Abstract
Using phragmoplastin as a bait, we isolated an Arabidopsis cDNA encoding a novel UDP-glucose transferase (UGT1). This interaction was confirmed by an in vitro protein–protein interaction assay using purified UGT1 and radiolabeled phragmoplastin. Protein gel blot results revealed that UGT1 is associated with the membrane fraction and copurified with the product-entrapped callose synthase complex. These data suggest that UGT1 may act as a subunit of callose synthase that uses UDP-glucose to synthesize callose, a 1,3-β-glucan. UGT1 also interacted with Rop1, a Rho-like protein, and this interaction occurred only in its GTP-bound configuration, suggesting that the plant callose synthase may be regulated by Rop1 through the interaction with UGT1. The green fluorescent protein–UGT1 fusion protein was located on the forming cell plate during cytokinesis. We propose that UGT1 may transfer UDP-glucose from sucrose synthase to the callose synthase and thus help form a substrate channel for the synthesis of callose at the forming cell plate.
INTRODUCTION
UDP-glucose is a substrate for the synthesis of both cellulose (1,4-β-glucan) and callose (1,3-β-glucan). UDP-glucose could bind directly to the catalytic subunit of cellulose synthases (CelSs) and callose synthases (CalSs), or its transfer to the catalytic site may be performed by a UDP-glucose transferase. It has been proposed that CelS and CalS might actually be the same enzyme, which changes linkage specificity and produces different products after being modified around the active site by an unknown mechanism (Jacob and Northcote, 1985; Delmer, 1999; Stasinopoulos et al., 1999). It has been difficult to test this hypothesis by biochemical approaches because CelS cannot be effectively purified and assayed in vitro. Furthermore, it is possible that CelS may synthesize 1,3-β-glucan under in vitro conditions (Brown et al., 1996). Isolation of a CalS gene (Hong et al., 2001) has demonstrated convincingly that CalS and CelS (Arioli et al., 1998; Taylor et al., 1999; Holland et al., 2000) are distinct enzymes.
Both CelS and CalS are large transmembrane proteins with similar topological arrangements in the membrane. However, they do not share homology in their primary amino acid sequence. CelS and the bacterial 1,3-β-glucan synthase contain a signature region that includes three highly conserved aspartate residues (the D, D, D motif) and a QXXRW motif (Saxena et al., 1995; Stasinopoulos et al., 1999). This region has been implicated in binding to UDP-glucose and transfer of the glucosyl group. The fact that plant CalS does not contain a similar signature suggests that CalS may have adopted a different mechanism to bind and transfer UDP-glucose. This function appears to be performed by a UDP-glucose transferase that may facilitate the transfer of UDP-glucose from sucrose synthase (SuSy), which has been localized at the cell plate (Amor et al., 1995) and suggested to be part of the CalS complex (Hong et al., 2001).
UDP-glycosyltransferases constitute a large family of enzymes that catalyze the transfer of the glycosyl group from a specific donor to a substrate/acceptor (Mackenzie et al., 1997). The common glycosyl donors include UDP-glucose and UDP-glucuronate, although UDP-galactose and UDP-rhamnose also are used by certain UDP-glycosyltransferases. The Arabidopsis genome alone encodes >120 members of the UDP-glycosyltransferase family. However, only a small number have been cloned and characterized to date. The maize Bronze-1 locus encoding UDP glucose:flavonol 3-O-glucosyltransferase was first cloned by transposon tagging (Fedoroff et al., 1984; Harborne and Williams, 1988), and since then, several genes from other plant species have been cloned based on homology (Sparvoli et al., 1994; Boss et al., 1996; Ford et al., 1998). Several UGTs involved in the glycosylation of various metabolites have been isolated using a variety of techniques (Szerszen et al., 1994; Moehs et al., 1997; Warnecke et al., 1997; Lee and Raskin, 1999; Martin et al., 1999; Yamazaki et al., 1999).
Here, we report a novel UDP-glucose transferase (UGT1) that appears to be a component of the CalS complex. The UGT1 cDNA was cloned in a two-hybrid interaction trap using phragmoplastin as a bait. In addition, UGT1 was found to interact with a cell plate–specific CalS1 (Hong et al., 2001) and the Rho1-like protein Rop1. The latter is known to regulate yeast 1,3-β-glucan synthase (Qadota et al., 1996). The green fluorescent protein (GFP)–UGT1 fusion protein was targeted to the forming cell plate during cytokinesis, suggesting that both CalS1 and UGT1 are colocalized in vivo and may form a functional CalS complex at the cell plate. This novel UGT appears to be conserved in higher plants, as shown by the cross-reactivity of the Arabidopsis UGT1 antibody with moth bean and tobacco membrane fractions.
RESULTS
Identification and Cloning of a Novel UGT1 cDNA
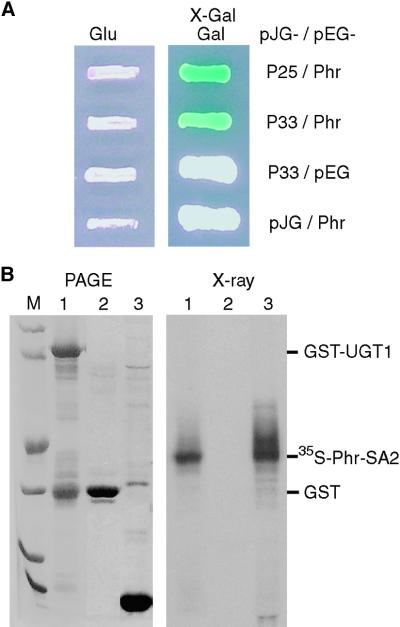
In an attempt to identify proteins that interact with phragmoplastin, a cell plate–associated protein (Gu and Verma, 1996, 1997), we took an interaction trap approach using phragmoplastin as a bait. The phragmoplastin cDNA was cloned in pEG202 in frame with the DNA binding protein LexA (Golemis et al., 1996) and used to screen an Arabidopsis cDNA library constructed in vector pJG4-5. Approximately two million yeast colonies were screened, and all positive clones were subjected to reconfirmation to eliminate any false positives. Two independent cDNA clones, pPIL33 and pPIL25, were isolated and found to be identical except that the former is one codon (three base pairs) longer than the latter, indicating that they represent the same gene. Cloning vectors (pEG202 and pJG4-5) were used as negative controls in combination with pPIL33 and pEG-Phr. Both controls were negative in the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) assay (Figure 1A), suggesting that the presence of both phragmoplastin and the UGT1 proteins encoded by pPIL33 is required for the observed interactions.
Figure 1.
Interaction between Phragmoplastin and UGT1.
(A) Interaction in the yeast two-hybrid system. Using soybean phragmoplastin (Phr; Gu and Verma, 1996) as a bait to screen an Arabidopsis cDNA library, two independent clones (pPIL25 [P25] and pPIL33 [P33]) were pulled out and later confirmed by sequencing to represent the same gene. Cloning vectors pEG202 (pEG) and pJG4-5 (pJG) were used as negative controls. Yeast colonies containing the plasmids were grown on SC-Ura-Trp-His+Leu medium in the presence of glucose (Glu) or galactose (Gal) and X-Gal.
(B) Pull-down assay of in vitro transcription/translation products. In lane 1, UGT1 was expressed as a glutathione S-transferase (GST)–tagged protein in Escherichia coli and purified using glutathione–agarose beads. The beads were incubated with 35S-labeled phragmoplastin, proteins bound to the beads were resolved by PAGE (left), and then the same gel was dried and exposed to x-ray film (right). Note that it is not unusual to see a GST band copurified with a GST-tagged recombinant protein. In lane 2, GST expressed from vector pGEX-KG served as a negative control and was treated as described for lane 1. In lane 3, plasmid pAGA3-SA2 expressing the SA2 domain of phragmoplastin (Phr-SA2; Zhang et al., 2000) was used to synthesize 35S-labeled phragmoplastin in an in vitro transcription/translation system. The positions of the proteins are marked. Lane M contains standard protein markers of (from bottom to top) 14, 21, 31, 45, 66, and 97 kD.
Database searches revealed that the insert in pPIL33 (∼1.0 kb long) encodes a new member of the UDP-glycosyltransferase superfamily (see below), but the sequence was not complete and lacked ∼600 bp at the 5′ terminus. To obtain a full-length clone of this UDP-glucose transferase (UGT1), we used the insert in pPIL33 as a probe to screen another Arabidopsis cDNA library, FL-1, constructed in λZapII vector (Seki et al., 1998). Several independent clones were found to contain the start codon (ATG), and the longest clone, pUGT1f, was used in further studies. The coding region of pUGT1f was cloned downstream from the glutathione S-transferase (GST) coding region, and the fusion protein was expressed in Escherichia coli and purified using glutathione–agarose beads. The SA2 domain of phragmoplastin was labeled with 35S-methionine in an in vitro transcription/translation system (Zhang et al., 2000). The radiolabeled phragmoplastin peptide was pulled down effectively by the recombinant fusion protein (Figure 1B), suggesting that the interaction between the phragmoplastin and UGT1 also occurs under in vitro conditions and that no additional protein is required for this interaction.
UGT1 Is a Novel Member of the UDP-Glycosyltransferase Family
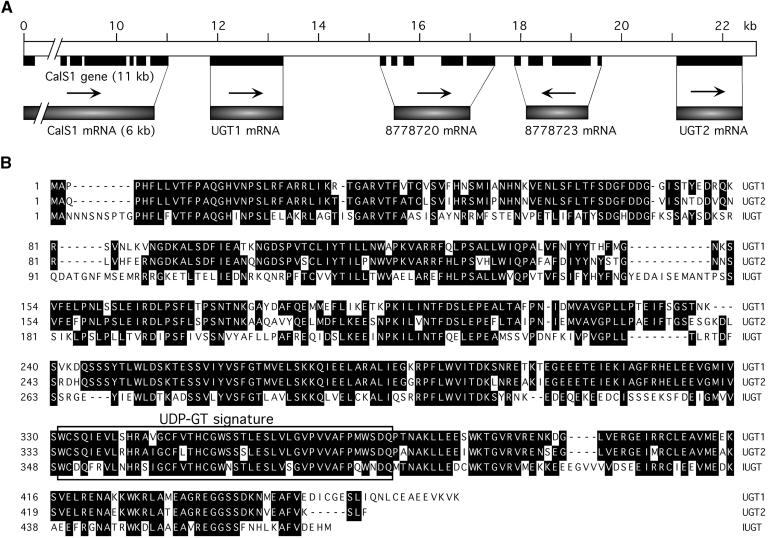
Analysis of the Arabidopsis genome database revealed that the UGT1 gene is contained in two overlapping bacterial artificial chromosomes, T25N20 and F23F20, which have been localized on chromosome 1. Upstream of the UGT1 locus is a CalS gene, designated CalS1 (Figure 2A) and characterized in the previous article (Hong et al., 2001). We showed that CalS1 interacts with UGT1 and is targeted to the forming cell plate during cytokinesis (Hong et al., 2001). A homolog of UGT1, designated UGT2, is found downstream of the UGT1 sequence. UGT2 shares high sequence homology with UGT1, 83.2% identity at the nucleotide level and 80.7% identity at the amino acid level (Figure 2B), suggesting that the UGT2 gene may have evolved recently from UGT1 via gene duplication and that these genes may be functionally similar. We have not studied the regulation of these genes (CalS1, UGT1, and UGT2), but they may be induced coordinately during cell plate formation. Because UGT1 interacts with CalS1 in the yeast two-hybrid system (Hong et al., 2001) and is associated with purified CalS (see below), we propose that UGT1 may function in the transfer of UDP-glucose to the catalytic subunit of CalS, thus representing a novel type of UGT in plants (see below). The UGT1 was assigned as UGT75B1 and UGT2 as UGT75B2 by the UGT Nomenclature Committee (Mackenzie et al., 1997).
Figure 2.
Chromosomal Location of UGT1 and Amino Acid Sequence Alignment of UGT1, UGT2, and Indoleacetic Acid–UGT.
(A) Genomic organization of the CalS1, UGT1, and UGT2 loci on chromosome 1. The open box represents the genomic fragment spanning the region from CalS1 to UGT2. The DNA length (kb) is indicated by numbers starting from the first ATG of CalS1. Exons are indicated by solid boxes. Arrows indicate the orientations of the genes from 5′ to 3′. Two genes with unknown functions are represented by their protein identification numbers from GenBank. The systematic nomenclature for UGT1 and UGT2 is UGT75B1 and UGT75B2, respectively. The GenBank accession number for UGT1 cDNA is AF196777.
(B) Alignment of the deduced amino acid sequences of UGT1, UGT2, and indoleacetic acid–UGT (IUGT). UGT1 was deduced from its cDNA as sequenced in this work. UGT2 and IUGT (Graham and Thornburg, 1997) were from GenBank. The UDP-glycosyltransferase (UDP-GT) signature is boxed.
UGT1 Interacts with the Rho-Like Protein Rop1
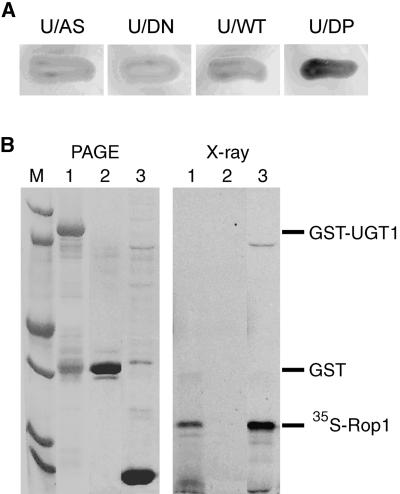
Yeast 1,3-β-glucan synthase requires the addition of GTP for its activity, but the enzyme preparation from yeast cells expressing constitutively active Rho1p does not require exogenous GTP for activity, suggesting that Rho1 is an activator of the yeast 1,3-β-glucan synthase (Qadota et al., 1996). We explored the possibility of whether a plant Rho homolog is part of the CalS/UGT1 complex. We coexpressed the Arabidopsis Rho-like protein Rop1 and UGT1 in the yeast two-hybrid system and performed X-Gal staining assays. Three forms of Rop1 created by Yang's group (Li et al., 1999) were used in this experiment: the dominant positive mutant (Rop1DP, GTP-bound form), the dominant negative mutant (Rop1DN, GDP-bound form), and the wild type (Rop1WT). The results showed that Rop1DP was able to interact with UGT1, whereas the interaction between Rop1DN and UGT1 was undetectable (Figure 3A). When Rop1WT was used, the interaction was either very weak or undetectable. These results suggest that UGT1 may interact with the GTP-bound form of Rop1 and that the GDP-bound form of Rop1 may dissociate from UGT1. We performed a pull-down experiment using UGT1 (as a GST-tagged protein) to interact with radiolabeled Rop1 (Figure 3B, lane 3). As a control (Figure 3B, lane 2), GST-bound agarose beads were not able to pull down 35S-labeled Rop1. When UGT1 was fused to GST and purified with the beads, it interacted quantitatively with 35S-labeled Rop1 and brought down the radiolabeled product to the pellet (Figure 3B, lane 1). This result confirmed the observation with the yeast two-hybrid system and provides further evidence for the interaction between UGT1 and Rop1. The fact that only the GTP-bound form interacted with UGT1 sheds light on its possible role in regulating the CalS complex as it does in the yeast β-1,3-glucan synthase (Qadota et al., 1996).
Figure 3.
Interaction between UGT1 and Rop1 Protein.
(A) Interaction in the yeast two-hybrid system. The UGT1 coding region (U) was subcloned into pACT2 to yield pACT-UGT1. Plasmids expressing wild-type Rop1 (WT), dominant negative Rop1 (DN), or dominant positive Rop1 (DP) in pAS1-CH2 vector (provided by Dr. Z. Yang, University of California, Riverside) were transformed to Y190 cells harboring pACT-UGT1. Vector plasmid pAS1-CH2 (AS) served as a negative control. Cells of transformants were streaked on medium containing X-Gal.
(B) Pull-down assay of in vitro transcription/translation products. In lane 1, purified GST-UGT1 was used to interact with 35S-labeled Rop1. In lane 2, GST alone served as a negative control. In lane 3, 35S-labeled Rop1 was used in an in vitro transcription/translation system. For more details, see the legend to Figure 1B.
UGT1 Is a Membrane-Associated Protein
Whereas most mammalian UGTs are endoplasmic reticulum– and Golgi-associated proteins, some bacterial and plant UGTs are cytosolic. To determine the subcellular location of UGT1, we raised antibodies against E. coli–expressed UGT1. The IgG fraction was affinity purified using nitrocellulose-bound UGT1 protein and used on a protein gel blot to react with soluble fraction and membrane pellet from Arabidopsis seedlings. Although the soluble fraction also contained some UGT1 antibody-reactive material, most immunoreactive material was found in the membrane fraction (Figure 4, lane 1). The membrane-bound UGT1 was composed of two bands, one of which was soluble in Triton X-100 and the other was insoluble (Figure 4, lanes 2 and 3). Both UGT1 bands were resistant to treatments with a chaotropic agent (NaI) and alkaline solution (Na2CO3, pH 11.0; Figure 4, lanes 4 and 5), suggesting that UGT1 is tightly associated with the membrane fraction. The antibody reacted with similar bands on protein gel blots by using membrane fractions from moth bean and tobacco (data not shown), suggesting that UGT1 is a conserved protein in higher plants. The apparent molecular mass of the in vivo forms was larger than that of the E. coli–expressed UGT1 (Figure 4, lane 6), indicating the possibility of glycosylation and/or other post-translational modifications of UGT1. Five consensus sites for N-glycosylation are present in the UGT1 sequence, but it is not known how many are glycosylated in planta, nor are the types of glycosyl residues present on the native UGT1 known.
Figure 4.
Association of UGT1 with Membranes.
Total membranes were isolated from extracts of Arabidopsis seedlings by centrifugation at 100,000g for 1 hr. The membrane pellet (lane 1) was treated with 1% Triton X-100, a chaotropic agent, or alkaline solution. Lane 2, Triton X-100–soluble fraction; lane 3, Triton X-100–insoluble fraction; lane 4, insoluble fraction after treatment with 0.5 M NaI; lane 5, insoluble fraction after treatment with 100 mM Na2CO3; lane 6, UGT1 peptide expressed in E. coli. The proteins were resolved by PAGE and transferred to a nitrocellulose membrane. Rabbit antibody raised against purified UGT1 was used as a primary antibody to probe the nitrocellulose membrane, followed by protein gel blotting. Two immunoreactive bands are indicated by arrowheads. Sizes (in kilodaltons) of the prestained protein markers (BioRad) are indicated at right.
UGT1 Is a Putative Subunit of the CalS Complex
We have demonstrated that CalS1 interacts with UGT1 in the yeast two-hybrid system, suggesting that UGT1 may be a subunit of the CalS complex (Hong et al., 2001). To determine if these two proteins exist as a complex in vivo, we purified the CalS complex using a product-entrapment procedure followed by sucrose gradient centrifugation. Total membranes (with a typical activity of 3000 fluorescence units mg−1 min−1) from seedling extracts were solubilized with a detergent mixture of 3-([3-cholamidopropyl]dimethylammonio)-1-propanesulfonic acid (Chaps) and cholesteryl hemisuccinate in the presence of 4 μM GTPγS. CalS was enriched by a product-entrapment procedure (Inoue et al., 1995). After incubation with UDP-glucose, CalS was purified from other proteins by centrifugation. The callose pellet containing entrapped CalS was dissolved in a buffer lacking UDP-glucose. This procedure resulted in a 90-fold increase in the specific activity (270,000 fluorescence units mg−1 min−1 ) of CalS, with ∼20% recovery of the total enzyme activity. The entrapped product was then subjected to centrifugation on a continuous sucrose density gradient (20 to 60%). The profile of CalS activity in a representative gradient is shown in Figure 5A. Two CalS activity peaks were observed, CalSLO at a low sucrose density of 40% and CalSHI at 52.5% sucrose. A similar profile of CalS activities from peanut seedlings has been reported (Kamat et al., 1992). Fractions from each peak were pooled, and proteins were resolved by SDS-PAGE. Anti-UGT1 antibody was used on a protein gel blot to determine if UGT1 was copurified with the CalS fractions. The two typical immunoreactive bands corresponding to UGT1 in total membranes were present in the purified CalS complexes, suggesting that UGT1 is a part of both the CalSLO and CalSHI complexes (Figure 5). We have shown in the preceding article (Hong et al., 2001) that product-entrapped CalS contains the CalS1 subunit, suggesting that CalS and UGT1 form a functional complex that comigrates in the sucrose density gradient. A significant difference in the densities of CalSLO and CalSHI suggests that other proteins may be part of these complexes and that they may be functionally different or located in different tissues.
Figure 5.
Copurification of UGT1 with the CalS Complex.
(A) Fractionation of the CalS complex by sucrose density gradient. A Chaps-solubilized fraction of membranes from moth bean root tips was incubated with the substrate for callose synthesis. Proteins remaining associated with the product-entrapped fraction were fractionated on a sucrose density gradient (20 to 60%). CalS activity (fluorescence units [FU] per 30-μL fraction) was measured in each fraction (see Methods). CalSHI, CalS present in high sucrose density (52%); CalSLO, CalS present in low sucrose density (40%).
(B) Presence of UGT1 with CalS as shown by protein gel blot analysis using UGT1 antibody. Fractions from each peak were pooled, and proteins were analyzed by protein gel blotting using UGT1 antibody. Two immunoreactive bands are indicated by arrowheads (see Figure 4). CalS1 was shown to be present with these fractions (Hong et al., 2001). Sizes (in kilodaltons) of the prestained protein markers (Bio-Rad) are indicated.
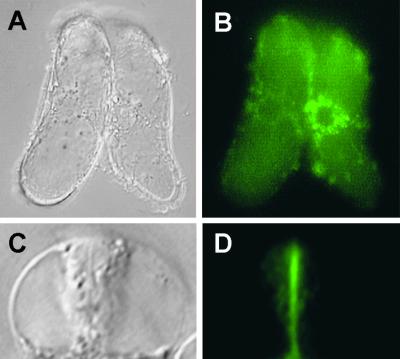
UGT1 Is Associated with the Cell Plate during Cytokinesis
A GFP–UGT1 fusion protein was created and expressed in transgenic tobacco BY-2 cells under the control of the cauliflower mosaic virus 35S promoter. Twenty independent transgenic lines were examined, and all were found to express the fusion protein. Cells expressing GFP alone were used as a control. In these cells, GFP was found to be distributed throughout the cytoplasm and nucleus and was excluded from the forming cell plate during cytokinesis (Gu and Verma, 1997; Hong et al., 2001). In interphase cells, the fluorescence of GFP–UGT1 was distributed in a punctate pattern in the perinuclear region (Figures 6A and 6B). In contrast, the fluorescence was found as a bright green line coinciding with the cell plate in cells undergoing cytokinesis (Figures 6C and 6D). In a parallel experiment, we cross-linked different fluorescent dyes, Alexa 594 and Alexa 488 (Molecular Probes, Eugene, OR), to IgG against UGT1 and used them to label wild-type BY-2 cells and Arabidopsis root tips. The localization data using Alexa-conjugated IgG (data not shown) were consistent with those obtained using the GFP–UGT1 fusion protein. These localization results are important for our understanding of the UGT1-interacting proteins at the forming cell plate, because two other proteins, phragmoplastin and CalS1, also have been shown to have the same distribution pattern (Gu and Verma, 1997; Hong et al., 2001). The colocalization of UGT1 and CalS1 at the forming cell plate and their copurification in the product-entrapped CalS complexes confirm that these two proteins are part of the CalS complex and that UGT1 may interact specifically with CalS1, making it a cell plate–specific complex that also interacts with phragmoplastin. The latter may not be a part of other CalS complexes, such as those induced as a result of pathogen infection. Because there are several CalS isoforms in plants (Hong et al., 2001), each may form a specific complex to be regulated spatially and temporally in response to developmental, biotic, and abiotic signals. It is possible that UGT1 and/or UGT2 are integral parts of all CalS complexes, which is suggested by the abundance of UGT1 in the membrane fraction.
Figure 6.
Fluorescence Localization of GFP-Tagged UGT1 in BY-2 Cells.
(A) and (B) Perinuclear distribution of GFP–UGT1 in nondividing cells.
(C) and (D) Association of GFP–UGT1 with the cell plate during cytokinesis.
Tobacco BY-2 cells expressing the GFP–UGT1 fusion protein were photographed in a bright field ([A] and [C]) or using a filter set for fluorescence ([B] and [D]).
DISCUSSION
UGT1 Is a Novel Glucose Transferase at the Forming Cell Plate
Using UGT1 sequence to search the database, we found that the Arabidopsis genome encodes many putative UDP-glycosyltransferases. Members of this superfamily can be identified by the presence of a highly conserved signature sequence (Figure 2B) (Mackenzie et al., 1997). On the basis of phylogenetic analysis of the Arabidopsis UGT family, we deduced that UGT1 and UGT2 belong to the subfamily that binds UDP-glucose and not UDP-glucuronate, UDP-galactose, or UDP-rhamnose as the glycosyl donor. The known compounds that form glucosides with UDP-glucose in plants include flavonoids, hormones (indoleacetic acid, zeatin, salicylic acids), secondary metabolites such as limonoid, and toxic compounds such as mandelonitrile (Szerszen et al., 1994; Warnecke et al., 1997; Lee and Raskin, 1999; Martin et al., 1999; Kita et al., 2000). Most UDP-glycosyltransferases comprise 450 to 500 amino acid residues with a calculated molecular mass of 50 to 60 kD, except for a subfamily that has an ∼100–amino acid extension at the C terminus and may have substrate specificity toward sterol compounds such as solanidine (Warnecke et al., 1997).
Although UGT1 interacted with phragmoplastin and CalS1 and may have a unique role in cell plate formation, this protein shares significant sequence homology with the known UGTs from other plants. However, unlike other UGTs that transfer glycosyl moiety to small organic compounds (Szerszen et al., 1994; Warnecke et al., 1997; Lee and Raskin, 1999; Martin et al., 1999), UGT1 may be involved in the synthesis of callose (see below). This conclusion is consistent with the observed interactions between UGT1 and phragmoplastin, Rop1, and CalS1 and with the subcellular localization of these proteins on the forming cell plate.
UGT1 Is Transported from the Golgi to the Forming Cell Plate during Cytokinesis
Glycosylation is one of the major functions of the Golgi apparatus, and many glycosyltransferases are localized in the Golgi bodies (Dupree and Sherrier, 1998). The distribution pattern of GFP–UGT1 fluorescence in nondividing cells (Figure 6A) is very similar to that of a GFP-fused α-mannosidase, a Golgi marker protein (Nebenführ et al., 1999, 2000). The fluorescence of GFP–UGT1 was clearly associated with the forming cell plate during cytokinesis (Figure 6B). We were able to follow the growth of the cell plate in living cells under a fluorescence microscope (Gu and Verma, 1997). The GFP–UGT1 fluorescence started to concentrate in the center of the phragmoplast, and it expanded centrifugally toward the parental cell wall (data not shown). Such a redistribution pattern corresponded well with that of GFP-phragmoplastin (Gu and Verma, 1997) and further suggests that UGT1 is associated with the growing cell plate. This localization is consistent with the presence of CalS, which synthesizes callose at the forming cell plate and with which UGT1 was found to interact (Hong et al., 2001).
Most mammalian and yeast Golgi enzymes are type II transmembrane proteins; each has a short cytoplasmic N terminus, a single membrane-spanning domain, a stalk region, and a catalytic domain (Machamer, 1993). The transmembrane region and the short cytoplasmic tail are key determinants for their localization. This rule applies to some plant type II Golgi proteins (Nebenführ et al., 1999). However, it is apparently not applicable to UGT1, because this protein does not contain any predicted transmembrane domain and may become membrane associated via interactions with other proteins, such as CalS1 (see below).
Rop1 May Regulate CalS Activity through Interaction with UGT1
The activity of yeast 1,3-β-glucan synthase requires the addition of GTP, and it has been demonstrated that the small GTP binding protein Rho1 is a subunit of the 1,3-β-glucan synthase complex (Qadota et al., 1996). We have cloned an Arabidopsis cDNA encoding a plant 1,3-β-glucan synthase (CalS1) that interacts with UGT1 and phragmoplastin in the yeast two-hybrid system and showed that CalS1 is targeted to the forming cell plate during cytokinesis (Hong et al., 2001). Partially purified CalS from plant extracts does not appear to require GTP for activity (Turner et al., 1998). However, the addition of GTP at low concentration (10 μM) increased CalS activity (20 to 30%) in our purified preparations from moth bean root tips (J. Olson and D.P.S. Verma, unpublished data). A difference in GTP requirement may exist between the yeast 1,3-β-glucan synthase and plant CalS. The latter appears to use a specific UGT to bind and transfer UDP-glucose for callose synthesis. Thus, Rop1, a plant Rho-like protein, may regulate CalS activity through interaction with UGT1, as illustrated in Figure 3. The fact that UGT1 interacts only with the GTP-bound configuration of Rop1 suggests the significance of Rop1 in controlling CalS activity via UGT1. Like other small GTP binding proteins, Rop1 may act as a molecular switch that controls CalS1 activity at the forming cell plate. Whether Rop1 is a part of all CalSs located in different tissues or is specific to cell plate CalS (Hong et al., 2001) remains to be determined.
The Cell Plate–Specific CalS1 Forms a Complex with UGT1, Rop1, and Possibly SuSy
Building the cell plate at cytokinesis is a rapid process during which copious amounts of callose need to be deposited in a few minutes after the tubulovesicular network is established (Samuels et al., 1995; Verma, 2001). We have shown that both CalS1 and UGT1 are localized at the cell plate during cytokinesis. SuSy also was reported to be associated with the forming cell plate (Amor et al., 1995), and overexpression of SuSy was found to enhance cellulose production in bacteria (Nakai et al., 1999). When expressed in BY-2 cells, a significant portion of Rop1 is also concentrated at the forming cell plate (Z. Yang, personal communication). We have demonstrated that CalS1 is a cell plate–specific enzyme (Hong et al., 2001). Together, these proteins may be targeted or relocated from other subcellular locations to the forming cell plate during cytokinesis.
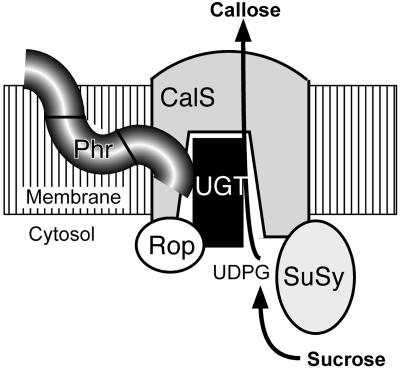
The available data suggest a model (Figure 7) in which several proteins associate to form a CalS complex. In this model, UGT1 acts in transferring UDP-glucose from SuSy to CalS for rapid synthesis of callose and Rop1 functions as a molecular switch to control CalS activity. Location of these two proteins, presumably in the central hydrophilic domain of the CalS1 peptide, may create a substrate channel to transfer UDP-glucose from sucrose. Moreover, UGT1, Rop1, and SuSy have no transmembrane domains, and if they form part of the CalS complex, they must interact directly with this protein or become associated with membrane after some post-translational modification. Such an association of these proteins may be essential to provide a rapid flux of substrate to synthesize the significant amounts of callose required to fill the large volume of growing cell plate in <60 min (Verma, 2001).
Figure 7.
Proposed Model of the CalS Complex at the Forming Cell Plate.
UGT1, SuSy, and Rop1 are possible subunits of the CalS complex. Phragmoplastin (Phr) is shown to interact with UGT1 (see Figure 1). This protein, however, may not be a part of CalS complexes present in other locations except the cell plate. Note that none of the other proteins besides CalS has transmembrane domains; hence, they must interact with CalS directly, most likely with the hydrophilic loop of CalS. UDPG, UDP-glucose.
Callose is synthesized at a precise time and location during cell plate formation (Samuels et al., 1995), which requires a mechanism for both temporal and spatial regulations of CalS activity. Phragmoplastin forms polymers (Zhang et al., 2000) that appear to facilitate the creation of vesicle-tubule-vesicles at the growing cell plate (Verma, 2001). The vesicle-tubule-vesicles are stabilized by the deposition of callose (Samuels et al., 1995; Verma, 2001). Rop1 may sense spatial or temporal signal(s) from the upstream proteins of the signal transduction pathway and thus regulate the activity of the CalS complex at the forming cell plate. The presence of two forms of CalS of different densities and the association of UGT1 with both forms suggest further complexity in the regulation of different forms of CalSs in plants. It is also possible that additional proteins are involved in making these complexes, which are very large, as indicated by the sedimentation data (Figure 5A). Further characterization of these complexes is essential to understanding how callose is deposited at different locations in plants under different developmental and physiological conditions as well as in response to external signals such as pathogen attacks and physical injuries.
METHODS
Bacterial and Yeast Strains
Plasmid propagation was performed in Escherichia coli strains Top10F′ (Invitrogen, Carlsbad, CA) and DH5α (Gibco BRL). Strains JM109 and BL21(DE3) were used for the expression of recombinant proteins. Saccharomyces cerevisiae strain EGY48 (MATα trp1 his3 ura3 leu2::6 LexAop-LEU2) was used for screening of an Arabidopsis thaliana cDNA library for interaction clones. Y190 (MATa ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3, 112 gal4Δ gal80Δ cyh2r LYS::GAL1UAS-HIS3TATA-HIS3 URA3::GAL1UAS-GAL1TATA-lacZ) was used to detect the interaction between UGT1 and Rop1. Yeast cells were selected on synthetic complete (SC) medium lacking specific amino acids and/or uracil supplemented with 2% glucose or 2% galactose (w/v).
Screening of the Arabidopsis cDNA Library for Interaction Clones
The Arabidopsis cDNA library, made in pJG4-5 from mRNAs of 6- to 8-day-old seedlings, was provided by Dr. H. Goodman (Massachusetts General Hospital, Boston). The library plasmids were amplified in E. coli Top10F′ cells and purified by CsCl gradient. The bait plasmid pEG-Phr was constructed by inserting the coding region of the soybean phragmoplastin cDNA (pSDL12; Gu and Verma, 1996) at the BamHI-XhoI sites of pEG202 (Golemis et al., 1996). Yeast EGY48 cells containing the reporter gene plasmid pSH18-34 were transformed by electroporation with the bait plasmid. The cells were selected in liquid SC medium without uracil and histidine for 12 hr and then propagated in yeast-peptone-dextrose medium to an OD600 of 0.6. Yeast cells were treated with lithium acetate (Golemis et al., 1996) and transformed with the pooled Arabidopsis cDNA library plasmids. Yeast colonies were first selected on SC-glucose medium without uracil, histidine, and tryptophan. The colonies (∼2 million) were mixed and then selected for putative interaction clones on SC-galactose medium followed by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) blue/white assay (Golemis et al., 1996). Plasmids were isolated from positive yeast colonies, and purified plasmids were reintroduced into EGY48 for confirmation of protein interaction. Inserts of the plasmids were sequenced using a Perkin-Elmer automatic DNA sequencer.
Isolation of a Full-Length UGT1-cDNA Clone
The plasmids pPIL33 and pPIL25, originally obtained from library screening in the yeast two-hybrid system, did not contain a full coding region of UGT1. The insert from pPIL33 was radiolabeled with 32P-α-dATP and used to screen an Arabidopsis FL-1 cDNA library constructed in λZapII (Seki et al., 1998; provided by Dr. K. Shinozaki, Institute of Physical and Chemical Research, Tsukuba, Japan). A full-length cDNA, pUGT1f, was obtained and used for protein expression in E. coli and for green fluorescent protein (GFP) fusion in BY-2 cells.
Interaction between UGT1 and Rop1 in the Yeast Two-Hybrid System
The coding region of pUGT1f was amplified by polymerase chain reaction (PCR) using primers 5′-CCGAATTCGTAGAAAAAAAATGGCGCC-3′ and 5′-AGCTCGAGACACCAACTCTTTTGTCC-3′. The PCR product was cloned at the EcoRI-XhoI sites of pACT2, giving rise to pACT2-UGT. Dominant positive and dominant negative mutants of Arabidopsis Rop1 cDNA (Li et al., 1999), cloned in pAS2 vector (Durfee et al., 1993), were made and provided by Dr. Z. Yang (University of California, Riverside). Plasmids were cotransferred into yeast Y190 cells by electroporation and selected on SC-Trp-Leu-His medium containing 30 mg/L 3-amino-1,2,4-triazole (Sigma). Colonies were lifted on 3 MM filter discs, permeated by liquid nitrogen, and incubated in a Z buffer containing X-Gal (120 mM sodium phosphate, pH 7.0, 10 mM KCl, 1 mM MgCl2, 0.2 mM β-mercaptoethanol, and 300 mg/L X-Gal).
Expression of the GST–UGT1 Fusion Protein
The coding region of UGT1 cDNA was released from pACT2-UGT by EcoRI-XhoI and cloned at the same sites of pGEX-KG (Frangioni and Neel, 1993), generating pGST-UGT. Cells expressing glutathione S-transferase (GST)–UGT fusion protein were washed in STE buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 1 mM EDTA) and lysed by incubation with lysozyme (100 μg/mL for 5 min) followed by sonication in the presence of 0.5% sarkosyl (Frangioni and Neel, 1993). The supernatant was diluted with Triton X-100 to a final concentration of 1% and incubated with glutathione–agarose beads (Sigma). The beads were washed extensively with STE buffer and used for both in vitro pull-down assay and purification of UGT1 for raising antibody.
Detection of Protein–Protein Interaction by in Vitro Pull-Down Assay
To confirm the interaction between UGT1 and phragmoplastin, we used agarose beads containing the GST–UGT1 fusion protein to pull down 35S-labeled phragmoplastin peptide. Plasmid pAGA3-SA2 expressing the SA2 domain of phragmoplastin (Zhang et al., 2000) was used with the TNT-T7 Quick-Coupled Transcription/Translation System (Promega) in the presence of 35S-methionine. The pull-down assay was performed as described previously (Zhang et al., 2000). To detect the interaction between UGT1 and Rop1, the coding region of Rop1 cDNA (Li et al., 1998) was released from pGEX-R1A (Li et al., 1999) by EcoRI-SacI digestion and subcloned into the same sites of pGEM-3Zf. The resulting plasmid, pGEM-Rop1, was cut with NcoI-SalI, and the insert was ligated to the same sites of pAGA3. The final plasmid, pAGA3-Rop1, was used to produce 35S-labeled Rop1 peptide, which then interacted with the GST–UGT1 fusion protein absorbed to the agarose beads.
Antibody Production and Purification
The GST–UGT1 fusion protein purified on the glutathione–agarose beads was digested with thrombin (2 units/mL; Sigma) in buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5 mM CaCl2, and 0.1% β-mercaptoethanol). The cleaved products were resolved by 10% SDS-PAGE. UGT1 bands from 10 gels were excised, lyophilized, and used to raise antibodies. For the purification of UGT1 IgG from serum, the GST–UGT1 fusion protein was resolved by PAGE and transferred to nitrocellulose membranes. The membrane was stained for protein with Ponceau S solution (0.2% Ponceau S in 3% trichloroacetic acid and 3% sulfosalicylic acid) for 5 min. The filter corresponding to the UGT1 band was cut and blocked with 5% nonfat milk in PBS (0.8% NaCl and 0.02% KCl in 12 mM Na2HPO4, pH 7.2). Rabbit serum against UGT1 was diluted with 2 volumes of PBS and incubated with the nitrocellulose filters containing UGT1 protein. After extensive washing with PBS, IgG bound to the membrane was eluted with 100 mM glycine, pH 2.5, and the eluate was neutralized immediately with 1 M Tris solution, pH 8.0.
Membrane Preparation, Product Entrapment, and Sucrose Gradient Analysis
Root tips of 4-day-old dark-grown moth bean (Vigna aconitifolia) seedlings or tobacco BY-2 cells were harvested into liquid nitrogen and homogenized in buffer (50 mM Hepes/KOH, pH 7.3, 5 mM EDTA, 1 mM DTT, and 20 μM phenylmethylsulfonyl fluoride [PMSF]) using a Brinkmann Instruments homogenizer. The homogenate was filtered through three layers of Miracloth and centrifuged at 10,000g for 20 min at 4°C. The supernatant was collected, and membranes were pelleted by ultracentrifugation at 100,000g for 1 hr at 4°C. The membranes were extracted with a detergent buffer (50 mM Tris-Cl, pH 7.5, 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid [Chaps], 0.1% cholesteryl hemisuccinate, 4 μM GTPγS, and 0.5 mM PMSF) on ice for 20 min. Supernatant was collected after centrifugation at 100,000g for 30 min, and reagents for callose production were added to a final concentration of 0.6 mM CaCl2, 10 mM cellobiose, and 5 mM UDP-glucose (Inoue et al., 1995). The reaction was incubated at 25°C for 1 hr, and callose polymer was collected by low-speed centrifugation (1500g for 5 min). The pellet was washed three times in buffer A (50 mM Tris-Cl, pH 7.5, 33% glycerol, 0.4% Chaps, 0.08% cholesteryl hemisuccinate, and 4 μM GTPγS) containing 5 mM UDP-glucose. The pellet was resuspended in buffer A without UDP-glucose, and the callose synthase (CalS) activity was recovered in the supernatant by ultracentrifugation at 200,000g for 10 min. The supernatant was layered onto a continuous sucrose density gradient (20 to 60% [w/v] sucrose in 10 mM Tris-Cl, pH 7.5). Centrifugation was performed at 100,000g for 4 hr at 4°C using a Beckman Instruments SW-55 rotor. Fractions were collected and assayed for CalS activity. The peak fractions were pooled, dialyzed, concentrated, and then analyzed on SDS gels and by protein gel blotting.
CalS Assay
CalS activity was measured using the microtiter-based fluorescence assay developed by Shedletzky et al. (1997). Enzyme assays (50 μL) in a flat-bottom, 96-well microtiter plate contained 50 mM Hepes/KOH, pH 7.3, 0.6 mM CaCl2, 10 mM cellobiose, 2 mM UDP-glucose, and 30 μL of sucrose gradient fraction material. Plates were incubated at 25°C for 1 hr, and the reactions were terminated by the addition of 10 μL of 6 N NaOH. Callose was solubilized by incubating the microtiter plate in a water bath at 80°C for 30 min. Callose was detected by the addition of 210 μL of aniline blue mix (40 volumes of 0.1% aniline blue in water, 21 volumes of 1 N HCl, and 59 volumes of 1 M glycine, pH 9.5) and incubated for 30 min at 50°C and then for 30 min at room temperature. Fluorescence was quantified using the Cytofluor 2350 Fluorescence Measurement System (Millipore, Bedford, MA) at an excitation wavelength of 365 nm, a bandwidth of 40 nm, and an emission wavelength of 456 nm.
Preparation of Plant Extracts and Protein Gel Blotting
One-week-old Arabidopsis seedlings frozen in liquid nitrogen were ground with mortar and pestle into a powder that was resuspended in extraction buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 2 mM β-mercaptoethanol, and 1 mM PMSF). The homogenate was filtered through two layers of Miracloth and centrifuged at 10,000g for 15 min. The supernatant (total protein extract) was further separated into soluble fraction and membrane pellet by ultracentrifugation at 100,000g for 45 min. The membrane pellet was extracted with detergent solution (1% Triton X-100 in extraction buffer), a chaotropic agent (0.5 M NaI in extraction buffer), or alkaline solution (100 mM Na2CO3, pH 11.0). Fractions were adjusted to 1 × SDS loading buffer (2% SDS, 80 mM Tris, pH 6.8, 10% glycerol, 100 mM DTT and 0.002% bromophenol blue) and resolved by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and probed with purified UGT1 IgG. Horseradish peroxidase–labeled goat antibody against rabbit IgG was used as a secondary antibody, and the signals were visualized on x-ray films using the enhanced chemiluminescence protein detection system (Amersham).
Construction of the GFP–UGT1 Fusion Protein
To facilitate the cloning and expression of GFP fusion proteins in plants, we removed the stop codon (TGA) of the GFP coding region in pMON30060 (Pang et al., 1996). The GFP coding region (NcoI-EcoRI fragment) was replaced with an NcoI-EcoRI restricted PCR product that was amplified using primers 5′-AACCATGGGCAAGGGCGAGG-3′ and 5′-CGGAATTCGAAGCTTGTAGAGTTCATCCATGCC-3′. The resulting plasmid, pMON-GFP, contains an enhanced cauliflower mosaic virus 35S promoter, an untranslated leader sequence from the GmHSP17.9 cDNA, the GFP coding sequence with HindIII and EcoRI for protein fusion, and a NOS-3′ terminator. The UGT1 coding region amplified by PCR as described above was cloned in pT-Adv (Clontech, Palo Alto, CA). The insert was released by EcoRI and subcloned into the same site of pMON-GFP. The entire cassette, including the promoter, the fusion protein coding region, and the terminator, was released by NotI and subcloned into the same site of the binary vector pMON18342 (Pang et al., 1996). The final plasmid, pMBIN-UGT1, was transferred to Agrobacterium tumefaciens (strain ABI) by electroporation.
Plant Transformation and Detection of GFP–UGT1
Tobacco BY-2 cells were transformed via A. tumefaciens–mediated procedures (Gu and Verma, 1997). Localization of the GFP fusion protein was performed using an epifluorescence microscope (Zeiss Axiophot, Jena, Germany) with appropriate filters for GFP (Pang et al., 1996).
NOTE ADDED IN PROOF
Recent phylogenetic and enzymatic analyses of all Arabidopsis glycosyltransferases placed UGT1 (UGT75B1) and UGT2 (UGT75B2) into Group L, which includes indole acetic acid UDP-glucose transferase (UGT84B1). UGT1 and UGT2 did not show significant enzymatic activity toward indoleacetic acid or other substrates examined (Li et al. [2001]. J. Biol. Chem. 276, 4338–4343; Rosamond et al. [2001]. J. Biol. Chem. 276, 4350–4356).
Acknowledgments
We thank Dr. Z. Yang for the provision of Arabidopsis Rop1 plasmids, Drs. H. Goodman and K. Shinozaki for the gifts of Arabidopsis cDNA libraries, and Dr. S. Pang for GFP plasmids. We also thank Dr. A. Delauney for his comments on this work. This work was supported by National Science Foundation Grant No. IBN-9724014.
References
- Amor, Y., Haigler, C.H., Johnson, S., Wainscott, M., and Delmer, D.P. (1995). A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. USA 92, 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., Davies, C., and Robinson, S.P. (1996). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111, 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.M., Jr., Saxena, I.M., and Kudlicka, K. (1996). Cellulose biosynthesis in higher plants. Trends Plant Sci. 1, 149–156. [Google Scholar]
- Delmer, D.P. (1999). Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276. [DOI] [PubMed] [Google Scholar]
- Dupree, P., and Sherrier, D.J. (1998). The plant Golgi apparatus. Biochim. Biophys. Acta 1404, 259–270. [DOI] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y., Kilburn, A.E., Lee, W.H., and Elledge, S.J. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V., Furtek, D.B., and Nelson, O.E. (1984). Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element activator (Ac). Proc. Natl. Acad. Sci. USA 81, 3825–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, C.M., Boss, P.K., and Hoj, P.B. (1998). Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J. Biol. Chem. 273, 9224–9233. [DOI] [PubMed] [Google Scholar]
- Frangioni, J.V., and Neel, B.G. (1993). Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210, 179–187. [DOI] [PubMed] [Google Scholar]
- Golemis, E.A., Gyuris, H., and Brent, R. (1996). Interaction trap/two hybrid system to identify interacting proteins. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley), pp. 20.1.1–20.1.28.
- Graham, R., and Thornburg, R. (1997). DNA sequence of UDP glucose:indole-3-acetate β-d-glucosyltransferase from Arabidopsis thaliana. Plant Physiol. 113, 1004. [Google Scholar]
- Gu, X., and Verma, D.P.S. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15, 695–704. [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P.S. (1997). Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell 9, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne, J.B., and Williams, C.A. (1988). Flavone and flavonol glycosides. In The Flavonoids, J.B. Harborne, ed (London: Chapman and Hall), pp. 303–328.
- Holland, N., Holland, D., Helentjaris, T., Dhugga, K.S., Xoconostle-Cazares, B., and Delmer, D.P. (2000). A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol. 123, 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., Delauney, A.J., and Verma, D.P.S. (2001). A cell plate–specific callose synthase and its interaction with phragmoplastin. Plant Cell 13, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S.B., Takewaki, N., Takasuka, T., Mio, T., Adachi, M., Fujii, Y., Miyamoto, C., Arisawa, M., Furuichi, Y., and Watanabe, T. (1995). Characterization and gene cloning of 1,3-β-d-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 231, 845–854. [DOI] [PubMed] [Google Scholar]
- Jacob, S.R., and Northcote, D.H. (1985). In vitro glucan synthesis by membranes of celery petioles: The role of the membrane in determining the type of linkage formed. J. Cell Sci. 2 (suppl.), 1.–11. [DOI] [PubMed] [Google Scholar]
- Kamat, U., Garg, R., and Sharma, C.B. (1992). Purification to homogeneity and characterization of a 1,3-β-glucan (callose) synthase from germinating Arachis hypogaea cotyledons. Arch. Biochem. Biophys. 298, 731–739. [DOI] [PubMed] [Google Scholar]
- Kita, M., Hirata, Y., Moriguchi, T., Endo-Inagaki, T., Matsumoto, R., Hasegawa, S., Suhayda, C.G., and Omura, M. (2000). Molecular cloning and characterization of a novel gene encoding limonoid UDP-glucosyltransferase in Citrus. FEBS Lett. 469, 173–178. [DOI] [PubMed] [Google Scholar]
- Lee, H.I., and Raskin, I. (1999). Purification, cloning, and expression of a pathogen inducible UDP-glucose:salicylic acid glucosyltransferase from tobacco. J. Biol. Chem. 274, 36637–36642. [DOI] [PubMed] [Google Scholar]
- Li, H., Wu, G., Ware, D., Davis, K.R., and Yang, Z. (1998). Arabidopsis Rho-related GTPases: Differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 118, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer, C.E. (1993). Targeting and retention of Golgi membrane proteins. Curr. Opin. Cell Biol. 5, 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, P.I., et al. (1997). The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7, 255–269. [DOI] [PubMed] [Google Scholar]
- Martin, R.C., Mok, M.C., and Mok, D.W. (1999). Isolation of a cytokinin gene, ZOG, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc. Natl. Acad. Sci. USA 96, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehs, C.P., Allen, P.V., Friedman, M., and Belknap, W.R. (1997). Cloning and expression of solanidine UDP-glucose glucosyltransferase from potato. Plant J. 11, 227–236. [DOI] [PubMed] [Google Scholar]
- Nakai, T., Tonouchi, N., Konishi, T., Kojima, Y., Tsuchida, T., Yoshinaga, F., Sakai, F., and Hayashi, T. (1999). Enhancement of cellulose production by expression of sucrose synthase in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA 96, 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Frohlick, J.A., and Staehelin, L.A. (2000). Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 124, 135–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, S.Z., DeBoer, D.L., Wan, Y., Ye, G., Layton, J.G., Neher, M.K., Armstrong, C.L., Fry, J.E., Hinchee, M.A., and Fromm, M.E. (1996). An improved green fluorescent protein gene as a vital marker in plants. Plant Physiol. 112, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota, H., Python, C.P., Inoue, S.B., Arisawa, M., Anraku, Y., Zheng, Y., Watanabe, T., Levin, D.E., and Ohya, Y. (1996). Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272, 279–281. [DOI] [PubMed] [Google Scholar]
- Samuels, A.L., Giddings, T.H., and Staehelin, L.A. (1995). Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, I.M., Brown, R.M., Jr., Fevre, M., Geremia, R.A., and Henrissat, B. (1995). Multidomain architecture of β-glycosyl transferases: Implications for mechanism of action. J. Bacteriol. 177, 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Carninci, P., Nishiyama, Y., Hayashizaki, Y., and Shinozaki, K. (1998). High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15, 707–720. [DOI] [PubMed] [Google Scholar]
- Shedletzky, E., Unger, C., and Delmer, D.P. (1997). A microtiter-based fluorescence assay for (1,3)-β-glucan synthases. Anal. Biochem. 249, 88–93. [DOI] [PubMed] [Google Scholar]
- Sparvoli, F., Martin, C., Scienza, A., Gavazzi, G., and Tonelli, C. (1994). Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol. Biol. 24, 743–755. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos, S.J., Fisher, P.R., Stone, B.A., and Stanisich, V.A. (1999). Detection of two loci involved in (1→3)-β-glucan (curdlan) biosynthesis by Agrobacterium sp. ATCC31749, and comparative sequence analysis of the putative curdlan synthase gene. Glycobiology 9, 31–41. [DOI] [PubMed] [Google Scholar]
- Szerszen, J.B., Szczyglowski, K., and Bandurski, R.S. (1994). iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265, 1699–1701. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, A., Bacic, A., Harris, P.J., and Read, S.M. (1998). Membrane fractionation and enrichment of callose synthase from pollen tubes of Nicotiana alata Link et Otto. Planta 205, 380–388. [DOI] [PubMed] [Google Scholar]
- Verma, D.P.S. (2001). Cytokinesis and building of the cell plate in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 751–784. [DOI] [PubMed] [Google Scholar]
- Warnecke, D.C., Baltrusch, M., Buck, F., Wolter, F.P., and Heinz, E. (1997). UDP-glucose:sterol glucosyltransferase: Cloning and functional expression in Escherichia coli. Plant Mol. Biol. 35, 597–603. [DOI] [PubMed] [Google Scholar]
- Yamazaki, M., Gong, Z., Fukuchi-Mizutani, M., Fukui, Y., Tanaka, Y., Kusumi, T., and Saito, K. (1999). Molecular cloning and biochemical characterization of a novel anthocyanin 5-O-glucosyltransferase by mRNA differential display for plant forms regarding anthocyanin. J. Biol. Chem. 274, 7405–7411. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Hong, Z., and Verma, D.P.S. (2000). Phragmoplastin polymerizes into spiral coiled structures via intermolecular interaction of two self-assembly domains. J. Biol. Chem. 275, 8779–8784. [DOI] [PubMed] [Google Scholar]