Abstract
The cyanobacterium Anabaena sp. strain PCC 7120 forms single heterocysts about every 10 to 15 vegetative cells along filaments. PatS is thought to be a peptide intercellular signal made by developing heterocysts that prevents neighboring cells from differentiating. Overexpression of the patS gene suppresses heterocyst formation. The hetL gene (all3740) was isolated in a genetic screen to identify genes involved in PatS signaling. Extracopy hetL allowed heterocyst formation in a patS overexpression strain. hetL overexpression from a heterologous promoter in wild-type Anabaena PCC 7120 induced multiple-contiguous heterocysts (Mch) in nitrate-containing medium. The predicted HetL protein is composed almost entirely of pentapeptide repeats with a consensus of A(D/N)L*X, where * is a polar amino acid. Thirty Anabaena PCC 7120 genes contain this repeat motif. A synthetic pentapeptide corresponding to the last 5 amino acids of PatS, which suppresses heterocyst formation in the wild type, did not suppress heterocyst formation in a hetL overexpression strain, indicating that HetL overexpression is affecting heterocyst regulation downstream of PatS production. The transcription regulator NtcA is required for the initiation of heterocyst formation. hetL overexpression allowed the initiation of heterocyst development in an ntcA-null mutant, but differentiation was incomplete. hetR and hetC mutations that block heterocyst development are epistatic to hetL overexpression. A hetL-null mutant showed normal heterocyst development and diazotrophic growth, which could indicate that it is not normally involved in regulating development, that it normally plays a nonessential accessory role, or perhaps that its loss is compensated by cross talk or redundancy with other pentapeptide repeat proteins.
The filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 forms heterocysts approximately 1 out of every 10 to 15 vegetative cells under diazotrophic growth conditions. Heterocysts are terminally differentiated cells specialized for nitrogen fixation (43). Heterocyst differentiation and pattern formation offers an important model of prokaryotic multicellular development. Several genes involved in heterocyst development have been identified (28, 43). A recent extensive review of heterocyst development presents updated models for the regulation in free-living and symbiotic growth states (28).
The patS gene encodes a small peptide that appears to control pattern formation through cell-to-cell signaling within a filament (46). The PatS peptide is thought to be a diffusible inhibitor produced by differentiating cells to suppress heterocyst formation in neighboring cells. A strain containing multiple copies of patS or in which patS is being overexpressed is unable to form heterocysts (46). In contrast, a patS-null mutant forms heterocysts in nitrate-containing medium and it forms an abnormally high percentage of heterocysts and multiple-contiguous heterocysts (Mch phenotype) in diazotrophic growth conditions. A synthetic pentapeptide corresponding to the last five patS-encoded amino acids (PatS-5) inhibits heterocyst development at submicromolar concentrations, which is consistent with its proposed role as an intercellular signal.
PatS is thought to control heterocyst pattern by lateral inhibition. In diazotrophic growth conditions, 60 nM PatS-5 in the medium restores a normal number of heterocysts in a patS-null mutant but the intervals between heterocysts do not show a normal pattern (46). However, expression of the patS open reading frame (ORF) from the heterocyst-specific hepA promoter complemented the patS-null mutant and restored the pattern to near normal. These data suggest that patS is involved in cell-to-cell signaling and that a gradient of PatS originating from differentiating cells is required to produce a normal pattern.
To examine the spatial expression pattern of PatS, a transcriptional gfp fusion was made to the patS promoter (46, 47). This strain showed that patS transcription increased after nitrogen step-down and was localized to small groups of cells by 8 to 10 h after induction. By 12 to 14 h, bright fluorescence was present in mostly single cells. Eighteen hours after nitrogen step-down, green fluorescent protein (GFP) fluorescence was found almost exclusively in proheterocysts. The temporal and spatial pattern of patS expression strongly supports the lateral inhibition model in which the patS product, possibly a processed C-terminal peptide, acts as an intercellular signal made by developing heterocysts to suppress heterocyst development in neighboring cells.
PatS is essential for establishing the initial heterocyst pattern and for the resolution of clusters of differentiating cells to a single cell that becomes committed to form a heterocyst. A second gene, hetN, has been proposed to be required for the maintenance of heterocyst spacing after the initial pattern is established (10, 11).
The general goal of this study was to identify genes involved in the production of the PatS signal and in the downstream signaling pathway by isolating bypass suppressor mutations. One approach was to screen a conjugal expression library for genes whose overexpression could bypass the heterocyst suppression caused by patS overexpression. One gene was identified in this screen, which we named hetL. To help identify which part of the heterocyst developmental pathway was affected by hetL overexpression, we determined the epistatic relationships between hetL and hetR, hetC, and ntcA, which are all required for important steps in heterocyst formation.
HetR plays a central role as a positive regulator of heterocyst formation. A hetR-null mutant shows no signs of heterocyst development, and multiple copies of hetR cause heterocyst development in nitrate- or ammonium-containing medium and an Mch phenotype in diazotrophic growth conditions (6, 7). hetR is positively autoregulated, and a high level of expression is localized to differentiating cells (3). The HetR protein has an auto-protease activity, but the mechanism by which HetR regulates development is still unknown (48, 49).
hetC is required for the formation of mature heterocysts (25). A hetC-null mutant produces a semiregular pattern of small cells that may represent an early stage of heterocyst differentiation (45). These small cells are weakly autofluorescent, a characteristic of proheterocysts and heterocysts that results from the degradation of phycobiliproteins (44, 45). More convincingly, the small cells express a hetR-gfp reporter (45). hetC is expressed most strongly in differentiating cells and encodes a predicted ABC protein exporter, but its role in heterocyst development has not been determined.
NtcA is a member of the Crp family of prokaryotic transcriptional regulators and regulates the expression of more than a dozen genes, many of which are involved in nitrogen metabolism (23). For example, the nirA-nrtABCD-narB operon is required for the uptake and assimilation of nitrate and its transcription increases immediately after nitrogen starvation through activation by NtcA (9, 18). NtcA's DNA binding and transcriptional activation activities are enhanced by 2-oxoglutarate, which reflects the carbon and nitrogen balance in cyanobacterial cells (31, 37, 40). NtcA is essential for the initiation of heterocyst development (17, 42) and is involved in the regulation of several heterocyst-specific genes. hetR expression fails to be induced in an ntcA mutant, although there is no evidence for NtcA directly interacting with the hetR promoter (23). NtcA directly activates the transcription of hetC and the devBCA operon (16, 30). NtcA also binds near genes required in the late stages of heterocyst development. NtcA binds to sequences near a site-specific recombination site in the region upstream of xisA, and it binds weakly to the upstream region of the nifHDK operon, which encodes nitrogenase (12, 33). Clearly, NtcA regulates multiple genes required for heterocyst development (23).
We report here on the identification of the Anabaena PCC 7120 hetL gene. hetL overexpression strongly stimulates the formation of heterocysts, even on media containing ammonium or PatS-5 pentapeptide, which normally suppresses heterocyst development.
MATERIALS AND METHODS
Strains and culture conditions.
A description of the bacterial strains and plasmids used in this study is presented in Table 1. Escherichia coli strains and culture conditions were similar to those previously described (20). Plasmids were maintained in E. coli strain DH10B. For selective growth, 50 μg of spectinomycin (Sp)/ml or 50 μg of kanamycin (Km)/ml was used. Anabaena PCC 7120 was grown in BG-11 medium, BG-110 medium, or BG-110 medium supplemented with 2 mM NH4Cl and 5 mM morpholinepropanesulfonic acid (MOPS) buffer, pH 8.0 (42). Plasmids were transferred into Anabaena cells by conjugation according to standard protocols (14, 21) with the following modifications. The mixed cells were placed directly onto the surface of 40 ml of agar-solidified (1.5%) BG-11 medium and grown overnight, and then the plates were underlaid with 400 μl of 100× antibiotic stock solution. Plates were returned to standard growth conditions, and colonies would typically appear in approximately 1 week. For selective growth in liquid, l μg each of Sp and streptomycin (Sm)/ml, 5 μg of erythromycin (Em)/ml, or 12.5 μg of neomycin (Nm)/ml was used. For selective growth on agar-solidified media, 2 μg each of Sp and Sm/ml, 5% sucrose, 5 μg of Em/ml, or 25 μg of Nm/ml was used. Light intensity was approximately 60 to 100 μE m−2 s−1 for Anabaena cultures.
TABLE 1.
Bacterial strains and plasmids
| Strain or, plasmid | Relevant characteristicsa | Source and/or reference |
|---|---|---|
| Strains | ||
| Anabaena sp. | ||
| 216 | hetR point mutant S179N, Het− | 6 |
| AMC236 | ntcA::ΩSpr/Smr cassette, Het− | 42 |
| AMC450 | PCC 7120 harboring pAM2474 (originally pAM1691), Het− | 46 |
| AMC451 | patS::ΩSpr/Smr cassette, Mch | 46 |
| AMC1043 | DR1653 harboring pAM2065 | This study |
| AMC1071 | PCC 7120 with pAM2836 containing the hetL-gfp translational fusion integrated into the chromosome by homologous single recombination | This study |
| AMC1131 | PCC 7120 harboring pAM1888, Het− | This study |
| DR1653 | hetC::luxAB-ΩSpr/Smr cassette, Het− | 25 |
| LD115 | PCC 7120 harboring pAM2065, which overexpresses hetL; accumulation of secondary mutations required this strain to be routinely remade | This study |
| PCC 7120 | Wild type | R. Haselkorn |
| E. coli | ||
| AM1358 | DH10B harboring pRL623, for triparental conjugations | This study |
| AM1359 | DH10B harboring pRL623 and pRL443, for biparental conjugations | 46 |
| AM1460 | HB101 harboring pRK2013, for triparental conjugations | J. Meeks (13) |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL nupG Smr | Life Technologies, GIBCO BRL |
| HB101 | recA13 mcrBC | 5 |
| Plasmids | ||
| pAM504 | Shuttle vector, Kmr/Nmr | 42 |
| pAM1011 | Shuttle vector, Kmr/Nmr, higher-copy-number | 34 |
| pAM1824 | pAM1011 containing rbcL promoter, used to construct expression library | This study |
| pAM1888 | pRL1272 containing 0.95-kb fragment with patS | This study |
| pAM1949 | pAM1824 library clone containing 2.2-kb insert with hetL and obg (missing 23 bases at 5′ end) | This study |
| pAM2045 | pAM1824 library clone containing 6-kb insert with hetL and obg | This study |
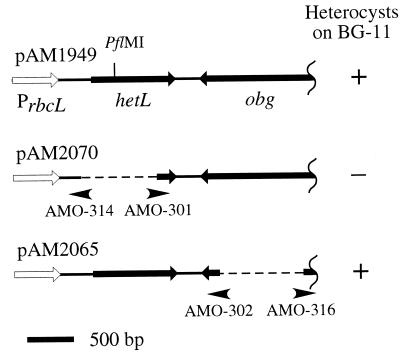
| pAM2065 | pAM1949 with obg deleted, hetL is expressed from rbcL promoter | This study |
| pAM2070 | pAM1949 with hetL deleted, obg is missing 23 bases at 5′ end | This study |
| pAM2110 | pAM2045 with hetL::ΩSpr/Smr cassette | This study |
| pAM2158 | pAM2110 insert cloned in pRL271; a suicide plasmid for hetL inactivation | This study |
| pAM2269 | hetL expressed from petE promoter in pAM504; Kmr/Nmr | This study |
| pAM2474 | patS expressed from glnA promoter, originally pAM1691; Kmr/Nmr | This study, reference 46 |
| pAM2836 | hetL-gfp translational fusion in suicide vector pRL277 | This study |
| pRK2013 | RK2 derivative; Kmr; colE1 oriV replaces RK2 oriV | 13 |
| pRL271 | Suicide vector containing sacB for the selection of double recombination events; Cmr Emr | 3 |
| pRL277 | Suicide vector containing sacB for the selection of double recombination events; Spr Smr | 3 |
| pRL443 | Conjugal plasmid; Apr Tcr; Kms derivative of RP-4 | 14 |
| pRL623 | Conjugation helper plasmid; Cmr; MobColK, M · AvaII, M · Eco47II, M · EcoT221 | 14 |
| pRL1272 | Shuttle vector derived from RSF1010; Cmr Emr | 50 |
Tc, tetracycline; Cm, chloramphenical; Ap, ampicillin
Expression library construction and screening.
Anabaena PCC 7120 chromosomal DNA was partially digested by TaqI and size fractionated by electrophoresis on a preparative 0.7% agarose gel. DNA fragments from 2 to 10 kb were purified and ligated into a dephosphorylated ClaI site in pAM1824. pAM1824 is a shuttle vector containing the Anabaena PCC 7120 rbcL promoter on an EcoRI-SalI fragment from pAM496 (33) cloned into the same sites of pAM1011 (34). pAM1011 contains an uncharacterized mutation resulting in acopy number in Anabaena PCC 7120 higher than that of the original pDU1-based shuttle vectors (J. Golden, unpublished data). Under standard growth conditions, a pDU1-based shuttle vector was found to be present at 17 copies per chromosome (M. Lee and J. Golden, unpublished data). The rbcL promoter is directed toward the ClaI site in pAM1824. E. coli strain AM1358, which is used for triparental conjugal mating with Anabaena, was transformed with ligated library DNA by electroporation, and approximately 80,000 primary clones were obtained. The colonies were washed off of plates, and this amplified library was frozen in several aliquots. Analysis of plasmids isolated from nine randomly selected colonies from the amplified pool showed that eight colonies contained inserts averaging about 2.5 kb in size. The pooled library clones in strain AM1358 were transferred into Anabaena strain AMC1131 by triparental conjugation with E. coli strain AM1460 by following standard protocols (13-15, 46). AMC1131 does not form heterocysts because it contains extra copies of the patS gene on plasmid pAM1888. Approximately 30,000 exconjugant colonies were screened for diazotrophic growth on BG-110 plates.
Plasmid constructions.
The obg and hetL genes were each deleted from pAM1949 by PCR to make pAM2065 and pAM2070, respectively. Two divergent primers were designed to amplify the whole plasmid except for the region to be deleted. The primers were phosphorylated by T4 polynucleotide kinase (Promega). The PCR was set up using 5 U of Pfu Turbo DNA polymerase (Stratagene), 50 ng of template DNA, 0.25 mM concentrations of each deoxynucleoside triphosphate, 10 pM concentrations of each primer, 2 mM MgCl2, and 2% dimethyl sulfoxide in 40-μl volumes. Each PCR cycle consisted of denaturation at 95°C for 30 s, annealing at 48°C for 20 s, and extension at 68°C for 6 min. This was repeated for 25 cycles. The PCR products were religated, and then the template plasmid DNA was digested with 0.3 μl (3 U) of DpnI. Primers AMO-316 and AMO-302 were used to delete obg, and AMO-314 and AMO-301 were used to delete hetL. The sequences of AMO-301, AMO-302, AMO-314, and AMO-316 were GTC GGA CAA TTC AGT GG, TTG CAA GCT TAC GGA CG, GGG TAT GAT ACC AAC TC, and AGA GGG CCC ACC GCA ACG AAA ATG, respectively.
hetL was placed under the control of the copper-inducible petE promoter in plasmid pAM2269. A PCR product containing the hetL ORF was made with primers AMO-364 (GAA TTA TTT ATG AAT GTG) and AMO-365 (GGG AGC TCT TGA CCA AGT AAA TGC). AMO-365 contains a SacI site in its 5′ end. The hetL ORF was inserted between the SmaI and SacI sites of pPet1 (7), and a SacI-ScaI fragment from the resulting plasmid was inserted into the SmaI-SacI sites in pAM504, a standard copy number shuttle vector, to produce pAM2269.
Alcian blue staining.
Alcian blue was used to stain the heterocyst-specific polysaccharide layer of the proheterocyst and proheterocyst envelope (22). A solution of 0.5% alcian blue (Sigma) in 50% ethanol-water was mixed with an equal volume of Anabaena culture before microscopic examination.
Inactivation of hetL in Anabaena PCC 7120.
The hetL gene in the Anabaena PCC 7120 chromosome was inactivated by sacB-mediated positive selection for double recombination (3, 8). The suicide plasmid pAM2158 was constructed in two steps. (i) A blunted HindIII fragment containing the ΩSpr/Smr cassette from pDW9 (21) was inserted into a blunted PflMI site in pAM2045 resulting in pAM2110. The HindIII ends were blunted by Klenow fragment, and the PflMI ends were blunted by T4 DNA polymerase. (ii) An XhoI-NcoI fragment from pAM2110 containing the interrupted hetL gene was inserted into the same sites of pRL271 (3), resulting in pAM2158.
The suicide plasmid pAM2158 was transferred into the wild-type strain by conjugation essentially according to standard methods (13-15, 46). E. coli conjugal donor strain AM1359 contains conjugal plasmid pRL443 and helper plasmid pRL623 (14). pAM2158 was transferred into AM1359 by electroporation. Cells from a 1.5-ml overnight culture of the resulting E. coli strain were washed twice by centrifugation to remove antibiotics and then mixed with the Anabaena recipient strain. The biparental mating mixture was transferred onto 40 ml of BG-11 agar-solidified medium. Plates were incubated overnight under low-light growth conditions and then underlaid with 600 μl of Em solution to produce a final concentration of 5 μg/ml. Plates were incubated under standard growth conditions for at least 7 to 10 days. Emr colonies representing single recombinants were grown in a liquid medium containing spectinomycin for 7 to 10 days. Double recombinants were selected for Spr, Smr, and sucrose resistance, and clones were then tested for loss of Em resistance (3, 8).
Construction of a strain carrying a hetL-gfp translational fusion.
The gfp ORF was amplified from pKEN2-GFPmut2 (46) by PCR with primers AMO-522 (CAG GTA CCA GTA AAG GAG AAG AAC TAT TCA CT) and AMO-523 (CAC TGA GAG CTC TTA TTT GTA TAG TTC ATC CAT GCC). The hetL ORF was amplified with primers AMO-521 (CAC TGG GGT ACC ATG AAT TGA ACC ATC AGG) and AMO-584 (CTG GAT CCT AAA ATG TGG GTG AAA TTC). AMO-522, AMO-523, AMO-521, and AMO-584 contain KpnI, SalI, KpnI, and BamHI sites near their 5′ ends, respectively. The gfp ORF and hetL ORF were inserted into the KpnI-SacI and BamHI-KpnI sites of pAM504 (42), respectively, such that the stop codon of hetL was replaced by the start codon of gfp. The BamHI-SacI fragment was cloned into the BglII-SacI sites of the suicide vector pRL277 (3), resulting in pAM2836, which was transferred into Anabaena PCC 7120 by conjugation with selection for Spr Smr single recombinants. The gfp ORF and its fusion to hetL in pAM2836 was confirmed by DNA sequencing. GFP fluorescence microscopy was performed as described previously (47).
Southern and Northern blot analysis.
DNA extraction and Southern blot analysis were performed as previously described (19, 20) with slight modifications. DNA samples were transferred to Magna-Charge nylon membranes (MSI) with a pressure blotter (Stratagene). RNA extraction and Northern blot analysis were performed as previously described (42). A 350-bp HpaI fragment from the hetL gene was used as a template for making a radioactive DNA probe with a random primer method.
RESULTS
hetL identification.
In an effort to identify genes involved in the PatS signaling pathway, we screened an expression library for plasmids that would enable the patS overexpression strain AMC1131 to form heterocysts and grow diazotrophically. The library plasmids in these strains were expected to contain genes or gene fragments that would bypass the PatS inhibition of heterocyst development. Forty clones were identified by their ability to grow and form green colonies on BG-110 plates. The library plasmids were isolated from each of the Het+ (Het, heterocyst formation) clones, subjected to restriction enzyme analysis, and retested by transfer back into strain AMC1131 by conjugation. Fourteen library plasmids with different inserts or insert orientation were confirmed to change the Het− strain AMC1131 to Het+. Restriction site mapping and DNA sequence analysis showed that a 2.2-kb chromosomal DNA region was common to all 14 plasmids (data not shown). The 2.2-kb DNA region was found in both orientations and with different lengths of flanking sequence, clearly indicating that extra copies of this region act as a bypass suppressor of the heterocyst suppression caused by patS overexpression. These results also show that the activity of the rbcL promoter on the library vector was not required for the phenotype. The isolation of 14 different clones all containing the same DNA region suggests both that the library contained good representation of clonable Anabaena PCC 7120 DNA and that other DNA regions capable of acting as patS bypass suppressors are unlikely to be identified with this particular screening procedure.
The 2.2-kb DNA fragment contained two obvious ORFs. One was similar to obg (38), and the other was similar to the C-terminal domain of the Anabaena PCC 7120 hglK gene (2). obg is an essential gene in Bacillus subtilis, Caulobacter crescentus, and Streptomyces coelicolor (27, 32, 38). obg is involved in sporulation stage zero in B. subtilis and in the regulation of cell differentiation in S. coelicolor. The hglK gene is required for heterocyst-specific glycolipid localization to the envelope. The ORF (all3740) similar to the hglK C-terminal domain was designated hetL because it was later found to strongly stimulate heterocyst formation when overexpressed.
pAM1949 contained the common 2.2-kb insert (Fig. 1). To determine which of the two ORFs, hetL or obg, caused bypass suppression of patS, each was deleted from pAM1949 with a PCR method. In plasmid pAM2070, bp −45 to 508 of the 714-bp hetL ORF were deleted (Fig. 1). Anabaena PCC 7120 carrying pAM2070 had a normal heterocyst formation and pattern. When pAM2070 was transferred into strain AMC1131, the resulting strain failed to bypass the patS inhibition of heterocyst development. Therefore, hetL is required to bypass patS overexpression.
FIG. 1.
Deletion analysis of the chromosomal insert in pAM1949. The open arrow indicates the rbcL promoter on the pAM1824 vector, and the black arrows indicate the obg and hetL ORFs. The obg ORF is missing 23 bp from the presumed 5′ end but contains an in-frame start codon at position 37. Deleted regions are shown as broken lines. Arrowheads indicate the primers used to make the deletions. Anabaena PCC 7120 carrying pAM1949 or pAM2065 formed multiple-contiguous heterocysts (an Mch phenotype) on BG-11 (N+) and BG-110 (N−) media. In contrast, a strain carrying pAM2070 showed a wild-type phenotype.
The obg ORF was intact in other isolated library clones, but in pAM1949, it was missing 23 bp at the 5′ end; however, an in-frame ATG start codon was present at position 37, which could possibly allow the expression of a slightly truncated polypeptide. To test the possible involvement of obg in the bypass phenotype, bp 150 to 786 of the 1,029-bp obg gene were deleted from pAM1949, resulting in plasmid pAM2065. pAM2065 was transferred into the wild type and strain AMC1131. The resulting strains showed multiple-contiguous heterocysts (an Mch phenotype) both in nitrate-containing medium and in diazotrophic growth conditions (Fig. 2). Both pAM2065 and its parental plasmid pAM1949 caused the wild-type strain and AMC1131 to show an Mch phenotype. Therefore, the obg ORF is not required to bypass the patS overexpression phenotype.
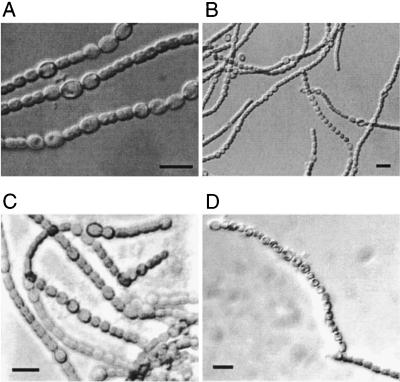
FIG. 2.
Micrographs of Anabaena PCC 7120 containing plasmid pAM2065 grown in BG-11 medium (A and B) and BG-110 medium (C and D), which lacks nitrate. Heterocysts are larger and have a thickened envelope compared to that for vegetative cells. hetL overexpression causes heterocysts to form in the presence of a reduced nitrogen source and produces an Mch phenotype. Scale bars, 10 μm.
hetL overexpression strongly stimulated heterocyst formation. When grown in BG-11 medium, which contains 17 mM nitrate, filaments wild-type Anabaena PCC 7120 typically form fewer than 1% heterocysts. Anabaena PCC 7120 carrying hetL on pAM2065 (strain LD115) formed 18% heterocysts on BG-11, which is approximately twice that of the wild-type strain growing diazotrophically on BG-110. When induced by nitrogen step-down, strain LD115 formed 20% heterocysts at 24 h after transfer to BG-110, which increased to 34% at 48 h after induction. Both strains LD115 and AMC1131 carrying pAM2065 possessed the Mch phenotype (Fig. 2). As many as 21 contiguous heterocysts were observed in filaments of strain LD115. There was no obvious difference in the frequency of heterocysts in these two strains, indicating that hetL overexpression completely bypasses heterocyst inhibition by PatS.
To confirm that transcription of the hetL gene was required to stimulate heterocyst formation, we placed the hetL ORF under the control of the copper-inducible petE promoter (7, 11, 46) in plasmid pAM2269. When pAM2269 was transferred into the patS overexpression strain AMC1131, the resulting strain formed heterocysts only after induction with 400 μM Cu2+ on BG-110. Wild-type Anabaena PCC 7120 containing pAM2269 showed no heterocyst development in copper-free BG-11 liquid medium, but the induction of PpetE-hetL expression with 400 μM Cu2+ caused the strain to form single and double heterocysts even in this nitrate-containing medium.
hetL overexpression in strain LD115 stimulated heterocyst formation and produced an Mch phenotype even when grown in ammonium-containing media, which generally suppresses heterocysts better than nitrate (data not shown). Several mutant strains will form heterocysts on nitrate-containing media, including patS (46), hetN (4), and mutants defective for nitrate transport or metabolism, such as moeA (35). However, heterocyst development is suppressed by ammonium in all these strains. In contrast, overexpression of hetR produces an Mch phenotype and stimulates heterocyst formation in the presence of nitrate or ammonium (7), which is similar to our results with hetL.
Predicted HetL protein.
The hetL ORF (all3740) is predicted to encode a 237-amino-acid polypeptide that is almost entirely composed of pentapeptide repeats (Fig. 3). HetL is homologous to a family of proteins containing pentapeptide repeats with the consensus A(D/N)L*X, where * represents a polar amino acid and X represents any amino acid (1, 24). Allowing for conservative amino acid substitutions, HetL contains approximately 40 pentapeptide repeats.
FIG. 3.
The predicted HetL polypeptide. HetL is a member of a family of pentapeptide repeat proteins with a consensus repeat sequence of A(D/N)L*X, where X represents any amino acid and * represents a polar amino acid.
Many pentapeptide repeat proteins are present in Anabaena PCC 7120 and other organisms; however, the function of the motif is not known. In the Anabaena PCC 7120 genome, 30 genes encode proteins with domains that appear to be homologous to HetL. In 13 of these genes, the entire ORF encodes pentapeptide repeats. The closest paralog is ORF all3256, which encodes 268 amino acids and shows 38% identity and 50% similarity to HetL. In the remaining 17 genes, the ORF encodes pentapeptide repeats and additional domains. Eleven of these contain predicted membrane spans (http://www.cbs.dtu.dk/services/TMHMM/) (26).
Proteins with pentapeptide repeat motifs homologous to HetL are also found in many other organisms. In the preliminary genome sequence of the heterocystous cyanobacterium Nostoc punctiforme, 40 proteins contain pentapeptide repeats similar to HetL. However, none of these can be clearly identified as a HetL ortholog and none are found next to the obg gene on the chromosome. In the sequenced genome of Synechocystis sp. strain PCC 6803, 16 proteins contain a pentapeptide repeat domain. Proteins with a HetL-like domain are found in at least 20 other bacteria, including E. coli, B. subtilis, Pseudomonas aeruginosa, C. crescentus, Salmonella enterica subsp. enterica, Mycobacterium tuberculosis, Sinorhizobium meliloti, S. coelicolor, and Legionella pneumophila. However, in no case has a function for the protein been established. Proteins showing some similarity to HetL are also found in plants and animals, including human, mouse, fruit fly, rice, and Arabidopsis thaliana genomes. In contrast to the cyanobacterial species, other bacteria, plants, and animals typically contain only a few proteins with apparent homology to the pentapeptide repeat domain.
hetL overexpression relieves inhibition by PatS-5 pentapeptide.
To determine whether hetL overexpression interrupted the production or the response to the PatS signal, we determined if the hetL overexpression strain LD115 would respond to the PatS-5 pentapeptide signal. PatS-5 (RGSGR) is a synthetic peptide that corresponds to the last 5 amino acids encoded by the patS ORF. A previous study showed that 1 μM PatS-5 suppresses heterocyst formation in wild-type Anabaena PCC 7120 (46). If LD115 is insensitive to the peptide signal, then the downstream portion of the signaling pathway must be affected by hetL overexpression. If the only effect of hetL overexpression is to block production of the PatS signal, then the addition of PatS-5 to filaments should suppress heterocyst formation in the hetL overexpression strain.
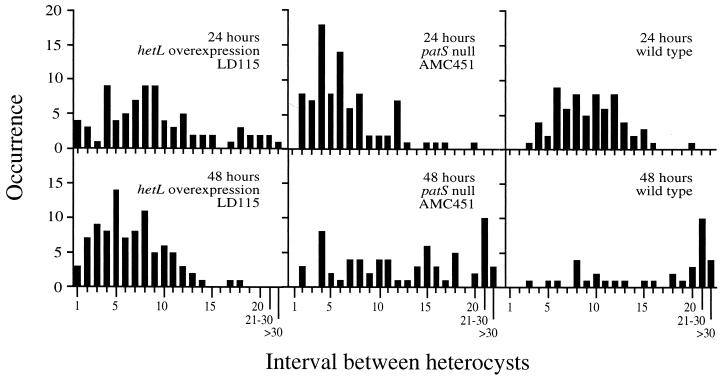
To determine if heterocyst formation was inhibited by PatS-5, the intervals between heterocysts were counted before and after the addition of the pentapeptide signal. This protocol, which tests for a reduction in heterocysts, was required because strains that overexpress hetL produce heterocysts under all tested growth conditions, even in the presence of a nitrogen source. Strain LD115, patS mutant AMC451, and the wild type were grown in BG-11 to exponential phase, and the cultures were then induced by transferring to BG-110 medium at time zero. At 24 h, PatS-5 was added to the cultures to a final concentration of 1 μM. The number of vegetative cells in the interval between heterocysts was counted at 24 and 48 h. If PatS-5 can suppress heterocyst formation in a strain, the number of vegetative cells between heterocysts should increase after one or two rounds of cell division. The results showed that the number of vegetative cells between heterocysts increased in the wild type and AMC451 but decreased in LD115 after PatS-5 addition (Fig. 4). Therefore, addition of PatS-5 signal failed to suppress heterocyst formation in the hetL overexpression strain. We conclude that HetL overexpression affects the downstream portion of the PatS signaling pathway, after the PatS signal is produced.
FIG. 4.
hetL overexpression results in insensitivity to PatS signaling. The heterocyst pattern was determined before and after the addition of PatS-5 pentapeptide. The hetL overexpression strain LD115, the patS-null mutant AMC451, and the wild type were grown in nitrate-containing medium to exponential phase. Cultures were induced by transferring filaments to BG-110 medium at time zero. At 24 h, PatS-5 was added to a final concentration of 1 μM. The numbers of vegetative cells in the intervals between heterocysts were counted at 24 and 48 h, and the number of occurrences of each interval length is shown. The data shown are representative of the results obtained from two independent experiments. The heterocyst pattern of the wild type carrying vector pAM1824 as a control was similar to that of the wild type alone (data not shown).
hetL inactivation.
The chromosomal hetL gene was inactivated by double recombination. The library plasmid pAM2045 contains hetL on a 6-kb fragment of Anabaena PCC 7120 genomic DNA. An ΩSpr/Smr cassette was inserted into a PflMI site within the hetL gene in pAM2045 (Fig. 1). The interrupted hetL gene was inserted into a suicide vector, and the resulting plasmid, pAM2158, was transferred into wild-type Anabaena PCC 7120 by conjugation. The exconjugants were selected for double recombination, and several clones that had the appropriate antibiotic resistance phenotype were obtained. PCR and Southern blot analysis confirmed the segregation of the inactivated hetL allele in these strains (data not shown).
Four independent hetL-null mutants were grown in BG-11 and BG-110 media. The hetL-null mutants all showed normal heterocyst formation and diazotrophic growth. They also showed normal growth and morphology in nitrate-containing medium. Therefore, the hetL gene is not required for heterocyst development. If HetL is normally involved in the regulation of heterocyst development, it must provide a nonessential function. This accessory role may not be required under our standardized laboratory growth conditions or could be too subtle to detect. Alternatively, hetL overexpression may produce a dominant-negative effect even though HetL may not normally be involved in heterocyst development.
hetL expression.
In wild-type Anabaena PCC 7120, hetL mRNA levels were very low in RNA samples isolated from vegetative cells and induced filaments. hetL mRNA was only weakly detectable by Northern blot analysis after a week-long exposure on a Fuji imaging plate, and no reliable pattern of temporal expression could be obtained (data not shown). A hetL-gfp translational fusion was constructed and recombined into the chromosome of Anabaena PCC 7120. Fluorescence from the GFP reporter was present in both vegetative cells and heterocysts but was very weak and did not the allow reliable measurement of a possible developmental expression pattern (data not shown).
hetL overexpression in hetR, ntcA, and hetC mutants.
To determine the epistatic relationship between hetL and other genes, hetL was overexpressed in hetR, ntcA, and hetC mutants defective for heterocyst formation. pAM2065, which contains hetL expressed from the strong rbcL promoter, was transferred into hetR mutant strain 216. The resulting strain formed some cells that were bigger than normal vegetative cells in both BG-11 and BG-110 media. However, the bigger cells failed to be stained by alcian blue, which stains heterocyst-specific polysaccharides in the heterocyst envelope (22). The intervals between the bigger cells did not resemble a normal heterocyst pattern. We conclude that hetR inactivation is epistatic to hetL overexpression.
ntcA is required for the earliest stages of heterocyst formation (17, 42). In strain AMC236, the ntcA gene is inactivated by an ΩSpr/Smr cassette. To determine whether hetL overexpression could cause heterocyst development in an ntcA-null mutant, pAM2065 was transferred into AMC236. Growth of AMC236 (pAM2065) required ammonium as a nitrogen source, which is the same as for the parental strain. On ammonium-containing medium and after transfer to BG-11 or BG-110 medium, strain AMC236 (pAM2065) showed signs of proheterocyst development (Fig. 5). Some cells were bigger than other cells; however, the filaments were highly fragmented, which made it difficult to determine their frequency or pattern. Many of the bigger cells had thicker cell walls, and all the bigger cells were stained by alcian blue (Fig. 5). Differentiating heterocysts degrade much of their photosynthetic pigments, which can be observed as a loss of fluorescence (28). Fluorescence microscopy showed that most of the bigger cells were dark compared with the bright vegetative cells, which indicates that these cells were attempting to differentiate (data not shown). Strain AMC236 (pAM2065) produced very short filaments (<20 cells) and many detached cells compared to the parental strain AMC236, which had much longer filaments (>200 cells) under the same growth conditions. After heterocyst induction, wild-type Anabaena PCC 7120 filaments tended to break at the more fragile connections between heterocysts and vegetative cells. We conclude that hetL overexpression forced the initiation of heterocyst development in the ntcA-null mutant background but that mature heterocysts could not form because of ntcA pleiotropy, which may reflect the involvement of NtcA in the regulation of many genes (23).
FIG. 5.
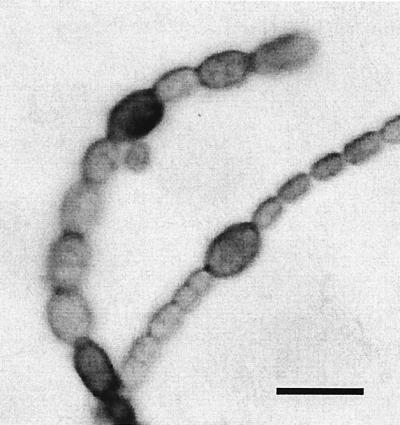
Micrograph of the ntcA-null mutant AMC236 containing the hetL overexpression plasmid pAM2065 in nitrate-containing BG-11 medium. The sample was stained with alcian blue, which stains the heterocyst-specific polysaccharide in the envelope of differentiating cells. Larger proheterocyst-like cells showed staining. Filaments were highly fragmented, presumably because of the weak connections between vegetative cells and partially differentiated cells. Scale bar, 10 μm.
hetC mutants appear to initiate development but cannot form mature heterocysts (25). Previous studies showed that, after heterocyst induction, the hetC-null mutant DR1653 forms weakly autofluorescent cells in a semiregular pattern along filaments (45). The weakly autofluorescent cells, which have degraded their phycobiliproteins (44), also showed increased expression of a hetR-gfp reporter, indicating the initiation of heterocyst differentiation (45). To test the epistasis between hetL and hetC, hetL on pAM2065 was transferred into strain DR1653, producing strain AMC1043. At 30 and 36 h after induction, there were no heterocysts and no obvious differences in the autofluorescence between AMC1043 and DR1653, showing that hetC is epistatic to hetL overexpression.
Interestingly, at 48 and 54 h after induction, the pattern of weakly fluorescent cells in strain AMC1043 was different from that in the parental strain DR1653, which had only a small fraction of weakly fluorescent cells. Most cells in AMC1043 filaments had weak autofluorescence at 54 h after induction (data not shown). The pattern of autofluorescence along filaments of DR1653 carrying vector pAM1824 as a control was similar to that of DR1653. Although hetL overexpression in the hetC-null mutant background increased the number of weakly fluorescent cells 2 days after induction, it is not clear if these cells had initiated heterocyst differentiation.
DISCUSSION
The hetL gene was identified by screening an expression library for plasmids that bypassed the inhibition of heterocyst formation produced by patS overexpression. Nearly the entire amino acid sequence of the predicted HetL protein is composed of pentapeptide repeats with a consensus of A(D/N)L*X (1, 24). Proteins containing this pentapeptide repeat motif have been identified in a wide variety of organisms, but they are particularly numerous in cyanobacteria. Bateman et al. have suggested that the pentapeptide repeats may form a parallel β-helical coil structure with 15 amino acids in each turn (1). This hypothetical structure is based on the crystal structure of a bacterial protein with hexapeptide repeats and other general protein-folding criteria (1, 24). The biochemical function of the pentapeptide repeat motif is not known.
Experimental data for several proteins containing pentapeptide repeat motifs are available, but in no case is the function of the motif known. In Anabaena PCC 7120, the predicted HglK protein has three membrane spans in the N-terminal region and pentapeptide repeats in the C-terminal region (2). hglK encodes a heterocyst-specific protein required for glycolipid localization in the heterocyst envelope. An hglK-null mutant is Fox− (defective for nitrogen fixation in the presence of oxygen) because of the defective heterocyst envelope.
The N-terminal region of the predicted IcmE protein from L. pneumophila contains pentapeptide repeats, and the C-terminal region is similar to a protein involved in DNA transfer (36, 41). An icmE-null mutant is defective in the ability to survive in host cells. The biochemical function of the pentapeptide repeat domains in HglK and IcmE is unclear.
Quinolones are antibacterial agents that target bacterial DNA gyrase and topoisomerase IV. The plasmid-encoded qnr gene provides quinolone resistance to E. coli by protecting DNA gyrase (39). Similar to HetL, Qnr is composed almost entirely of 39 pentapeptide repeats. Purified Qnr protected E. coli DNA gyrase, but not topoisomerase IV, from ciprofloxacin; however, the mechanism of the protection is not known.
Leucine-rich repeats, which are found in proteins with diverse functions, tend to show similarity to the pentapeptide repeat proteins. For example, the N-terminal region of the human follicle-stimulating hormone receptor (FSHR) (29) shows 19% identity and 50% similarity to HetL in its amino acid sequence, even though FSHR does not have pentapeptide repeats. The N-terminal region of FSHR contains leucine-rich repeats and is believed to form a pocket to bind follicle-stimulating hormone. Qnr also shows similarity to leucine-rich repeat proteins (39). Generally, leucine-rich repeat motifs are thought to be involved in protein-protein interactions. However, it is thought that leucine-rich repeat motifs and bacterial pentapeptide repeat motifs fold into different structures and therefore could be expected to have different functions (24).
Our results showed that synthetic PatS-5 pentapeptide could not inhibit heterocyst formation in the hetL overexpression strain. These results indicate that hetL overexpression stimulates heterocyst formation downstream of PatS production. However, hetL overexpression cannot simply relieve inhibition by the PatS signaling pathway because hetL overexpression causes a stronger heterocyst stimulation phenotype than a patS-null mutant. For example, in nitrate-containing medium, a patS-null mutant forms 4% heterocysts (47) but a hetL overexpression strain forms nearly 20% heterocysts and forms multiple-contiguous heterocysts. Therefore, it seems likely that hetL overexpression affects the regulatory pathway at or downstream of the point that integrates multiple signals including PatS and nitrogen limitation.
After nitrogen step-down, hetC mutants produce a pattern of weakly autofluorescent cells that are thought to have initiated heterocyst differentiation (25, 45). When hetL is overexpressed in the hetC-null mutant, most cells showed weak autofluorescence 2 days after nitrogen step-down. It is possible that these cells are all attempting to form heterocysts because of the hetL overexpression. Alternatively, hetL overexpression could be altering the response of vegetative cells to nitrogen starvation.
hetL overexpression in an ntcA-null mutant caused proheterocyst development, which was detected with alcian blue staining of the proheterocyst envelope. hetL overexpression is epistatic to the ntcA-null mutation for the initiation of heterocyst development; however, the proheterocysts do not mature. NtcA is known to be required for the expression of several genes involved in heterocyst development (23). Our results show that NtcA is required for the progression of heterocyst differentiation past the stage reached by the above strain, but the specific nature of the block is unknown.
There are several parallels between the phenotype produced by the overexpression of hetL and the overexpression of hetR, which suggests that HetL overexpression may stimulate heterocyst development by altering HetR activity or abundance. Overexpression of hetL or hetR increases the frequency of heterocysts and produces an Mch phenotype even on media containing nitrate or ammonium. The stimulation of heterocyst development by hetL or hetR overexpression is stronger than the absence of either nitrogen supply or PatS signaling alone, indicating that HetL and HetR affect the signaling pathway downstream of where these signals are integrated.
It is not clear if HetL is normally involved in the heterocyst regulatory pathway or if only its overexpression affects the pathway. The lack of a phenotype for the hetL mutant shows that it is not essential for normal heterocyst development. It is possible that HetL is normally not involved in the process or it could function as a nonessential component such as an accessory or scaffold protein. In either case, hetL overexpression could affect development through dominant-negative interactions with components of the heterocyst regulatory pathway. These interactions could be between HetL and its normal partners, or they could be cross talk resulting from the similarity between HetL and another member of the pentapeptide repeat family of proteins. These different possibilities cannot be resolved until a more complete picture of the heterocyst regulatory pathway has been determined.
We propose a model in which overexpression of hetL stimulates heterocyst development downstream of NtcA and PatS and upstream of HetR and HetC. PatS and HetL could interact with unknown factor(s) between NtcA and HetR in the regulatory pathway; however, it is possible that PatS, HetL, or both could directly influence the activity of HetR. HetC is thought to be downstream of HetR because hetC mutants appear to initiate development whereas hetR mutants do not; however, different models would be consistent with available data.
Acknowledgments
Martha Sprague isolated some of the patS overexpression bypass strains. Ivan Khudyakov helped in epistasis and gene inactivation experiments. Michael Scherer constructed the hetL-gfp fusion strain AMC1071. Beth Givens helped with plasmid sequencing. Xiaoqiang Wu constructed a plasmid containing PpetE-hetL to verify the hetL overexpression phenotype. Jin Xiong helped with bioinformatic searches. We thank all other colleagues in the laboratory for beneficial discussions and critical review.
This work was supported by Public Health Service grant GM36890 and by Texas Advanced Research Program grant 010366-0010-1999.
REFERENCES
- 1.Bateman, A., A. G. Murzin, and S. A. Teichmann. 1998. Structure and distribution of pentapeptide repeats in bacteria. Protein Sci. 7:1477-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, K., W. J. Buikema, and R. Haselkorn. 1995. The hglK gene is required for localization of heterocyst-specific glycolipids in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:6440-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 4.Black, T. A., and C. P. Wolk. 1994. Analysis of a Het− mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol. 176:2282-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321-330. [DOI] [PubMed] [Google Scholar]
- 7.Buikema, W. J., and R. Haselkorn. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. USA 98:2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, Y., and C. P. Wolk. 1997. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J. Bacteriol. 179:258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callahan, S. M., and W. J. Buikema. 2000. Suppression of heterocyst formation by hetN. 10th International Symposium on Phototrophic Prokaryotes, Barcelona, Spain.
- 11.Callahan, S. M., and W. J. Buikema. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40:941-950. [DOI] [PubMed] [Google Scholar]
- 12.Chastain, C. J., J. S. Brusca, T. S. Ramasubramanian, T.-F. Wei, and J. W. Golden. 1990. A sequence-specific DNA-binding factor (VF1) from Anabaena sp. strain PCC 7120 vegetative cells binds to three adjacent sites in the xisA upstream region. J. Bacteriol. 172:5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, M. F., J. C. Meeks, Y. A. Cai, and C. P. Wolk. 1998. Transposon mutagenesis of heterocyst-forming filamentous cyanobacteria. Methods Enzymol. 297:1-17. [Google Scholar]
- 14.Elhai, J., A. Vepritskiy, A. M. Muro-Pastor, E. Flores, and C. P. Wolk. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 16.Fiedler, G., A. M. Muro-Pastor, E. Flores, and I. Maldener. 2001. NtcA-dependent expression of the devBCA operon, encoding a heterocyst-specific ATP-binding cassette transporter in Anabaena spp. J. Bacteriol. 183:3795-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frias, J. E., E. Flores, and A. Herrero. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14:823-832. [DOI] [PubMed] [Google Scholar]
- 18.Frias, J. E., E. Flores, and A. Herrero. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179:477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golden, J. W., S. J. Robinson, and R. Haselkorn. 1985. Rearrangement of nitrogen fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature (London) 314:419-423. [DOI] [PubMed] [Google Scholar]
- 20.Golden, J. W., L. L. Whorff, and D. R. Wiest. 1991. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173:7098-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golden, J. W., and D. R. Wiest. 1988. Genome rearrangement and nitrogen fixation in Anabaena blocked by inactivation of xisA gene. Science 242:1421-1423. [DOI] [PubMed] [Google Scholar]
- 22.Hebbar, P. B., and S. E. Curtis. 2000. Characterization of devH, a gene encoding a putative DNA binding protein required for heterocyst function in Anabaena sp. strain PCC 7120. J. Bacteriol. 182:3572-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajava, A. V. 2001. Proteins with repeated sequence—structural prediction and modeling. J. Struct. Biol. 134:132-144. [DOI] [PubMed] [Google Scholar]
- 25.Khudyakov, I., and C. P. Wolk. 1997. hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J. Bacteriol. 179:6971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 27.Maddock, J., A. Bhatt, M. Koch, and J. Skidmore. 1997. Identification of an essential Caulobacter crescentus gene encoding a member of the Obg family of GTP-binding proteins. J. Bacteriol. 179:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meeks, J. C., and J. Elhai. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 66:94-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyle, W. R., and R. K. Campbell. 1995. Gonadotropins, p. 230-241. In L. J. DeGroot (ed.), Endocrinology. WB Saunders Company, Philadelphia, Pa.
- 30.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 1999. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J. Bacteriol. 181:6664-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muro-Pastor, M. I., J. C. Reyes, and F. J. Florencio. 2001. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 276:38320-38328. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto, S., and K. Ochi. 1998. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol. Microbiol. 30:107-119. [DOI] [PubMed] [Google Scholar]
- 33.Ramasubramanian, T. S., T.-F. Wei, and J. W. Golden. 1994. Two Anabaena sp. strain PCC 7120 DNA-binding factors interact with vegetative cell- and heterocyst-specific genes. J. Bacteriol. 176:1214-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramaswamy, K. S., C. D. Carrasco, T. Fatma, and J. W. Golden. 1997. Cell-type specificity of the Anabaena fdxN-element rearrangement requires xisH and xisI. Mol. Microbiol. 23:1241-1249. [DOI] [PubMed] [Google Scholar]
- 35.Ramaswamy, K. S., S. Endley, and J. W. Golden. 1996. Nitrate reductase activity and heterocyst suppression on nitrate in Anabaena sp. strain PCC 7120 require moeA. J. Bacteriol. 178:3893-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanigawa, R., M. Shirokane, S. Maeda Si, T. Omata, K. Tanaka, and H. Takahashi. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 99:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trach, K., and J. A. Hoch. 1989. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171:1362-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez-Bermudez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512:71-74. [DOI] [PubMed] [Google Scholar]
- 41.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 42.Wei, T.-F., T. S. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176:4473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolk, C. P. 2000. Heterocyst formation in Anabaena, p. 83-103. In Y. V. Brun, and L. J. Shimkets (ed.), Prokaryotic development. American Society of Microbiology, Washington, D.C.
- 44.Wood, N. B., and R. Haselkorn. 1980. Control of phycobiliprotein proteolysis and heterocyst differentiation in Anabaena. J. Bacteriol. 141:1375-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, X., and C. P. Wolk. 2001. Role for hetC in the transition to a nondividing state during heterocyst differentiation in Anabaena sp. J. Bacteriol. 183:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon, H. S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 47.Yoon, H.-S., and J. W. Golden. 2001. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 183:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, R., Z. Cao, and J. Zhao. 1998. Characterization of HetR protein turnover in Anabaena sp. PCC 7120. Arch. Microbiol. 169:417-423. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, R., X. Wei, N. Jiang, H. Li, Y. Dong, K. L. Hsi, and J. Zhao. 1998. Evidence that HetR protein is an unusual serine-type protease. Proc. Natl. Acad. Sci. USA 95:4959-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, J., R. Kong, and C. P. Wolk. 1998. Regulation of hepA of Anabaena sp. strain PCC 7120 by elements 5′ from the gene and by hepK. J. Bacteriol. 180:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]