Abstract
Pasteurella multocida was grown in iron-free chemically defined medium supplemented with hemoglobin, transferrin, ferritin, and ferric citrate as iron sources. Whole-genome DNA microarrays were used to monitor global gene expression over seven time points after the addition of the defined iron source to the medium. This resulted in a set of data containing over 338,000 gene expression observations. On average, 12% of P. multocida genes were differentially expressed under any single condition. A majority of these genes encoded P. multocida proteins that were involved in either transport and binding or were annotated as hypothetical proteins. Several trends are evident when the data from different iron sources are compared. In general, only two genes (ptsN and sapD) were expressed at elevated levels under all of the conditions tested. The results also show that genes with increased expression in the presence of hemoglobin did not respond to transferrin or ferritin as an iron source. Correspondingly, genes with increased expression in the transferrin and ferritin experiments were expressed at reduced levels when hemoglobin was supplied as the sole iron source. Finally, the data show that genes that were most responsive to the presence of ferric citrate did not follow a trend similar to that of the other iron sources, suggesting that different pathways respond to inorganic or organic sources of iron in P. multocida. Taken together, our results demonstrate that unique subsets of P. multocida genes are expressed in response to different iron sources and that many of these genes have yet to be functionally characterized.
Pasteurella multocida is a gram-negative, rod-shaped, facultative anaerobe that has been isolated from a wide range of mammals and birds throughout the world. This organism is the etiologic agent of a variety of economically significant diseases, including fowl cholera in chickens and turkeys, hemorrhagic septicemia in cattle and buffalo, atrophic rhinitis in swine, and snuffles in rabbits (17, 21). P. multocida is also the most common human infection resulting from small-mammal bites (22). Previously identified P. multocida virulence factors include a capsule, hemagglutinins, lipopolysaccharide, the filamentous hemagglutinin PfhB, and a dermonecrotic toxin in certain toxigenic strains (7). However, the molecular mechanisms by which P. multocida can survive in the environment and successfully infect and cause disease in various hosts remain unclear.
Iron is an essential nutrient for most organisms because of its importance in metabolic electron transport chains. Iron has very low solubility at neutral pH, and hence, it must be associated with specialized protein carriers in order to remain accessible. In mammals and birds, these protein carriers can include transferrin in blood, lactoferrin in secretory fluids, and ferritin within cells. It has been determined that because of the activity of these protein carriers, the concentration of free iron normally present in mammals and birds is not enough to support the growth of bacteria (3, 4). The low availability of iron within host species is one of the first barriers to infection that microorganisms must overcome. Bacteria have developed several strategies for obtaining iron from their hosts. Among the best-studied bacterial systems for iron acquisition are siderophores, which are low-molecular-weight iron ligands that are able to compete with host protein carriers for ferric iron binding (29). Bacteria can also acquire iron when it is associated with other molecules, including heme, hemoglobin, and ferric citrate.
Whole-genome sequencing has revealed that P. multocida has many iron-related genes, some of which are duplicated (16) (Table 1). Previous investigations of P. multocida have identified iron-regulated outer membrane proteins that are produced in response to iron limitation (8, 13, 18, 25, 27, 30); however, the genes encoding most of these proteins have not been identified. Importantly, Ruffolo and coworkers have shown that P. multocida grown under low-iron conditions can provide cross-protection against subsequent infections (23). More recently, we utilized microarrays to monitor gene expression in P. multocida grown under low-iron conditions formed by the addition of an iron chelator (20). The results of these investigations showed that 135 genes were significantly altered in expression during the first 2 h of growth under iron-limiting conditions. These results led us to conclude that a variety of P. multocida genes are transcriptionally responsive to environmental iron and that many of the genes have not yet been functionally characterized and their mechanism of regulation is unknown.
TABLE 1.
Iron acquisition-related genes identified by homology in P. multocida
| ORF | Gene | Description |
|---|---|---|
| 40 | pfhR | Outer membrane heme receptor |
| 42 | hugZ | Heme utilization protein |
| 51 | fbpA | Iron-binding protein FbpA precursor |
| 128 | fecE | Membrane-bound iron(III) dicitrate transport protein |
| 129 | fecD | Iron(III) dicitrate transport system permease |
| 130 | fecC | Iron(III) dicitrate transport system permease |
| 131 | fecB | Iron(III) dicitrate periplasmic binding protein |
| 203 | afuA | AfuA protein |
| 299 | hugZ | Heme utilization protein |
| 352 | fur | Ferric uptake regulation protein |
| 397 | yfeD | Iron (chelated) transporter, permease protein |
| 398 | yfeC | Iron (chelated) transporter, permease protein |
| 399 | yfeB | Iron (chelated) transporter, ATP-binding protein |
| 400 | yfeA | Iron (chelated) transporter, periplasmic-binding protein |
| 576 | hemR | Heme-hemopexin utilization protein C |
| 592 | hbpA | Heme-binding protein A precursor |
| 666 | rsgA | Ferritin-like protein 1 |
| 667 | rsgA | Ferritin-like protein 2 |
| 953 | afuA | AfuA protein |
| 954 | afuA | AfuA protein |
| 955 | afuA | AfuA protein |
| 956 | afuB | AfuB protein |
| 957 | afuC | Ferric transport protein |
| 1186 | exbB | Biopolymer transport protein |
| 1187 | exbD | Biopolymer transport protein |
| 1188 | tonB | TonB protein |
| 1308 | hemU | ABC transporter, hemin permease homolog |
| 1455 | afuB | AfuB homolog |
| 1622 | hasR | Heme receptor |
Although the function of many iron transport systems present in P. multocida has been well studied in other organisms, the exact role these genes play in the uptake of iron from different biological sources has not been established. To identify the transcriptional response of P. multocida to defined sources of iron, we have used whole-genome DNA microarrays representing all of the 2,014 identified open reading frames (ORFs) (16). The gene expression patterns of P. multocida grown in iron-free chemically defined medium with and without supplemental iron sources were then compared to identify genes that respond to specific sources of iron.
Details of the methods used are described on our website (http://www.agac.umn.edu/Microarray/definedirondata/index.htm). In brief, P. multocida PM70 was grown to log phase, split into two equal volumes, and resuspended in chemically defined medium (CDM) or CDM supplemented with ferric citrate (0.01 mM), hemoglobin (turkey; 0.01 mg/ml), transferrin (bovine; 0.01 mg/ml), or ferritin (horse; 0.01 mg/ml) (Sigma, St. Louis, Mo.). Samples were taken 1, 5, 10, 15, 30, 60, and 90 min after resuspension, and total RNA was extracted. Gene expression analysis with DNA microarrays was performed as described at our website (http://www.agac.umn.edu/microarray/protocols/protocols.htm). Replicate hybridizations with reverse dye labeling did not reveal any systematic biases. Quality filtering of the replicate spots was achieved by applying the Shapiro-Wilk normality test (W test) (24) to the sets of data, and replicates not falling within a normal distribution were discarded. A minimum of three data points was needed to calculate the average ratio. Data visualization and analysis were performed by using Spotfire DecisionSite 7.0 software.
Effect of supplemental hemoglobin.
A complete set of all of our results is available at http://www.agac.umn.edu/Microarray/definedirondata/index.htm. Hemoglobin functions as an oxygen carrier by reversibly binding oxygen and transporting it to tissues throughout the body. It is composed of four polypeptide chains, each of which contains a heme group complexed to an iron atom. Many bacteria are able to remove these iron atoms from hemoglobin and use them as an iron source (15, 28). The expression of 7% of all P. multocida genes was significantly elevated over the course of the experiment, and 5% of the genes were expressed at reduced levels when hemoglobin was added to the medium, as determined by significance analysis of microarrays (SAM) software (26).
Several genes involved in the use of tetrathionate for electron transport were elevated in expression an average of 2.1-fold in the presence of hemoglobin. They included ttrAC and the two-component regulatory system ttrRS. Interestingly, the ttr genes are under the global control of fnr, which is located adjacent to two ferritin-like genes that were expressed at reduced levels during the experiment (10). Several other genes thought to be under the regulatory control of Fnr were transcribed at elevated levels in response to hemoglobin, including cydB, nrfF, focA, frdD, and glpACTQ (1.5- to 2.4-fold). The involvement of Fnr regulation may indicate that the presence of hemoglobin drives P. multocida toward anaerobic metabolism. Other genes involved in electron transport that were expressed at elevated levels included PM0973 (cytochrome D oxidase), PM1329 (ubiquinone oxidoreductase), and PM1598 (cytochrome c-type protein), whose expression was an average of twofold higher. Interestingly, four genes involved in glycolysis (eno, tpiA, pfkA, and gapdh) were expressed at an average maximum of 2.1-fold reduced levels when hemoglobin was added to the minimal medium used.
The gene encoding the filamentous hemagglutinin, PfhB2, was expressed at 2.6-fold elevated levels when hemoglobin was added to the medium. This gene has previously been identified as a P. multocida virulence factor and was associated with in vivo survival in a mouse model (10, 16). Several genes thought to participate in cell surface biosynthesis were also expressed at elevated levels. They included ponC (2.2-fold), gmhA (2-fold), and type 4 fimbrial subunit PM0084 (2-fold). PM0885 was also expressed at twofold lower levels for the first 30 min of the experiment and encodes an acyl transferase involved in lipid A biosynthesis. These data are consistent with the presence of a general sensor-effector system in P. multocida that responds to stimuli, indicative of in vivo growth.
Several regulatory genes were expressed at lower levels in the presence of hemoglobin. Aerobic respiration control protein ArcA is encoded by PM0219 and was expressed at reduced levels throughout the experiment, including a sixfold reduction in expression at 10 min. The genes encoding the chemotaxis sensor protein CpxA and universal stress protein A were expressed at 2.6-fold reduced levels during the experiment, as was sigma factor RpoH, which peaked at approximately 3-fold reduced levels at 5 and 90 min.
Many hypothetical proteins (n = 47) were also expressed at altered levels in the presence of hemoglobin. Genes encoding the hypothetical proteins PM1880 to PM1884 were expressed at 2.3-fold elevated levels when hemoglobin was added to the medium. Genes that were expressed at reduced levels in the presence of hemoglobin included PM1566 and PM0980, which were expressed at nearly twofold lower levels throughout the experiment.
A large number (n = 49) of transport and binding genes were expressed at altered levels in response to hemoglobin. PM0068 encodes a putative transport permease protein and was expressed at nearly twofold higher levels for the entire experiment. ORFs PM0335 to PM0337 were expressed at a level an average of 1.6-fold higher and encode a proton-glutamate symport, a TonB-dependent receptor, and a hemoglobin-binding protein, respectively. The genes encoding the peptide ABC transport proteins SapBCD and C4-dicarboxylate permease proteins (PM1252, PM1253, and PM1254) were all maximally elevated in expression an average of 1.7-fold. The ABC transport periplasmic binding protein AfuA encoded by PM0953 and the DNA-binding protein AfuC (PM0957) were both transcribed at twofold higher levels. Several other genes homologous to cation transport genes were expressed at elevated levels, including PM0576 (HemR) and PM0428 (antiporter). PM0331 encodes the outer membrane protein OmpW and was expressed at more than twofold reduced levels for the first 30 min of the experiment. PM0666 and PM0667 encode ferritin-like proteins and were expressed at twofold reduced levels in the presence of hemoglobin. This intuitively makes sense, as cells would not want to store iron if it were present in limited amounts. The putative arginine transport genes artP and artI were also expressed at twofold reduced levels during most of the experiment. These genes have also been previously shown to be responsive to nutrient limitation and warrant further study as putative stress response regulatory genes in P. multocida (19).
Effect of supplemental transferrin.
Transferrins are metal-binding proteins that have a high affinity for iron in vivo and are capable of reversibly binding two ferric ions. They function to reduce the concentration of extracellular iron, thereby preventing the formation of oxygen radicals and making it unavailable for use by bacterial pathogens. Bacteria may acquire iron from transferrin by binding and unloading transferrin at the cell surface or secreting their own iron-binding proteins (9). Ten percent of all P. multocida genes were significantly elevated over the course of the experiment, and 8% of the genes were expressed at reduced levels over the course of the experiment when transferrin was added to the medium.
PM0352 encodes the ferric uptake regulator protein Fur and was transcribed at levels elevated greater than twofold from 5 to 10 min after the addition of transferrin to the medium. Interestingly, fur expression was not dramatically altered under any of the other experimental conditions tested. Other genes encoding regulatory proteins that were expressed at elevated levels included the tryptophan biosynthesis repressor gene trpR (1.6-fold), the ribose utilization repressor gene rbsR (1.7-fold), and the stringent response modulator gene spoT (1.7-fold). Correspondingly, genes involved in tryptophan biosynthesis and ribose transport and utilization were expressed at reduced or undetectable levels. Among the regulatory genes expressed at reduced levels were the nitrogen regulatory gene glnB (1.9-fold), the glycerol-3-phosphate repressor gene glpR (1.4-fold), and the arginine biosynthesis repressor gene argR (1.4-fold). Genes under the control of these repressors were expressed at relatively higher levels than the corresponding regulatory genes.
There were a large number (n = 75) of genes homologous to genes encoding hypothetical proteins that were significantly altered in expression. The genes encoding hypothetical proteins PM1125 and PM1637 were initially expressed at twofold higher levels within the first 5 min. Genes expressed at reduced levels included PM0133, PM1095, and PM1125.
The iron transport protein energy-transducing genes exbB, exbD, and tonB were expressed at 2.1-fold elevated levels when transferrin was added to iron-free medium. Some of the other genes that were elevated in expression included those encoding the tryptophan transport protein Mtr, the transmembrane permease FbpB, and the branched-chain amino acid transporter AzlD. Interestingly, the gene encoding outer membrane protein OmpW was expressed at 1.7-fold elevated levels in response to transferrin but was expressed at significantly (2.9-fold) reduced levels when hemoglobin was present. Conversely, PM0953 encodes one of the afuA homologs and was expressed at elevated levels in both transferrin and hemoglobin but afuC expression was reduced 1.4-fold in response to transferrin and its expression was elevated in hemoglobin. Together, these data suggest that the expression of these genes occurs in response to specific, specialized signals in P. multocida that are coregulated.
Effect of supplemental ferritin.
Ferritin proteins form hollow spheres that are capable of storing iron atoms, thereby making them available for use by cells, but protect the cells from the toxic effects of iron accumulation. They are commonly found inside bacterial and eukaryotic cells but may also be found circulating in plasma (1, 12). The expression of 4% of all P. multocida genes was significantly elevated over the course of the experiment, and 7% of the genes were expressed at reduced levels over the course of the experiment when ferritin was added to the medium.
Regulatory genes that were expressed at elevated levels included the nitrogen regulator gene ptsN (1.8-fold), as well as the ribose operon repressor gene rbsR (1.9-fold), which was also highly expressed in response to transferrin. Genes expressed at levels reduced 1.6-fold or more in response to ferritin included those encoding the regulatory proteins PM0417 (putative transcriptional regulator), Mtl (mannitol biosynthesis repressor), and ArgR (arginine biosynthesis repressor). Interestingly, genes encoding the transcription factors NusA and NusG were expressed at 1.9-fold elevated levels during the experiment. These proteins have been shown to have counteracting, although noncompeting, effects (5).
As in the other experiments discussed thus far, a relatively large number (n = 43) of genes homologous to genes encoding hypothetical proteins were expressed at altered levels when cells were exposed to ferritin.
PM0241 and PM0242 were expressed at twofold elevated levels in response to ferritin and encode a putative membrane protein and an ATP-binding protein with homology to ABC transport proteins. Other transport genes whose expression was elevated included PM0050 (transmembrane permease FbpB), PM0435 and PM0436 (phosphate), PM0914 (peptide), and PM1971 and PM1972 (carbohydrate and an unknown product, respectively). Genes expressed at lower levels in the presence of ferritin included a sodium-dependent transport gene (PM0718), the magnesium and cobalt transport gene corA, and lctP, which encodes a lactase permease.
Effect of supplemental ferric citrate.
Citrate is a chemical compound found in most cells and is important for its role in the metabolism of carbohydrates and fats. It is capable of binding ferric iron, thereby making it soluble. Some bacteria are able to utilize Fe3+ when it is delivered to the cell surface as a citrate complex (2). Five percent of all P. multocida genes were significantly elevated over the course of the experiment, and 5% of the genes were expressed at reduced levels over the course of the experiment when ferric citrate was added to the medium. Overall, stimulation with ferric citrate resulted in the lowest transcriptional response level.
Interestingly, transcripts encoding a putative alpha-hemolysin (PM1164) and competence factor (comD) were present at 2.1- and 2.6-fold higher levels during the first 10 min of the experiment, respectively. The regulatory genes uspA and fnr were transcribed at 1.9-fold lower levels in response to ferric citrate. Several genes encoding transport proteins, including the S-independent protein transporter TatB and a putative permease protein (PM471), were expressed at reduced levels when ferric citrate was present in the medium.
Confirmatory RT-PCR analyses.
To confirm the results obtained from the microarray analyses, we used real-time reverse transcription (RT)-PCR to compare RNA levels between the samples obtained at 10 min after hemoglobin was added to iron-free medium. The 10 genes analyzed were found to have a strong correlation (r2 = 0.88) to the microarray results.
General trends.
Several trends become apparent when results from multiple experiments are compared. Figure 1 presents an overall view of the experiments, with the genes clustered according to the similarity of their expression profiles. Cluster A contains several cell surface and iron transport protein-encoding genes that were expressed at elevated levels in response to transferrin and ferritin, including the filamentous hemagglutinin PfhB2, the iron-binding protein FbpA, and the rod shape-determining proteins MreBD. Genes that were generally expressed at elevated levels in all experiments except hemoglobin fell into cluster B and include three involved in cell division (ftsQAZ) and menaquinone biosynthesis (menCBG), fur, and uspA. Cluster D contains genes that were generally expressed at elevated levels in hemoglobin only. This cluster includes genes encoding proteins involved in competence (ComBCE), oligopeptide transport (OppACF), and tryptophan biosynthesis (TrpBCDG). Genes generally expressed in all of the experiments are found in cluster E and include one that encodes a tryptophan transporter (mtr) and one of the afuA copies found in the P. multocida genome.
FIG. 1.
Hierarchical clustering of microarray expression data. The ratio of fluorescence intensities between control and experimental populations across four experiments (hemoglobin, transferrin, ferritin, and ferric citrate) was log transformed and normalized between experiments by subtracting the mean change (n-fold). Red and green represent the increase (n-fold) and the decrease (n-fold), respectively, in gene expression. Data from an experiment using BHI medium as the iron source are presented for reference only and were not used for clustering. Unweighted hierarchical clustering using correlation as a similarity measure was performed by using Spotfire DecisionSite 6.3 software. Tables display selected genes from the clusters highlighted by the corresponding colors. Genes encoding hypothetical proteins are designated by “hyp.” The cluster tables contain genes that were generally expressed at elevated levels in response to transferrin and ferritin (A), expressed at elevated levels in response to all conditions except hemoglobin (B), expressed at reduced levels in all experiments (C), expressed at elevated levels in response to hemoglobin only (D), and expressed at elevated levels in all experiments (E).
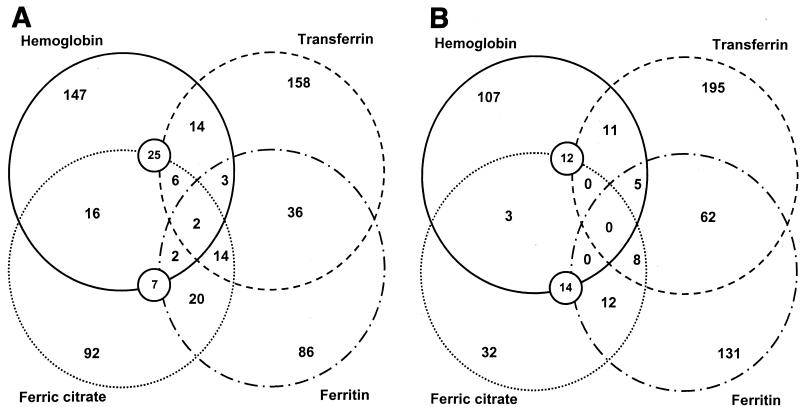
The numbers of genes altered in expression across multiple experiments are shown in Fig. 2. Transferrin and ferritin have the most coregulated genes (n = 98). Genes expressed at altered levels in the presence of hemoglobin had the least in common with the other conditions tested, although there were many similarities between cells grown in hemoglobin and those grown in brain heart infusion (BHI) medium (Fig. 1), which intuitively makes sense, as BHI medium contains hemoglobin (19). Forty genes were expressed at altered levels in response to three or more conditions. For cells growing in the presence of hemoglobin, ferritin, and ferric citrate, genes whose expression was elevated included those encoding the putative regulatory protein PtsN (6), the transcriptional antitermination factor NusA, and the ATP-binding proteins YebM and SapD. The genes encoding PtsN and SapD were also expressed at elevated levels when transferrin was added to the medium. Genes expressed at reduced levels in response to transferrin, ferritin, and ferric citrate included the mannitol operon repressor gene mtl, the glycerol-3-phosphate regulon repressor gene glpR, and the putative permease gene PM0471.
FIG. 2.
Numbers of P. multocida genes that were significantly altered in expression. Significance was determined by using SAM software as described in Materials and Methods. The numbers enclosed by overlapping circles represent the numbers of genes whose expression was elevated (A) or lowered (B) under the conditions indicated.
The gene encoding tryptophan repressor protein TrpR was expressed at elevated levels when transferrin and ferritin were added to the medium. It is worth noting that the gene encoding the arginine repressor ArgR was expressed at reduced levels during these times. Throughout all of the experiments, whenever trpR was transcribed at elevated levels, argR transcription was reversed and vice versa. This supports previous observations indicating an inverse relationship between the expressions of genes under the control of these regulators (14, 19).
PM1020, encoding a putative ADP-ribose pyrophosphatase, was expressed at reduced levels when hemoglobin, transferrin, and ferritin were added to the medium. Also, the gene encoding the catabolite repressor protein GlpR and the ribosomal subunit L21 were generally transcribed at lower levels when any of the iron sources was added to the culture. PM0242 (yebM) codes for a putative zinc uptake ATP-binding protein, and its expression was elevated at least twofold when each of the iron sources was added to iron-free medium.
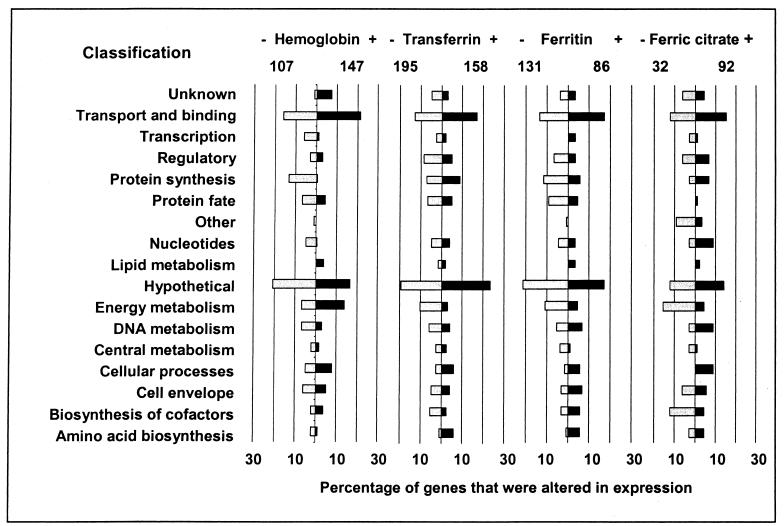
The percentage of genes from different functional categories whose expression was altered during the experiments is presented in Fig. 3. P. multocida expressed a higher percentage of energy metabolism genes (14%) when grown in the presence of hemoglobin than when grown in any of the other iron sources. Additionally, growth of cells in the presence of ferric citrate resulted in the decreased expression of only 3% of the genes related to protein synthesis and fate. Another interesting feature of genes expressed at reduced levels in ferric-citrate-supplemented medium is the large percentage (13%) of genes encoding cofactor biosynthesis proteins.
FIG. 3.
Percentages of P. multocida genes whose expression was significantly altered upon exposure to defined iron sources. The total numbers of genes that were expressed at significant levels throughout the experiments are displayed below the supplemented iron sources. Genes are grouped by functional category, and the values shown are the percentages of the total number of genes that were transcriptionally altered.
Concluding comments.
Acquisition of iron from host animals is one of the first hurdles bacteria must overcome in order to establish an infection. The absolute requirement of most bacteria for iron, coupled with the inherently scarce iron supply in healthy animals, means bacteria must produce proteins that are capable of acquiring iron from the environment. These proteins are ideal candidates for further study, as they are likely essential for bacterial growth and pathogenesis. Our present study focused on the identification of genes in P. multocida that are expressed in response to specific biological iron sources in vitro. By utilizing whole-genome microarrays, we hoped to classify genes on the basis of their transcriptional activity, as well as to identify hypothetical genes that may play an important role in iron acquisition. The results of this type of analysis can then be used to generate new hypotheses and identify genes that require further study and characterization. For each of the iron sources tested, some of the most strongly transcriptionally altered genes were homologous only to genes encoding proteins with hypothetical or unknown functions.
The expression of many known iron acquisition genes was altered during exposure to different iron sources. The iron transport genes afuABC appear to be regulated by Fur, as they were transcribed at elevated levels only when fur expression was reduced. The bacterial heme biosynthesis and utilization genes hemBGL and hemR were expressed at elevated levels only in the presence of hemoglobin. Interestingly, the genes encoding iron transport proteins FpbB and TonB were expressed at similar levels throughout all of the experiments and did not appear to be under Fur regulation. Many of the genes known to be involved in iron acquisition were not altered in transcription during growth in the presence of defined iron sources. This may indicate that these genes do not respond to the presence of extracellular iron and are instead used as generic transporters of free iron that has been acquired by specialized iron-specific genes.
One notable feature of the data was an overall lack of dramatic changes in gene expression. None of the genes varied more then sixfold between the experimental and control populations. Several factors may have contributed to this. P. multocida was initially grown to log phase in BHI medium; therefore, the cells may have had a reserve of iron available for utilization immediately following resuspension in minimal medium. Additionally, although several steps were taken to ensure that the medium and glassware did not contain free iron, a trace amount of contaminating iron may be enough to weaken the transcriptional response to supplemental iron sources. It is also likely that the bacteria were growing at a generally reduced rate because of the nutrient limitations imposed by the minimal medium. Finally, some of the observed transcriptional changes may have been due to species-specific effects. Supplemental iron sources were obtained from several different species (avian, bovine, and equine), and it is likely that unique transcriptional responses would be observed if iron sources from alternate species were used. The preceding caveats notwithstanding, the experimental design used in these experiments provides us with a useful system for studying the initial events of infection in vitro and provides strong evidence for the existence of specialized responses to defined sources of iron in P. multocida.
Acknowledgments
M.P. is supported by an NIH NIGMS Training for Future Biotechnology Development grant (T32 GM08347). The work of D.C. and D.B. was supported in part by NSF grant IIS-9811229. Research in the laboratory of V.K. is supported by grants from the Minnesota Turkey Growers Association, the Minnesota Agricultural Experimentation Station, the University of Minnesota Academic Health Center, the National Institutes of Health, and the U.S. Department of Agriculture's National Research Initiative.
REFERENCES
- 1.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 2.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109. [DOI] [PubMed] [Google Scholar]
- 3.Bullen, J. J. 1981. The significance of iron in infection. Rev. Infect. Dis. 3:1127-1138. [DOI] [PubMed] [Google Scholar]
- 4.Bullen, J. J., H. J. Rogers, and E. Griffiths. 1978. Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol. 80:1-35. [DOI] [PubMed] [Google Scholar]
- 5.Burns, C. M., L. V. Richardson, and J. P. Richardson. 1998. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J. Mol. Biol. 278:307-316. [DOI] [PubMed] [Google Scholar]
- 6.Cases, I., J. A. Lopez, J. P. Albar, and V. de Lorenzo. 2001. Evidence of multiple regulatory functions for the PtsN (IIANtr) protein of Pseudomonas putida. J. Bacteriol. 183:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanter, N. 1990. Molecular aspects of the virulence of Pasteurella multocida. Can. J. Vet. Res. 54:S45-S47. [PubMed] [Google Scholar]
- 8.Choi-Kim, K., S. K. Maheswaran, L. J. Felice, and T. W. Molitor. 1991. Relationship between the iron regulated outer membrane proteins and the outer membrane proteins of in vivo grown Pasteurella multocida. Vet. Microbiol. 28:75-92. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen, C. N., and P. F. Sparling. 1994. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol. Microbiol. 14:843-850. [DOI] [PubMed] [Google Scholar]
- 10.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 11.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 12.Harrison, P. M., and P. Arosio. 1996. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275:161-203. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda, J. S., and D. C. Hirsh. 1988. Antigenically related iron-regulated outer membrane proteins produced by different somatic serotypes of Pasteurella multocida. Infect. Immun. 56:2499-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, B. C. 1995. Quelling the red menace: haem capture by bacteria. Mol. Microbiol. 18:383-390. [DOI] [PubMed] [Google Scholar]
- 16.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morishita, T. Y., L. J. Lowenstine, D. C. Hirsh, and D. L. Brooks. 1996. Pasteurella multocida in raptors: prevalence and characterization. Avian Dis. 40:908-918. [PubMed] [Google Scholar]
- 18.Ogunnariwo, J. A., J. Alcantara, and A. B. Schryvers. 1991. Evidence for non-siderophore-mediated acquisition of transferrin-bound iron by Pasteurella multocida. Microb. Pathog. 11:47-56. [DOI] [PubMed] [Google Scholar]
- 19.Paustian, M. L., B. May, and V. Kapur. 2002. Transcriptional response of Pasteurella multocida to nutrient limitation. J. Bacteriol. 184:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rimler, R. B., R. D. Angus, and M. Phillips. 1989. Evaluation of the specificity of Pasteurella multocida somatic antigen-typing antisera prepared in chickens, using ribosome-lipopolysaccharide complexes as inocula. Am. J. Vet. Res. 50:29-31. [PubMed] [Google Scholar]
- 22.Rimler, R. B., and K. R. Rhoades. 1989. Solubilization of membrane-associated cross-protection factor(s) of Pasteurella multocida. Avian Dis 33:258-263. [PubMed] [Google Scholar]
- 23.Ruffolo, C. G., B. H. Jost, and B. Adler. 1998. Iron-regulated outer membrane proteins of Pasteurella multocida and their role in immunity. Vet. Microbiol. 59:123-137. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro, S. S., and M. B. Wilk. 1965. An analysis of variance test for normality (complete samples). Biometrika 52:591-611. [Google Scholar]
- 25.Snipes, K. P., L. M. Hansen, and D. C. Hirsh. 1988. Plasma- and iron-regulated expression of high molecular weight outer membrane proteins by Pasteurella multocida. Am. J. Vet. Res. 49:1336-1338. [PubMed] [Google Scholar]
- 26.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veken, J. W., N. H. Shah, P. Klaasen, B. Oudega, and F. K. de Graaf. 1996. Binding of host iron-binding proteins and expression of iron-regulated membrane proteins by different serotypes of Pasteurella multocida causing haemorrhagic septicaemia. Microb. Pathog. 21:59-64. [DOI] [PubMed] [Google Scholar]
- 28.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 29.Weitl, F. L., W. R. Harris, and K. N. Raymond. 1979. Sulfonated catecholamide analogues of enterobactin as iron sequestering agents. J. Med. Chem. 22:1281-1283. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, G., C. Pijoan, K. Choi, S. K. Maheswaran, and E. Trigo. 1995. Expression of iron-regulated outer membrane proteins by porcine strains of Pasteurella multocida. Can. J. Vet. Res. 59:46-50. [PMC free article] [PubMed] [Google Scholar]