Abstract
Leinamycin (LNM), produced by Streptomyces atroolivaceus, is a thiazole-containing hybrid peptide-polyketide natural product structurally characterized with an unprecedented 1,3-dioxo-1,2-dithiolane moiety that is spiro-fused to a 18-member macrolactam ring. LNM exhibits a broad spectrum of antimicrobial and antitumor activities, most significantly against tumors that are resistant to clinically important anticancer drugs, resulting from its DNA cleavage activity in the presence of a reducing agent. Using a PCR approach to clone a thiazole-forming nonribosomal peptide synthetase (NRPS) as a probe, we localized a 172-kb DNA region from S. atroolivaceus S-140 that harbors the lnm biosynthetic gene cluster. Sequence analysis of 11-kb DNA revealed three genes, lnmG, lnmH, and lnmI, and the deduced product of lnmI is characterized by domains characteristic to both NRPS and polyketide synthase (PKS). The involvement of the cloned gene cluster in LNM biosynthesis was confirmed by disrupting the lnmI gene to generate non-LNM-producing mutants and by characterizing LnmI as a hybrid NRPS-PKS megasynthetase, the NRPS module of which specifies for l-Cys and catalyzes thiazole formation. These results have now set the stage for full investigations of LNM biosynthesis and for generation of novel LNM analogs by combinatorial biosynthesis.
The five-member heterocycles of thiazole or oxazole or their reduced structures of thiazoline and thiazolidine or oxazoline and oxazolidine are signature pharmacophores common to many clinically important natural products (44), such as bacitracin (26) and pristinamycin IIB (4) (antibacterial) and bleomycin (12) and epothilone (35, 49) (anticancer) (Fig. 1A). They are also common structural motifs for many microbial pathogenesis factors (10), such as pyochelin from Pseudomonas aeruginosa (9, 39), vibriobactin from Vibrio cholerae (23, 52), yersiniabactin from Yersinia pestis (17, 40), mycobactin from Mycobacterium tuberculosis (41), and microcin B17 from Escherichia coli (24, 29) (Fig. 1B).
FIG. 1.
Examples of five-member, heterocycle (boxed)-containing, natural products: antibiotics (A), pathogenesis factors (B), and leinamycin (C).
Biosynthetically these heterocycles result from cyclization (Cy) of the cysteine (Cys), serine, or threonine side chain onto the proceeding carbonyl group of the peptide substrates, and two mechanisms are known for this process. One is exemplified by the maturation of microcin B17, where the heterocycle-forming steps occur posttranslationally, catalyzed by the microcin B17 synthetase (29). The other is observed for nonribosomal peptide biosynthesis where the peptide elongation and heterocycle-forming steps proceed processively, catalyzed by nonribosomal peptide synthetases (NRPS). The latter enzymes are characterized by unique catalytic domains, such as the Cy, oxidization (Ox), and reduction domains, which act on the nascent peptidyl-S-NRPS intermediate to furnish the heterocycles into the resultant natural products (11, 27, 44, 51).
Leinamycin (LNM) is a novel thiazole-containing natural product produced by several Streptomyces atroolivaceus species (18, 20, 36). Its structure was established by spectroscopic (18, 20) and X-ray crystallographic analyses (21) and was confirmed by total synthesis (15, 22). LNM is characterized by an unusual 1,3-dioxo-1,2-dithiolane moiety that is spiro-fused to a 18-member macrolactam ring, a molecular architecture that has not been found to date in any other natural product (Fig. 1C). LNM exhibits a broad spectrum of antimicrobial activity against gram-positive and gram-negative bacteria but not against fungi. It shows potent antitumor activity in murine tumor models in vivo, most significantly against tumors that are resistant to clinically important anticancer drugs, such as cisplatin, doxorubicin, mitomycin, and cyclophosphamide (18, 36). LNM preferentially inhibits DNA synthesis, resulting in interference with the growth of susceptible cells. It alkylates DNA to cause single-strand scission of DNA in the presence of a thiol as a reducing cofactor, and the presence of a sulfoxide group in the dithiolane moiety is essential for the DNA cleavage activity (19). Although simple 1,3-dioxo-1,2-dithiolanes are also thiol-dependent DNA cleavage agents in vitro (16, 34), the mechanisms for DNA cleavage by simple 1,3-dioxo-1,2-dithiolanes and LNM are distinct—oxidative cleavage by the former mediated by DNA-cleaving oxygen radicals and alkylative cleavage by the latter mediated by an episulfonium ion intermediate (1, 5, 19).
The biosynthetic origin of LNM has not been established by in vivo isotope-labeling experiment. The structural similarity of LNM to macrolides in general and to thiazole-containing hybrid peptide-polyketide metabolites, such as bleomycin (12) and epothilone (35, 49), in particular, however, supports the hypothesis that it is of hybrid peptide-polyketide origin. Thus, on the basis of the emerging paradigm for hybrid peptide-polyketide biosynthesis (6, 7, 13), we postulated that a hybrid NRPS-polyketide synthase (PKS) system would be responsible for the biosynthesis of the 18-member LNM macrolactam ring and that an NRPS module characterized by a Cys-specific adenylation (A) domain and the thiazole-forming Cy and Ox domains would specifically furnish the thiazole moiety of LNM. We set to clone this NRPS gene by a PCR approach as the starting point toward cloning the entire lnm biosynthetic gene cluster. The availability of the lnm gene cluster should greatly aid our effort to investigate LNM biosynthesis and to produce novel LNM analogs for anticancer drug development by manipulation of genes governing LNM biosynthesis. Here we report the amplification of a Cy domain sequence from the genome of an LNM-producing S. atroolivaceus strain S-140 by PCR, the cloning and characterization of a thiazole-forming NRPS module and confirmation of this NRPS module in LNM biosynthesis, and the subsequent identification and localization of the entire lnm biosynthetic gene cluster.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli DH5α (45), E. coli XL1-Blue MRF' (Stratagene, La Jolla, Calif.), E. coli S17-1 (25), and E. coli BL21(DE3) (Novagen, Madison, Wis.) were used in this work. Vectors pGEM-T, pGEM-5zf, pGEM-9zf (Promega, Madison, Wis.), pBluescript II SK (Stratagene), and pET-37b (Novagen) were from commercial sources, and pOJ260, pKC1139, and pOJ446 were described previously (3). The S. atroolivaceus strains and other plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. atroolivaceus strains | ||
| S-140 | Wild-type, LNM producing | 18 |
| SB3001 | lnmI-disrupted mutant resulted from homologous integration of pBS3012 into the S-140 strain; Apr, non-LNM producing | This work |
| Plasmids | ||
| pBS3001 | 1.1-kb PCR product from S. atroolivaceus S-140 with Cy-FP and Cy-RP primers in pGEM-T | This work |
| pBS3002 | 0.3-kb PCR product from S. atroolivaceus S-140 with Ox-FP and Ox-RP primers in pGEM-T | This work |
| pBS3003 | 5.2-kb PCR product from S. atroolivaceus S-140 with Cy-FP and Ox-RP primers in pGEM-T | This work |
| pBS3004 | pOJ446-derived S. atroolivaceus S-140 genomic library cosmid | This work |
| pBS3005 | pOJ446-derived S. atroolivaceus S-140 genomic library cosmid | This work |
| pBS3006 | pOJ446-derived S. atroolivaceus S-140 genomic library cosmid | This work |
| pBS3007 | pOJ446-derived S. atroolivaceus S-140 genomic library cosmid | This work |
| pBS3008 | pOJ446-derived S. atroolivaceus S-140 genomic library cosmid | This work |
| pBS3009 | 4.6-kb NcoI fragment from pBS3006 into the same site of pGEM-5zf | This work |
| pBS3010 | 2.8-kb NcoI fragment from pBS3006 into the same site of pGEM-5zf | This work |
| pBS3011 | 5.1-kb BamHI fragment from pBS3006 into the same site of pBluescript II SK | This work |
| pBS3012 | 1.1-kb EcoRI fragment from pBS3001 into the same site of pOJ260 | This work |
| pBS3013 | 1.1-kb EcoRI fragment from pBS3001 into the same site of pKC1139 | This work |
| pBS3014 | PCR-amplified 5′ end (a 372-bp EcoRI-SpeI fragment) and 3′ end (a 293-bp SpeI-HindIII fragment) of lnmI (A-PCP) into the EcoRI and HindIII sites of pSP72 | This work |
| pBS3015 | 1.2-kb NcoI-BamHI internal fragment of lnmI (A-PCP) from pBS3010 into the same sites of pBS3014 | This work |
| pBS3016 | 1.9-kb NdeI-HindIII fragment of lnmI (A-PCP) from pBS3015 into the same sites of pET-37b | This work |
Biochemicals, chemicals, and media.
LNM standard was a gift from Kyowa Hakko Kogyo Co. Ltd. (Tokyo, Japan). Unless specified otherwise, other common biochemicals and chemicals were from standard commercial sources. E. coli strains carrying plasmids were grown in Luria-Bertani (LB) medium and were selected with appropriate antibiotics (45). All media for Streptomyces growth were prepared according to standard protocols (25). DNB, ISP-2, ISP-4, and tryptic soy broth (TSB) were from Difco Laboratories (Detroit, Mich.), and modified ISP-4 is ISP-4 supplemented with 0.05% yeast extract and 0.1% tryptone (30).
DNA isolation, manipulation, and sequencing.
Plasmid preparation and DNA extraction were carried out by using commercial kits (Qiagen, Valencia, Calif.). Total S. atroolivaceus DNA was isolated according to protocols from the literature (25, 42). Restriction endonuclease digestion and ligation were done by standard methods (45). For Southern analysis, digoxigenin labeling of DNA probes, hybridization, and detection were performed according to the protocols provided by the manufacturer (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Automated DNA sequencing was carried out on an ABI Prism 377 DNA sequencer (Perkin-Elmer/ABI, Foster City, Calif.), and this service was provided by Davis Sequencing Inc. (Davis, Calif.). Data were analyzed by the ABI Prism Sequencing 2.1.1 software and the Genetics Computer Group program (Madison, Wis.).
PCR.
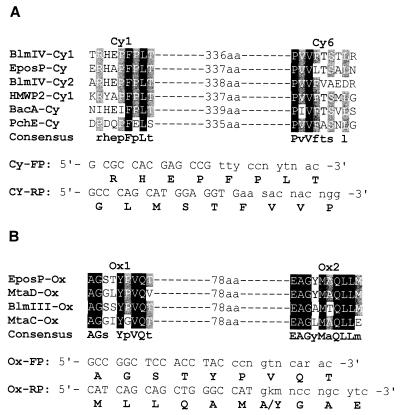
PCR primers for Cy (Cy-FP and Cy-RP) and Ox (Ox-FP and Ox-RP) domain amplification (Fig. 2) were designed by using the COnsensus-DEgenerate Hybrid Oligonucleotide Primer (CODEHOP) strategy of PCR primer design (43). They were synthesized at the Macromolecular Structure Facility, University of California, Davis. PCRs were performed on a GeneAmp 2400 thermocycler (Perkin-Elmer/ABI). A typical PCR (50 μl) consists of 1.5 mM MgCl2, 5 ng of S. atroolivaceus genomic or cosmid DNA as the template, 7.5% dimethyl sulfoxide, 100 μM deoxynucleoside triphosphates, 25 pmol of each primer, and 2.5 U of Taq DNA polymerase in 1× PCR buffer. For Cy domain amplification, the program was as follows: prerun denaturation at 95°C for 3 min, followed by 10 cycles of ramp amplification (45 s at 94°C, 1 min of annealing with ramping from 65 to 55°C, and 1.5 min at 72°C), then 25 cycles of amplification with a constant annealing temperature of 55°C, and finally postrun extension for 10 min at 72°C. A distinctive product with the predicted size of 1.1 kb was obtained with the Cy-FP and Cy-RP pair of primers and was cloned into pGEM-T to yield pBS3001. For Ox domain amplification, the program was the same except that the ramp temperature was from 60 to 50°C and that the extension time was 45 s. With the Ox-FP and Ox-RP pair of primers, no product was amplified from S. atroolivaceus genomic DNA but a distinctive product with the predicted size of 0.3 kb was amplified from several Cy-containing cosmid clones. This product was subsequently cloned in pGEM-T to yield BS3002. For amplification of the entire NRPS module, the PCR program was the same as for Cy domain amplification except that extension time was 5 min. With the Cy-FP and Ox-RP pair of primers, a distinctive product of 5.2 kb was amplified from S. atroolivaceus cosmid library DNA and cloned in pGEM-T to afford pBS3003.
FIG. 2.
The conserved core motifs and PCR primers designed for them according to the CODEHOP strategy: Cy domain (A) and Ox domain (B). Protein accession numbers (GenBank) are given in parentheses: BacA (O68006), BlmIII (AAG02365), BlmIV (AAG02364), EposP (AAF26925), HMWP2 (P48633), MtaC (AAF19811), MtaD (AAF19812), and PchE (AAD55800). The numbers between the motifs indicate the distance in amino acids (aa). K, G/T; M, A/C; N, G/A/T/C; R, A/G; S, C/G; and Y, C/T.
S. atroolivaceus genomic library construction and screening.
A genomic library of S. atroolivaceus S-140 was constructed in the pOJ446 cosmid according to protocols from the literature (25, 42). Gigapack III XL packaging extract (Stratagene) was used for library construction according to the manufacturer's instructions. The titer of the primary library was approximately 2.2 × 105 PFU per μg of DNA. The primary library was amplified once; aliquots of it were stored at −80°C. The titer for the amplified library was approximately 2 × 105 CFU/μl.
To screen the genomic library, cosmid clones from four plates (with approximately 1,000 colonies per plate) were transferred to Immobilon-Ny+ nylon membranes (Millipore Corp., Bedford, Mass.) and were hybridized with the digoxigenin-labeled 1.1-kb EcoRI fragment of the Cy domain from pBS3001 as a probe (Fig. 3A, Probe-1). Resultant positive cosmid clones were rescreened by PCR with primers specific for Cy (Cy-FP and Cy-RP), Ox (Ox-FP and Ox-RP), and the thiazole-forming NRPS module (Cy-FP and Ox-RP) and were confirmed by Southern hybridization. Further restriction endonuclease mapping of and chromosomal walking from these overlapping cosmids lead to the genetic localization of a 172-kb contiguous DNA region, as represented by pBS3004, pBS3005, pBS3006, pBS3007, and pBS3008 (Fig. 3A). A total of 11 kb of DNA from pBS3006 that hybridized to the PCR-amplified NRPS fragment from pBS3003 was subcloned as three overlapping clones: a 4.6-kb NcoI fragment into pGEM-5zf to yield pBS3009, a 2.8-kb NcoI fragment into pGEM-5zf to yield pBS3010, and a 5.1-kb BamHI fragment into pBluescript II SK to yield pBS3011; their DNA sequences were subsequently determined.
FIG. 3.
(A) A 172-kb DNA region from S. atroolivaceus S-140 harboring the lnm biosynthetic gene cluster as represented by five overlapping cosmid clones. Probes used to clone and map the lnm gene cluster were marked. (B) Genetic organization of the lnmG, lnmH, and lnmI genes and domain organization of the LnmI hybrid NRPS-PKS protein. Cy, condensation/cyclization. A, adenylation; KS, ketoacyl synthase; B, BamHI; N, NcoI.
Construction of the lnmI mutant by gene disruption.
The 1.1-kb internal fragment of lnmI amplified by PCR with the Cy-FP and Cy-RP pair of primers was cloned as an EcoRI fragment from pBS3001 into the same site of pOJ260 and pKC1139 to yield pBS3012 and pBS3013, respectively. Introduction of pBS3012 and pBS3013 into S. atroolivaceus S-140 was carried out by conjugation, following a procedure from the literature (30) with minor modifications. In brief, S. atroolivaceus S-140 spores were heat shocked in TSB medium at 42°C for 20 min, followed by incubation at 30°C for up to 6 h. Spore germination was monitored microscopically every 30 min after 4 h of incubation at 30°C. Germinated spores were pelleted and resuspended in LB medium as S. atroolivaceus recipients. E. coli S17-1(pBS3012) or E. coli S17-1(pBS3013) was grown in LB medium with appropriate antibiotics for selection to cell density of 0.3 to 0.4 at 600 nm. Cells from 2-ml culture were pelleted, washed with LB medium, and resuspended in 100 μl of LB medium as the E. coli donors. For conjugation, the donors (100 μl) and recipients (100 μl, 108 spores) were mixed and spread onto a modified ISP-4 plate freshly supplemented with 10 mM MgCl2. The plates were incubated at 28°C for 16 to 22 h. After removal of most of the E. coli S17-1 donors from the plates by washing the surface with sterile water, each plate was overlaid with 3 ml of soft LB medium (0.7% agar) containing apramycin (Apr) at a final concentration of 50 μg/ml and nalidixic acid at a final concentration of 50 μg/ml. Incubation continued at 28°C until exconjugants appeared (in about a week).
Production, isolation, and analysis of LNM.
S. atroolivaceus strains were fermented under conditions taken from the literature (18, 20) with modifications. In brief, the seed medium (25 ml in a 250-ml baffled flask), consisting of 10 g of glucose, 10 g of soluble starch, 3 g of beef extract, 5 g of yeast extract, 5 g of Bacto-Tryptone, and 2 g of CaCO3 per liter, pH 7.2, was inoculated with 10 μl of spore suspension and incubated at 25°C, 250 rpm, for 48 h. The resultant seed inoculum (5 ml) was then used to inoculate the production medium (50 ml in a 250-ml baffled flask), consisting of 40 g of soluble starch, 5 g of soybean meal, 2.5 g of corn steep liquor, 0.5 g of KH2PO4, 0.25 g of MgSO4, 40 mg of ZnSO4 · 7H2O, 0.1 g of methionine, 0.1 mg of vitamin B12, and 5 g of CaCO3 per liter, pH 7.0. The production culture was fermented at 28°C, 250 rpm, for 72 h. Sterile, moisturized Diaion HP-20 resins (5% [wt/vol]) (Supelco, Bellefonte, Pa.) were added to the production culture 18 h after inoculation. For LNM isolation, the production culture was acidified to pH 2 with H2SO4. The HP-20 resins were recovered by filtration through two layers of cheesecloth and were then lyophilized to dryness. Crude LNM was eluted off the HP-20 resins with methanol (10 resin volumes), and the methanol elute was concentrated in vacuo. High-performance liquid chromatography (HPLC) analysis of LNM was achieved through a two-step procedure with a Microsorb-MV C18 column (5 μM, 100 Å, 4.6 mm by 25 cm; Varian, Walnut Creek, Calif.) on a ProStar-210 HPLC system with a PDA-330 UV-VIS detector at 320 nm (Varian). In the first step, partially pure LNM was collected upon isocratic elution with 50% methanol in 25 mM KH2PO4, pH 4.0, at a flow rate of 0.6 ml/min. In the second step, the partially purified LNM was baseline resolved by a linear gradient elution from 20 to 80% acetonitrile in 10 mM KH2PO4, pH 4.0, in 30 min at a flow rate of 0.6 ml/min. The identity of LNM was confirmed by coelution with authentic LNM standard and electrospray ionization-mass spectrometry (ESI-MS) analysis. The latter was performed on an LCQ-Decca mass spectrometer (ThermoQuest, San Jose, Calif.) at the Molecular Structure Facility, University of California, Davis.
Overexpression of lnmI (A-peptidyl carrier protein [PCP]) in E. coli and purification of the LnmI (A-PCP) protein.
The 5′ end of lnmI (A-PCP) was amplified from pBS3006 by PCR using a forward primer of 5′-G GAA TTC CAT ATG GAG CTG CAC AAG ATC CTG CAC-3′ (both EcoRI and NdeI sites are underlined) and a reverse primer of 5′-CC ACT AGT GCG CCT CCT CGA GGA TGA C-3′ (the SpeI site is underlined) as a 372-bp fragment. The 3′ end of lnmI (A-PCP) was amplified from pBS3006 by PCR using a forward primer of 5′-GG ACT AGT CGC GCG ACG AGC TGT GCG-3′ (the SpeI site is underlined) and a reverse primer of 5′-CG CGC AAG CTT CGC GGT CTC CGC CTC GGC GGC-3′ (the HindIII site is underlined) as a 293-bp fragment. These two fragments were digested with EcoRI-SpeI and SpeI-HindIII, respectively, and were cloned together into the EcoRI-HindIII sites of pSP72 to afford pBS3014. The 1.2-kb NcoI-BamHI internal fragment of lnmI (A-PCP) from pBS3010 was then cloned into the same sites of pBS3014 to yield pBS3015. DNA sequencing confirmed that pBS3015 contains the complete lnmI (A-PCP) sequence. The lnmI (A-PCP) fragment was finally moved as a 1.9-kb NdeI-HindIII fragment from pBS3015 into the same sites of pET-37b to yield the expression construct pBS3016. Introduction of pBS3016 into E. coli BL21(DE3) under standard conditions resulted in the production of LnmI (A-PCP) as a C-terminal His8-tagged fusion protein. To improve the solubility, the incubation temperature was lowered to 25°C with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) induction for 4 h at 250 rpm. The soluble fraction of LnmI (A-PCP) was subsequently purified by affinity chromatography on Ni-nitrilotriacetic acid resin according to the manufacturer's protocols (Qiagen). Purification was monitored by sodium polyacrylamide gel electrophoresis (SDS-PAGE), and the yield of the purified protein was 92 mg/liter. The purified protein was dialyzed against the storage buffer (0.1 mM EDTA and 10% glycerol in 25 mM Tris-HCl, pH 8.0) and was stored at −80°C for an in vitro assay.
Amino acid specificity assay of LnmI (A-PCP).
Amino acid-dependent ATP-sodium pyrophosphate (PPi) assays were performed according to procedures in the literature (14, 28). A typical assay reaction (100 μl) contained 100 nM LnmI (A-PCP), 1 mM ATP, and 0.1 mM PPi with 0.5 μCi of [32P]PPi (40.02 Ci/mmol; NEN Life Science Products, Boston, Mass.), 1 mM MgCl2, 0.1 mM EDTA, and 0.5 mM amino acid in 50 mM sodium phosphate buffer, pH 7.8. After 30 min of incubation at 30°C, the assays were stopped by addition of 0.9 ml of cold 1% (wt/vol) activated charcoal in 3% (vol/vol) perchloric acid. The precipitates were collected on glass fiber filters (2.4 cm, G-4; Fisher Scientific, Pittsburgh, Pa.), washed successively with 10 ml of 40 mM sodium phosphate buffer, pH 8.0, 10 ml of water, and 5 ml of ethanol, and dried briefly in air. The filters were mixed with 7 ml of scintillation fluid (ScintiSafe Gel; Fisher Scientific) and were counted on a Tri-Carb 2900TR liquid scintillation counter (Packard Instrument Co., Meriden, Conn.) to determine the radioactivity.
In vitro phosphopantetheinylation and aminoacylation of holo-LnmI (A-PCP) were carried out essentially according to methods from the literature (12, 14, 46). For in vitro phosphopantetheinylation, a typical assay solution (100 μl) consisted of 3 μM LnmI (A-PCP), 1 μM Svp (46), 25 μM coenzyme A, 10 mM MgCl2, and 5 mM dithiothreitol in 75 mM Tris-HCl buffer, pH 8.0, and the reaction mixture was incubated at 30°C for 30 min. For in vitro aminoacylation, to the holo-LnmI (A-PCP) reaction mixture were added 0.1 μM l-[35S]Cys (103 Ci/mmol; NEN Life Science) and 3 mM ATP, and the reaction was continued at 30°C for additional 30 min. The reaction was finally stopped by addition of 0.9 ml of cold 7% (vol/vol) trichloroacetic acid. The precipitated proteins were collected by centrifugation and resolved by SDS-PAGE on a 4 to 20% gradient gel (Bio-Rad Laboratories Inc., Richmond, Calif.). To determine the radioactivity incorporated into the holo-LnmI (A-PCP)-l-[35S]Cys species, the gel was dried between two sheets of cellulose membrane (KOH Development Inc., Ann Arbor, Mich.) and visualized by a PhosphorImager (Molecular Dynamics/Amersham Pharmacia Biotech, Piscataway, N.J.).
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the GenBank database under accession number AF484556.
RESULTS
Cloning of a thiazole-forming NRPS module from S. atroolivaceus S-140 by PCR and mapping of the lnm gene cluster.
Seven conserved motifs for the Cy domain (Cy1 to Cy7) and two conserved motifs for the Ox domain (Ox1 and Ox2) are known for thiazole-forming NRPS (Fig. 4). PCR primers for the Cy and Ox domains were based on the Cy1 (for Cy-FP) and Cy6 (Cy-RP) motifs and Ox1 (Ox-FP) and Ox2 (Ox-RP) motifs, respectively. Initial PCR attempts with conventionally designed degenerate primers, taking into the GC bias at the wobble position for G/C-rich Streptomyces DNA, failed to amplify any specific products. We then adopted the CODEHOP strategy of PCR primer design (43) and designed the two pairs of primers as shown in Fig. 2. With S. atroolivaceus S-140 genomic DNA as the template, while products appeared to be nonspecific when the Ox-FP and Ox-RP pair of primers was used, a distinct band with the expected size of 1.1 kb was readily amplified with the Cy-FP and Cy-RP pair of primers. This product was cloned (pBS3001), and 12 randomly selected clones were subjected to sequencing. Eight yielded an identical sequence (except differences resulting from primer utilization), which shows significant homology to Cy domains of known thiazole-forming NRPS (Fig. 4B), and four were nonspecific sequences. This PCR-amplified Cy fragment was then recovered as a 1.1-kb EcoRI fragment from pBS3001 and used as a probe (Fig. 3A, Probe-1) to screen an S. atroolivaceus genomic library that was constructed in the Streptomyces-E. coli shuttle cosmid, pOJ446. Of the 4,000 colonies screened, 12 positive clones were identified, 11 of which were confirmed by PCR and Southern analysis to contain the same fragment hybridizing to the Cy probe. To ensure that we have a full coverage of the entire lnm gene cluster, additional chromosomal walking from this locus was carried out with probe-2, probe-3, and probe-4 (Fig. 3A), eventually leading to the localization of a 172-kb continuous DNA region covered by overlapping cosmids as exemplified by pBS3004, pBS3005, pBS3006, pBS3007, and pBS3008 (Fig. 3A).
FIG. 4.
Conserved motifs from LnmI-NRPS module and alignments of the A, Cy, Ox, and PCP domains with known NRPS domains. Protein accession numbers (GenBank) are given in parentheses: AcvS (B61212), BacA (O68006), BlmIII (AAG02365), BlmIV (AAG02364), EposP (AAF26925), GrsA (P14687), HMWP2 (P48633), MtaC (AAF19811), MtaD (AAF19812), LnmI-NRPS (AF484556), PchE (AAD55800), TycC (O30409), and VibF (AAG00566).
To examine if these cosmids also harbor an Ox domain, we carried out PCRs using the Ox-FP and Ox-RP pair of primers with the cosmid clone DNA as templates. Among the 11 Cy domain-containing cosmids tested, 9 yielded a distinct band with the predicted size of 0.3 kb. This fragment was cloned (pBS3002) and sequenced, revealing that it is highly homologous to known Ox domains (Fig. 4C). Finally, to determine if the amplified Cy and Ox domains constitute the same thiazole-forming NRPS module, we carried out additional PCRs using the Cy-FP and Ox-RP pair of primers with one of the cosmids, pBS3006, as the template. A distinct band with the predicted size of 5.2 kb was amplified and cloned (pBS3003). DNA sequencing of the cloned fragment revealed that it contained an additional Cy domain, an A domain, and a PCP domain, flanked by the Cy and Ox domains that had been amplified individually (Fig. 3B). The type of domains and organization identified from the PCR product is exactly what would be predicted for a thiazole-forming NRPS module (11, 27, 44, 51).
Sequence analysis of the lnmI locus.
Restriction mapping and Southern analysis mapped the PCR-amplified 5.2 kb in pBS3003 to three overlapping fragments, a 4.6-kb NcoI fragment (pBS3009), a 2.8-kb NcoI fragment (pBS3010), and a 5.1-kb BamHI fragment (pBS3011), from pBS3006. These three fragments constitute an 11-kb contiguous region. DNA sequencing of this region (10,881 bp) revealed one completed open reading frame (ORF), lnmH, and two incomplete ORFs, lnmG and lnmI, all of which are transcribed in the same direction (Fig. 3B). The incomplete lnmG gene starts out of the sequenced region and ends with a TGA codon 1,643 bp into the sequence. The deduced product of lnmG, the 546-amino acid C terminus, shows significant homology to a class of putative enoyl reductase domain of PKSs, such as the Orf8 protein from a Shewanella sp. (GenBank accession number T30186) and the OrfB protein from Schizochytrium sp. strain ATCC 20888 (GenBank accession number AAK72880) (33). The Orf8 and OrfB proteins are components of the newly discovered PKSs involved in the biosynthesis of very-long-chain polyunsaturated fatty acids. Immediately downstream (70 bp) of lnmG is the complete ORF, lnmH, which starts with an ATG codon and ends with a TGA codon. The deduced product of lnmH consists of 274 amino acids but shows no significant homology to any protein in the databases. Separated from lnmH by 132 bp, the other incomplete ORF, lnmI, starts with an ATG codon and ends out of the sequenced region (10,881 bp).
The sequenced lnmI portion contains all three PCR-amplified fragments, and the deduced product of lnmI, the 3,623-amino-acid N terminus, clearly consists of domains characteristic for both known NRPS and PKSs (6, 7, 13). Located at the N termini of LnmI is a complete thiazole-forming NRPS module composed of two-tandem Cy domains, an A domain, a PCP domain, and an Ox domain. All the domains contain the known conserved motifs (12, 27), which is indicative that they likely are functional (Fig. 4B). It is very unusual to find two-tandem Cy domains in a single NRPS module, but a precedent has been observed recently in VibF (GenBank accession number AAG00566) for vibriobactin biosynthesis (31). Using site-directed and domain-deletion mutagenesis, Marshall and coworkers mapped the specific functions of the two-tandem Cy domains in VibF—the second Cy domain for condensation and the first Cy domain for cyclization and dehydration. The two Cy domains in LnmI could play a similar role in LNM biosynthesis. The amino acid specificity of an NRPS module is determined by the A domain, which could be predicted according to the so-called “specificity-conferring codes” (8, 47). The specificity-conferring codes for the A domain were identified as follows: D-235, L-236, F-239, N-278, F-299, S-301, L-322, V-330, W-331, and K-517, suggesting that LnmI-A specifies for l-Cys. This is consistent with the fact that the A domain exhibits the highest sequence homology to A domains of NRPS known to specify for l-Cys (Fig. 4A). The PCP domain is only modestly conserved at the signature LGGXS motif (Fig. 4D). In order to be functional, PCP must be posttranslationally modified by the covalent attachment of the 4′-phosphopantetheine group to the Ser residue of this motif (S, underlined) (6, 46). The Ox domain has only been found in thiazole- and oxazole-forming NRPS modules but not in thiazoline- and oxazoline- or thiazolidine- and oxazolidine-forming NRPS modules (10, 11, 27, 44). This is consistent with the fact that the biosynthesis of the latter does not require an oxidation step. Two locations within an NRPS module have been observed for the Ox domain (11). The Ox domain could be located between the A8 and A9 motifs of an A domain, as for EposP (GenBank accession number AAF26925) and MtaD (GenBank accession number AAF19812), or could be located downstream of PCP, as for BlmIII (GenBank accession number AAG02365) and MtaC (GenBank accession number AAF19811). The Ox domain in the LnmI-NRPS module apparently falls into the latter group, being located downstream of the PCP domain.
Following the NRPS module throughout the rest of the sequenced LnmI are two tandem KS domains (the second KS domain is incomplete), indicative of an incomplete PKS module. Both KS domains are highly homologous to known KS domains (6, 7, 48). To our knowledge, tandem KS domains within a PKS module are unprecedented, although it is not known if both KS domains are functional. The LnmI-PKS module would start with a KS domain consisting of 420 amino acids. This KS domain is characterized by the well-defined signature TSCSSSFAAL motif, the Cys residue of which (C, underlined) has been established to be the active site for all known KS domains (48). Although incomplete (only 78 amino acids), the sequenced 3′ end of lnmI clearly encodes a KS domain. Hybrid NRPS-PKS has been defined as megasynthetases that can incorporate biosynthetic building blocks of both amino acids and short carboxylic acids into the resultant hybrid peptide-polyketide product—an NRPS-bound growing peptidyl intermediate is further elongated by a PKS module or vice versa (6, 7, 13). The interacting NRPS and PKS modules could either be covalently linked with all domains arranged in a linear order on the same protein, known as type I hybrid, or could physically reside on separate proteins, known as type II hybrid (14). Pending the completion of its C terminus, LnmI would be a hybrid NRPS-PKS megasynthetase with a type I hybrid architecture.
Development of a genetic system and disruption of the lnmI gene in S. atroolivaceus.
No genetic study of S. atroolivaceus had been reported prior to our investigation. We set out to establish a genetic system for this strain in order to confirm that the cloned gene cluster encodes LNM biosynthesis, setting the stage for production of novel LNM analogs by manipulating LNM biosynthesis in vivo. S. atroolivaceus S-140 grows optimally between 28 and 30°C but cannot grow at or beyond 37°C. Among the 10 media (AS-1, DNB, GMP, ISP-2, ISP-4, R2YE, SGGP, SR12, TSB, and YEME) (25) tested, TSB medium was found to be most suitable for vegetative growth of S. atroolivaceus. However, for protoplast preparation and genomic DNA isolation, it is necessary to supplement TSB with 25 mM MgCl2 (final concentration) to facilitate lysozyme digestion, whereas supplementing glycine at a concentration as low as 0.1% (wt/vol) has adverse effects on both protoplast preparation and DNA isolation. S. atroolivaceus S-140 sporulates very well on ISP-4 agar at 28°C, and the color of spores ranges from grayish to dark blue, depending on the age of the culture. S. atroolivaceus S-140 is highly sensitive to both Apr and thiostrepton (Thi) (complete inhibition of growth at 10 and 3 μg/ml, respectively); therefore, 50 μg of Apr and 25 μg of Thi per ml were used throughout this study for selection.
Although S. atroolivaceus S-140 can be protoplasted in a respectable yield (30%), they cannot be regenerated efficiently under all conditions tested. The best regeneration frequency (0.6%) was obtained when the protoplasts were made from cells grown in YEME medium and regenerated on R2YE plates supplemented with 10 mM MgCl2. Due to the low regeneration frequency, protoplast-mediated transformation for S. atroolivaceus S-140 was not very successful. With nonreplicative plasmids such as pBS3012, the transformation frequency was only about 10−9 per μg of DNA. In contrast, we were successful in developing a protocol for conjugation between E. coli S17-1 and S. atroolivaceus S-140. Following a procedure taken from the literature (30) with modifications, we found that conjugation between E. coli S17-1 and S. atroolivaceus S-140 occurred with the highest frequency on modified ISP-4 agar freshly supplemented with 10 mM MgCl2. The exconjugant frequencies for replicating plasmids such as pBS3013 were approximately 10−4 to 10−5 and for nonreplicating plasmids such as pBS3012 were 10−5 to 10−6.
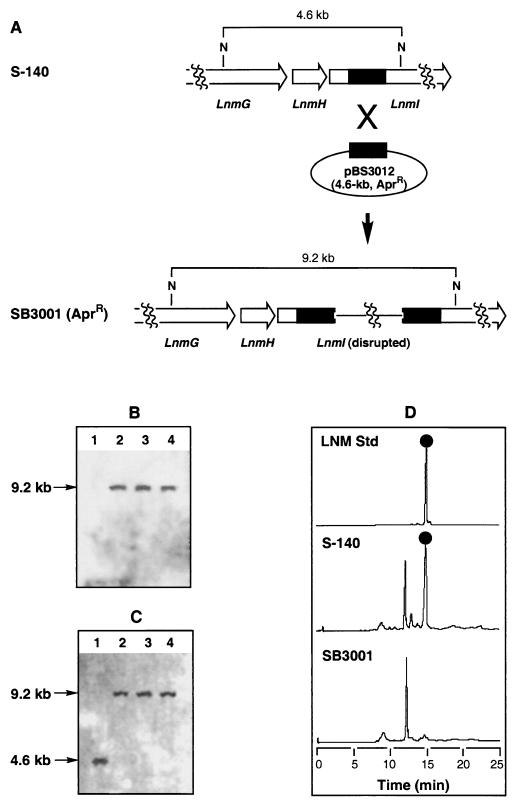
To confirm that the cloned gene cluster encodes LNM biosynthesis, we inactivated lnmI by insertional disruption via a single-crossover homologous recombination event (Fig. 5). Construct pBS3012 was selected for this experiment, which contains an 1.1-kb internal fragment of lnmI. After introduction of pBS3012 into S. atroolivaceus S-140 by conjugation, exconjugants that were resistant to Apr were isolated. Since pBS3012 is derived from the Streptomyces nonreplicating plasmid pOJ260, those exconjugants must have resulted from integration of pBS3012 into the S. atroolivaceus chromosome by homologous recombination and were named S. atroolivaceus SB3001 mutant strains (Fig. 5A).
FIG. 5.
Disruption of lnmI by single-crossover homologous recombination. (A) Restriction maps of the S. atroolivaceus S-140 wild-type and SB3001 mutant strains showing predicted fragment sizes upon NcoI digestion. Southern analysis of S-140 (lane 1) and SB3001 strain (lanes 2 to 4 for three individual isolates) genomic DNAs digested with NcoI by using the pOJ260 vector (B) and the 1.1-kb lnmI fragment from pBS3012 (C) as a probe, respectively. (D) HPLC analysis with UV detection at 320 nm of partially purified LNM from the S-140 wild-type and SB3001 mutant strain with authentic LNM standard as a control. LNM, •.
Characterization of the lnmI-disrupted S. atroolivaceus SB3001 strain as a non- LNM-producing mutant.
To confirm that lnmI disruption had occurred by a single-crossover homologous event, Southern analysis of the DNA from three randomly selected isolates of the SB3001 mutant strain was carried out with either pOJ260 or the 1.1-kb internal fragment of lnmI from pBS3012 as a probe. As shown in Fig. 5B, a distinctive band of the predicted size of 9.2 kb was detected with the pOJ260 vector as a probe in all three isolates; this band was absent from the wild-type strain. Complementarily, when the 1.1-kb internal fragment of lnmI was used as a probe, the 4.6-kb band in the wild-type strain was shifted to the 9.2-kb band in the mutant isolates (Fig. 5C), as would be expected for disruption of lnmI by a single-crossover homologous recombination event (Fig. 5A).
No apparent difference in growth characteristics and morphologies between the wild-type S-140 and mutant SB3001 strains was observed. S. atroolivaceus fermentation was carried out according to protocols from the literature (18) with modifications. The LNM produced was distributed among the mycelia (<15%), broth (<5%), and Diaion HP-20 resins (>80%); therefore, LNM isolation was focused only on the resins in this study. The isolated yield for LNM from S. atroolivaceus S-140 is 3 to 5 mg/liter. The LNM identity was confirmed by coelution with the authentic standard and ESI-MS analysis. Upon ESI-MS analysis under positive ionization mode, the purified product yielded a characteristic (M+H)+ ion at an m/z of 511, consistent with the molecular formula of C22H26N2O6S3 for LNM (calculated molecular weight, 510). LNM production was completely abolished in all three isolates of the lnmI mutant strain of S. atroolivaceus SB3001, confirming that lnmI and therefore the cloned lnm gene cluster encode LNM biosynthesis (Fig. 5D).
Specific incorporation of l-Cys by the LnmI NRPS module.
To verify the sequence-based prediction that the LnmI NRPS module specifies for l-Cys, we overexpressed lnmI (A-PCP) in E. coli and purified the resultant LnmI (A-PCP) as a fusion protein with the His8 tag at the C terminus (Fig. 6A). By variation of the expression conditions, lnmI (A-PCP) was very well expressed, resulting in the production of a significant quantity of pure protein (92 mg/liter). After SDS-PAGE, LnmI (A-PCP) migrates with an Mr of 73 kDa, consistent with the calculated Mr of 73,231 Da. The purified LnmI (A-PCP) protein was active as measured by the amino acid-dependent ATP-PPi exchange assay. Among the 22 amino acids tested, LnmI (A-PCP) specifically activates l-Cys, an amino acid that would be required for the biosynthesis of the thiazole moiety of LNM (Fig. 6B). The fact that the l-Cys-specific, thiazole-forming LnmI NRPS module is followed by a PKS module would agree well with the hybrid peptide-polyketide hypothesis for LNM biosynthesis.
FIG. 6.
(A) Expression and purification of LnmI (A-PCP) as analyzed by electrophoresis on an SDS-12% polyacrylamide gel (the calculated molecular weight for LnmI [A-PCP] is 73.231 kDa). Lane 1, molecular weight markers; lane 2, total protein before IPTG induction; lane 3, total protein after IPTG induction; lane 4, total soluble proteins; and lane 5, purified LnmI (A-PCP). (B) Amino acid substrate specificity as determined by the ATP-PPi exchange assay (100% relative activity corresponds to 8 × 105 cpm). In vitro aminoacylation of holo-LnmI (A-PCP) with l-[35S]Cys as a substrate was performed; results of SDS-polyacrylamide gel analysis on a 4 to 20% gradient gel (C) and phosphorimaging (D) are shown. Lane 1, molecular weight markers; lane 2, complete assay with no ATP; lane 3, complete assay with no Svp; and lane 4, complete assay.
To confirm that the activated l-Cys could be loaded to the PCP domain of LnmI, we carried out the in vitro aminoacylation of LnmI (A-PCP). Since PCPs heterologously overproduced in E. coli are often in the apo-form or only partially modified into the holo-form (46) (these PCPs are poor substrates for the endogenous E. coli phosphopantetheinyl transferases [PPTases]), we first carried out the in vitro phosphopantetheinylation of the purified LnmI (A-PCP) to ensure that it is fully in the holo form. We chose the Svp PPTase for its broad carrier protein specificity (46). In vitro aminoacylation was then carried out by incubating the holo-LnmI (A-PCP) in the presence of l-[35S]Cys and ATP and was visualized by phosphorimaging. As shown in Fig. 6C and D, aminoacylation of LnmI (A-PCP) is very efficient and specific. Removal of either ATP (lane 2) or the Svp PPTase (lane 3) from the complete assay solution completely abolished or significantly reduced l-Cys labeling of LnmI (A-PCP).
DISCUSSION
Methods for PCR cloning of both NRPS and PKS genes have been reported, relying on the amino acid sequence conservation of the A domain in NRPS (50) and the KS domain in PKSs (32, 37), respectively. While very effective, these methods suffer from the multiple nonribosomal peptide or polyketide pathways commonly existing in the genome of a given microorganism, especially in the Streptomyces species (2, 38). The amplified A or KS fragments often represent a pool of heterologous products from different nonribosomal peptide or polyketide biosynthetic gene clusters. To determine which PCR product and subsequently which gene cluster are the desired ones requires unambiguous genetic, biochemical, or chemical evidence. If a genetic system is available to the given strain, it is possible but still time consuming to test individual PCR products by a series of gene inactivation experiments. However, most strains were discovered by their ability to produce a biologically active metabolite and are, therefore, poorly characterized physiologically and genetically. Thus, an amenable genetic system is often not available, which presents a serious challenge for deciding which gene cluster is to be further pursued.
We recognized, on the basis of its structural features, that LNM is a hybrid peptide-polyketide macrolactam containing a thiazole moiety. Taking advantage of the emerging paradigm for NRPS-templated thiazole biosynthesis, we reasoned that the choice of amplifying a Cy and/or Ox domain of a thiazole-forming NRPS module rather than the KS domain of a PKS module or A domain of an NRPS module as a probe would greatly increase our chances of specifically isolating the lnm biosynthetic gene cluster. Realizing that the amino acid sequence conservations among the known Cy and Ox domains are limited to a few very short core motifs, we adopted the CODEHOP strategy of PCR primer design, which allowed us to design hybrid primers consisting of a short 3′degenerate core region (for selectivity) and a longer 5′consensus clamp region (for specificity) (Fig. 2) (43). We successfully amplified a Cy domain from the S. atroolivaceus S-140 genome, and, with it as a probe, subsequently cloned a thiazole-forming NRPS module, thereby localizing the lnm biosynthetic gene cluster. The involvement of the cloned DNA in LNM biosynthesis has been unambiguously established by insertional inactivation of the lnmI gene (Fig. 5) and biochemical characterization of the LnmI protein (Fig. 6). Although we were unable to amplify the Ox domain directly from the S. atroolivaceus genome, we were successful in amplifying the Ox domain from cosmid DNA, validating the usefulness of these primers. The latter should not come as a surprise, retrospectively, because the Ox1 and Ox2 motifs of the LnmI-Ox domain are less conserved than those used to design the Ox-FP and Ox-RP primers (Fig. 4C). It should be pointed out that, while the PCR products appear to be very specific in this study, this approach could not discriminate among different heterocycle-forming NRPS genes if the organism produces multiple heterocycle-containing metabolites. Using this PCR cloning strategy, we recently amplified a thiazoline-forming NRPS module and a thiazole-forming NRPS module from Streptomyces flavoviridis ATCC 21892 that produces the antitumor antibiotic phleomycin (Fig. 1) and mapped them to the phleomycin biosynthetic gene cluster (Y. Fan and B. Shen, unpublished data). This method should now be taken into consideration in formulating a strategy for cloning biosynthesis gene clusters of other heterocycle-containing natural products.
On the basis of the hybrid NRPS-PKS paradigm, we could propose the following model for the early steps of LNM biosynthesis involving two NRPS modules. A yet-to-be-identified loading NRPS module, consisting of at least A and PCP domains, would initiate LNM biosynthesis by selecting, activating, and loading a d-alanine (Ala) to its PCP. (It cannot be predicted a priori if d-Ala is incorporated directly or derived from l-Ala via an epimerization step. The latter process would require an epimerization domain within the loading NRPS module [27, 51].) The NRPS module, residing at the N terminus of LnmI reported in this study, would represent the second NRPS module that would select, activate, and load a Cys to its PCP. Condensation between the aligned d-Ala-S-PCP and Cys-S-PCP and subsequent Cy and Ox would furnish the thiazole moiety for LNM biosynthesis. The characteristic Cy-Cy-A-PCP-Ox domain organization of the LnmI NRPS module agrees well with the enzyme functions that would be required for the biosynthesis of the thiazole moiety of LNM according to the proposed mechanism. The amino acid specificity of the LnmI NRPS module has been confirmed biochemically in vitro to be l-Cys, providing direct evidence for the proposed pathway. Thus, in a mechanistic analogy to VibF (31), we propose that the second Cy domain is responsible for the condensation step, yielding the dipeptidyl-S-PCP intermediate that is subsequently cyclized by the first Cy domain to afford the thiazolinyl-S-PCP intermediate. The latter is finally oxidized by the Ox domain to yield the thiazonyl-S-PCP species. At this point, the growing peptidyl-S-PCP intermediate is switched to the PKS machinery. Further elongations of thiazonyl-S-PCP by PKS modules complete the biosynthesis of the LNM hybrid peptide-polyketide carbon backbone. The identification of the incomplete PKS module residing immediately after the NRPS module on LnmI supports the hybrid NRPS-PKS hypothesis for LNM biosynthesis.
Since we have identified overlapping cosmids that cover a total of 172 kb of the DNA region (Fig. 3), we believe that we have localized the entire lnm gene cluster. In addition to the two NRPS modules, at least six PKS modules would be needed for the incorporation of the six C-2 units from malonyl coenzyme A to form the LNM macrolactam structure. Although we cannot predict a priori the biosynthetic origin of the 1,3-dioxo-1,2-dithiolane moiety due to its unprecedented structural features, genetic analysis of the complete lnm gene cluster could shed light on its biosynthesis. The localization of the lnm gene cluster, the confirmation of the LNM biosynthesis by a hybrid NRPS-PKS megasynthetase, and development of an efficient genetic system for in vivo manipulation of LNM biosynthesis in S. atroolivaceus have now set up an excellent stage for future investigations of LNM biosynthesis. Given LNM's potent antitumor activity, in particularly against tumors that are resistant to clinically important anticancer drugs, we envisage applying the methodologies of combinatorial biosynthesis to LNM for the production of novel analogs, some of which could lead to the discovery of novel anticancer drugs.
Acknowledgments
We thank Kyowa Hakko Kogyo Co. Ltd. for an authentic sample of leinamycin and the wild-type S. atroolivaceus S-140 strain; César Sánchez for assistance in the designing of PCR primers; and Liangcheng Du for advising on the ATP-PPi assays.
This work is supported in part by a grant from the University of California BioSTAR Program (Bio99-10045) and Kosan Biosciences, Inc., Hayward, Calif. B.S. is the recipient of a CAREER Award from NSF (MCB9733938) and an Independent Scientist Award from NIH (AI51689).
REFERENCES
- 1.Asai, A., H. Saito, and Y. Saitoh. 1997. Thiol-independent DNA cleavage by a leinamycin degradation product. Bioorg. Med. Chem. 4:723-729. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-69. [DOI] [PubMed] [Google Scholar]
- 4.Blanc, V., D. Lagneaux, P. Didier, P. Gil, P. Lacroix, and J. Crouzet. 1995. Cloning and analysis of structural genes from Streptomyces pristinaespiralis encoding enzymes involved in the conversion of pristinamycin IIB to pristinamycin IIA (PIIA): PIIA synthase and NADH:riboflavin 5′-phosphate oxidoreductase. J. Bacteriol. 177:5206-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breydo, L., H. Zang, K. Mitra, and K. S. Gates. 2001. Thiol-independent DNA alkylation by leinamycin. J. Am. Chem. Soc. 123:2060-2061. [DOI] [PubMed] [Google Scholar]
- 6.Cane, D. E., and C. T. Walsh. 1999. The parallel and convergent universes of polyketide synthases and non-ribosomal peptide synthases. Chem. Biol. 6:R319-325. [DOI] [PubMed] [Google Scholar]
- 7.Cane, D. E., C. T. Walsh, and K. Khosla. 1998. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282:63-68. [DOI] [PubMed] [Google Scholar]
- 8.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 9.Cox, C. D., K. L. Rinehart, Jr., M. L. Moore, and J. C. Cook, Jr. 1981. Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 78:4256-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, L., M. Chen, C. Sánchez, and B. Shen. 2000. An oxidation domain in the BlmIII non-ribosomal peptide synthetase probably catalyzing thiazole formation in the biosynthesis of the antitumor drug bleomycin in Streptomyces verticillus ATCC15003. FEMS Microbiol. Lett. 189:171-175. [DOI] [PubMed] [Google Scholar]
- 12.Du, L., C. Sánchez, M. Chen, D. J. Edwards, and B. Shen. 2000. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 7:623-642. [DOI] [PubMed] [Google Scholar]
- 13.Du, L., C. Sánchez, and B. Shen. 2001. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab. Eng. 3:78-95. [DOI] [PubMed] [Google Scholar]
- 14.Du, L., and B. Shen. 1999. Identification and characterization of a type II peptidyl carrier protein from the bleomycin producer Streptomyces verticillus ATCC 15003. Chem. Biol. 6:507-517. [DOI] [PubMed] [Google Scholar]
- 15.Fukuyama, T., and Y. Kanda. 1994. Total synthesis of (+)-leinamycin. J. Synth. Org. Chem. Jpn. 52:888-899. [Google Scholar]
- 16.Gates, K. S. 2000. Mechanisms of DNA damage by leinamycin. Chem. Res. Toxicol. 13:953-956. [DOI] [PubMed] [Google Scholar]
- 17.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 18.Hara, M., K. Asano, I. Kawamoto, T. Takiguchi, S. Katsumata, K. Takahashi, and H. Nakano. 1989. Leinamycin, a new antitumor antibiotic from Streptomyces: producing organism, fermentation and isolation. J. Antibiot. 42:1768-1774. [DOI] [PubMed] [Google Scholar]
- 19.Hara, M., Y. Saitoh, and H. Nakano. 1990. DNA strand scission by the novel antitumor antibiotic leinamycin. Biochemistry 29:5676-5681. [DOI] [PubMed] [Google Scholar]
- 20.Hara, M., I. Takahashi, M. Yoshida, K. Asano, I. Kawamoto, M. Morimoto, and H. Nakano. 1989. DC 107, a novel antitumor antibiotic produced by a Streptomyces sp. J. Antibiot. 42:333-335. [DOI] [PubMed] [Google Scholar]
- 21.Hirayama, N., and E. S. Matsuzawa. 1993. Molecular structure of a novel antitumor antibiotic leinamycin. Chem. Lett. 11:1957-1958. [Google Scholar]
- 22.Kanda, Y., and T. Fukuyama. 1993. Total synthesis of (+)-leinamycin. J. Am. Chem. Soc. 115:8451-8452. [Google Scholar]
- 23.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry 39:15522-15530. [DOI] [PubMed] [Google Scholar]
- 24.Khmel, I. A. 1999. Microcins—peptide antibiotics of enterobacteria: genetic control of the synthesis, structure, and mechanism of action. Genetika 35:5-16. [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 26.Konz, D., A. Klens, K. Schörgendorfer, and M. A. Marahiel. 1997. The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: molecular characterization of three multi-modular peptide synthetases. Chem. Biol. 4:927-937. [DOI] [PubMed] [Google Scholar]
- 27.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R39-R48. [DOI] [PubMed] [Google Scholar]
- 28.Ku, J., R. G. Mirmira, L. Liu, and D. V. Santi. 1997. Expression of a functional non-ribosomal peptide synthetase module in Escherichia coli by coexpression with a phosphopantetheinyl transferase. Chem. Biol. 4:203-207. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y. M., J. C. Milne, L. L. Madison, R. Kolter, and C. T. Walsh. 1996. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science 274:1188-1193. [DOI] [PubMed] [Google Scholar]
- 30.Liu, W., and B. Shen. 2000. Genes for production of the enediyne antitumor antibiotic C-1027 in Streptomyces globisporus are clustered with the cagA gene that encodes the C-1027 apoprotein. Antimicrob. Agents Chemother. 44:382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall, C. G., N. J. Hillson, and C. T. Walsh. 2002. Catalytic mapping of the vibriobactin biosynthetic enzyme VibF. Biochemistry 41:244-250. [DOI] [PubMed] [Google Scholar]
- 32.Metsa-Ketela, M., V. Salo, L. Halo, A. Hautala, J. Hakala, P. Mantsala, and K. Ylihonko. 1999. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 180:1-6. [DOI] [PubMed] [Google Scholar]
- 33.Metz, J. G., P. Roessler, D. Facciotti, C. Levering, F. Dittrich, M. Lassner, R. Valentine, K. Lardizabal, F. Domergue, A. Yamada, K. Yazawa, V. Knauf, and J. Browse. 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290-293. [DOI] [PubMed] [Google Scholar]
- 34.Mitra, K., W. Kim, J. S. Daniels, and K. S. Gates. 1997. Oxidative DNA cleavage by the antitumor antibiotic leinamycin and simple 1,2-dithiolan-3-one 1-oxides: evidence for thiol-dependent conversion of molecular oxygen to DNA-cleaving radicals mediated by polysulfides. J. Am. Chem. Soc. 119:11691-11692. [Google Scholar]
- 35.Molnar, I., et al. 2000. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 7:97-109. [DOI] [PubMed] [Google Scholar]
- 36.Nakano, H., and T. Tamaoki. 1992. Mechanism-based screens for natural products lead as sources for antitumor drugs, p. 72-75. In M. R. Ladisch and A. Bose (ed.), Harnessing biotechnology for the 21st century. Proceedings of the Ninth International Biotechnology Symposium and Exposition. American Chemical Society, Washington, D.C.
- 37.Nicholson, T. P., B. A. Rudd, M. Dawson, C. M. Lazarus, T. J. Simpson, and R. J. Cox. 2001. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 8:157-178. [DOI] [PubMed] [Google Scholar]
- 38.Omura, S., et al. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, H. M., and C. T. Walsh. 2001. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry 40:9023-9031. [DOI] [PubMed] [Google Scholar]
- 40.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quadri, L. E. N., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 42.Rao, R. N., M. A. Richardson, and S. Kuhstoss. 1987. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 153:166-198. [DOI] [PubMed] [Google Scholar]
- 43.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy, R. S., A. M. Gehring, P. J. Belsham, and C. T. Walsh. 1999. Thiazole and oxazole peptides: biosynthesis and molecular machinery. Nat. Prod. Rep. 16:249-263. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Sánchez, C., L. Du, D. J. Edwards, M. D. Toney, and B. Shen. 2001. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem. Biol. 8:725-738. [DOI] [PubMed] [Google Scholar]
- 47.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 48.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]
- 49.Tang, L., S. Shah, L. Chung, J. Carney, L. Katz, C. Khosla, and B. Julien. 2000. Cloning and heterologous expression of the epothilone gene cluster. Science 287:640-642. [DOI] [PubMed] [Google Scholar]
- 50.Turgay, K., and M. A. Marahiel. 1994. A general approach for identifying and cloning peptide synthetase genes. Peptide Res. 7:238-241. [PubMed] [Google Scholar]
- 51.Walsh, C. T., H. Chen, T. A. Keating, B. K. Hubbard, H. C. Losey, L. Luo, C. G. Marshall, D. A. Miller, and H. M. Patel. 2001. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 5:525-534. [DOI] [PubMed] [Google Scholar]
- 52.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]