Abstract
The major structural components of the P2 contractile tail are encoded in the FETUD tail gene operon. The sequences of genes FI and FII, encoding the major tail sheath and tail tube proteins, have been reported previously (L. M. Temple, S. L. Forsburg, R. Calendar, and G. E. Christie, Virology 181:353-358, 1991). Sequence analysis of the remainder of this operon and the locations of amber mutations Eam30, Tam5, Tam64, Tam215, Uam25, Uam77, Uam92, and Dam6 and missense mutation Ets55 identified the coding regions for genes E, T, U, and D, completing the sequence determination of the P2 genome. Inspection of the DNA sequence revealed a new open reading frame overlapping the end of the essential tail gene E. Lack of an apparent translation initiation site and identification of a putative sequence for a programmed translational frameshift within the E gene suggested that this new reading frame (E′) might be translated as an extension of gene E, following a −1 translational frameshift. Complementation analysis demonstrated that E′ was essential for P2 lytic growth. Analysis of fusion polypeptides verified that this reading frame was translated as a −1 frameshift extension of gpE, with a frequency of approximately 10%. The arrangement of these two genes within the tail gene cluster of phage P2 and their coupling via a translational frameshift appears to be conserved among P2-related phages. This arrangement shows a striking parallel to the organization in the tail gene cluster of phage lambda, despite a lack of amino acid sequence similarity between the tail gene products of these phage families.
Bacteriophage P2 belongs to a large, environmentally widespread family of temperate double-stranded DNA phages that are unrelated to the lambdoid phages in gene organization and lytic regulation (for reviews, see references 3 and 8). The similarity in contractile tail structure between the P2-related phages, P1, and the T-even phages led to the classification of this family under the general heading of Myoviridae (11). The P2 tail, 135 nm in length, is composed of an inner tube and a contractile sheath of subunits in an ordered array that gives the appearance of regular horizontal striations (24). A baseplate at the distal end has a slender spike and six tail fibers. Complementation studies, DNA sequence analysis, and antibody staining of phage particles have identified a total of 16 genes, in three operons, essential for P2 tail assembly (13, 14, 28, 44, 47). T4, which has served as the prototype for studies of contractile tail assembly, requires 22 genes for tail assembly (for a review, see reference 7). P2 provides an attractive alternative to phage T4 for detailed molecular studies of tail assembly, because fewer genes are involved. In addition, it provides a model for assembly of the R-type pyocins of Pseudomonas aeruginosa, which are closely related to P2-like contractile tails (36). The work reported here was undertaken to complete sequence analysis of the P2 genome and lay the framework for future isolation and molecular analyses of the proteins involved in assembly of the P2 contractile tail.
In the course of this study, a site was discovered that results in a programmed translational frameshift. DNA sequence analysis led to the identification of an additional open reading frame between the previously identified essential tail genes E and T that overlaps the end of gene E in the −1 reading frame. Work described here demonstrates that this reading frame encodes an essential P2 function and that ribosomes translating gene E undergo a programmed frameshift near the 3′ end of the gene about 10% of the time and enter the −1 reading frame. The resulting 15.4-kDa protein shares 85 N-terminal amino acids (aa) with gpE and contains a C-terminal extension encoded by the overlapping reading frame. This extended protein has been designated gpE+E′.
MATERIALS AND METHODS
Bacterial and bacteriophage strains and growth conditions.
Bacterial and bacteriophage strains used in this study are listed in Table 1. P2 phage stocks were propagated in derivatives of Escherichia coli strain C by the method of Kahn et al. (21). Suppressor strains C-1757 and C-1792 were used for propagation of phages carrying amber mutations. The medium used was Luria broth (LB) (21), supplemented as appropriate; antibiotics were added to final concentrations of 100 μg of ampicillin per ml and 60 μg of kanamycin per ml.
TABLE 1.
Bacterial and bacteriophage strains used in this study
| E. coli or phage strain | Characteristic(s) | Reference or source |
|---|---|---|
| E. coli C derivatives | ||
| C-1a | F− prototrophic | 41 |
| C-1757 | rpsL (Smr) supD | 42 |
| C-1792 | supF | 42 |
| C-2420 | F− Δ(argF-lac)U169 | 20 |
| C-6518 | argE(Am) btuB::Tn10 placUV5 lacZ::T7 gene 1 Knr | Ian Molineux |
| C-6519 | argE(Am) btuB::Tn10 supD placUV5 lacZ::T7 gene 1 Knr | Ian Molineux |
| E. coli K-12 derivative | ||
| DH5α | F− φ80dlacZ ΔM15 Δ(lacZYA-argF)U169 deoR supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 phoA | GibcoBRL |
| Bacteriophages | ||
| P2 vir1 | Clear plaques | 2 |
| P2 Eam30 vir1 | Conditional-lethal, tail defective | 42 |
| P2 Ets55 | Temperature-sensitive | 26 |
| P2 Tam64 vir1 | Conditional-lethal | 42 |
| P2 Tam5 | Conditional-lethal, tail defective | 27 |
| P2 Tam215 vir1 | Conditional-lethal | 42 |
| P2 Uam25 vir1 | Conditional-lethal, tail defective | 42 |
| P2 Uam77 vir1 | Conditional-lethal | 42 |
| P2 Uam92 vir1 | Conditional-lethal | 42 |
| P2 Dam6 | Conditional-lethal, tail defective | 27 |
| P2 Dam22 vir1 | Conditional-lethal | 42 |
| P2 Dam44 vir1 | Conditional-lethal | 42 |
| P2 Dam45 vir1 | Conditional-lethal | 42 |
DNA manipulations.
New plasmids constructed for this study are described in Table 2. Oligonucleotide primers used for cloning or mutagenesis are listed in Table 3. Cloning vectors used were pUC18 and pUC19 (37), pKK232-8 (5), pT7-5 and pT7-7 (43), pMAL-c2 (New England Biolabs), pCRII (Invitrogen), and p138 (45). Restriction and DNA modification enzymes were obtained from commercial sources and used as recommended by the suppliers. Other DNA manipulations were performed by standard procedures (39). P2 DNA was isolated from phage particles by phenol extraction and ethanol precipitation. Plasmid DNA was prepared by the minilysate procedure of Hattori and Sakaki (16) or a Wizard (Promega, Inc.) or Qiagen kit. PCR amplification of DNA from plasmid-containing bacterial colonies and from phage lysates was performed as described previously (47). Amplified fragments were gel purified and eluted as described previously (28) or by chromatography on a QIAquick Spin column (Qiagen, Inc.). All cloned fragments generated by PCR amplification were sequenced to verify that no errors were introduced by Taq polymerase.
TABLE 2.
P2-containing plasmids for marker rescue, sequencing, and expression
| Plasmid | Descriptiona | Reference or source |
|---|---|---|
| pBB1 | PstI (20997)-KpnI (22065) fragment in pUC19/PstI + KpnI | This work |
| pBB2 | KpnI (22065)-EcoRV (22710) fragment in pUC19/KpnI + HincII | This work |
| pBB3 | EcoRV (22710)-NruI (24033) fragment in pUC18/HincII | This work |
| pBBT64 | PstI (20997)-KpnI (22065) fragment from Tam64 in pUC19/KpnI + PstI | This work |
| pEE260 | PstI (20997-25733) fragment from Uam25 in pBR322/PstI | 29 |
| pEE269 | PstI (20997-25733) fragment from Dam6 in pBR322/PstI | 29 |
| pGCE10 | AccI (19000)-EcoRV (19951) in pBR322/EcoRV + PvuII | This work |
| pGCE30 | BglII (17242)-PstI (20997) fragment from Eam30 in pUC18/BamHI + PstI | 44 |
| pKA1 | PstI (20997-25733) fragment in pBR322/PstI | 4 |
| pKL2 | HincII (19403-19891) fragment (E+E′) from P2 vir1 in pT7-5/SmaI | This work |
| pNB59 | BstEII (23399)-BglII fragment from P2 del15 in pKK232-8 | N. K. Birkeland |
| pTG203 | 567-bp NsiI-TaqI U gene fragment in pT7-7/PstI + ClaI | This work |
| pTG260 | HincII (19403-19891) fragment (E+E′) from Eam30 in pT7-5/SmaI | This work |
| pTG267 | 482-bp P2 fragment by PCR using GG2 and GG3 in pCRII | This work |
| pTG316 | HindIII-ApaI fragment from PCR using EH3 and EA1 in p138/HindIII + ApaI | This work |
| pTG373 | HindIII-ApaI fragment from PCR using EH3 and EA1.1 in p138/HindIII + ApaI | This work |
| pTG414 | P2 E+E′ by PCR using Ema1 and Ema12 in pMAL-c2/EcoRI + BamHI | This work |
| pTG502 | P2 E+E′ am by PCR using E1, E7, and T1b in pUC19/SmaI | This work |
| pTG546 | HincII fragment (E+E′ am) from pTG502 in pT7-5/SmaI | This work |
Numbers in parentheses are nucleotide positions. pUC19/PstI + KpnI, PstI- and KpnI-digested pUC19.
TABLE 3.
P2-specific oligonucleotide primers used for cloning and mutagenesis
| Oligonucleotide primer | Sequence (5′ to 3′)a | Location (nt)b |
|---|---|---|
| Wild type | ||
| E1 | CCGGCGAAACGGTGGCCG | 19144-19161 |
| GG2 | CGCAATCTTGCACATGA | 20246-20262 |
| GG3 | TCTTCCATTGTGCGACG | 20712-20728 |
| T1b | TTCAGCTCGCGCAGTGAT | 19869-19986 |
| Mutagenic | ||
| E7 | GGTCATCGACCTAGAGATTT | 19716-19735 |
| EA1 | GGCAGGGCCCGTCACTGCACCGAGTTCGG | 19688-19707 |
| EA1.1 | GGCAGGGCCCGTCAGCTGCACCGAGTTCGG | 19688-19707 |
| EH3 | GTGAAAGCTTGGCCGGTAAGGTGGTCG | 19659-19676 |
| EMal1 | CCCGAATTCAGAAGTGAGAAAACC | 19411-19429 |
| EMal2 | GCGGATCCAGCCCTGAGCAATAC | 19874-19890 |
Primers shown in italics are complementary to the strand reported in GenBank accession no. NC_001895. Nucleotides in the mutagenic oligonucleotides that differ from those in the wild-type sequence are underlined.
Location from the left end of P2. For primers adding restriction sites at the ends, numbering corresponds only to the region identical to P2 sequence.
An amber mutation in E′, the open reading frame giving rise to the gpE′ frameshift extension, was introduced by incorporation of a phosphorylated mutant oligonucleotide during PCR amplification (32). DNA was amplified from a P2 vir1 phage lysate, using 3 μl of a 1:50 dilution of the phage as the template and 100 ng (each) of primers E1 and T1b and 800 ng of phosphorylated mutagenic primer E7 (Table 3). PCR was performed using Vent DNA polymerase in the presence of Taq DNA ligase (both from New England Biolabs). The full-length PCR product was gel purified, ligated with pUC19 that had been cleaved with SmaI, and introduced into competent E. coli DH5α cells. The desired mutation in the resulting plasmid, pTG502, was verified by sequence analysis prior to subsequent subcloning.
Marker rescue and complementation analysis.
P2 amber mutations were localized by marker rescue. Phage lysates were treated with UV light (approximately 300 erg/mm2) from a General Electric germicidal lamp. Titers of the UV-irradiated lysates were determined on a nonsuppressing strain (C-1a) containing a plasmid with a fragment of wild-type P2 DNA and on C-1a alone. An increase of at least 200-fold in the plating efficiency of the amber mutant on the plasmid-bearing strain indicated the presence of the wild-type allele on the cloned fragment. Results of the marker rescue are summarized below (see Fig. 1B).
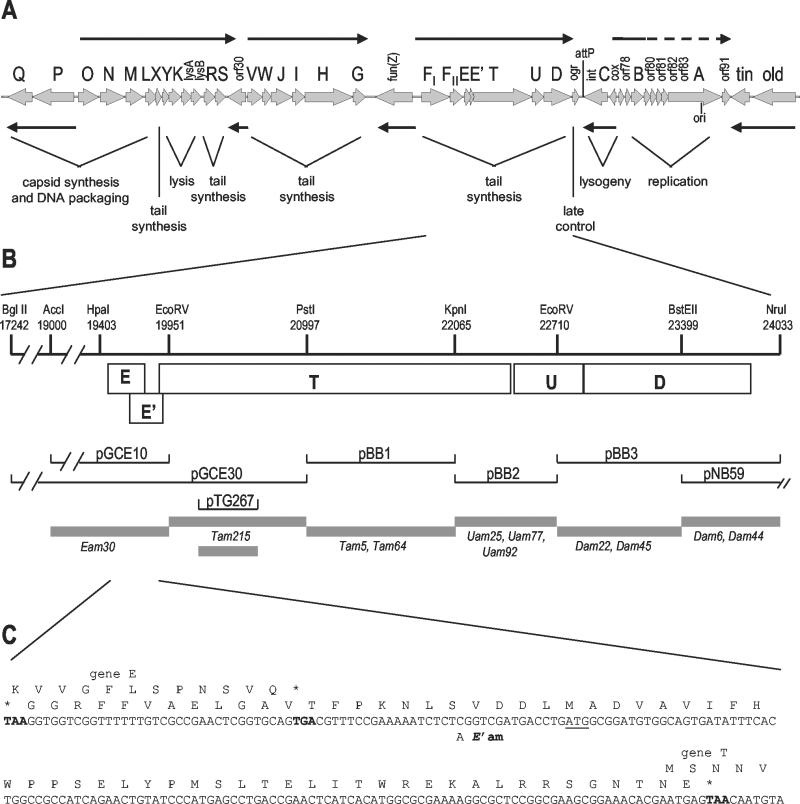
FIG. 1.
Genetic map of P2 and physical map of the tail gene region reported in this paper. (A) Linear map of the P2 genome, with cosL at the left. Thin black arrows indicate the direction and extent of the known transcription units. orf, open reading frame. (B) Expanded view of the physical map of the part of the P2 genome characterized in this report and the location of amber mutations in this region. Selected restriction sites used in subcloning and marker rescue are indicated, with coordinates shown in nucleotides from the left end of the P2 genome. Boxed regions delineate the coding regions for each of the genes indicated in the physical map. The extent of DNA carried by each plasmid subclone is aligned below the relevant restriction sites or position on the physical map; gray bars indicate the intervals to which amber mutations were mapped by marker rescue. (C) DNA sequence of the region between the end of gene E and the beginning of gene T, showing the −1 (E′) open reading frame between these two genes. The TGA stop codon for gene E and the two TAA stop codons defining the boundaries of the −1 reading frame are shown in boldface type. The first in-frame Met codon in the −1 reading frame is underlined, and the location of the C-to-A change introduced to create an amber mutation in E′ is indicated.
Complementation of P2 amber mutants by plasmids derived from pT7-5 and pT7-7 and expressing P2 genes under the control of the T7 φ10 promoter was assayed in strain C-2420 carrying the compatible plasmid pGP1-2 (43). This plasmid carries T7 gene 1 under control of a temperature-sensitive λ repressor. Expression of T7 RNA polymerase was sufficiently leaky at 33°C to allow complementation of P2 amber mutants by plasmids expressing the corresponding wild-type genes from the T7 promoter. Plating efficiencies of P2 amber mutants on strains carrying complementing plasmids were equivalent to those obtained on a supD strain. Complementation of P2 Ets55 by plasmids carrying wild-type and mutant copies of the E+E′ region was examined in strains C-6518 (Su−) and C-6519 (supD), which express T7 RNA polymerase from the lacUV5 promoter. Expression in the absence of induction was sufficiently leaky to allow complementation, which was tested at 42°C.
DNA sequence determination and computer analysis.
DNA sequences were determined by the method of Sanger et al. (40) using a Sequenase kit (U.S. Biochemicals, Cleveland, Ohio) and 5′[α-35S]thio-dATP or using a Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, Calif.). Plasmids carrying cloned DNA from this region of the P2 genome (Table 2) were used as templates. The sequence changes in amber mutants were determined from cloned fragments or from PCR-amplified phage DNA of the regions identified by marker rescue. Sequencing reactions using the fluorescent dideoxynucleotides were analyzed on an ABI automated sequencer in the Nucleic Acids Core Facility, Massey Cancer Center, Medical College of Virginia, Virginia Commonwealth University. At least two independent sequence determinations were performed for each strand, and oligonucleotide primers were used to obtain sequences spanning all restriction sites used in subcloning. All sequences obtained from PCR-amplified DNA were determined from the products of two independent amplification reactions. M13 universal sequencing primers were purchased from commercial suppliers; additional oligonucleotide primers were synthesized by the Nucleic Acids Core Facility, Massey Cancer Center, Medical College of Virginia, Virginia Commonwealth University, or purchased from Oligos, Etc. (Wilsonville, Oreg.). Details about primers used in sequence analysis will be provided upon request.
Sequence data were stored and analyzed using the GCG (Genetics Computer Group) Wisconsin Package (Accelrys, Madison, Wis.). The sequence reported in this paper corresponds to coordinates 19384 to 23954 in the sequence of the entire P2 genome and is in the GenBank database.
Assay of β-galactosidase.
Cultures of strain C-2420 carrying pTG316 or pTG373 were grown overnight in LB plus ampicillin. Cultures grown overnight were diluted 20-fold in LB supplemented with ampicillin in 96-well microtiter plates and incubated for 90 min with shaking at 37°C. The cultures were then diluted into fresh medium with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubation was continued at 37°C for another 90 min. After the optical densities at 600 nm (OD600) of the cultures were read, growth was halted by mixing aliquots (130 μl) of each culture with 30 μl of chloroform. The β-galactosidase activity of a 20-μl aliquot of each culture was determined by the method of Menzel (31). Six independent colonies of each construct in the presence and absence of IPTG were assayed, and activities are expressed as the mean ± standard error of the induced activity in arbitrary units. Background activity in this assay was essentially undetectable.
Purification of fusion proteins.
The gpE-β-galactosidase fusion protein was prepared from strain C-2420 carrying pTG316 and the compatible lacI plasmid pRG1 (R. Garcea, unpublished data). A 1-liter culture of cells was grown at 37°C in LB supplemented with ampicillin and kanamycin to an OD600 of 0.55. IPTG was added to a final concentration of 1 mM, and incubation was continued overnight. Cells were pelleted and resuspended in 100 ml of 50 mM Tris-HCl [pH 7.5]-0.5 mM EDTA. Lysozyme was added to a final concentration of 1 mg/ml, and the cells were incubated on ice for 1 h. NaCl and MgCl2 were added to final concentrations of 50 mM and 1 mM, respectively, and the lysate was treated with 2 mg of DNase I for 30 min on ice followed by a 10-min incubation at 37°C. Cell debris was removed by centrifugation at 10,000 × g for 15 min. The gpE-β-galactosidase fusion protein was precipitated by the addition of ammonium sulfate to 40%, followed by an additional centrifugation at 10,000 × g for 20 min. The β-galactosidase activity, which was present in the pellet fraction, was resuspended in 20 ml of 20 mM Tris-HCl. Insoluble material was removed by centrifugation for 10 min at 10,000 rpm in a Sorvall SS-34 rotor, and the supernatant was applied to a Promega Protosorb lacZ immunoaffinity column. The fusion protein was eluted under the high pH conditions recommended by the manufacturer, dialyzed overnight against water, and dried. Protein was redissolved in Laemmli sample buffer for further analysis.
For purification of MalE fusion proteins, a 1-liter culture of DH5α cells containing pTG414 or the vector pMALc-2 was grown at 37°C in LB supplemented with ampicillin to an OD600 of 0.55. IPTG was added to a final concentration of 0.5 mM, and incubation was continued for 2 h. Cells were pelleted, weighed, and resuspended in 10 ml of lysis buffer (25 mM morpholinepropanesulfonic acid [MOPS] [pH 7.1], 100 mM NaCl, 20 μM EDTA, 5% [vol/vol] glycerol, 1 mM β-mercaptoethanol) plus 1 mM phenylmethylsulfonyl fluoride for each gram of cell paste. Cells were lysed in a chilled French pressure cell at 12,000 lb/in2, and the lysate was centrifuged at 41°C for 30 min at 16,000 rpm in a Sorvall SS-34 rotor. The supernatant was applied to a column containing 5 ml of amylose resin (New England Biolabs) at a flow rate of 15 ml/h. The column was washed with approximately 40 ml of lysis buffer, and then the fusion protein was eluted with lysis buffer containing 10 mM maltose. One-milliliter fractions were collected, and aliquots from fractions containing the protein peak were analyzed by electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels.
N-terminal sequence determination.
The N-terminal sequence of purified β-galactosidase fusion protein was obtained by automated Edman degradation on a Hewlett-Packard model G1005A protein sequencer, by Commonwealth Biotechnologies, Inc., Richmond, Va.
Nucleotide sequence accession number.
The sequence reported in this paper, corresponding to coordinates 19384 to 23954 in the sequence of the entire P2 genome, was used to complete the P2 genome sequence. The complete genome has been deposited in the GenBank database under accession number NC_001895.
RESULTS
The sequence reported here completes the characterization of the P2 genome. This sequence spans P2 nucleotides (nt) 19384 to 23954, as measured from the left cohesive end of the phage genome. Identification of the known genes that have been mapped previously to this region was accomplished by marker rescue of mutations in genes E, T, U, and D. In addition to these known genes, we discovered an additional small reading frame overlapping the end of gene E that is translated as a frameshifted extension of gpE. A physical and genetic map of this region of the P2 genome and a summary of the marker rescue data are presented in Fig. 1. The detailed characterization of each gene in this region is described below.
Identification of gene E and the −1 frameshift extension E′.
Gene E was originally defined by the ts mutation 55 (26); nonsense mutation am30 lies in the same complementation group and was shown by Lengyel et al. (24) to result in a lack of tails. A polypeptide corresponding to the product of gene E has not been identified in infected cells or phage particles. Both the Ets55 (C to A at nt 19473) and Eam30 (C to T at nt 19619) mutations lie in an open reading frame just distal to FII that encodes an acidic polypeptide of 91 aa (Fig. 1 and 2). The role of gpE is unknown; Lengyel et al. (24) reported that the product of gene T appears to be unstable in the absence of gpE and suggested that gpE plays a role in stabilizing gpT.
FIG. 2.
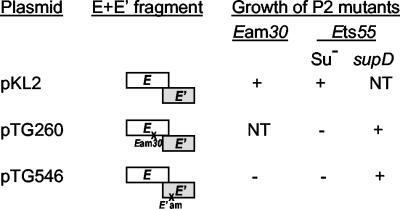
Complementation of P2 E mutants by cloned fragments carrying the E-E′ region. P2 Ets55 was plated at 42°C. Complementation was shown by growth of P2 mutants as follows: +, growth; −, no growth; NT, not tested.
Between gene E and gene T is a small open reading frame (E′) that overlaps the start of gene T and extends 37 nt back into gene E in the −1 reading frame (Fig. 1C). The most 5′ in-frame Met codon for this open reading frame is 33 nt beyond the end of E but lacks a potential ribosome binding sequence. A possible GUG start overlaps the termination codon for E; translation from this site (presumably via translational coupling with E to compensate for the poor translation initiation signals) would yield a polypeptide of 51 aa. About 20 nt upstream of this GUG, within the region overlapping gene E, is a run of six T residues that resembles sites used for −1 translational frameshifting (10, 12). To explore this possibility and determine whether E′ represented a bona fide gene, several strategies were pursued. A single nucleotide change was introduced 19 bp downstream of gene E (C to A at nt 19724 [Fig. 1]); this change creates an amber codon in place of a Ser in the −1 reading frame between the end of E and the first in-frame Met codon in E′. A HincII fragment including E+E′(am) was ligated into the SmaI site of pT7-5, placing expression of both open reading frames under control of the T7 φ10 promoter. The resulting plasmid, pTG546, failed to complement P2 Eam30 in a nonsuppressing strain expressing low levels of T7 RNA polymerase (Fig. 2). An otherwise identical plasmid carrying the corresponding wild-type fragment (pKL2) did complement P2 Eam30 in the same background. These results demonstrate that E′ is essential for P2 lytic growth and indicate that the Eam30 mutation also affects expression of E′. The pTG546 plasmid also failed to complement P2 Ets55 at 42°C in a nonsuppressing strain, although complementation was obtained in an otherwise isogenic supD strain. The Ets55 missense mutation should not be polar on E′, so this result is consistent with expression of the E′ reading frame as a frameshifted extension of E.
Further evidence in support of an essential role for E′ comes from studies of coliphage 186, a close relative of P2. The homologous 186 tail gene cluster contains a similar pair of overlapping reading frames (38); the upstream one encodes a polypeptide 67% identical to gpE, while the downstream −1 reading frame encodes a polypeptide 74% identical to gpE′. Two 186 amber mutations have been mapped to the downstream reading frame, which was identified as a tail gene, H, by Hocking and Egan (19). Lysates of 186 Ham56-infected cells showed an accumulation of apparently normal tails (19). This finding suggests that the longer frameshifted gene product either is a minor protein that must be added to completed tails, such as a collar component, or is involved directly in the process of head-tail attachment. The P2-related Pseudomonas aeruginosa phage φCTX (35) encodes a similar potential protein generated by a −1 frameshift (see Discussion), while the P. aeruginosa R2 pyocin gene cluster does not (36). This observation is also consistent with a role for gpE+E′ in head-tail attachment, since the R-type pyocins are completed tail structures that are not attached to heads.
Since all mutations that affect P2 gpE also affect gpE+E′, there is no direct evidence that the shorter gpE protein is in fact essential, as originally reported. A strong case can be made, however, based on the difference in reported phenotypes caused by Eam30 and the amber mutation affecting just the downstream reading frame in the homologous gene from phage 186, Ham56. If the essential role of gpE were just to permit proper expression of gpE+E′, the defects conferred by Eam30 (which makes neither protein) and Ham56 (which makes the protein equivalent to gpE but not the longer frameshifted polypeptide) should be the same. However, as described above, no tails were observed in cells infected with P2 Eam30 (24), while apparently complete but unattached tails accumulated in cells infected with 186 Ham56 (19). If one accepts the hypothesis that these homologous genes encode proteins that have equivalent functions in P2 and 186, this evidence strongly supports the conclusion that both gene products are essential for tail assembly.
Confirmation of a programmed translational frameshift between E and E′.
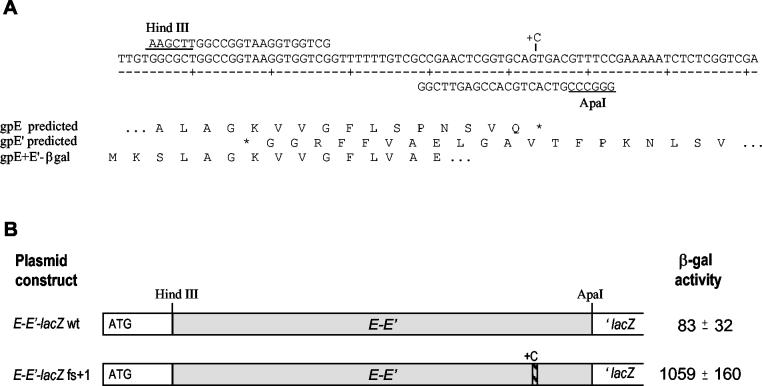
To test directly the hypothesis that the region of overlap between the gene E reading frame and the E′ reading frame included determinants for a −1 translational frameshift, we employed a plasmid designed to allow expression of a reporter gene only if such a frameshift occurs. Plasmid p138, described by Weiss et al. (45), carries a modified lacZ gene in which DNA between codons 2 and 5 is replaced with a short “stuffer” region flanked by HindIII and ApaI sites. Expression of lacZ is controlled by Ptac and the E. coli lpp translation start site. A fragment containing the putative frameshift site near the end of gene E and the beginning of E′ was generated by PCR using primers EH3 and EA1, which introduced HindIII and ApaI sites, respectively (Fig. 3). Insertion of this fragment between the ApaI and HindIII sites of p138 generated plasmid pTG316. Production of active β-galactosidase from this construct can occur only if a frameshifted ribosome bypasses the termination codon at the end of E and enters the downstream lacZ coding region in the correct frame. We also created a parallel construct, pTG373, by inserting a fragment generated by PCR with primers EA1.1 and EH3. This plasmid contains an additional nucleotide just upstream of the stop codon of gene E; this places the lacZ gene in frame with the initiating AUG and allows measurement of translation that bypasses the frameshifting site without changing reading frame. A comparison of the levels of β-galactosidase produced by these two plasmids (Fig. 3) indicates that the ratio of ribosomes that shift reading frame to those that bypass the frameshift is about 1:13. The fraction of ribosomes that shift reading frame during translation of the E gene is therefore on the order of 7 to 8%.
FIG. 3.
Analysis of the region containing a programmed translational frameshift. (A) The nucleotide sequence spanning the end of gene E and the beginning of open reading frame E′ is shown, along with the oligonucleotides used to clone this region into the frameshift detection plasmid p138. The position where a C residue was inserted to allow translation of lacZ by ribosomes that did not shift reading frame is indicated. Below the DNA sequence the predicted amino acid sequences encoded by E and the −1 open reading frame E′ are shown, along with the actual N-terminal sequence determined from the β-galactosidase (βgal) fusion protein purified from cells carrying pTG316. (B) Measurement of frameshifting efficiency in vivo. The β-galactosidase (β-gal) activity was determined in cells carrying a plasmid with the wild-type (wt) E-E′ fragment, in which lacZ must be translated by ribosomes that have undergone a −1 frameshift, and from cells with a plasmid carrying the equivalent fragment in which a C was inserted just upstream of the stop codon for gene E (fs+1), so that lacZ is translated by ribosomes that did not shift reading frame into E′.
To determine precisely where the frameshift occurs, we purified the β-galactosidase fusion protein produced in cells carrying plasmid pTG316 and obtained the amino acid sequence of the N terminus (Fig. 3). Residues 1 to 3 corresponded to amino acids encoded by the start of lacZ in the vector, spanning the HindIII site; residues 4 to 12 matched those predicted by the coding sequence for gene E. At residue 13, a strong signal for Val was detected; this corresponds to the residue encoded by the −1 reading frame. There was also a possible weak signal for Ser at this position; it is unclear whether this was due to background in the sample or whether some fraction of the protein was translated in the unshifted reading frame for one additional codon. Beyond residue 13, the sequence matched that predicted for the −1 reading frame.
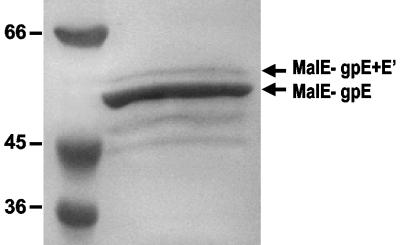
We wished to demonstrate that the frameshift observed in the context of the lacZ fusion also occurred during normal translation of gene E. Unlike the relatively abundant λ gpG and gpG-T proteins, the products of gene E and its frameshifted extension (hereafter referred to as gpE+E′) have never been identified in P2-infected cells. A number of attempts were made to overexpress these proteins from plasmid constructs using either the T7 expression system or an N-terminal hexahistidine tag. No proteins with a predicted molecular mass corresponding to that of either gpE or gpE+E′ were observed, nor were there any identifiable proteins whose appearance was affected if the constructs carried the Eam30 mutation. It may be that these presumptive tail assembly proteins are unstable in the absence of other P2 tail components. As a final attempt, we constructed plasmid pTG414, which placed the coding region for genes E and E′ at the end of the malE gene in plasmid pMAL-c2. Amylose affinity purification of the fusion proteins yielded a major product with an apparent molecular mass of 52.6 kDa (Fig. 4), in good agreement with the predicted size of 52.4 kDa for the MalE-gpE fusion protein. In addition, a small amount of a larger 57.8-kDa fusion protein was observed; this is the predicted size for the MalE-gpE+E′ frameshifted product. Several smaller products, presumably arising from degradation, were also apparent. Densitometric analysis of the relative proportions of the two largest species indicates that the larger protein represents approximately 12% of the total, in reasonably good agreement with the frequency of frameshifting measured in the β-galactosidase assay.
FIG. 4.
Synthesis of a MalE fusion protein carrying the frameshifted gpE+E′polypeptide. Protein was affinity purified as described in Materials and Methods and analyzed by electrophoresis on a 10% SDS-polyacrylamide gel stained with Gelcode blue (Pierce). Protein bands corresponding to the expected sizes for the MalE-gpE fusion protein and the MalE-gpE+E′ frameshifted protein are indicated, as are the sizes (in kilodaltons) of molecular mass standards. Densitometry and digital imaging of the gel was performed on a ChemiImager 4000 (Alpha Innotech Corp.), and the figure was compiled using Microsoft PowerPoint.
Identification of gene T.
Distal to gene E+E′ is an open reading frame encoding a polypeptide of 815 aa, with a predicted molecular mass of 86.5 kDa, which correlates reasonably well with the 94-kDa mass (±2.2 kDa) determined by SDS-polyacrylamide gel electrophoresis for the product of gene T (24). Three T amber mutations, Tam5 (C to T at nt 21101), Tam64 (G to A at nt 21528), and Tam215 (C to T at nt 20669), were located within this reading frame, confirming its assignment. The presumptive start codon of this gene overlaps the very end of E′ (Fig. 1C) and is preceded by a potential ribosome binding site, GGA. The ability of a plasmid encoding just E+E′ to complement P2 Eam30 indicates that the E amber mutation is not polar on expression of T, so there is no obligatory translational coupling between E′ and T.
Lengyel et al. (24) suggested that gpT was the P2 tail fiber, based on its large size and the fact that it was present in approximately six copies per phage particle. Subsequent work, however, established gpH as the P2 tail fiber protein (13). The most likely role for gpT is that of tail length determination. It is the only P2 tail protein that is large enough to span the length of the P2 tail shaft. Both λ (18, 22) and T4 (1) encode proteins that have been shown to act as tape measures for tail shaft polymerization. The sizes of these proteins correspond to a fairly constant 0.15 nm of tail length per amino acid residue, suggesting that their structure is that of an extended α-helix. The secondary structure predicted for gpT (6) is also largely α-helical. In addition, a BLAST homology search revealed multiple regions of similarity between gpT and myosin heavy chains, which are proteins with known extended α-helical structure. The tail length-to-amino acid ratio for gpT is 0.17 nm per amino acid residue, suggesting that this protein might be a bit more extended than in the lambdoid phages.
A comparison of P2 gpT with the homologous proteins encoded by other P2-related phages provides further support for assigning the role of tail length determination to this gene product. The coliphage 186 gpG is 78% identical to P2 gpT and virtually the same size (812 aa compared to 814 aa [38]), and the tails of phage 186 and P2 are indistinguishable in length. The P2-related Pseudomonas phage φCTX encodes a 904-aa protein, predicted to be rich in α-helix, that is 29% identical to P2 gpT (35). The product of this gene has the predicted N-terminal sequence but apparently migrates to the position of an 67-kDa protein, rather than to the position of a 95.8-kDa protein predicted by the DNA sequence, suggesting that the C-terminal portion of the protein is removed by processing. Consistent with the proposed role of this protein in tail length determination, the φCTX tail is 105 nm in length (17) compared to 135 nm for P2. While C-terminal processing of tail proteins during the assembly process has been documented for λ gpH and the analogous proteins from several other lambdoid phages (18), there is no evidence to suggest proteolytic cleavage of gpT during tail assembly, since the protein identified in phage particles is roughly the same size predicted by the coding sequence.
Identification of gene U.
Beginning with the TAA termination codon for gene T is a potential ribosome binding site with extensive homology to the 3′ end of 16S rRNA, i.e., TAAGGAGGTGA. Seven base pairs beyond this is the first of two tandem ATG codons, which begins the next open reading frame. We characterized three U amber mutations: Uam25 (C to T at nt 22363) and Uam77 and Uam92 (identical mutations that change A to T at nt 22501). Both changes lie in this next open reading frame, which encodes a protein of 160 aa with a predicted molecular mass of 17.45 kDa. Although gpU was not found in phage particles (24), Ljungquist and Bertani (29) identified a protein that appeared to be synthesized in maxicells from a plasmid carrying a wild-type P2 PstI fragment, but not from pEE260, which carries the same fragment from P2 Uam25. This protein, which was 28 kDa, was thought to be the product of gene U. The open reading frame predicted by sequence determination encodes a considerably smaller protein. To ensure that we had correctly identified the entire coding sequence, a 567-bp fragment generated by cleavage with NsiI and TaqI (P2 coordinates 22286 to 22853) was cloned into pT7-7. The resulting plasmid, pTG203, complemented P2 Uam25, indicating that a functional U gene was contained within this fragment. Further confirmation is provided by the paralogous gene from φCTX, which encodes a predicted protein of similar size (16.2 kDa) that is 51% identical to P2 gpU (35). N-terminal microsequencing of φCTX virion proteins indicates that this protein is present in the phage particle in small amounts, suggesting that P2 gpU is likely to be in P2 virions as well and may have escaped detection in earlier studies (24) due to its small size and low abundance.
Identification of gene D.
The product of the D gene was also not identified in phage particles (24) but was expressed in minicells from the cloned PstI fragment examined by Ljungquist and Bertani (29). They identified a protein with a molecular mass of about 46 kDa that was replaced by a 32-kDa protein encoded by a plasmid carrying the Dam6 mutation. Overlapping the termination codon of gene U is a start codon for an open reading frame of 388 aa, encoding a protein with a molecular mass of 42.8 kDa. The Dam6 mutation (C to T at nt 23592) is in this reading frame and would generate a protein of 267 aa (29.4 kDa), consistent with the reported reduction in size (29). The gpD homologue of φCTX, 45.4% identical to P2 gpD, is present in phage virions (35). It is likely, therefore, that gpD is present in P2 virions as well. The failure to detect gpD in phage particles or infected cells (24) can be explained by the similarity in size between gpD and the relatively abundant gpFI (42.8 versus 43.1 kDa), which would make these proteins difficult to resolve on polyacrylamide gels. The polar effect of the Fam4 mutation on gene D also prevented detection of gpD in the absence of gpFI. Antibodies to gpD will be required to confirm the presence of gpD in P2 virions.
DISCUSSION
In recent years, a number of programmed frameshifts have been characterized in retroviruses and in several phage and bacterial genes (reviewed in references 10 and 12). Most of the natural −1 frameshifts occur within a run of nucleotides in which simultaneous slippage of the tRNAs in the P and A sites can occur while preserving the identity of the first 2 (nonwobble) bp in the codon-anticodon interaction. Accordingly, we anticipated that if a −1 frameshift occurred in the region where gene E overlapped the beginning of open reading frame E′, it would be within the run of six T residues (i.e., slipping back from the ..T TTT TTG Phe-Leu to a TTT TTT G.. Phe-Phe). Surprisingly, though, the sequence of the frameshifted β-galactosidase fusion protein indicated that the frameshift actually occurred one codon further downstream. Slippage of the tRNALeu at the P site would maintain the codon-anticodon pairing of the UU dinucleotide, but the new tRNAVal occupying the A site in the −1 frame would have codon-anticodon mismatches at all three positions relative to the Ser codon in the zero reading frame. This suggests that a simultaneous slippage mechanism may not be involved at this site. A slowing or pausing of the ribosome during translation has been proposed to enhance tRNA-message realignment during a programmed frameshift. This can be affected by codon usage and/or RNA secondary structures. Potential downstream secondary structures, including stem-loops and pseudoknots, have been found in a number of frameshift sites. Base pairing between the 3′ end of 16S rRNA and the mRNA just upstream of the coding region has been implicated in both −1 and +1 frameshifting in E. coli (23, 46). Analysis of the sequence 3′ of the E-E′ frameshifting site does reveal a weak potential hairpin distal to the frameshift. Furthermore, a region complementary to the 3′ end of 16S rRNA lies 9 nt upstream of the site where the frameshift occurs (Fig. 5A), suggesting a possible role for the ribosome in promoting frameshifting at this site.
FIG. 5.
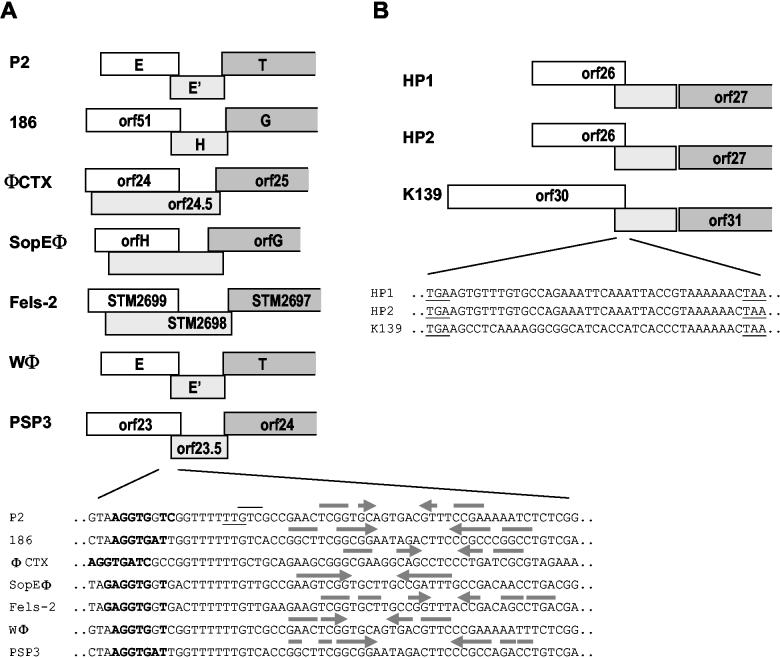
Potential translational frameshift sites in P2-related phages. (A) Alignment of sequences from the region corresponding to E-E′ in related phages that parallel P2 in genome organization. Sequences are from the following phages (GenBank accession numbers are shown in parentheses): E. coli phages 186 (NC_001317) and Wφ (Esposito et al., unpublished); S. enterica phages SopEφ (AF153829), Fels-2 (sequenced as a prophage in the S. enterica serovar Typhimurium LT2 genome; (AE006468) and PSP3 (Christie et al., unpublished); and P. aeruginosa phage 102 φCTX (NC_003278). Boxes indicate the extent of the open reading frame (orf) and overlap in the genes equivalent to P2 E (white) and the −1 reading frame E′ (light gray), as well as show the beginning of T (dark gray, which in all cases is in the same frame as E). The DNA sequences from this region are aligned below the sequence alignment. The codons for the last amino acid read in the gpE reading frame and the first amino acid read in the gpE+E′ reading frame are indicated by an underline and overline, respectively. Nucleotides shown in boldface type are complementary to the 3′ end of 16S rRNA. Arrows indicate complementary sequence encoding potential RNA hairpins predicted by the GCG program MFold. In the case of P2 and Wφ, the most stable of several predicted structures is shown. However, the extensive similarity between these two sequences might also allow formation of a hairpin equivalent to that of P2 in Wφ and vice versa. (B) A similar comparison of open reading frames upstream of the putative tail tape measure protein in the more distantly related P2-like phages from H. influenzae (HP1 [NC_001697] and HP2 [NC_003315]) and V. cholerae (K139 [NC_003313]) (GenBank accession numbers shown in brackets). The DNA sequence of the region of overlap between the genes in locations equivalent to E and E is shown. The TAA stop codon for open reading frame 26 (orf26) (HP1 and HP2) or orf30 (K139) is underlined, as is the TGA stop codon that defines the extent of overlap in the −1 reading frame.
A potential for a similar translational frameshift exists in the equivalent genes of the other characterized P2-related phages. These include not only the highly homologous late gene region of phage 186 (38) but also similar regions in the P. aeruginosa phage φCTX (36), Salmonella enterica phages SopEφ(15, 33) and Fels-2 (30), and two additional phages whose genomes have been determined recently, Wφ (D. Esposito, B. J. Schmidt, F. R. Bloom, and G. E. Christie, unpublished data) and PSP3 (G. E. Christie, P. Xu, P. Vitazka, and G. A. Buck, unpublished data). As illustrated in Fig. 5A, the extent of potential overlap in the −1 open reading frame is virtually identical for P2 and 186, as well as for Wφ and PSP3 (which have sequences in this region identical to P2 and 186, respectively). The other three phages have a much longer region in which a frameshift could occur. The gpT homologue is encoded in the same reading frame as the gpE homologue in all cases, and its reading frame overlaps the end of the reading frame for the E′ homologue by 8 nt except in φCTX, where the overlap is 11 nt. A comparison of the DNA sequence of these seven phages in the region surrounding the E-E′ frameshift in P2 shows absolute conservation of the T6G at the upstream boundary of the frameshift site (Fig. 5A). Another feature shared by all of these sites is a sequence complementary to the 3′ end of 16S rRNA, just upstream of the T6G. These sites all have potential hairpins 3′ of the frameshift site as well, but the strength and position of each of these predicted RNA secondary structures is somewhat variable, suggesting that this feature may be less important. Genetic or biochemical evidence supporting the existence of a frameshift is available only for P2 and 186 at present, but on the basis of conservation of sequence and features in this region, it seems likely that a similar frameshift occurs at this position in all of these closely related phages. Three additional phages have been classified as P2-related phages on the basis of amino acid sequence similarities of some of their genes. These include the Haemophilus influenzae phages HP1 (9) and HP2 (B. J. Williams, M. Golomb, M. V. Olson, and A. L. Smith, GenBank entry NC_003315), and the Vibrio cholerae phage K139 (D. Kapfhammer, J. Nesper, J. Blass, and J. Reidl, GenBank entry NC_003313). The capsid gene clusters of these three phages are clearly homologous to those of the other P2-related phages, but the gene organization and amino acid sequences of the tail genes are much less similar. The tail gene clusters of these three phages are closely related to each other, however. No homologue of P2 E has been identified in these phages, but all possess a putative tail tape measure gene that does show similarity to that of P2 T. Inspection of the sequence upstream of the T gene homologue reveals two small overlapping reading frames in a location similar to that of E and E′ (Fig. 5B). All three phages have an overlap of the same length between the putative E and E′ equivalent genes, and the extended open reading frame that could be generated by a −1 frameshift ends just upstream of the beginning of the putative tape measure gene, which is translated in the same reading frame as the potential frameshifted polypeptide. The sequences of HP1 and HP2 in the region of overlap are identical; the sequence of K139 differs from the HP1 and HP2 sequence, except for conservation of a potential “slippery sequence,” A6C, preceding the stop codon in the upstream reading frame (Fig. 5B). No potential Shine-Dalgarno sequence or RNA secondary structures are evident in this region. While there are no amino acid similarities between these small open reading frames and those in the P2 E-E′ region or any genetic evidence suggesting a similar role for these genes, the parallel in gene organization and the potential for a translational frameshift are suggestive of a similar mechanism in these three more distant relatives of P2 as well.
Translational frameshifting in tail assembly genes was first reported in bacteriophage lambda (25). The virion proteins of P2-related phages and lambdoid phages show little amino acid sequence similarity. The roles of the two polypeptides encoded by the overlapping reading frames have not been clearly established for lambda or P2, but in both cases they play a role in tail assembly. The apparent frequency of frameshifting in P2 is two- to threefold higher than that reported for lambda gpG-T, and features of the frameshift sites suggest differences in the mechanism involved in promoting frameshifting. Nevertheless, the arrangement of the cluster of tail assembly genes that includes the programmed translational frameshift is strikingly parallel. The overlapping open reading frames are preceded in both cases by the gene (or in the case of P2, with a contractile tail, genes) encoding the major structural component(s) of the phage tail. Distal to the frameshifted gene is the gene encoding the protein that determines tail length. A similar organization of tail genes and a putative site for a translational frameshift have recently been reported for bacteriophage Mu and several of its relatives as well (34). While it is possible that this remarkable similarity in gene organization is accidental, it is tempting to speculate that the frameshift may play a biological function (beyond regulating the relative molar ratios of the two polypeptides encoded by the overlapping reading frames) that has been conserved during phage evolution.
Acknowledgments
We are grateful to many individuals for providing strains and plasmids and sharing unpublished data, including Richard Calendar, Elisabeth Haggård-Ljungquist, Bryan Julien, Rainer Ziermann, Erich Six, Mel Sunshine, Nils Kåre Birkeland, Roger Hendrix, Ian Molineux, Tetsuya Hayashi, and Ian Dodd. Undergraduate students Karin Lee and Kate Bacon constructed some of the plasmids used in the course of this study, and Rodney King provided a detailed protocol for the automated β-galactosidase assay.
This work was supported in part by grants (to G.E.C.) from the National Institutes of Health (GM34651), American Cancer Society (NP869A), and the A. D. Williams Foundation, Medical College of Virginia Campus, Virginia Commonwealth University.
REFERENCES
- 1.Abuladze, N. K., M. Gingery, J. Tsai, and F. A. Eiserling. 1994. Tail length determination in bacteriophage T4. Virology 199:301-310. [DOI] [PubMed] [Google Scholar]
- 2.Bertani, L. E. 1957. The effect of the inhibition of protein synthesis on the establishment of lysogeny. Virology 4:53-71. [DOI] [PubMed] [Google Scholar]
- 3.Bertani, L. E., and E. W. Six. 1988. The P2-like phages and their parasite, P4, p. 73-143. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Publishing Corp., New York, N.Y.
- 4.Birkeland, N. K., and B. H. Lindqvist. 1986. Coliphage P2 late control gene ogr: DNA sequence and product identification. J. Mol. Biol. 188:487-490. [DOI] [PubMed] [Google Scholar]
- 5.Brosius, J. 1984. Plasmid vectors for the selection of promoters. Gene 27:151-160. [DOI] [PubMed] [Google Scholar]
- 6.Chou, P. Y., and G. D. Fasman. 1978. Empirical prediction of protein conformation. Annu. Rev. Biochem. 47:251-276. [DOI] [PubMed] [Google Scholar]
- 7.Coombs, D. H., and F. Arisaka. 1994. T4 tail structure and function, p. 259-281. In J. Karam (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 8.Dodd, I. B., and J. B. Egan. 1999. P2, 186, and related phage, p. 1087-1094. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, 2nd ed. Academic Press, New York, N.Y.
- 9.Esposito, D., W. P. Fitzmaurice, R. C. Benjamin, S. D. Goodman, A. S. Waldman, and J. J. Scocca. 1996. The complete nucleotide sequence of bacteriophage HP1. Nucleic Acids Res. 24:2360-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farabaugh, P. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60:103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraenkel-Conrat, H. 1985. Phages of prokaryotes (bacteria and cyanobacteria), p. 171-222. In H. Fraenkel-Conrat and R. R. Wagner (ed.), The viruses: catalogue, characterization, and classification. Plenum Press, New York, N.Y.
- 12.Gesteland, R. F., and J. F. Atkins. 1996. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 65:741-768. [DOI] [PubMed] [Google Scholar]
- 13.Haggård-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haggård-Ljungquist, E., K. Jacobsen, S. Rishovd, E. W. Six, O. Nilssen, M. G. Sunshine, B. H. Lindqvist, K.-J. Kim, V. Barreiro, E. V. Koonin, and R. Calendar. 1995. Bacteriophage P2: genes involved in baseplate assembly. Virology 213:109-121. [DOI] [PubMed] [Google Scholar]
- 15.Hardt, W.-D., H. Urlaub, and J. E. Galán. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hattori, M., and Y. Sakaki. 1986. Dideoxy sequencing method using denatured plasmid templates. Anal. Biochem. 152:232-238. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, T., T. Baba, H. Matsumoto, and Y. Terawaki. 1990. Phage conversion of cytotoxin production in Pseudomonas aeruginosa. Mol. Microbiol. 4:1703-1709. [DOI] [PubMed] [Google Scholar]
- 18.Hendrix, R. W. 1988. Tail length determination in double-stranded DNA bacteriophages. Curr. Top. Microbiol. Immunol. 136:21-29. [DOI] [PubMed] [Google Scholar]
- 19.Hocking, S. M., and J. B. Egan. 1982. Genetic studies of coliphage 186. I. Genes associated with phage morphogenesis. J. Virol. 44:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien, B., and R. Calendar. 1995. The purification and characterization of the bacteriophage P4 δ protein. J. Bacteriol. 177:3743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahn, M. L., R. Ziermann, G. Dehò, D. W. Ow, M. G. Sunshine, and R. Calendar. 1991. Bacteriophage P2 and P4. Methods Enzymol. 204:264-280. [DOI] [PubMed] [Google Scholar]
- 22.Katsura, I. 1990. Mechanism of length determination in bacteriophage lambda tails. Adv. Biophys. 26:1-18. [DOI] [PubMed] [Google Scholar]
- 23.Larsen, B., N. M. Wills, R. F. Gesteland, and J. F. Atkins. 1994. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 176:6842-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lengyel, J. A., R. N. Goldstein, M. Marsh, and R. Calendar. 1974. Structure of the bacteriophage P2 tail. Virology 62:161-174. [DOI] [PubMed] [Google Scholar]
- 25.Levin, M. E., R. W. Hendrix, and S. J. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage λ tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl, G. 1969. Genetic map of bacteriophage P2. Virology 39:839-860. [DOI] [PubMed] [Google Scholar]
- 27.Lindahl, G. 1971. On the control of transcription in bacteriophage P2. Virology 46:620-633. [DOI] [PubMed] [Google Scholar]
- 28.Linderoth, N. A., B. Julien, K. E. Flick, R. Calendar, and G. E. Christie. 1994. Molecular cloning and characterization of bacteriophage P2 genes R and S involved in tail completion. Virology 200:347-359. [DOI] [PubMed] [Google Scholar]
- 29.Ljungquist, E., and L. E. Bertani. 1983. Properties and products of the cloned int gene of bacteriophage P2. Mol. Gen. Genet. 192:87-94. [DOI] [PubMed] [Google Scholar]
- 30.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 31.Menzel, R. 1989. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for β-galactosidase activity. Anal. Biochem. 181:40-50. [DOI] [PubMed] [Google Scholar]
- 32.Michael, S. F. 1994. Mutagenesis by incorporation of a phosphorylated oligo during PCR amplification. BioTechniques 16:410-412. [PubMed] [Google Scholar]
- 33.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W.-D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama, K., S. Kanaya, M. Ohnishi, Y. Terawaki, and T. Hayashi. 1999. The complete nucleotide sequence of φCTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 31:399-419. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama, K., K. Takashima, H. Ishihara, T. Shinomiya, M. Kageyama, S. Kanaya, M. Ohnishi, T. Murata, H. Mori, and T. Hayashi. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213-231. [DOI] [PubMed] [Google Scholar]
- 37.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 38.Portelli, R., I. B. Dodd, Q. Xue, and J. B. Egan. 1998. The late-expressed region of the temperate coliphage 186 genome. Virology 248:117-130. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasaki, I., and G. Bertani. 1965. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J. Gen. Microbiol. 40:365-376. [DOI] [PubMed] [Google Scholar]
- 42.Sunshine, M. G., M. Thorn, W. Gibbs, R. Calendar, and B. Kelly. 1971. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology 46:691-702. [DOI] [PubMed] [Google Scholar]
- 43.Tabor, S. 1990. Expression using the T7 RNA polymerase/promoter system, p. 16.2.1-16.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates/Wiley Interscience, New York, N.Y.
- 44.Temple, L. M., S. L. Forsburg, R. Calendar, and G. E. Christie. 1991. Nucleotide sequence of the genes encoding the major tail sheath and tail tube proteins of bacteriophage P2. Virology 181:353-358. [DOI] [PubMed] [Google Scholar]
- 45.Weiss, R. B., D. M. Dunn, J. F. Atkins, and R. F. Gesteland. 1987. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, +6 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 52:687-693. [DOI] [PubMed] [Google Scholar]
- 46.Weiss, R. B., D. M. Dunn, A. E. Dahlber, J. F. Atkins, and R. F. Gesteland. 1988. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 7:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziermann, R., B. Bartlett, R. Calendar, and G. E. Christie. 1994. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J. Bacteriol. 176:4974-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]