Abstract
Production of complex extracellular polysaccharides (EPSs) by the nitrogen-fixing soil bacterium Sinorhizobium meliloti is required for efficient invasion of root nodules on the host plant alfalfa. Any one of three S. meliloti polysaccharides, succinoglycan, EPS II, or K antigen, can mediate infection thread initiation and extension (root nodule invasion) on alfalfa. Of these three polysaccharides, the only symbiotically active polysaccharide produced by S. meliloti wild-type strain Rm1021 is succinoglycan. The expR101 mutation is required to turn on production of symbiotically active forms of EPS II in strain Rm1021. In this study, we have determined the nature of the expR101 mutation in S. meliloti. The expR101 mutation, a spontaneous dominant mutation, results from precise, reading frame-restoring excision of an insertion sequence from the coding region of expR, a gene whose predicted protein product is highly homologous to the Rhizobium leguminosarum bv. viciae RhiR protein and a number of other homologs of Vibrio fischeri LuxR that function as receptors for N-acylhomoserine lactones (AHLs) in quorum-sensing regulation of gene expression. S. meliloti ExpR activates transcription of genes involved in EPS II production in a density-dependent fashion, and it does so at much lower cell densities than many quorum-sensing systems. High-pressure liquid chromatographic fractionation of S. meliloti culture filtrate extracts revealed at least three peaks with AHL activity, one of which activated ExpR-dependent expression of the expE operon.
The soil bacterium Sinorhizobium meliloti fixes atmospheric dinitrogen to ammonia in symbiotic association with the host plant Medicago sativa (alfalfa). A successful symbiosis is the result of a complex series of interactions between the host and the symbiont (10, 18, 48, 55, 78). A broad range of plant compounds can function to influence the production of Nod factors by S. meliloti. Nod factors stimulate root hair curling and root nodule formation. S. meliloti cells colonize curled root hairs and invade developing root nodules via extended invaginations of the root hair cell membrane called infection threads. The S. meliloti cells are individually surrounded by host cell membrane and released into the host nodule cells, where they differentiate into bacteroids, the nitrogen-fixing form of the bacteria.
Extracellular polysaccharides (EPSs) produced by S. meliloti are crucial for establishing a successful nitrogen-fixing symbiosis with alfalfa. S. meliloti mutants that are unable to produce symbiotically active polysaccharides are defective in nodule invasion and primarily induce the formation of symbiotically ineffective root nodules that are devoid of bacteria and bacteroids (34, 45, 62). Any one of three S. meliloti polysaccharides, succinoglycan, EPS II, or K antigen, can mediate alfalfa root nodule invasion. By using green fluorescent protein-expressing S. meliloti strains, we have recently demonstrated that each of these three polysaccharides functions to mediate infection thread initiation and extension on alfalfa (16, 56). However, under laboratory conditions, there are quantitative and qualitative differences in the manners in which succinoglycan, EPS II, and K-antigen function (56). This suggests that certain polysaccharides are able to function more efficiently under different conditions, thus providing strains that produce multiple polysaccharides with a selective symbiotic advantage under variable conditions.
Succinoglycan, EPS II, and K antigen are structurally diverse polysaccharides. The succinoglycan repeating unit is composed of one galactose and seven glucose residues with pyruvyl, acetyl, and succinyl modifications (2, 61). EPS II has a galactoglucan repeating unit modified with acetyl and pyruvyl moieties (34, 39). The K-antigen repeating unit is a disaccharide containing glucuronic acid and 5,7-diamino-3,5,7,9-tetradeoxynonulosonic acid (63). Despite the structural diversity of these three polysaccharides, genetic and biochemical evidence strongly suggests that particular low-molecular-weight forms of each polysaccharide are the symbiotically active species (i.e., the forms able to promote nodule invasion) (6, 35, 64, 77, 81).
Under nonstarvation conditions, wild-type laboratory S. meliloti strain Rm1021, whose genome has recently been sequenced (4, 11, 27, 32), produces measurable quantities of succinoglycan but does not produce detectable EPS II and does not make symbiotically active K antigen. EPS II production in Rm1021 can be induced by the expR101 mutation (34), by a null allele of the mucR gene (42, 88), or by growth under very low phosphate conditions (89). However, only the expR101 mutation, a spontaneous mutation in Rm1021 that results in mucoid colony morphology, stimulates production of symbiotically active EPS II. Both the mucR::Tn5 mutation and growth under very low phosphate conditions result in production of high-molecular-weight EPS II, a form that is symbiotically inactive (36, 52), whereas expR101-induced EPS II ranges from a high molecular weight to a low molecular weight and includes EPS II molecules with 15 to 20 disaccharide repeating units, the fraction active in promoting nodule invasion (36).
To date, molecular analysis of the genetics of EPS II production has revealed a cluster of genes (the exp gene cluster) required for EPS II production (34). The exp gene cluster has been sequenced (7), revealing 21 exp genes that are organized into five operons, including the expA (9 genes), expC (1 gene), expD (2 genes), expE (8 genes), and expG (1 gene) transcriptional units. However, among the predicted exp gene products are several proteins whose roles in EPS II production are not easily explained. Furthermore, there are more predicted glycosyltransferases (five) present than might have been expected to synthesize a polysaccharide with only two sugars in the repeating unit. Thus, it is unclear exactly what roles these multiple exp gene products have in the synthesis of EPS II. In addition, it is not clear how the molecular weight distribution of EPS II is controlled and which gene products are responsible for adding the pyruvyl and acetyl modifications to the EPS II galactoglucan backbone, as genes predicted to encode these functions appear to be absent. A regulatory gene, mucR, has also been subjected to molecular analysis. MucR is 80% identical to the Ros protein from Agrobacterium tumefaciens and is a negative regulator of EPS II production and a positive regulator of succinoglycan production in S. meliloti (42).
In a previous study, the expR101 locus was mapped to the ∼3.4-Mb S. meliloti main chromosome (34). Cotransduction analyses showed that the expR locus was ∼66% transductionally linked to one allele of the ndvB gene and ∼7% transductionally linked to the trp-33 locus (33). However, prior to this work, the nature of the expR101 mutation had not been elucidated. In order to gain insight into how the production of symbiotically active EPS II by Rm1021 is controlled, we have cloned and characterized the expR101 mutation.
We have found that the production of symbiotically active EPS II in S. meliloti strain Rm1021 is dependent on the presence of a functional copy of the expR gene. The predicted expR gene product is a member of the LuxR family of proteins, many of which are receptors for N-acylhomoserine lactones (AHLs) and are transcriptional regulators involved in the control of gene expression in response to changes in population density, a process known as quorum sensing (30, 84). ExpR controls the transcription of the exp genes (and hence production of symbiotically active EPS II) in a density-dependent manner. Compared to many other LuxR family systems, ExpR-dependent activation of exp transcription is induced at relatively low cell densities.
MATERIALS AND METHODS
Bacterial strains and culture media.
The S. meliloti strains used in this study are listed in Table 1. Strains were grown at either 30°C (S. meliloti) or 37°C (Escherichia coli) in LBMC medium (Luria-Bertani [LB] [70] liquid or agar supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2) or in TYC medium (TY [9] liquid or agar supplemented with 12 mM CaCl2). In the conditioned-medium experiments, LBMC medium was buffered with 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) to a pH of 6.75 (LBMCP). Antibiotics were used at the following concentrations: streptomycin, 500 μg/ml; neomycin, 200 μg/ml; gentamicin, 50 μg/ml for S. meliloti and 5 μg/ml for E. coli; spectinomycin, 100 μg/ml; tetracycline, 10 μg/ml; kanamycin, 25 μg/ml.
TABLE 1.
S. meliloti strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| Rm1021 | SU47 str-21 expR102::ISRm2011-1 | 51 |
| Rm7210 | Rm1021 exoY210::Tn5 | 45 |
| Rm8530 | Rm1021 expR+ (formerly expR101) | 34 |
| Rm10006 | Rm1021 expR+ ΩTn5-233 #3-15 | 34 |
| Rm9000 | Rm1021 expR+exoY210::Tn5 | 35 |
| Rm10002 | Rm1021 expA3::Tn5 | 34 |
| Rm9025 | Rm1021 expR103::lacZ-Gm | This study |
| Rm9026 | Rm8530 expR103::lacZ-Gm exoY210::Tn5 | This study |
| Rm9028 | Rm8530 expR103::lacZ-Gm | This study |
| RmAR1014 | Rm2011 expA1::lacZ-Gm | 7 |
| RmAR1016 | Rm2011 expC::lacZ-Gm | 7 |
| RmAR1018 | Rm2011 expG::lacZ-Gm | 7 |
| RmAR1020 | Rm2011 expD1::lacZ-Gm | 7 |
| RmAR1022 | Rm2011 expE2::lacZ-Gm | 7 |
| Rm9030-2 | Rm1021 expR+expA1::lacZ-Gm | This study |
| Rm9031 | Rm1021 expR+expC::lacZ-Gm | This study |
| Rm9034 | Rm1021 expR+expG::lacZ-Gm | This study |
| Rm9032 | Rm1021 expR+expD1::lacZ-Gm | This study |
| Rm9033 | Rm1021 expR+expE2::lacZ-Gm | This study |
For AHL extraction, S. meliloti strain Rm1021 was grown in a defined NM salts medium (65) supplemented with 0.028 M glucose and Gotz vitamins and adjusted to a pH between 6.6 and 6.8. To prevent precipitation, MgSO4 · 7H2O, CaCl2, and FeSO4-citric acid stocks were autoclaved separately. Extracts were prepared from the defined-medium-grown culture to facilitate future chromatographic studies and AHL identification.
The AHL reporter strains used were E. coli HB101/pSB401 (luxRI′::luxBCDAE) (85), E. coli HB101/pSB1075 (lasRI′::luxBCDAE) (85), E. coli HB101/pSB536 (ahyRI′::luxBCDAE) (75), Chromobacterium violaceum CV026 (50), and Pseudomonas aureofaciens 30-84I (86). These strains were maintained as frozen glycerol stocks and subcultured into the appropriate media as necessary. For the AHL bioassays, luminescent E. coli reporter strains were first grown overnight in LB broth with appropriate antibiotics. These cultures were then diluted 100-fold in fresh LB, cultured for 2 h, again diluted 100-fold in fresh LB, and then allowed to grow to an optical density at 600 nm (OD600) of 0.2. The culture was then spun down, and the pellet was resuspended in 10 volumes of fresh LB and vortexed for the bioassays as described below.
Alfalfa nodulation assays.
Alfalfa nodulation assays were performed as previously described (45). Each plant was inoculated with 1 ml of a cell suspension with an OD600 of 0.05. Plants were scored 4 weeks after inoculation for foliage condition, plant height, and the presence of pink, nitrogen-fixing nodules.
Molecular cloning of the expR101 mutation.
The molecular cloning of the expR101 mutation was accomplished by preparing genomic DNA (3) from S. meliloti strain Rm10006 (Rm1021 expR101 ΩTn5-233 #3-15) (34), partially restricting the genomic DNA with EcoRI (New England Biolabs, Beverly, Mass.), and ligating 20- to 35-kb genomic DNA fragments to EcoRI-restricted pLAFR1 (29) by using T4 DNA ligase (New England Biolabs). The resultant recombinant cosmid DNA was delivered into E. coli strain HB101 (Gibco BRL, Gaithersburg, Md.) with the Gigapack III-XL kit (Stratagene, La Jolla, Calif.). Tetracycline-resistant HB101 cells (those carrying recombinant cosmids) were screened for the ability to confer resistance to spectinomycin and kanamycin, the antibiotic resistance markers located on Tn5-233 (19) that function in E. coli. The insertion target for ΩTn5-233 #3-15 was found to be the 9-bp sequence including nucleotides 3997 to 4105 in the ndvB open reading frame (ORF) (nucleotides 4229 to 4237 in the total published sequence [40]). This ndvB allele confers the expected symbiotic defect (22) but not a hypoosmotic adaptation defect (21) (data not shown).
Diagnostic PCR analysis of the expR region and DNA sequencing.
Both strands of the expR region were sequenced with two templates, pBSKSII+ (Stratagene) subclones of the expR region and PCR products generated from genomic DNA templates with oligonucleotide primers (Gibco BRL) specific to the expR region. DNA sequencing was performed by the Molecular Biology Core Facility at Dartmouth College with reaction products generated by following the recommended reaction and purification protocol.
The two primers used to amplify the expR region both for DNA sequencing and for the diagnostic PCR were RmndvA5′out (5′-GCGAGGAGATCCTGCCCGAG-3′) and Rmpyc5′out (5′-AGAGTGGCGTGAACATTCGG-3′). We used 2.5 U of Pfu enzyme (Stratagene) and the manufacturer's recommended buffer conditions. The template consisted of 1 μl of a cell suspension consisting of a small scoop of cells from a 10% dimethyl sulfoxide frozen permanent strain stock suspended in 100 μl of water. Primers were used at a concentration of 1 μM, and deoxynucleoside triphosphates (Pharmacia Biotech, Piscataway, N.J.) were used at a concentration of 200 μM. The PCR program used was as follows: (i) 95°C for 5 min, (ii) 94°C for 30 s, (iii) 65°C for 30 s, (iv) 72°C for 5 min, (v) go to step ii 29 times, (vi) hold at 4°C.
Construction of an expR-null allele and a lacZ transcriptional fusion.
The genomic expR-null allele and transcriptional fusion was generated as follows. A 2.2-kb PCR product with an XbaI restriction site engineered at the expR 5′ end was generated with the expR101 locus from Rm8530 genomic DNA as the template and the primer and deoxynucleoside triphosphate concentrations listed above. The manufacturer's recommended buffer and amplification conditions for Platinum Taq Hifi (Gibco BRL) were used. The PCR product generated was purified with a PCR purification kit (Qiagen, Valencia, Calif.). Following restriction with XbaI and HindIII, the PCR product was ligated to XbaI/HindIII-restricted pK19mobGII (41) with T4 DNA ligase (New England Biolabs), generating pK19expR. The knockout construct was generated by inserting a 4.0-kb lacZ-Gm SphI cassette from pAB2001 (8) into the unique SphI site in the expR ORF on pK19expR and screening for an insertion with the lacZ-Gm cassette in the proper orientation to generate a transcriptional fusion. The resultant knockout plasmid, pK19expR::lacZ-Gm, was mobilized into Rm8530, and homologous recombinant candidates were identified by gentamicin resistance, kanamycin sensitivity, and white color on plates containing 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid. The expR::lacZ-Gm allele was transduced into Rm8530 (generating Rm9028) or Rm1021 (generating Rm9025) before use in subsequent assays. This analysis verified that the expected colony morphology phenotype cotransduced with the antibiotic resistance marker. The proper insertion of the expR::lacZ-Gm allele in the S. meliloti genome was confirmed by both PCR analysis and Southern blot analysis.
Effects of conditioned medium and culture filtrate extracts.
S. meliloti cultures grown in unbuffered LBMC medium have a basic terminal pH of ∼8. Because AHL molecules are unstable at a basic pH (71), conditioned medium was buffered with 50 mM PIPES (pH 6.75). Conditioned medium was generated by growing cultures of Rm1021 to an OD600 of ∼3.0 in LBMCP medium and then filter sterilizing the culture supernatant. Before use in quorum-sensing experiments, conditioned LBMCP medium was supplemented with 0.05 volume of a filter-sterilized solution containing 100 g of Bacto Tryptone per liter and 50 g of yeast extract per liter. Addition of this concentrate to fresh LBMCP medium did not influence the transcriptional activity of our fusions. Cells from mid-log-phase cultures (OD600 between 0.6 and 0.8) were diluted to an OD600 of 0.002 and allowed to grow for 8 h in either fresh LBMCP medium, conditioned LBMCP medium, or LBMCP medium supplemented with dilutions of culture filtrate extract. After the growth period, cultures had reached an OD600 of between 0.010 and 0.020, at which point β-galactosidase activity was measured. β-Galactosidase activity assays were performed as previously described (53), with cells from 2 ml of each culture.
To prepare crude AHL extracts, cell-free culture supernatant from a 1-liter NM glucose-nitrate culture of S. meliloti Rm1021 (OD600 of 1.1 to 1.3) was extracted twice with 500 ml of ethyl acetate acidified with 1 ml of acetic acid per liter. The extracts were pooled, dried over anhydrous sodium sulfate, and then filtered and rotary evaporated to dryness. The dry residue was dissolved in acetonitrile.
AHL detection and fractionation.
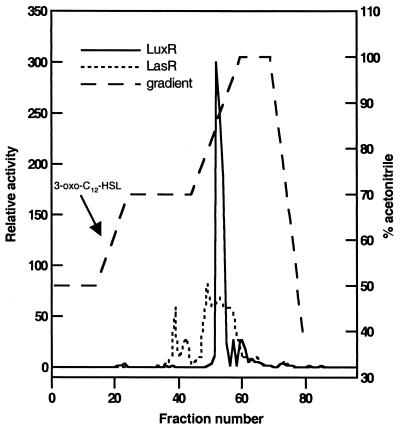
For reverse-phase high-pressure liquid chromatography (HPLC) of S. meliloti AHLs, the residue from 6 liters of bacterial culture was dissolved in 1 ml of 50% acetonitrile-water, injected onto a 50% acetonitrile-water-equilibrated semipreparatory C18 column (Whatman Partisil 10 ODS-3), and then eluted for 80 min with a step gradient (see Fig. 5) at a flow rate of 2 ml/min. Absorbance was monitored at 210 nm. Sequential 1-min fractions were collected.
FIG. 5.
Reverse-phase HPLC fractionation of AHLs present in the ethyl acetate culture filtrate extracts of S. meliloti strain Rm1021. One-minute fractions from a C18 HPLC column were collected, and serial dilutions were assayed with the AHL reporters as described in Materials and Methods. Luminescence was measured with a Wallac Victor-2 microtiter plate reader. The acetonitrile concentration in the elution gradient was 50 to 100%. The retention time of a synthetic 3-oxo-C12-HSL standard is indicated. Similar AHL activity profiles were obtained with culture filtrate extracts of an expR+ strain (Rm8530).
Aliquots of each fraction were bioassayed with the AHL reporters listed above. Serial dilutions of the HPLC fractions were dried in 96-well microtiter plates (Life Sciences Inc., Denver, Colo.) in a laminar-flow hood and then inoculated with 80 μl of the AHL reporter suspension in LB. After 3 h of incubation, the luminescence of the E. coli reporters was measured with a Wallac Victor-2 microtiter plate reader (Perkin-Elmer Inc., Gaithersburg, Md.). Pigment production by C. violaceum CV026 and P. aureofaciens was assayed as previously described (50, 76, 86). For the bioassays with S. meliloti exp::lacZ fusions, aliquots of the fractions were dried in Eppendorf tubes and then mixed with the cultures as described above.
Synthetic 3-oxo-C12-L-HSL (3-oxo-C12-L-homoserine lactone), C4-L-HSL, C6-L-HSL, C8-L-HSL, and C14-L-HSL were purchased from Aurora Biosciences (Coralville, Iowa).
RESULTS
Molecular cloning of the expR101 mutation.
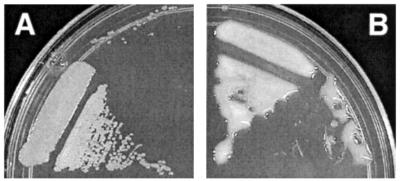
The expR101 mutant was originally noticed during a screen of S. meliloti Rm1021 Tn5-derived mutants because of its mucoid colony morphology (Fig. 1). The mucoid colony phenotype proved to be unlinked to the Tn5 insertion, indicating that the expR101 mutation had arisen spontaneously (34). Attempts to complement or suppress the mucoid colony morphology of the expR101-containing strain with cosmids from an Rm1021 pLAFR1 genomic library (29) were unsuccessful (J. Glazebrook and G. C. Walker, unpublished results; B. J. Pellock and G. C. Walker, unpublished results), suggesting that the expR101 mutation was dominant.
FIG. 1.
The expR101 mutation in S. meliloti strain Rm1021 results in a mucoid colony morphology. Colonies formed by wild-type strain Rm1021 (A), which does not produce EPS II, are dry, whereas colonies formed by Rm9000 (Rm1021 expR+ [formerly expR101] exoY210:: Tn5) (B), which produces EPS II, are mucoid. The colonies pictured were grown on LBMC agar.
To clone the expR101 mutation, we utilized a direct cloning approach that exploited a transposon insertion (termed ΩTn5-233 #3-15 [34] and located in the ndvB ORF [see Materials and Methods for details]) that was ∼95% linked (5 to 10 kb) to the expR101 mucoid colony phenotype in generalized transductions with φM12 (26). We used genomic DNA from Rm10006 (Rm1021 expR101 ΩTn5-233 #3-15) to construct a cosmid library (see Materials and Methods for details) and isolated p1-37, a cosmid that carried Tn5-233 #3-15 and more than 10 kb of chromosomal DNA flanking the transposon on each side (data not shown).
To determine whether the expR101 mutation was present on p1-37, we constructed an Rm1021/p1-37 transconjugant and examined its colony morphology. On both LBMC agar and TYC agar plates (see Materials and Methods) Rm1021/p1-37 had a mucoid colony morphology (similar to that shown in Fig. 1B) whereas Rm1021 carrying the vector control had a dry colony morphology (similar to that shown in Fig. 1A). p1-37 also induced a mucoid colony morphology when present in Rm7210 (Rm1021 exoY210::Tn5), a strain incapable of producing succinoglycan. However, p1-37 failed to induce a mucoid colony morphology in Rm10002 (Rm1021 expA3::Tn5), a strain incapable of producing EPS II. Taken together, our results strongly suggested that we had cloned the expR101 mutation on p1-37, that the expR101 mutation is dominant over the expR allele in wild-type strain Rm1021, and that the mucoid colony phenotype induced by p1-37 is a result of EPS II production, not succinoglycan production.
The predicted S. meliloti expR gene product is a LuxR homolog.
By using the ability to induce a mucoid colony morphology in Rm7210 as an assay for the presence of the expR101 mutation, we subcloned p1-37 into pSW213, a broad-host-range IncP vector (15), and examined the colony morphologies of Rm7210 subclone transconjugants on both LBMC and TYC plates. We isolated two overlapping subclones, pK5 and pH 6, that induced a mucoid colony phenotype in Rm7210. The ca. 3.2-kb KpnI-HindIII fragment common to pK5 and pH 6 (subcloned to generate pKH3.2) was also able to induce a mucoid colony phenotype in Rm7210.
We determined the DNA sequence of this 3,274-bp KpnI-HindIII fragment and compared the DNA sequences and predicted protein sequences to the GenBank database with a BLAST search (1). Sequence analysis revealed the 5′ ends of the ndvA (74) ORF and the 5′ end of an ORF predicted to encode pyruvate carboxylase (pyc) flanking one complete ORF, which we have designated expR. The expR protein product was predicted to be 246 amino acids long and showed significant homology to the Rhizobium leguminosarum bv. viciae RhiR protein (17), the E. coli SdiA protein (82), and a number of other Vibrio fischeri LuxR (24, 25) homologs. The predicted ExpR sequence contained all seven of the amino acid residues conserved in most LuxR homologs (31).
To test whether the expR ORF on the 3,274-bp KpnI-HindIII fragment was responsible for the ability of this fragment to induce a mucoid colony phenotype in Rm7210, we deleted the 3′ terminus of the expR ORF by removing a 1.4-kb SphI fragment internal to the 3,274-bp KpnI-HindIII fragment, creating pKH3.2ΔSphI. This deletion eliminated the ability of pKH3.2 to induce a mucoid colony phenotype.
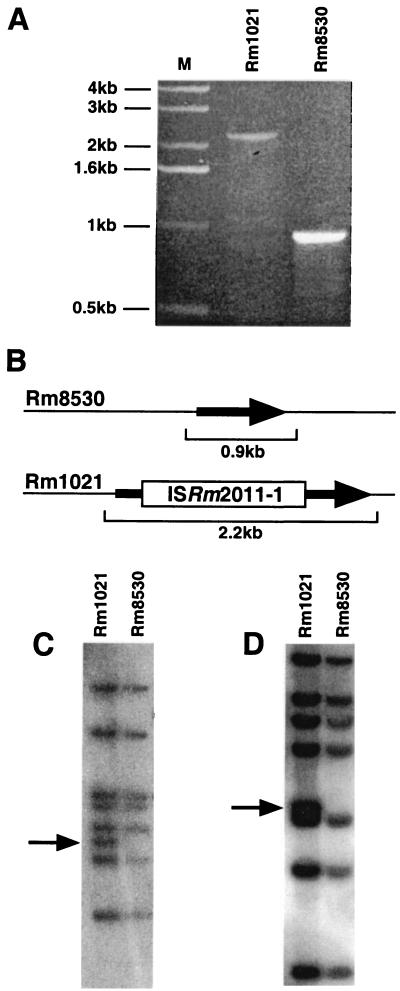
The expR ORF in S. meliloti Rm1021 is disrupted by an insertion sequence (IS) element.
When we PCR amplified the expR region (located on the S. meliloti main chromosome) from the wild-type and expR101 strains for sequence analysis, we were surprised to find that, although the PCR product from the expR101 mutant was of the expected size (0.9 kb), the PCR product from wild-type Rm1021 with the same primer pair was 2.2 kb (Fig. 2A). Sequence analysis of this 2.2-kb PCR product indicated that the expR ORF was disrupted in Rm1021 by a copy of ISRm2011-1 (Fig. 2B), a previously described 1,319-bp IS element (43, 72). This result was corroborated by the published sequence of the S. meliloti main chromosome (11). Previous Southern blot analyses suggested that eight copies of ISRm2011-1 exist in the Rm1021 genome (72), although it is now clear from the composite genome sequence that, including the copy that disrupts the expR ORF, Rm1021 has nine copies of this IS element (http://sequence.toulouse.inra.fr/meliloti.html). We observed eight ISRm2011-1-hybridizing bands in Rm1021 with both EcoRI-restricted genomic DNA and EcoRV-restricted genomic DNA (Fig. 2C and D). However, in Rm8530 (Rm1021 expR101), we detected only seven copies of ISRm2011-1 (Fig. 2C and D), suggesting that the expR-disrupting copy of ISRm2011-1 in Rm8530 had been eliminated from the genome.
FIG. 2.
The expR ORF in S. meliloti strain Rm1021 is disrupted by a 1,319-bp IS element. (A) A 0.8% agarose gel showing the expR region PCR products from Rm1021 (2.2 kb) and from Rm8530 (Rm1021 expR+ [formerly expR101]) (0.9 kb). The marker (M) lane contains the Gibco BRL 1-kb ladder. (B) Schematic representation of the expR ORFs from Rm1021 and Rm8530. The scale bars represent the diagnostic PCR products produced from each strain. (C and D) Autoradiographs of Southern blots of S. meliloti DNA probed with a sequence specific for ISRm2011-1. (C) ISRm2011-1 fingerprint of EcoRI-restricted genomic DNA from Rm1021 and Rm8530 (Rm1021 expR+ [formerly expR101]). (D) ISRm2011-1 fingerprint of EcoRV-restricted genomic DNA from Rm1021 and Rm8530 (Rm1021 expR+ [formerly expR101]). The arrows indicate the ISRm2011-1-hybridizing bands present in Rm1021 that are missing in Rm8530.
ISRm2011-1 is a member of the ISRm1 family of IS elements. Previous work indicated that members of this IS element family create 5-bp target site duplications upon insertion (69, 83). The ISRm2011-1 insertion in expR appears to have created a 5-bp target site duplication in the expR coding sequence. Precise excision of the IS element and the duplicated 5-bp target site recreated a functional expR ORF. Thus, it is highly likely that the expR101 mutation is a return to the state of the expR locus prior to the insertion of the IS element. Since the expR101 mutation appears to have recreated a functional expR allele, we have renamed the expR101 allele expR+. The expR allele present in strain Rm1021 will be designated expR102::ISRm2011-1.
Disruption of the expR+ allele in the expR101 mutant eliminates EPS II production.
To demonstrate unequivocally that it is the presence of an intact expR ORF in the expR101 mutant that is responsible for EPS II production, we disrupted the expR+ allele in strain Rm8530 (Rm1021 expR+ [formerly expR101]) (see Materials and Methods). In contrast to strain Rm8530, which has a mucoid colony phenotype, strain Rm9028 (Rm8530 expR103::lacZ-Gm) had a dry colony phenotype (data not shown), confirming that expR+ is required for the mucoid colony morphology of strain Rm8530. As expected, introduction of the expR103::lacZ-Gm allele into Rm1021 (which already has a dry colony morphology) did not impact its colony phenotype (data not shown).
To confirm that a strain carrying the expR103::lacZ-Gm allele was incapable of producing any EPS II that could function in symbiosis, we generated Rm9026 (expR103::lacZ-Gm exoY210::Tn5), a derivative of strain Rm9028 that is unable to produce succinoglycan. Rm9026 induced only white, ineffective nodules when inoculated onto alfalfa, whereas Rm9000 (Rm1021 expR+ exoY210::Tn5), which is able to produce symbiotically active EPS II, had the ability to induce pink, symbiotically effective nodules on alfalfa (data not shown). This indicated that disruption of the expR+ allele prevented production of symbiotically active EPS II. In addition, we could not detect any low-molecular-weight EPS II (35) in Rm9026 culture supernatants by Dionex high-performance anion-exchange chromatography coupled with highly sensitive pulsed amperometric detection (data not shown).
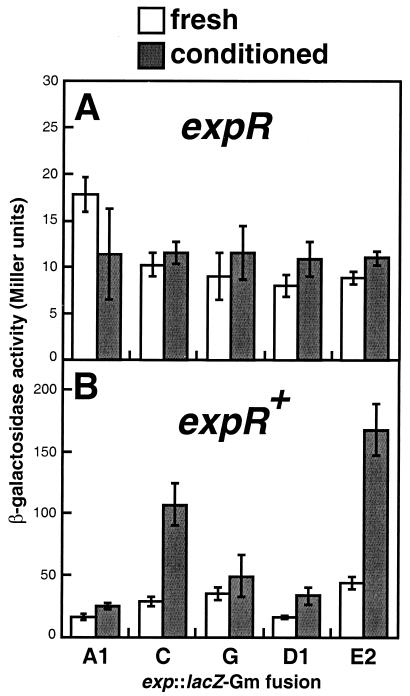
Extracellular signals control ExpR-dependent transcription of expC and expE.
Because the predicted protein sequence of ExpR has significant homology to both the AHL- and DNA-binding domains of canonical AHL receptors, we were interested in knowing whether ExpR functions to activate target gene transcription in a density-dependent fashion. To determine whether ExpR activates transcription of exp genes in response to an extracellular factor(s) produced by S. meliloti, we measured the activity of lacZ transcriptional fusions to expA1, expC, expG, expD1, and expE2 in both fresh medium and medium conditioned by growth of Rm1021 (see Materials and Methods). We performed these measurements in both the Rm1021 expR102::ISRm2011-1 and expR+ strain backgrounds. In the absence of a functional expR ORF, the levels of transcription of the five exp::lacZ fusions were relatively low (usually ∼10 Miller units) and were very similar in fresh and conditioned media (Fig. 3A). In the expR+ background in fresh medium, expression of each of the exp::lacZ fusions was equal to (for expA1 and expD1) or two- to fourfold higher (for expG, expC, and expE2) than that observed in the Rm1021 background (Fig. 3B). Overall, however, the activity of these fusions in fresh medium (at less than 50 Miller units) was modest. In contrast, in conditioned medium in the expR+ background, we observed substantial activity by both the expC and expE2 fusions (Fig. 3B). The expA1 and expG transcriptional fusions were not induced by conditioned medium, and the expD1 fusion showed only a modest increase in activity in conditioned medium (Fig. 3B). Taken together, these results strongly suggested that some density-dependent signal produced by Rm1021 activated the transcription of exp genes in an ExpR-dependent fashion.
FIG. 3.
Results of conditioned-medium experiments. The transcription of lacZ transcriptional fusions to five exp genes (expA1, expC, expG, expD1, and expE2) was measured in both fresh and conditioned media (see Materials and Methods). The activity of each fusion was determined in the expR mutant (A) and expR+ (B) backgrounds. Assays for each strain tested were performed on at least three independent cultures and repeated with independent preparations of conditioned medium. Error margins represent 1 standard deviation. The β-galactosidase activities of Rm1021 and Rm8530 (Rm1021 expR+) were nearly identical in both fresh and conditioned media (∼5 Miller units).
Intriguingly, ExpR is tuned to activate exp gene transcription at low cell densities (Fig. 4). To observe density-dependent transcription of the exp genes by ExpR required that we extensively dilute mid-log-phase cultures and allow the cells to grow for several generations prior to the transcription assay (see Materials and Methods). This allowed exp transcription to return to baseline levels. Following cellular growth in fresh medium, we consistently observed that expC and expE transcription began to be activated at an OD600 between 0.02 and 0.03 (which corresponds to approximately 107 CFU/ml for S. meliloti [data not shown]) and was strongly activated before the cells reached an OD600 of 0.07 (Fig. 4). Consistent with this observation, use of medium conditioned by growth of S. meliloti to a relatively low cell density (OD600 of ∼0.1) produced results similar to those shown in Fig. 3 (data not shown). This was an interesting result because many LuxR homologs are tuned to respond to cell densities much higher than those that activate ExpR (see Discussion).
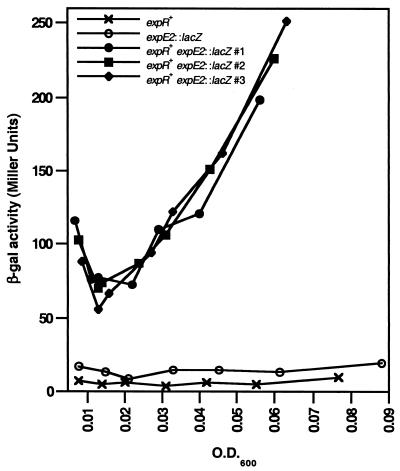
FIG. 4.
ExpR mediates density-dependent transcriptional activation of exp genes at low cell densities. Mid-log-phase cultures of Rm8530 (×), RmAR1022 (○), and three independent cultures of Rm9033 (•, ▪, and ♦) were diluted to an OD600 of 0.002 in fresh LBMCP and allowed to grow for 14 h. Samples were analyzed for β-galactosidase (β-gal) activity at 6.5, 8, 9.5, 11, 12, 13, and 14 h. Following dilution, β-galactosidase activity in Rm9033 cultures (expR+ expE2::lacZ) falls until an OD600 of 0.015 to 0.02 is reached and then begins increasing again at an OD600 of 0.02 to 0.03. The results shown are typical of those seen in multiple iterations of similar experiments. Similar results were obtained with the expC::lacZ fusion.
Transcription of expE and expC was also activated in an ExpR-dependent manner by ethyl acetate extracts of S. meliloti strain Rm1021 culture supernatants. Addition of this extract activated ExpR-dependent expC and expE transcription 2- to 2.5-fold (data not shown). Our ethyl acetate extracts also activated the LuxR- and LasR-based AHL reporter strains (data not shown), which are responsive to AHLs with long, 3-oxo-alkanoyl side chains (85). We further separated compounds present in these extracts by reverse-phase HPLC (Fig. 5). S. meliloti compounds that activated both the LuxR and LasR reporters eluted at high acetonitrile concentrations, and HPLC fractions 52 to 55 also activated ExpR-dependent expE expression (data not shown). Commercially available AHLs, including C4-L-HSL, C6-L-HSL, and C8-L-HSL (Aurora Biosciences), did not activate the ExpR-dependent fusions, although we consistently observed a very modest (1.2- to 1.4-fold) stimulation of expE transcription by C14-L-HSL.
Under our culture conditions, the presence of a functional copy of ExpR did not seem to change the AHL profile (Fig. 5) we have observed for S. meliloti; AHL activity profiles of expR+ cultures were generally similar to those from Rm1021 (data not shown). No additional AHL activities were detected in the expR+ culture filtrates with expR+ expE::lacZ or with AHL biosensors. Neither the culture filtrate extracts from S. meliloti strain Rm1021 or S. meliloti strain Rm8530 (Rm1021 expR+) nor any of the HPLC fractions activated CviR-, AhyR-, and PhzR-based reporters (C. violaceum CV026, E. coli pSB536, and P. aureofaciens 30-84I, respectively), all of which respond most strongly to AHLs with alkanoyl (C4 to C8) side chains (50, 75, 86). Taken together, our results suggest that the putative AHL molecule responsible for ExpR-dependent stimulation of exp transcription is not an AHL with a short (C4 to C8) alkanoyl side chain and that the ExpR receptor is very specific in the type of molecule that it recognizes.
Analysis of the expR ORF in other S. meliloti strains.
During the course of our analyses of the expR101 mutation, we became curious about the status of the expR ORF in a number of other S. meliloti strains that have been used in genetic laboratory studies and performed diagnostic PCRs on the expR regions of a number of S. meliloti strains. The results of these analyses are summarized in Table 2. The PCR product from strain SU47 (the parent strain of Rm1021, Rm2011, Rm5000, and RCR2011) and those from Rm2011, Rm5000, and RCR2011 were 2.2 kb, implying that these strains, like Rm1021, had an ISRm2011-1-disrupted expR ORF. This was confirmed by sequence analysis for SU47 and Rm2011. Strain YE-2Sl, an independently isolated strain that constitutively produces both succinoglycan and EPS II, had an intact expR ORF. Strain Rm41, which is more mucoid than Rm1021 but much less mucoid than Rm8530 and YE-2Sl, also had an intact expR ORF. Another independently isolated S. meliloti strain that has been genetically analyzed, strain 102F34, gave a 0.9-kb expR PCR product, suggesting that this strain does not have ISRm2011-1 inserted in the expR ORF. However, when we sequenced the expR region from strain 102F34, we discovered that, although no IS element is present, the 102F34 expR ORF has an 11-bp deletion in its coding sequence, a deletion predicted to result in premature translational termination of the predicted expR gene product in strain 102F34. This is consistent with the dry colony morphology of strain 102F34, and our sequence data agree completely with the previously published ndvA region sequence data (74).
TABLE 2.
Status of the expR locus in S. meliloti strains
| Strain | Reference | Size (kb) of expR PCR product | Status of expR ORF |
|---|---|---|---|
| Rm1021 | 51 | 2.2 | expR102::ISRm2011-1 |
| Rm8530 | 34 | 0.9 | Intact (expR+) |
| SU47 | 79 | 2.2 | expR102::ISRm2011-1a |
| Rm2011 | 12 | 2.2 | expR102::ISRm2011-1a |
| Rm5000 | 26 | 2.2 | expR102::ISRm2011-1b |
| RCR2011 | 68 | 2.2 | expR102::ISRm2011-1b |
| Rm41 | 59 | 0.9 | Intactc |
| YE-2S1 | 87 | 0.9 | Intactd |
| 102F34 | 20 | 0.9 | Disrupted by 11-bp deletion |
Genotype confirmed via DNA sequencing.
This genotype is inferred from the relatedness of these strains to SU47 and the size of the PCR product.
Rm41 expR has a sequence distinct from those of Rm1021 and YE-2S1.
YE-2S1 expR has a sequence distinct from those of Rm1021 and Rm41.
DISCUSSION
The only known circumstance under which S. meliloti strain Rm1021 synthesizes EPS II in a symbiotically active form is when it carries a mutation originally designated expR101. In this report, we have shown that widely used laboratory strain Rm1021 (whose genome has recently been sequenced [32]) and several related strains have a copy of ISRm2011-1 (43, 72) within the coding region of expR, a gene that encodes a LuxR homolog. The expR101 mutation evidently resulted from precise excision of this IS, which restored the reading frame of the expR gene, with the excised copy of ISRm2011-1 having been lost from this strain. The presence of a functional expR ORF on a plasmid or in the genome is sufficient to promote the production of symbiotically active EPS II, and disruption of the expR101 allele eliminates EPS II production. We have therefore renamed the expR101 allele expR+. Rm1021 and related strains carry the expR102::ISRm2011-1 allele.
The expR gene product, which is a LuxR homolog, significantly activates transcription of the expC and expE operons in conditioned medium and in response to specific HPLC fractions that also activate two AHL reporter systems. This result is consistent with our observation that, of the five exp::lacZ fusions tested, expC and expE are by far the most strongly expressed in an expR+ background (B. J. Pellock, J. Lloret, and G. C. Walker, unpublished results). To the best of our knowledge, this is the first description of quorum-sensing regulation in S. meliloti. AHL-dependent quorum-sensing systems that impact nodulation efficiency have been described in R. leguminosarum (17, 37, 47, 66) and R. etli (67). However, it is not clear exactly how density-dependent gene expression mediated by these R. etli and R. leguminosarum systems is involved in the nodulation process. Previous studies have also shown that a number of distinct plant-associated and free-living bacteria use quorum-sensing systems to regulate EPS synthesis and colony mucoidy (for example, see references 28, 58, 80, and 90), and in some cases, nonmucoid derivatives are impaired in the ability to infect or colonize their plant hosts (80, 90).
A striking feature of ExpR-mediated density-dependent production of EPS II is that exp transcription is stimulated at much lower cell densities than in many of the other described LuxR-type systems (Fig. 4). For example, substantial activation of the V. fischeri LuxR system occurs when the OD600 of a culture is between 0.1 and 0.3, depending on the strain tested (23, 54). Additionally, the Pseudomonas aeruginosa LasR and RhlR systems induce target gene expression at much higher densities than S. meliloti ExpR (57). R. leguminosarum, which has a multitiered regulatory network involving multiple AHL molecules and several LuxI and LuxR homologs (17, 37, 47, 66), also contains a quorum-sensing system (CinR) that is tuned to respond to low cell densities. In fact, we initially overlooked the density-dependent regulation of the exp genes by ExpR, and it was this R. leguminosarum work that prompted us to explore the possibility that expR is tuned to activate exp gene transcription at low cell densities. However, ExpR is more closely related to many other LuxR family members than it is to CinR (data not shown), suggesting that S. meliloti ExpR and R. leguminosarum CinR are distinct systems tuned to respond to low cell densities.
The results presented here have changed our understanding of how S. meliloti controls the production of symbiotically active EPS II. Prior to this work, it was thought that expR+ strains constitutively produce EPS II. However, we now know that this is not the case. We had previously overlooked the density-dependent production of EPS II because the cell densities used in typical laboratory experiments are much greater than those at which ExpR begins activating transcription of the exp genes. Since the densities of S. meliloti found in soil are typically less than 107 cells per gram of soil (73), it may be that expression of the exp genes in an expR+ strain in the field will be at low levels until the cells are in an environment where the local density is above the threshold for activation of exp gene expression. This could act to conserve cell resources until synthesis of symbiotic polysaccharides is required, for example, inside colonized curled root hairs and infection threads. However, further studies are needed to determine the quantity and molecular weight distribution of EPS II produced at various cell densities and to determine the precise roles of the exp gene products in EPS II synthesis.
The expC- and expE-activating substances present in the conditioned medium and the ethyl acetate culture filtrate extracts have not been chemically identified. However, because our ethyl acetate extracts and HPLC fractions activate both ExpR-dependent transcription of some exp genes and long-chain AHL reporter systems, it seems likely that ExpR also responds to an AHL. The side chains of the expE-active Rm1021 AHL(s) are likely to be very nonpolar. Gray et al. have described purification of a LasR-activating compound from S. meliloti Rm1021 culture supernatants that eluted from a C18 column later than R. leguminosarum 3OH,C14:1-HSL (37). This suggests that the putative AHL(s) produced by strain Rm1021 is structurally distinct from R. leguminosarum 3OH,C14:1-HSL and has carbon chains at least as long as C14. Independent chromatographic analyses also indicate that the AHLs produced by S. meliloti strain Rm1021 are very hydrophobic (13, 58). Further experimentation is required to determine the structures of the Rm1021 AHL molecules and to identify which of these Rm1021 AHLs activate ExpR-mediated transcription of the exp genes.
Interestingly, the S. meliloti genome (http://sequence.toulouse.inra.fr/meliloti.html) contains a number of genes that may have roles in quorum sensing. In addition to expR (the intact gene is SMc03899 plus SMc03896), five ORFs (SMc00170, SMc00877, SMc00878, SMc00658, and SMc04032) in the S. meliloti genome are predicted to encode possible LuxR family members. Two candidate AHL synthase genes are also present: one gene (SMc00168) predicted to encode a LuxI homolog and one gene (SMc00714) predicted to encode a homolog of the Pseudomonas fluorescens HdtS protein (44). It is not clear whether all of these predicted genes function in density-dependent gene expression, but recent work indicates that SMc00170 and SMc00168 may be part of an S. meliloti quorum-sensing network similar to that found in R. leguminosarum (49).
In a number of bacteria, IS elements modulate polysaccharide-dependent phase variation but it is not clear whether this occurs in S. meliloti. Some examples of this include Neisseria meningitidis switching from a capsule-producing form to a capsule-deficient form (38), IS-mediated reversible production of EPS in Pseudoalteromonas atlantica (5), and phase variation of Xanthomonas oryzae pv. oryzae with regard to xanthan gum production (60). It is possible that, in the field, S. meliloti uses the ISRm2011-1 insertion in expR as a genetically plastic switch to control EPS II production. However, under laboratory conditions, both the mucoid colony phenotype of expR+ strains and the nonmucoid colony phenotype of expR::ISRm2011-1 strains are highly stable (Pellock and Walker, unpublished). Additionally, it has been reported that S. meliloti IS element genomic fingerprint patterns are highly stable over long periods of time under laboratory conditions (72). Consistent with this observation, our ISRm2011-1 fingerprinting analysis of SU47 and its daughters (data not shown) revealed a hybridization pattern identical to that reported previously (72). Since the expR ORF is disrupted by ISRm2011-1 in all of the daughters of SU47 that we tested, insertion of the IS element almost certainly occurred before these daughters of SU47 were isolated. However, it is not known whether the expR ORF was already disrupted in the original SU47 field isolate or whether the IS insertion occurred later under laboratory conditions and a SU47 isolate with a less mucoid colony phenotype was chosen for further study.
ISRm2011-1 is a member of the IS1 family of IS elements, which are thought to transpose by a replicative mechanism (14). Since the copy of ISRm2011-1 that disrupts expR has been eliminated from the genome (Fig. 2C and D), it seems likely that this precise excision resulted from a microhomologous recombination event between the 5-bp target site and its duplicate. Precise excisions of IS elements occur with a frequency of 10−6 to 10−10 per element per generation (46), which is consistent with the high stability of the ISRm2011-1 insertion in expR. The fact that the expR+ allele is also very stable suggests that the expR gene is not a hot spot for ISRm2011-1 insertion, as appears to be the case for insertions of ISRm1 in the nif gene region (69).
The results of this study raise a number of intriguing questions about the regulation of the production of EPS II and other symbiotically active S. meliloti polysaccharides. The identity of the cell density signal(s) that stimulates ExpR-dependent gene expression remains to be determined. It will also be of interest to determine whether the production of succinoglycan and/or symbiotically active K antigen is controlled in a density-dependent fashion, whether any non-exp genes are regulated by ExpR, and how the expression of expR itself is regulated. In addition, further biochemical and genetic studies are needed to determine the exact mechanism of ExpR action. The question of how the molecular weight distribution of EPS II is controlled also remains open.
Acknowledgments
We thank Anke Becker, Allan Downie, Clay Fuqua, and the members of the Walker laboratory for helpful suggestions and discussions. We are indebted to Jim Metzger and Anatol Eberhard for help and advice with the HPLC separations of the AHLs. We are grateful to Paul Williams, F.-C. Gong, and S. Swift for the generous gifts of the AHL reporter strains.
This work was supported by Public Health Service grant GM31030 from the National Institutes of Health to G.C.W., National Institutes of Health predoctoral training grant T32GM07287 (B.J.P.), the Massachusetts Institutes of Technology Undergraduate Research Opportunities Program (R.B.), the Ohio Agricultural Research and Development Center (W.D.B.), and an Ohio Agricultural Research and Development Center Graduate Research Enhancement grant (M.T.).
REFERENCES
- 1.Altschul, S., T. Madden, A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aman, P., M. McNeil, L.-E. Franzen, A. G. Darvill, and P. Albersheim. 1981. Structural elucidation, with HPLC-MS and GLC-MS, of the acidic exopolysaccharide secreted by Rhizobium meliloti strain Rm1021. Carbohydr. Res. 95:263-282. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology, vol. 1. Wiley, New York, N.Y.
- 4.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, D. H., and M. Silverman. 1989. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J. Bacteriol. 171:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battisti, L., J. C. Lara, and J. A. Leigh. 1992. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc. Natl. Acad. Sci. USA 89:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, A., S. Rüberg, H. Küster, A. A. Roxlau, M. Keller, T. Ivashina, H. Cheng, G. C. Walker, and A. Pühler. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 179:1375-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker, A., M. Schmidt, W. Jäger, and Pühler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 9.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 10.Brewin, N. J. 1991. Development of the legume root nodule. Annu. Rev. Cell Biol. 7:191-226. [DOI] [PubMed] [Google Scholar]
- 11.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casse, F., C. Boucher, S. Julliot, M. Michel, and J. Dénarié. 1979. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J. Bacteriol. 113:229-242. [Google Scholar]
- 13.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 14.Chandler, M., and O. Fayet. 1993. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 7:497-503. [DOI] [PubMed] [Google Scholar]
- 15.Chen, C.-Y., and S. C. Winans. 1991. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J. Bacteriol. 173:1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cubo, M. T., A. Economou, G. Murphy, A. W. Johnston, and J. A. Downie. 1992. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarum biovar viciae. J. Bacteriol. 174:4026-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dénarié, J., and J. Cullimore. 1993. Lipo-oligosaccharide nodulation factors: a new class of signaling molecules mediating recognition and morphogenesis. Cell 74:951-954. [DOI] [PubMed] [Google Scholar]
- 19.De Vos, G. F., G. C. Walker, and E. R. Signer. 1986. Genetic manipulations in Rhizobium meliloti using two new transposon Tn5 derivatives. Mol. Gen. Genet. 204:485-491. [DOI] [PubMed] [Google Scholar]
- 20.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dylan, T., D. R. Helinski, and G. S. Ditta. 1990. Hypoosmotic adaptation in Rhizobium meliloti requires β-(1→2)-glucan. J. Bacteriol. 172:1400-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dylan, T., L. Ielpi, S. Stanfield, L. Kashyap, C. Douglas, M. Yanofsky, E. Nester, D. R. Helinski, and G. Ditta. 1986. Rhizobium meliloti genes required for nodule development are related to chromosomal virulence genes in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 83:4403-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberhard, A. 1972. Inhibition and activation of bacterial luciferase synthesis. J. Bacteriol. 109:1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 25.Engebrecht, J., and M. Silverman. 1987. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 15:10455-10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finan, T. M., E. K. Hartwieg, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flavier, A. B., S. J. Clough, M. A. Schell, and T. P. Denny. 1997. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26:251-259. [DOI] [PubMed] [Google Scholar]
- 29.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Biukema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 30.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 31.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 32.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 33.Glazebrook, J., G. Meiri, and G. C. Walker. 1992. Genetic mapping of symbiotic loci on the Rhizobium meliloti chromosome. Mol. Plant-Microbe Interact. 5:223-227. [DOI] [PubMed] [Google Scholar]
- 34.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the Calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661-672. [DOI] [PubMed] [Google Scholar]
- 35.González, J. E., B. L. Reuhs, and G. C. Walker. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 93:8636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.González, J. E., G. M. York, and G. C. Walker. 1996. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene 179:141-146. [DOI] [PubMed] [Google Scholar]
- 37.Gray, K. M., J. P. Pearson, J. A. Downie, B. E. Boboye, and E. P. Greenberg. 1996. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J. Bacteriol. 178:372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 39.Her, G.-R., J. Glazebrook, G. C. Walker, and V. N. Reinhold. 1990. Structural studies of a novel exopolysaccharide produced by a mutant of Rhizobium meliloti strain Rm1021. Carbohydr. Res. 198:305-312. [DOI] [PubMed] [Google Scholar]
- 40.Ielpi, L., T. Dylan, G. S. Ditta, D. R. Helinski, and S. W. Stanfield. 1990. The ndvB locus of Rhizobium meliloti encodes a 319-kDa protein involved in the production of β-(1→2)-glucan. J. Biol. Chem. 265:2843-2851. [PubMed] [Google Scholar]
- 41.Katzen, F., A. Becker, M. V. Ielmini, C. G. Oddo, and L. Ielpi. 1999. New mobilizable vectors suitable for gene replacement in gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl. Environ. Microbiol. 65:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keller, M., A. Roxlau, W. M. Weng, M. Schmidt, J. Quandt, N. Karsten, D. Jording, W. Arnold, and A. Pühler. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant-Microbe Interact. 8:267-277. [DOI] [PubMed] [Google Scholar]
- 43.Labes, G., and R. Simon. 1990. Isolation of DNA insertion elements from Rhizobium meliloti which are able to promote transcription of adjacent genes. Plasmid 24:235-239. [DOI] [PubMed] [Google Scholar]
- 44.Laue, B. E., Y. Jiang, S. R. Chhabra, S. Jacob, G. S. Stewart, A. Hardman, J. A. Downie, F. O'Gara, and P. Williams. 2000. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology 146:2469-2480. [DOI] [PubMed] [Google Scholar]
- 45.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewin, B. 1994. Genes V. Oxford University Press, Oxford, United Kingdom.
- 47.Lithgow, J. K., A. Wilkinson, A. Hardman, B. Rodelas, F. Wisniewski-Dye, P. Williams, and J. A. Downie. 2000. The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 37:81-97. [DOI] [PubMed] [Google Scholar]
- 48.Long, S. R. 1989. Rhizobium-legume nodulation: life together in the underground. Cell 56:203-214. [DOI] [PubMed] [Google Scholar]
- 49.Marketon, M. M., and J. E. González. 2002. Identification of two quorum sensing systems in Sinorhizobium meliloti. J. Bacteriol. 184:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 51.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendrygal, K. E., and J. E. González. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 54.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niehaus, K., and A. Becker. 1998. The role of microbial surface polysaccharides in the Rhizobium-legume interaction. Subcell. Biochem. 29:73-116. [DOI] [PubMed] [Google Scholar]
- 56.Pellock, B. J., H.-P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puskas, A., E. P. Greenberg, S. Kaplan, and A. L. Schaefer. 1997. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179:7530-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putnoky, P., E. Grosskopf, D. T. Ha, G. B. Kiss, and A. Kondorosi. 1988. Rhizobium fix genes mediate at least two communication steps in symbiotic nodule development. J. Cell Biol. 106:597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajeshwari, R., and R. V. Sonti. 2000. Stationary-phase variation due to transposition of novel insertion elements in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 182:4797-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reinhold, B. B., S. Y. Chan, T. L. Reuber, A. Marra, G. C. Walker, and V. N. Reinhold. 1994. Detailed structural characterization of succinoglycan, the major symbiotically important exopolysaccharide of Rhizobium meliloti strain Rm1021. J. Bacteriol. 176:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reuhs, B. L., R. W. Carlson, and J. S. Kim. 1993. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-D-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 175:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reuhs, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. Kolli, and S. G. Pueppke. 1998. Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64:4930-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reuhs, B. L., M. N. V. Williams, J. S. Kim, R. W. Carlson, and F. Côté. 1995. Suppression of the Fix− phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J. Bacteriol. 177:4289-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson, J. B., O. H. Tuovinen, and W. D. Bauer. 1992. Role of divalent cations in the subunit associations of complex flagella from Rhizobium meliloti. J. Bacteriol. 174:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodelas, B., J. K. Lithgow, F. Wisniewski-Dye, A. Hardman, A. Wilkinson, A. Economou, P. Williams, and J. A. Downie. 1999. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 181:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosemeyer, V., J. Michiels, C. Verreth, and J. Vanderleyden. 1998. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J. Bacteriol. 180:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosenburg, C., P. Boistard, J. Dénarié, and F. Casse-Delbart. 1981. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol. Gen. Genet. 184:326-333. [DOI] [PubMed] [Google Scholar]
- 69.Ruvkun, G. B., S. R. Long, H. M. Meade, R. C. van den Bos, and F. M. Ausubel. 1982. ISRm1: a Rhizobium meliloti insertion sequence that transposes preferentially into nitrogen fixation genes. J. Mol. Appl. Genet. 1:405-418. [PubMed] [Google Scholar]
- 70.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 71.Schaefer, A. L., B. L. Hanzelka, M. R. Parsek, and E. P. Greenberg. 2000. Detection, purification, and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 305:288-301. [DOI] [PubMed] [Google Scholar]
- 72.Simon, R., B. Hötte, B. Klauke, and B. Kosier. 1991. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J. Bacteriol. 173:1502-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spaink, H. P., A. Kondorosi, and P. J. J. Hooykaas (ed.). 1998. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 74.Stanfield, S. W., L. Ielpi, D. O'Brochta, D. R. Helinski, and G. S. Ditta. 1988. The ndvA gene product of Rhizobium meliloti is required for β-(1→2)glucan production and has homology to the ATP-binding export protein HlyB. J. Bacteriol. 170:3523-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. Macintyre, and G. S. A. B. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teplitski, M., J. B. Robinson, and W. D. Bauer. 2000. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microbe Interact. 13:637-648. [DOI] [PubMed] [Google Scholar]
- 77.Urzainqui, A., and G. C. Walker. 1992. Exogenous suppression of the symbiotic deficiencies of Rhizobium meliloti exo mutants. J. Bacteriol. 174:3403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Rhijn, P., and J. Vanderleyden. 1995. The Rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vincent, J. M. 1941. Serological properties of the root-nodule bacteria. I. Strains of Rhizobium meliloti. Proc. Linn. Soc. N. S. W. 66:145-154. [Google Scholar]
- 80.von Bodman, S. B., D. R. Majerczak, and D. L. Coplin. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. USA 95:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang, L.-X., Y. Wang, B. J. Pellock, and G. C. Walker. 1999. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J. Bacteriol. 181:6788-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, X. D., P. A. de Boer, and L. I. Rothfield. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10:3363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watson, R. J., and R. Wheatcroft. 1991. Nucleotide sequence of Rhizobium meliloti insertion sequence ISRm1: homology to IS2 from Escherichia coli and IS426 from Agrobacterium tumefaciens. DNA Seq. 2:163-172. [DOI] [PubMed] [Google Scholar]
- 84.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 85.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 86.Wood, D. W., and L. S. Pierson III. 1996. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene 168:49-53. [DOI] [PubMed] [Google Scholar]
- 87.Zevenhuizen, L. P. T. M., and P. Faleschini. 1991. Effect of the concentration of sodium chloride in the medium on the relative proportions of poly- and oligo-saccharides excreted by Rhizobium meliloti strain YE-2Sl. Carbohydr. Res. 209:203-209. [DOI] [PubMed] [Google Scholar]
- 88.Zhan, H., S. B. Levery, C. C. Lee, and J. A. Leigh. 1989. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc. Natl. Acad. Sci. USA 86:3055-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhan, H. J., C. C. Lee, and J. A. Leigh. 1991. Induction of the second exopolysaccharide (EPSb) in Rhizobium meliloti SU47 by low phosphate concentrations. J. Bacteriol. 173:7391-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang, Z., and L. S. Pierson III. 2001. A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 67:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]