiological changes (47). A peculiar characteristic of this aminoalcohol replacement is the inhibition of cell separation (chain formation), a property originally ascribed to a defective autolytic system of EA-grown pneumococci since LytA has an absolute requirement for choline in the cell wall teichoic acid for activity (49). The addition of GFP-LytB to M31B cells grown in EA-containing medium revealed that the fusion protein was capable of recognizing the same polar zones (Fig. 8B, images g and h), but these cells remained as long chains. We have also prepared a similar fusion of LytA with GFP (Fig. 8A); this protein predominantly targeted the equatorial zone (septum at the midcell) when added to R6B cells. Some fluorescence was also seen at the cell poles, although the chain length remained apparently unaltered. These observations are consistent with previous findings by using immunochemical techniques (6) and suggested spatial distribution of carbohydrates in bacterial cell walls as reported recently for other murein hydrolases of Listeria monocytogenes (27).
DISCUSSION
The presence of repeated motifs in the lytic enzymes of several gram-positive bacteria and their bacteriophages is well documented in the recent literature (reference 19 and references therein). We have demonstrated that, for S. pneumoniae and its bacteriophages, the ChBD, comprising a variable number of repeated motifs, recognizes the choline present in teichoic acid (28). This result has been further corroborated recently by the elucidation of the crystal structure of the ChBD of LytA, revealing a novel solenoid fold that is maintained by choline molecules (11). Interestingly, it has also been reported that a different set of sequence repeats located in the N-terminal region of the CwlB amidase and CwlG glucosaminidase of Bacillus subtilis plays a direct role in cell wall binding (24). In L. monocytogenes, four tandemly arranged units are supposed to be involved in wall binding (53). A peculiar case of domain organization is provided by the Atl autolysin of Staphylococcus aureus, a bifunctional protein with an N-terminal amidase domain and a C-terminal glucosaminidase domain; here, a three-repeat sequence located between the two catalytic regions is the binding domain (2). However, the domain organization of LytB, as well as that reported recently for the LytC lysozyme of S. pneumoniae (19), contrasts with the structural organization documented for all other cell wall lytic enzymes of the pneumococcal system (28). The ChBD and the catalytic domain for these enzymes are located at the N- and C-terminal moieties in LytB and LytC, respectively. These examples of modular shuffling in the ChBPs of pneumococcus might imply recombination between bacteria and also between phages and bacteria, as illustrated by the preparation of functional chimeric lytic enzymes (7, 12). In this sense, it has been suggested that gene rearrangement in bacteria has occurred continually during evolution (10).
We have now cloned and expressed the lytB gene in E. coli. The failure to clone the DNA fragment coding for the unprocessed form of the LytB in E. coli suggests that the signal peptide might be processed in E. coli and that the mature protein, then located in the outer side of the cytoplasmic membrane, could degrade the peptidoglycan and cause the lysis of the culture. Similar events were previously reported for the Ejl amidase and the Cpl1 lysozyme, two lytic enzymes of pneumococcal phages EJ-1 and Cp-1, respectively (9, 19). A similar situation was documented recently when we failed to clone the lytC gene coding for the intact form of the pneumococcal lysozyme in E. coli (19). The set of murein hydrolases reported so far in pneumococcus consists of an amidase (LytA), a lysozyme (LytC), and the glucosaminidase (LytB) described here. Additionally, Pce (phosphorylcholine esterase) is also involved in remodeling the outer surface by releasing phosphorylcholine residues from teichoic acid (5, 50). All these enzymes are ChBPs and, consequently, dependent on the presence of choline in teichoic acid for maximum activity. However, it is not clear whether the total number of pneumococcal murein hydrolases has been already determined. In contrast to findings for other bacterial systems we have not detected in pneumococcus the presence of several enzymes displaying the same specificity, although we cannot rule out the possibility of other choline-independent or choline-dependent murein hydrolases.
The purified LytB enzyme liberated, in in vitro assays, up to 25% of the choline residues present in purified pneumococcal cell walls. This limited hydrolysis of the cell walls is reminiscent of that recently reported for Pce (50). This esterase is also a ChBP which degrades only some of the phosphorylcholine residues present in the teichoic acid of the cell wall without any known implication for autolysis (50). The inability of some bacterial cell wall enzymes to efficiently degrade the homologous substrate has been best illustrated in the case of the Atl of S. aureus. The Atl bifunctional enzyme plays a precise role in cell septation, which requires the specific recognition of some zones of the cell wall. This limited degradation of the walls of S. aureus hindered the precise biochemical characterization of these murein hydrolases when using homologous cell walls for in vitro assays. However, the use of cell walls prepared from Micrococcus luteus allowed the characterization of these enzymes (44). This alternative procedure has not been feasible for the lytic enzymes of the pneumococcal system, most probably due to the absolute requirement of these enzymes for choline in the cell wall substrate for activity. However, gel filtration analysis of the degradation products of [3H]choline- and [3H]lysine-labeled pneumococcal cell walls together with bioinformatic sequence comparison strongly suggested that LytB is an endo-N-acetylglucosaminidase.
The new murein hydrolase of S. pneumoniae described here was capable of dispersing, in a dose-dependent way, the long chains formed by mutants R6B and M31B. Daughter cell separation, the final event of the cell division cycle, has been demonstrated in S. aureus to be the result of the synergistic activity of two enzymes at the equatorial surface ring (2). Targeting these two murein hydrolases to a specific zone of the division ring is carried out by the three-repeat domain located at the center of pro-Atl prior to its proteolytic processing (2, 54). Inactivation of the atl gene led to the formation of clusters of cells (44). On the other hand, long-chain formation in B. subtilis requires the inactivation of several autolysins (30), suggesting that different lytic enzymes might replace each other or are capable of cooperative action. In contrast, for S. pneumoniae, the LytB glucosaminidase is the main enzyme responsible for the separation process and LytA amidase appears to have a minor role in cell separation; deletion of the lytA gene only provokes the formation of short chains (37). It has been previously demonstrated that LytA amidase targets the bacterial surface via binding to choline, and other surface proteins of the pneumococcal system use a similar mechanism to anchor to the cell envelope, e.g., PspA, a lactoferrin-binding protein (16). This mechanism of anchoring proteins to the cell wall is considered unique (4, 11). We therefore anticipate that the 18 repeat motifs identified in the N-terminal domain of LytB might also play a role similar to that demonstrated for other pneumococcal ChBPs. For the pro-Atl protein of S. aureus, where different experimental approaches, including preparation of chimeric proteins (2) and immunogold-labeling techniques, have been used, the repeat motifs direct the two murein hydrolases to the equatorial surface ring (2, 54). It has been postulated that these particular lytic enzymes do not hydrolyze the peptidoglycan randomly, but instead separate dividing cells by cleaving the cell wall at designed sites (2).
Several observations reported in this work provide strong support for the view that the pneumococcal glucosaminidase might recognize only certain regions of the cell wall for catalytic activity. Acetylation may play a role in selective control of the activities of the two murein hydrolases involved in daughter cell separation in S. aureus (44). In this regard, heterogeneity within the cell wall of pneumococcus (51) may influence the activity of lytic enzymes, suggesting that acetylated zones of the pneumococcal cell wall could allow control of the activity of these potentially autolytic enzymes. The activity of a peptidoglycan N-acetylglucosamine deacetylase (PgdA) found in S. pneumoniae modifies over 80% of the glucosamine residues and 10% of the muramic acid residues; these modifications provide protection against the enzymatic action of some lysozymes (50). Interestingly, we report here that full acetylation of the peptidoglycan does not affect the in vitro activity of LytB, whereas LytA amidase was drastically reduced, as already shown (51). On the other hand, the preparation of enzymatically active fusion protein GFP-LytB provided valuable information about cell polarity and the biological role of LytB. Combined observation of lytB mutants and R6 diplococci treated with GFP-LytB revealed that the fusion protein remains active in cell separation and is localized at the poles of the cell. The finding that GFP-LytB also binds specifically to the polar zones of EA-grown pneumococcal cells, although the long chains remain uncut, suggests that LytB presents a wider range of substrate recognition, in contrast to the absolute requirement of LytA for choline to bind and hydrolyze the cell walls. During the preparation of this work, fusions between C-terminal cell wall binding domains of two murein hydrolases (CBP500 and CBP118) coded by phages of L. monocytogenes were used to show that the domain of CBP500 targeted the entire cell surface whereas CBP118 bound to a ligand predominantly present at septal regions and cell poles. The authors of this study concluded that the uneven distribution of the ligand in CBP118 was due to asymmetric display or to some modification that alters or masks the molecule along the lateral walls (27). On the other hand, examination of E. coli peptidoglycan composition has also revealed that the polar murein composition differs from that of the lateral walls; localization data suggest that old poles remain rather static when new material is being incorporated. It has been proposed that poles provide a mechanism by which proteins, once inserted, are maintained at this location (29). Apparently, only the old wall (poles) appears brilliantly fluorescent when treated with fusion protein GFP-LytB. This results from the binding of the protein to the specific target (3), to which it remains bound for a long time. In S. pneumoniae, the presence of choline is essential for the successful cell septation by LytB, but initial binding to other aminoalcohol-containing substrates can be also achieved. In this regard, the different blocks might play a role in cell substrate recognition and degradation (Fig. 1A and 8A).
It has been suggested that murein hydrolases may exert their selective advantage only in the in vivo environment (21). Moreover, it has been proposed that bacterial chain formation limits the dissemination of the bacteria during infection (33). Since a single murein hydrolase appears to be fundamental for cell separation in S. pneumoniae, the LytB glucosaminidase might be an interesting target for the design of a future conjugated vaccine or new antibiotics. In fact, it has been very recently reported that an antiserum raised against LytB significantly protected mice from lethal challenge with some pneumococcal strains (52). Finally, we have found that several clinical isolates of S. pneumoniae form long chains and that these isolates recovered the diplococcus form after treatment with LytB (unpublished observations). Our forthcoming studies on these isolates will explore the role of the LytB glucosaminidase in natural environments.
Acknowledgments
We are indebted to E. García for his critical reading of the manuscript and many fruitful discussions and S. Lapole for getting ultrathin sections. The valuable advice of P. López for the preparation of GFP fusion proteins is gratefully acknowledged. We appreciate the skillful guidance of M. A. Ollacarizqueta and O. Ahrazem with the fluorescence studies and the technical assistance of E. Cano and M. Carrasco. We also thank V. Muñoz and M. Fontenla for the artwork and W. Sanders for correcting the English version of the manuscript.
This work was supported by grant BCM2000-1002 from the Dirección General de Investigación Científica y Técnica. B.R. was the recipient of a grant from Comunidad Autónoma de Madrid.
REFERENCES
- 1.Arrecubieta, C., E. García, and R. López. 1995. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene 167:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 17:4639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, R. M. 1965. Symposium on the fine structure and replication of bacteria and their parts. III. Bacterial cell wall replication followed by immunofluorescence. Bacteriol. Rev. 29:326-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cossart, P., and R. Jonquières. 2000. Sortase, a universal target for therapeutic agents against gram-positive bacteria? Proc. Natl. Acad. Sci. USA 97:5013-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Las Rivas, B., J. L. García, R. López, and P. García. 2001. Molecular characterization of the pneumococcal teichoic acid phosphorylcholine esterase. Microb. Drug Res. 7:213-222. [DOI] [PubMed] [Google Scholar]
- 6.Díaz, E., E. García, C. Ascaso, E. Méndez, R. López, and J. L. García. 1989. Subcellular localization of the major pneumococcal autolysin: a peculiar mechanism of secretion in Escherichia coli. J. Biol. Chem. 264:1238-1244. [PubMed] [Google Scholar]
- 7.Díaz, E., R. López, and J. L. García. 1990. Chimeric phage-bacterial enzymes: a clue to the modular evolution of genes. Proc. Natl. Acad. Sci. USA 87:8125-8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Díaz, E., R. López, and J. L. García. 1992. Role of the major pneumococcal autolysin in the atypical response of a clinical isolate of Streptococcus pneumoniae. J. Bacteriol. 174:5508-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Díaz, E., M. Munthali, H. Lünsdorf, J.-V. Höeltje, and K. Timmis. 1996. The two-step lysis system of pneumococcal bacteriophage EJ-1 is functional in gram-negative bacteria: triggering of the major pneumococcal autolysin in Escherichia coli. Mol. Microbiol. 19:667-681. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle, R. F. 1995. Convergent evolution: the need to be explicit. Annu. Rev. Biochem. 64:287-314. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Tornero, C., R. López, E. García, G. Giménez-Gallego, and A. Romero. 2001. A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 8:1020-1024. [DOI] [PubMed] [Google Scholar]
- 12.García, E., J. L. García, P. García, A. Arrarás, J. M. Sánchez-Puelles, and R. López. 1988. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc. Natl. Acad. Sci. USA 85:914-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García, E., J. M. Rojo, P. García, C. Ronda, R. López, and A. Tomasz. 1982. Preparation of an antiserum against the pneumococcal autolysin. Inhibition of autolysin activity and some autolytic processes by the antibody. FEMS Microbiol. Lett. 14:133-136. [Google Scholar]
- 14.García, J. L., E. García, A. Arrarás, P. García, C. Ronda, and R. López. 1987. Cloning, purification, and biochemical characterization of the pneumococcal bacteriophage Cp-1 lysin. J. Virol. 61:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García, J. L., E. García, and R. López. 1987. Overproduction and rapid purification of the amidase of Streptococcus pneumoniae. Arch. Microbiol. 149:52-56. [DOI] [PubMed] [Google Scholar]
- 16.García, J. L., A. R. Sánchez-Beato, F. J. Medrano, and R. López. 2000. Versatility of choline-binding domain, p. 231-244. In A. Tomasz (ed.), Streptococcus pneumoniae. Molecular biology and mechanisms of disease. Mary Ann Liebert, Inc., Larchmont, N.Y.
- 17.García, P., J. L. García, E. García, and R. López. 1986. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene 43:265-272. [DOI] [PubMed] [Google Scholar]
- 18.García, P., J. L. García, E. García, J. M. Sánchez-Puelles, and R. López. 1990. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene 86:81-88. [DOI] [PubMed] [Google Scholar]
- 19.García, P., M. P. González, E. García, J. L. García, and R. López. 1999. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33:128-138. [DOI] [PubMed] [Google Scholar]
- 20.García, P., M. P. González, E. García, R. López, and J. L. García. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1277. [DOI] [PubMed] [Google Scholar]
- 21.Handberger, S., and A. Tomasz. 1985. Antibiotic tolerance among clinical isolates of bacteria. Rev. Infect. Dis. 7:368-386. [DOI] [PubMed] [Google Scholar]
- 22.Höltje, J. V., and A. Tomasz. 1976. Purification of the pneumococcal N-acetylmuramyl-L-alanine amidase to biochemical homogeneity. J. Biol. Chem. 251:4199-4207. [PubMed] [Google Scholar]
- 23.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda, A., and J. Sekiguchi. 1991. Molecular cloning and sequencing of the major Bacillus subtilis autolysin gene. J. Bacteriol. 173:7304-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme activity in Pneumococcus. Biochim. Biophys. Acta 39:508-517. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 28.López, R., E. García, P. García, and J. L. García. 1997. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 3:199-211. [DOI] [PubMed] [Google Scholar]
- 29.Lybarger, S. R., and J. R. Maddock. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margot, P., M. Pagni, and D. Karamata. 1999. Bacillus subtilis 168 gene lytF encodes a γ-D-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, σD. Microbiology 145:57-65. [DOI] [PubMed] [Google Scholar]
- 31.Miller, W. G., and S. E. Lindow. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149-153. [DOI] [PubMed] [Google Scholar]
- 32.Mosser, J. L., and A. Tomasz. 1970. Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J. Biol. Chem. 245:287-298. [PubMed] [Google Scholar]
- 33.Musher, D. M., R. F. Breiman, and A. Tomasz. 2000. Streptococcus pneumoniae: at the threshold of the 21st century, p. 485-491. In A. Tomasz (ed.), Streptococcus pneumoniae. Molecular biology and mechanism of disease. Mary Ann Liebert, Inc., Larchmont, N.Y.
- 34.Paton, J. C., P. W. Andrew, G. J. Boulnois, and T. J. Mitchell. 1993. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu. Rev. Microbiol. 47:89-115. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds, S. 1963. The use of lead citrate at high pH as electron-opaque stain in electron microscopy. J. Cell Biol. 17:200-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sánchez-Puelles, J. M., C. Ronda, J. L. García, P. García, R. López, and E. García. 1986. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur. J. Biochem. 158:289-293. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez-Puelles, J. M., J. M. Sanz, J. L. García, and E. García. 1992. Immobilization and single-step purification of fusion proteins using DEAE-cellulose. Eur. J. Biochem. 203:153-159. [DOI] [PubMed] [Google Scholar]
- 39.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shockman, G. D., and J.-V. Höltje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 41.Speicher, D. W. 1994. Methods and strategies for the sequence analysis of proteins of PVDF membranes. Methods 6:569-577. [Google Scholar]
- 42.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microsocpy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 43.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 44.Sugai, M., H. Komatsuzawa, T. Akiyama, Y.-M. Hong, T. Oshida, Y. Miyake, T. Yamaguchi, and H. Suginaka. 1995. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J. Bacteriol. 177:1491-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabor, S. 1990. Expression using T7 RNA polymerase/promoter system, p. 16.2.1-16.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates Inc. and John Wiley & Sons, New York, N.Y.
- 46.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Redune, E. Holtzapple, H. Khouri, A. M. Wof, T. R. Utterback, C. L. Hansen, L. A. MacDonald, T. V. Feldblyum, S. Angiouli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 47.Tomasz, A. 1968. Biological consequences of the replacement of choline by ethanolamine in the cell wall of pneumococcus: chain formation, loss of transformability and loss of autolysis. Proc. Natl. Acad. Sci. USA 59:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomasz, A. 1967. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in pneumococcus. Science 157:694-697. [DOI] [PubMed] [Google Scholar]
- 49.Tomasz, A., M. Westphal, E. B. Briles, and P. Fletcher. 1975. On the physiological functions of teichoic acids. J. Supramol. Struct. 3:1-16. [DOI] [PubMed] [Google Scholar]
- 50.Vollmer, W., and A. Tomasz. 2001. Identification of the teichoic acid phosphorylcholine esterase in Streptococcus pneumoniae. Mol. Microbiol. 39:1610-1622. [DOI] [PubMed] [Google Scholar]
- 51.Vollmer, W., and A. Tomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]
- 52.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barash, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wuenscher, M. D., S. Köhler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, P60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamada, S., M. Sugai, H. Komatsuzawa, S. Nakhasima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]