Abstract
The Aux/IAA genes are rapidly and specifically induced by the plant hormone auxin. The proteins encoded by this gene family are short-lived nuclear proteins that are capable of homodimerizing and heterodimerizing. Molecular, biochemical, and genetic data suggest that these proteins are involved in auxin signaling. The pleiotropic morphological phenotype and altered auxin responses of the semidominant axr3-1 mutant of Arabidopsis result from a single amino acid change in the conserved domain II of the Aux/IAA protein IAA17. Here, we show that the biochemical effect of this gain-of-function mutation is to increase the half-life of the iaa17/axr3-1 protein by sevenfold. Intragenic mutations that suppress the iaa17/axr3-1 phenotype have been described. The iaa17/axr3-1R3 revertant contains a second site mutation in domain I and the iaa17/axr3-1R2 revertant contains a second site mutation in domain III. Transient expression assays show that the mutant forms of IAA17/AXR3 retain the ability to accumulate in the nucleus. Using the yeast two hybrid system, we show that the iaa17/axr3-1 mutation does not affect homodimerization. However, the iaa17/axr3-1 revertants counteract the increased levels of iaa17/axr3-1 protein by decreasing the capacity of the mutant protein to homodimerize. Interestingly, heterodimerization of the revertant forms of IAA17/AXR3 with IAA3/SHY2, another Aux/IAA protein, and ARF1 or ARF5/MP proteins is affected only by changes in domain III. Collectively, the results provide biochemical evidence that the revertant mutations in the IAA17/AXR3 gene affect the capacity of the encoded protein to dimerize with itself, other members of the Aux/IAA protein family, and members of the ARF protein family. By extension, these findings may provide insight into the effects of analogous mutations in other members of the Aux/IAA gene family.
INTRODUCTION
The plant hormone auxin, typified by indoleacetic acid, regulates a broad range of cellular and physiological processes in response to abiotic and biotic stimuli (Davies, 1995). Auxin transcriptionally activates a select set of immediate early genes that are thought to mediate processes ranging from embryo formation to tropic responses. Data gathered from several species have resulted in the classification of three groups of auxin early response genes: the Aux/IAA gene family, the GH3 gene family, and the SAUR gene family (Abel and Theologis, 1996). Of these, the protein products of the Aux/IAA genes are the best characterized. Biochemical, molecular, and genetic studies suggest that the Aux/IAA proteins play a central role in auxin signaling and plant development.
The large family of Aux/IAA genes encode short-lived, nucleus-localized proteins that contain four highly conserved domains (I, II, III, and IV) (Abel et al., 1994, 1995; Abel and Theologis, 1995; Kim et al., 1997). Domain III of these proteins contains a predicted βαα protein fold that is found in the prokaryotic transcriptional repressors Arc and MetJ (Abel and Theologis, 1995). In the prokaryotic proteins, this domain is involved in dimerization and DNA binding (Raumann et al., 1994). Although specific DNA binding by the Aux/IAA proteins has not been demonstrated, domains III and IV mediate homodimerization and heterodimerization among members of this protein family (Kim et al., 1997).
Most members of the auxin response factor (ARF) family of proteins also contain domains III and IV at their C termini (Guilfoyle et al., 1998). The Aux/IAA proteins can heterodimerize with the ARF proteins through interactions mediated by these conserved domains (Kim et al., 1997; Ulmasov et al., 1997b; Guilfoyle et al., 1998). In addition, the ARFs are capable of binding synthetic and natural auxin responsive promoter elements through a VP1-B3 DNA binding domain located at their N termini (Ulmasov et al., 1997a, 1997b, 1999).
Although the Aux/IAA proteins were initially identified by molecular and biochemical methods, three semidominant Arabidopsis mutants, axr3, axr2, and shy2, have been shown to carry lesions in IAA17, IAA7, and IAA3, respectively (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000). The iaa17/axr3, iaa7/axr2, and iaa3/shy2 plants exhibit altered auxin responses and pleiotropic morphological phenotypes. The mutations in these three genes alter amino acids in the conserved qvVGWPPvrsyRkN motif found in domain II of all Aux/IAA proteins. In addition, intragenic mutations that suppress the mutant phenotypes to varying degrees have been described (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000). Of the five intragenic revertant alleles of iaa17/axr3-1 (Rouse et al., 1998), three contain a second site mutation that causes a single amino acid substitution. The iaa17/axr3-1R1 revertant leads to a P to S substitution in domain III, the iaa17/axr3-1R2 revertant causes a D to N substitution in domain III, and the iaa17/axr3-1R3 revertant leads to an L to F change in domain I. The other two revertants affect splice sites and result in either increased spacing between domains III and IV (iaa17/axr3-1R5) or deletion of half of domain IV (iaa17/axr3-1R4) (Rouse et al., 1998).
The gain-of-function phenotype seen in the iaa17/axr3, iaa7/axr2, and iaa3/shy2 mutants suggests that changes in domain II are likely to be hypermorphic because they increase the stability of these proteins (Leyser et al., 1996; Tian and Reed, 1999; Nagpal et al., 2000). Recent experiments have shown that domain II of the pea PS-IAA4/5 confers instability on the luciferase (LUC) reporter protein in transient assays, providing additional evidence that domain II may fulfill an important regulatory role in controlling the half-lives of Aux/IAA proteins (Worley et al., 2000). Here, we examine the in vivo effects of mutations in iaa17/axr3 on IAA17/AXR3 protein function. Using the three assays that have been used to characterize the Aux/IAA proteins, we show that the iaa17/axr3-1 mutation has a dramatic effect on the stability of IAA17/AXR3 but does not abolish its accumulation in the nucleus or its ability to form protein–protein interactions. In contrast, the capacity of the revertant forms of iaa17/axr3 protein to homodimerize and heterodimerize is dramatically affected. These experiments show that the fine control of Aux/IAA protein levels and protein–protein interactions is critical for normal plant development and proper auxin responses.
RESULTS
Effect of the iaa17/axr3-1 Mutation on IAA17/AXR3 Transcription
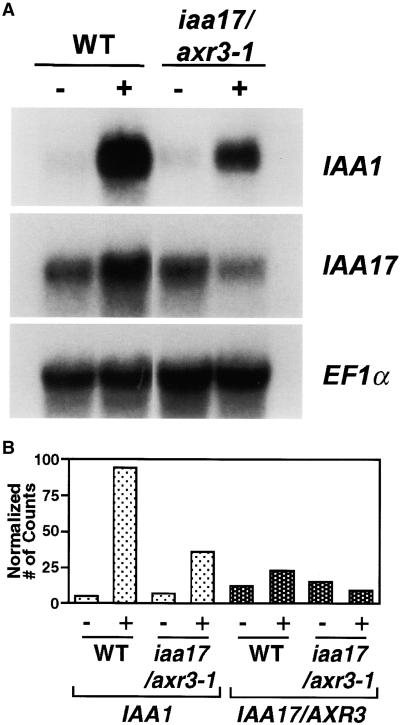
To determine the effect of the iaa17/axr3-1 mutation on Aux/IAA gene expression, RNA gel blot analysis was performed using RNA from etiolated seedlings that had been mock treated or treated with 20 μM IAA. Figure 1 shows that in untreated seedlings the level of IAA17/AXR3 mRNA is higher than the level of IAA1 mRNA. When wild-type seedlings are treated with auxin, the mRNA of both genes accumulates. Because the amount of IAA1 mRNA in untreated tissue is lower than IAA17/AXR3 and the amount of IAA1 mRNA in auxin-treated tissue is higher than IAA17/AXR3, the magnitude of the auxin response is much greater for IAA1 than IAA17/AXR3.
Figure 1.
RNA Gel Blot Analysis of IAA17/AXR3 Expression in Wild-Type and iaa17/axr3-1 Plants.
(A) Total RNA was isolated from 7-day-old wild-type (WT) or iaa17/axr3-1 etiolated seedlings that had been mock treated (−) or treated with 20 μM IAA for 2 hr (+). Twenty micrograms of glyoxalated RNA was separated by electrophoresis, blotted onto a GeneScreen membrane, and probed with gene-specific probes for IAA1 or IAA17/AXR3. Hybridization with elongation factor 1α probe (EF1α) was used to ensure equal loading.
(B) Relative intensity of IAA1 and IAA17/AXR3 transcript levels as shown in (A). The IAA1 and IAA17/AXR3 mRNA levels were normalized to the EF1α transcript levels after quantification using the STORM phosphorimager system. The normalized number of counts was calculated by dividing the number of counts present in the IAA1 or IAA17/AXR3 bands by the number of counts present in the corresponding EF1α band. These values were then multiplied by 100 and used to generate the graph.
In untreated iaa17/axr3-1 seedlings, the mRNA levels of IAA1 and IAA17/AXR3 are similar to untreated wild-type seedlings. When iaa17/axr3-1 seedlings are treated with auxin, however, IAA1 and IAA17/AXR3 show very different responses. The IAA1 mRNA level is still inducible by auxin, but compared with wild type, the amplitude of the response is less (Figure 1). This attenuation of auxin-inducible expression of Aux/IAA genes has been observed in other auxin response mutants (Abel et al., 1995). In contrast, the level of IAA17/AXR3 mRNA is repressed slightly by auxin treatment of the iaa17/axr3-1 seedlings (Figure 1). The differential response of these two genes suggests that only one of multiple pathways leading to auxin-induced transcription is affected by the iaa17/axr3-1 mutation.
The iaa17/axr3-1 Mutation Affects Protein Stability
The pleiotropic effects of the iaa17/axr3-1 mutation on auxin-regulated gene expression and plant development (Figure 1) (Leyser et al., 1996) led us to examine the biochemical nature of lesions in IAA17/AXR3. To specifically and directly assess the level and stability of IAA17/AXR3 in planta, anti-peptide antibodies were generated against a unique region of the protein that lies between domains I and II. Immunoblot analysis of recombinant (HIS)6-tagged IAA17/AXR3 or iaa17/axr3-1 showed that both the wild-type and mutant forms of the protein were detected with equal affinity by these antibodies (data not shown). Although the antibodies could detect ⩽20 ng of protein, they could not detect IAA17/AXR3 on an immunoblot of total soluble Arabidopsis proteins isolated from either mock-treated or IAA-treated tissues. These results indicate that IAA17/AXR3 is a very low abundance protein.
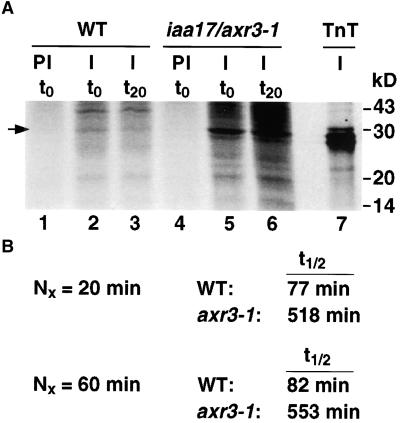
Because of the low level of IAA17/AXR3, in vivo labeling of proteins followed by immunoprecipitation was used as a more sensitive assay. The specificity and utility of the antibody was verified by immunoprecipitation of in vitro transcribed and translated protein. Figure 2A shows a representative result of the pulse–chase experiments. The anti-IAA17/AXR3 peptide antibody (lanes 2, 3, 5, and 6) immunoprecipitates a protein of the same size as the in vitro transcribed and translated IAA17/AXR3 (lane 7). The preimmune serum does not precipitate any proteins in this size range (lanes 1 and 4). The amount of IAA17/AXR3 remaining after a 20- or 60-min chase period was determined using a phosphorimager. In wild-type plants, the half-life of IAA17/AXR3 is ∼80 min (Figure 2B). In contrast, the half-life of the mutant iaa17/axr3-1 is ∼550 min (Figure 2B). This sevenfold increase in half-life demonstrates directly that the mutation in domain II of IAA17/AXR3 increases the stability of the protein.
Figure 2.
Pulse–Chase Analysis of the IAA17/AXR3 Protein in Wild-Type and iaa17/axr3-1 Plants.
(A) Intact 7-day-old etiolated seedlings were labeled with 35S-methionine for 2 hr. The tissue was rinsed and either harvested (t0, lanes 1, 2, 4, and 5) or allowed to incubate in a buffer containing a 1000-fold excess of cold methionine and cysteine for 20 min (t20, lanes 3 and 6). The tissue was frozen in liquid N2, ground, and lyophilized. Proteins were extracted and immunoprecipitated with protein A–purified preimmune serum (PI, lanes 1 and 4) or with affinity-purified anti-IAA17/AXR3 antibody (I, lanes 2, 3, 5, 6, and 7). The protein sample in lane 7 was generated by a coupled in vitro transcription and translation (TnT) reaction in the presence of 35S-methionine using the IAA17/AXR3 cDNA as template. The precipitated proteins were separated by SDS-PAGE and detected using the STORM phosphorimager system. The arrow indicates the position of IAA17/AXR3. Numbers at the right denote molecular mass in kilodaltons (kD).
(B) Determination of the half-life (t1/2) of IAA17/AXR3 and iaa17/axr3-1. In two separate experiments, the amount of radioactivity present in the immunoprecipitated IAA17/AXR3 bands at t0 or after either a 20- min (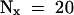 ) or 60-min (
) or 60-min (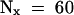 ) chase period was determined using the STORM phosphorimager system. These values were used to calculate the half-lives of IAA17/AXR3 and iaa17/axr3-1 based on the formula
) chase period was determined using the STORM phosphorimager system. These values were used to calculate the half-lives of IAA17/AXR3 and iaa17/axr3-1 based on the formula 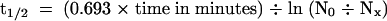 , where N0 is the amount of protein at t0 and Nx is the amount of protein remaining after a given chase time.
, where N0 is the amount of protein at t0 and Nx is the amount of protein remaining after a given chase time.
The Intragenic Suppressors of iaa17/axr3-1 Accumulate in the Nucleus
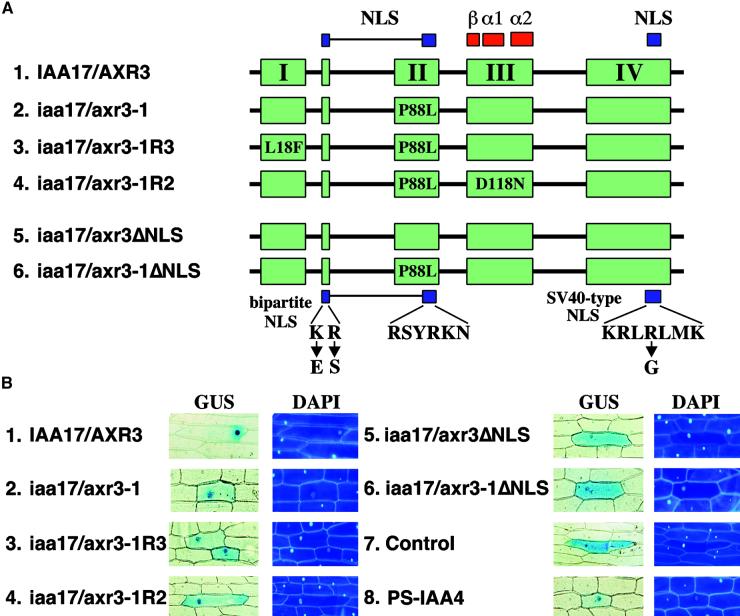
Because the intragenic revertants suppress the iaa17/axr3-1 phenotype, the second site mutations must counteract the effects of enhanced protein stability conferred by the mutation in domain II. Given that the Aux/IAA proteins normally accumulate in the nucleus, one mechanism for suppressing the activity of the stabilized protein would be to prevent the collection of iaa17/axr3-1 in the nucleus. To test this possibility we used transient expression in onion epidermal cells to assess the ability of wild-type and mutant IAA17/AXR3 to accumulate in the nucleus. Figure 3A shows a scheme of the conserved domains of IAA17/AXR3; the conserved clusters of basic amino acids that function as nuclear localization signals (NLS) are highlighted in blue (Abel et al., 1994; Abel and Theologis, 1995; Kim et al., 1997).
Figure 3.
Nuclear Accumulation of Wild-Type and Mutant Forms of the IAA17/AXR3 Protein.
(A) Scheme of the proteins analyzed. The four conserved domains (I, II, III, and IV) present in the Aux/IAA protein family are shown in green. The two NLS found in these proteins are shown in blue. The location of the prokaryotic βαα protein motif is shown in red. The positions and substitutions of amino acids found in the iaa17/axr3-1 and revertant proteins are shown. The changes corresponding to the mutations were introduced into the IAA17/AXR3 cDNA using the QuikChange method (Stratagene). Shown at the bottom are the substitutions introduced to disrupt the NLS (ΔNLS) of IAA17/AXR3 and iaa17/axr3-1. The different cDNAs were fused in frame with the 3′ end of the GUS cDNA.
(B) Intracellular distribution of GUS fusion proteins. Onion epidermal cells were transformed by microprojectile bombardment using gold particles coated with plasmid DNA. After recovery and staining, cells showing GUS activity were photographed using bright-field optics (GUS); the fluorescence micrographs show the positions of the nuclei in the same cells after staining with 4′-6-diamidino-2-phenylindole (DAPI). A plasmid expressing GUS without a functional NLS (Control) was used as a negative control, and a plasmid expressing the nucleus-localized GUS::PS-IAA4 fusion (PS-IAA4) was used as a positive control.
Plasmids expressing different forms of the IAA17/AXR3 fused to the C terminus of β-glucuronidase (GUS) were introduced by particle bombardment followed by staining to ascertain the subcellular location of the chimeric protein. The pRTL2-GUS plasmid, which encodes a GUS protein lacking an NLS, and the pGUS::PS-IAA4 plasmid were used as controls (Figure 3B, panels 7 and 8) (Carrington et al., 1991; Abel and Theologis, 1995). Both the IAA17/AXR3 and iaa17/axr3-1 proteins accumulate in the nucleus (Figure 3B, panels 1 and 2). The accumulation of iaa17/axr3-1 in the nucleus shows that the mutation in domain II does not abolish the subcellular localization of the mutant protein, although more cytoplasmic staining is seen.
IAA17/AXR3 or iaa17/axr3-1 lacking a functional NLS accumulates throughout the cell (Figure 3B, panels 5 and 6). As such, the accumulation of iaa17/axr3-1 in the nucleus is not the result of its association with an endogenous Aux/IAA protein containing a functional NLS. Together, these data show that IAA17/AXR3 and iaa17/axr3-1 accumulate in the nucleus and that this targeting is dependent on the conserved clusters of basic amino acids found in the Aux/IAA protein family.
To determine whether the second site mutations affect the nuclear accumulation of iaa17/axr3-1, the IAA17/AXR3 cDNA was modified to express the substituted amino acids found in the iaa17/axr3-1R3 and iaa17/axr3-1R2 proteins (Figure 3A). Figure 3B (panels 3 and 4) shows that both iaa17/axr3-1R3 and iaa17/axr3-1R2 retain the ability to collect in the nucleus, demonstrating that the reverting effects of the second site mutation are not caused by preventing protein accumulation in the nucleus. The data shown are representative of several experiments and numerous transformed cells stained under a variety of conditions including changes in time and temperature. The minor increase in cytoplasmic staining seen with the iaa17/axr3-1 and revertant proteins (Figure 3B, panels 2, 3, and 4) may reflect a slight decrease in targeting efficiency, but is unlikely to be sufficient to explain the revertant phenotype.
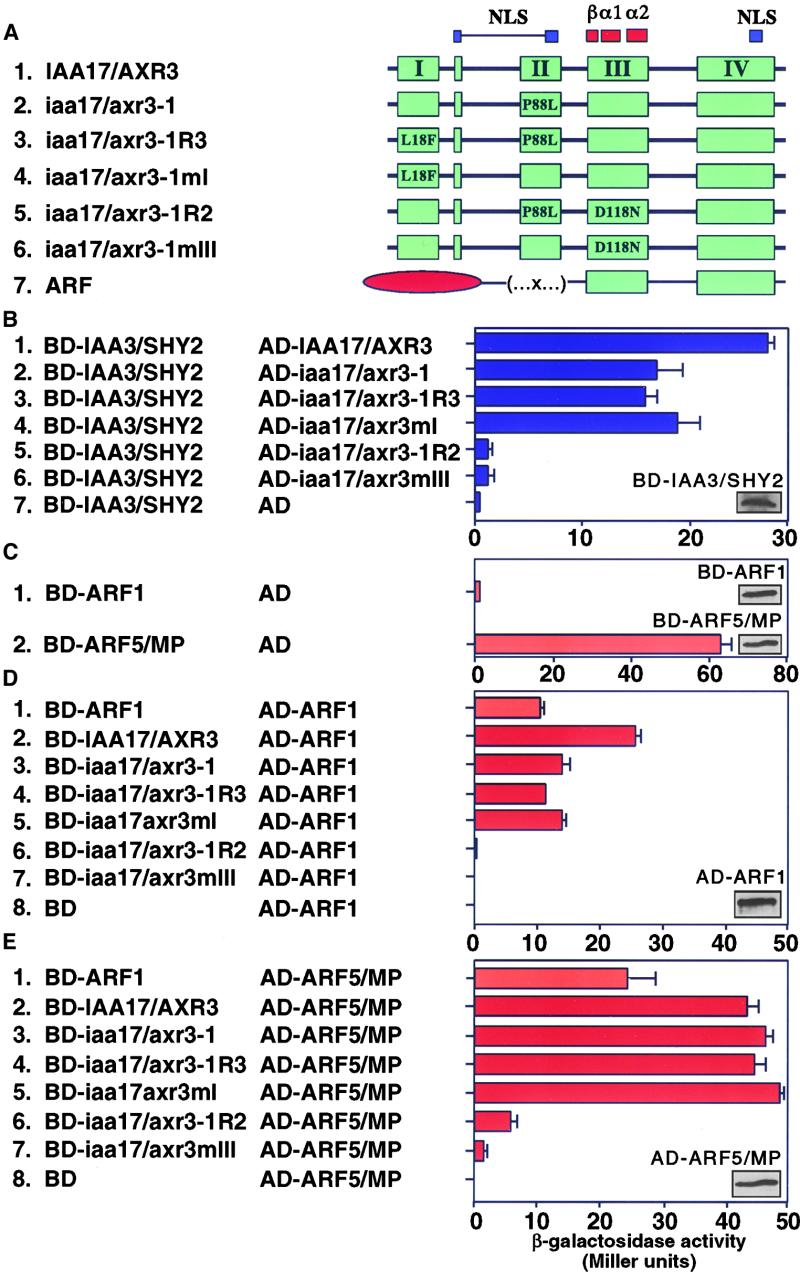
Suppression of the iaa17/axr3-1 Phenotype Is Mediated by Altered Protein–Protein Interactions
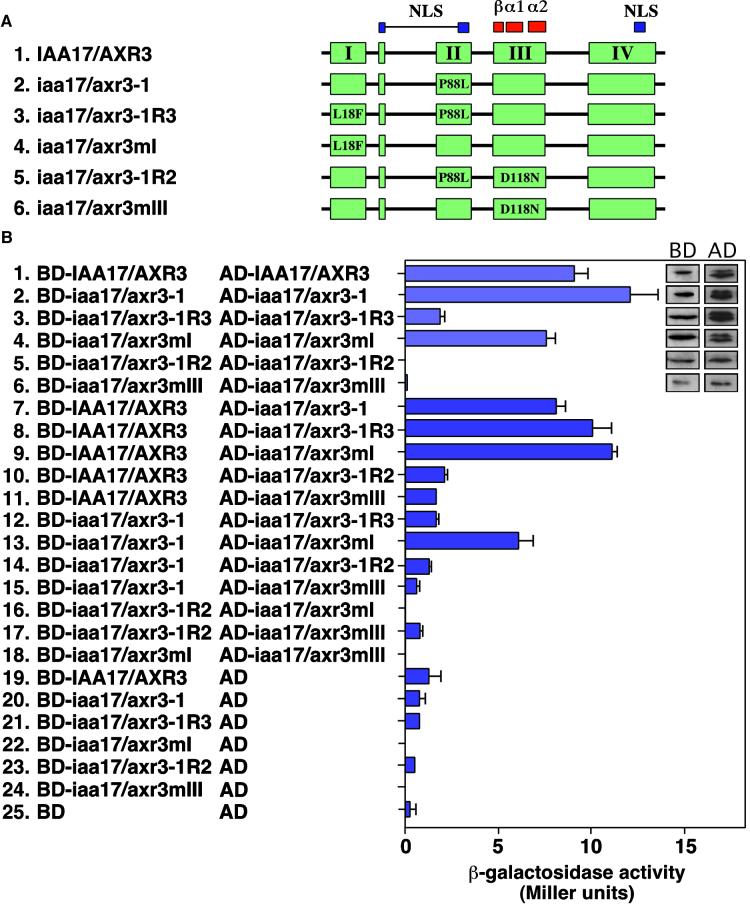
The capacity of Aux/IAA proteins to form intermolecular contacts raises the possibility that the revertant mutations could counteract the increased stability of IAA17/AXR3 by altering its capacity to interact with itself, with other Aux/IAA proteins, or with the ARF proteins. To test these possibilities, we used the yeast two-hybrid system. Figure 4A shows a scheme of the IAA17/AXR3 cDNAs that were fused independently to the C terminus of either the GAL4 activation domain (AD) or the GAL4 DNA binding domain (BD). AD- and BD-containing plasmids were transformed into the yeast strain Y190, which harbors the β-galactosidase and HIS3 reporter genes under the control of the GAL1 promoter. The capacity of each of these BD and AD fusion proteins to interact was determined by monitoring the activation of the two reporter genes. In each case, the ability of the transformed yeast to grow on synthetic medium lacking histidine and supplemented with 3-aminotriazole, a competitive inhibitor of the HIS3 gene product, correlated well with the level of β-galactosidase activity (data not shown). In addition, similar patterns of growth and β-galactosidase activity were observed when the AD and BD were swapped.
Figure 4.
Homodimerization of Wild-Type and Mutant Forms of IAA17/AXR3.
(A) Scheme of the constructs used. The coloring and designation of conserved elements are the same as in Figure 3A. The amino acid changes and their positions in the mutant proteins are indicated. The different cDNAs were fused in frame at the 3′ end of either the GAL4 AD or the GAL4 BD.
(B) Analysis of IAA17/AXR3 homodimerization using the yeast two-hybrid system. Y190 cells transformed with the indicated plasmids were analyzed for the level of β-galactosidase activity (Miller units). The two columns of inset sections show immunoblots demonstrating the expression of different forms of the IAA17/AXR3 protein expressed as fusions with either the GAL4 DNA BD or the GAL4 AD. The level of β-galactosidase activity was determined using orthonitrophenyl-β-d-galactopyranoside as a substrate. The values shown are averages of triplicate assays performed with at least two independent yeast colonies. Error bars represent the standard deviation. AD, plasmid with the GAL4 activation domain alone; BD, plasmid with the GAL4 binding domain alone.
Similar to results obtained with IAA1 and IAA2 (Kim et al., 1997), IAA17/AXR3 is capable of forming homodimers and the iaa17/axr3-1 mutation does not affect this interaction (Figure 4B, panels 1, 2, and 7). Furthermore, proteins with substitutions in domains I and II (iaa17/axr3-1R3) or domain I alone (iaa17/axr3mI) retain the capacity to interact with IAA17/AXR3 (Figure 4B, panels 8 and 9). In contrast, proteins that contain a mutation in domain I show differing capacities to interact with proteins that also contain a mutation in domain II. For instance, if the mutation in domain I is present alone (iaa17/axr3mI), the protein can interact with itself, IAA17/AXR3, or iaa17/axr3-1 (Figure 4B, panels 4, 9, and 13). However, if the mutation in domain I is present as a second site change (iaa17/axr3-1R3), the protein can no longer interact with itself or iaa17/axr3-1 (Figure 4B, panels 3 and 12). Collectively, these results suggest that the reverting mutation in domain I inhibits the homodimerization of proteins that also contain a mutation in domain II. In contrast, IAA17/AXR3 proteins that contain substitutions in domain III alone (iaa17/axr3mIII) or domains II and III (iaa17/axr3-1R2) are unable to interact with IAA17/AXR3, iaa17/axr3-1, or themselves (Figure 4B, panels 5, 6, 10, 11, 14 to 18). These results support the hypothesis that domain III forms a critical contact point for interactions between and among Aux/IAA proteins.
Because the BD fusion proteins alone contain no reporter gene activity (Figure 4B, panels 19 to 25), the data do not merely reflect differences in the capacity of the fusion proteins to activate transcription on their own. Similarly, the inset sections in Figure 4B show that each of the fusion proteins is expressed at a similar level in yeast. These control experiments therefore indicate that the changes in reporter gene activation shown in Figure 4 reflect changes in the ability of the different forms of IAA17/AXR3 to interact.
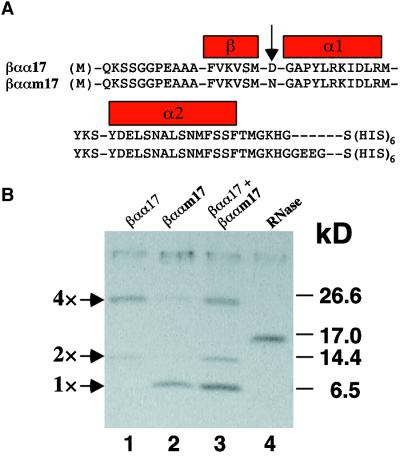
As an independent means to assess whether the mutation in domain III disrupts protein–protein interactions, the βαα domain of IAA17/AXR3 (βαα17) and iaa17/axr3-1R2 (βααm17) were expressed in Escherichia coli as (HIS)6-tagged fusion proteins. This approach has been used to show that the βαα domain contains secondary structure and can form homotypic and heterotypic interactions in vitro (Morgan et al., 1999). Figure 5A shows the amino acid sequence of the polypeptides used and highlights the position of the reverting amino acid substitution between the β sheet and the α1 helix. The (HIS)6-tagged peptides were purified under denaturing conditions, renatured, and analyzed by SDS-PAGE. Figure 5B shows that the renatured wild-type βαα17 polypeptide is capable of forming stable multimers (lane 1). In contrast, the βααm17 polypeptide (lane 2) is present only as a monomer, like the control protein, RNase A (lane 4). Because the recombinant peptides differ in size (Figure 5A), the two proteins were mixed to determine if an additional complex could be detected. The failure to observe a newly migrating complex indicates that these two polypeptides do not interact (lane 3). Together, the yeast two-hybrid and in vitro multimerization data indicate that the iaa17/axr3-1R2 mutation disrupts the ability of domain III to mediate protein–protein interactions.
Figure 5.
In Vitro Dimerization of Domain III Polypeptides.
(A) The cDNAs encoding domain III of IAA17/AXR3 (βαα17; 62 amino acids) and iaa17/axr3-1R2 (βααm17; 66 amino acids) were amplified by polymerase chain reaction and cloned in frame with a methionine codon and a histidine hexapeptide (HIS)6 in the pQE vector series (Qiagen). The arrow points to the single amino acid substitution present in iaa17/axr3-1R2.
(B) After isopropyl-β-d-thiogalactopyranoside induction of protein expression, bacteria were lysed and the proteins were purified under denaturing conditions on a nickel–nitrilotriacetic acid agarose column. After renaturation, the proteins were resolved on a 16.5% Tris–Tricine gel and stained with Coomassie Blue. Numbers at the right denote molecular mass in kilodaltons (kD).
A second set of interactions that might be affected by the amino acid changes introduced by the revertants involves contact between different Aux/IAA proteins. To determine if the mutant forms of IAA17/AXR3 retain the ability to interact with another Aux/IAA protein, we tested their interaction with IAA3/SHY2 using the yeast two-hybrid system. Both IAA17/AXR3 and iaa17/axr3-1 can form heterodimers with IAA3/SHY2 (Figure 6B, panels 1 and 2). Similarly, both iaa17/axr3-1R3 and iaa17/axr3mI are able to interact with IAA3/SHY2 (Figure 6B, panels 3 and 4). In contrast, neither iaa17/axr3-1R2 nor iaa17/axr3mIII is able to interact with IAA3/SHY2 (Figure 6B, panels 5 and 6), suggesting that mutations in domain III eliminate heterodimerization. The BD-IAA3/SHY2 construct by itself does not stimulate any reporter gene expression (Figure 6B, panel 7), even though the inset shows that the protein is expressed. Because each form of IAA17/AXR3 is expressed at similar levels in yeast (Figure 4B), the differences seen in Figure 6B reflect the ability of these proteins to interact rather than differences in protein levels.
Figure 6.
Heterodimerization of the Wild-Type and Mutant Forms of IAA17/AXR3 with IAA3/SHY2, ARF1, or ARF5/MP.
(A) Scheme of the constructs used. The coloring and designation of conserved elements are the same as in Figure 3A. The conserved DNA BD found in the ARF proteins is shown as a red oval. The amino acid changes and their positions in the mutant protein are indicated. The different cDNAs were fused in frame at the 3′ end of either the GAL4 AD or the GAL4 DNA BD.
(B) Analysis of the ability of wild-type and mutant forms of IAA17/AXR3 to heterodimerize with IAA3/SHY2. The inset section shows an immunoblot demonstrating the expression of BD-IAA3/SHY2.
(C) Analysis of the ability of BD-ARF1 and BD-ARF5/MP to activate transcription in Y190 cells. The inset sections show immunoblots demonstrating that the BD-ARF1 and BD-ARF5/MP proteins are expressed at similar levels.
(D) Analysis of the capacity of ARF1 to homodimerize and heterodimerize with the wild-type and mutant forms of IAA17/AXR3. The inset section shows an immunoblot demonstrating the expression of AD-ARF1.
(E) Analysis of the capacity of ARF5/MP to heterodimerize with ARF1 or the wild-type and mutant forms of IAA17/AXR3. The inset shows an immunoblot demonstrating the expression of AD-ARF5/MP.
The level of β-galactosidase activity was determined using orthonitrophenyl-β-d-galactopyranoside as a substrate. The values shown are averages of triplicate assays performed with at least two independent yeast colonies. Error bars represent the standard deviation. AD, plasmid with the GAL4 AD alone; BD, plasmid with the GAL4 BD alone.
A third set of interactions that might be affected by the amino acid changes present in the revertants involves the interaction of IAA17/AXR3 with the ARF proteins. ARF1 was originally identified as a protein that could interact with the synthetic auxin-responsive P3 element (Ulmasov et al., 1997a). Subsequently, the C-terminal region of ARF1 containing the conserved Aux/IAA domains III and IV was shown to mediate interaction with ARF2, IAA4, IAA12, IAA13, and Aux/IAA proteins from soybean (Ulmasov et al., 1997a, 1997b). In control experiments in which BD-ARF1 or BD-ARF5/MP were introduced into Y190, similar levels of BD fusion protein were seen, but vast differences in the level of reporter gene expression were detected (Figure 6C). The BD-ARF5/MP on its own strongly activates reporter gene expression (Figure 6C, panel 2), which prevents the use of this construct in the yeast two-hybrid assay. In contrast, the BD-ARF1 construct shows a much reduced capacity to interact with the yeast transcriptional machinery (Figure 6C, panel 1). Because of the reduced activity of the BD-ARF1 protein, we were able to assess the capacity of the full-length ARF1 to homodimerize and heterodimerize. The full-length ARF1 is able to form a homodimer (Figure 6D, panel 1) and has the capacity to heterodimerize with ARF5/MP (Figure 6E, panel 1).
To test the interaction of the ARFs with wild-type and mutant forms of IAA17/AXR3, we used the yeast two-hybrid system. Both IAA17/AXR3 and the iaa17/axr3-1 can form heterodimers with either ARF1 or ARF5/MP (Figure 6D, panels 2 and 3, and Figure 6E, panels 2 and 3). Similar to the interactions with IAA3/SHY2, both iaa17/axr3-1R3 and iaa17/axr3mI are able to interact with ARF1 and ARF5/MP (Figure 6D, panels 4 and 5, and Figure 6E, panels 4 and 5). In contrast, neither iaa17/axr3-1R2 nor iaa17/axr3mIII are able to heterodimerize with ARF1 or ARF5/MP (Figure 6D, panels 6 and 7, and Figure 6E, panels 6 and 7). Together, these results show that IAA17/AXR3–ARF interactions are mediated through domain III and that the revertants alter the capacity for this interaction to occur. Because the AD-ARF1 and AD-ARF5/MP proteins are expressed at similar levels in yeast (Figures 6D and 6E), the differences in reporter gene activity seen in Figure 6 reflect the capacity of these proteins to interact rather than differences in protein levels.
DISCUSSION
In addition to sharing extensive amino acid similarity, the Aux/IAA proteins have been defined by three criteria: they are short-lived, they are localized in the nucleus, and they have the capacity to form homodimers and heterodimers (Abel et al., 1994; Abel and Theologis, 1995; Oeller and Theologis, 1995; Kim et al., 1997). In this report, we have used these characteristics as functional assays to examine the effect of the iaa17/axr3-1 and intragenic revertant mutations on IAA17/AXR3 protein function.
Pulse–chase experiments show that the half-life of iaa17/axr3-1 is sevenfold longer than the wild-type IAA17/AXR3. The subsequent increase in IAA17/AXR3 protein levels is consistent with the semidominant, hypermorphic nature of the iaa17/axr3-1 mutation (Leyser et al., 1996; Rouse et al., 1998). Semidominant mutations in IAA3/SHY2 (shy2-2 and shy2-3) and in IAA7/AXR2 (axr2-1) also alter invariant amino acids in domain II (Tian and Reed, 1999; Nagpal et al., 2000), and it has been shown that the shy2-2 gain-of-function mutation increases the steady-state level of iaa3/shy2-2 protein (Cólon-Carmona et al., 2000).
Further support for the hypothesis that domain II regulates the stability of Aux/IAA proteins comes from the work of Worley et al. (2000). Using chimeric genes consisting of the PS-IAA4/5 or IAA1 open reading frame fused to the LUC reporter coding sequence, these authors show that the N-terminal region of the Aux/IAA proteins can serve as a transferable signal to decrease the half-life of LUC in transgenic plants or in transient assays. If LUC is fused to domain II of PS-IAA6 containing the iaa17/axr3-1 mutation, however, the LUC half-life is increased 50-fold. The differences in the degree of stabilization may be due to a number of differences in experimental design, including the use of a fusion protein, the use of protein domains, the stability of LUC alone, and the level of LUC expression. Nevertheless, these results suggest that domain II functions to mediate the short half-lives of the Aux/IAA proteins. How the mutations in domain II alter interaction with the machinery that targets the Aux/IAA proteins for rapid proteolysis is unknown.
The hypothesis that protein turnover may be critical for controlling auxin-induced gene expression is supported by several lines of direct and indirect evidence. First, early auxin-responsive genes are induced by blocking de novo protein synthesis with cycloheximide, suggesting the presence of a short-lived nuclear repressor(s) of auxin-inducible gene expression (Ballas et al., 1993, 1995; Koshiba et al., 1995). Second, the cloning of the auxin-resistant mutant axr1 suggested that this gene product is involved in the targeting of select proteins for ubiquitin-mediated degradation (Leyser et al., 1993). Subsequent studies have shown that AXR1 forms a heterodimer with ECR1 and that this complex is capable of activating the ubiquitin-like RUB polypeptide (del Pozo et al., 1998). Finally, cloning of the auxin transport inhibitor–resistant mutant tir1 further implicated protein turnover in auxin signaling (Ruegger et al., 1998). TIR1 is the F-box component of an E3 ubiquitin–ligase complex known as Skp1•cdc53/cullin•F-boxTIR1 (SCFTIR). In yeast and Drosophila, proteins targeted for ubiquitination and subsequent degradation are first phosphorylated before being recruited by the SCF complexes (Maniatis, 1999). There are several reports implicating kinase cascades in auxin-mediated responses (Kovtun et al., 1998, 2000; Christensen et al., 2000; Mockaitis and Howell, 2000). Although several lines of evidence have emerged concerning the function of ubiquitination in auxin response (Gray and Estelle, 2000), a number of important questions remain to be answered: (1) Do the protein phosphorylation and protein degradation pathways converge on a common effector? (2) Are the activities of these distinct pathways influenced by auxin? (3) Are the Aux/IAA proteins targets of these pathways? (4) Are the ARF proteins targets of these pathways?
The identification of intragenic revertants of iaa17/axr3-1 suggests that the gain-of-function effects of the original mutation are negated by the biochemical properties of the revertant proteins. The Aux/IAA proteins share two highly conserved NLS elements that are necessary and sufficient to mediate nuclear localization (Abel and Theologis, 1995). As expected, both IAA17/AXR3 and iaa17/axr3-1 accumulated in the nucleus (Figure 3). Furthermore, proteins containing the amino acid substitutions that suppress the iaa17/axr3-1 phenotype accumulated in the nucleus. These results exclude the possibility that the iaa17/axr3-1 or revertant phenotypes are simply the result of altering the subcellular localization of the mutant proteins.
The Aux/IAA proteins have the capacity to form homodimers and heterodimers (Kim et al., 1997). Furthermore, the Aux/IAA proteins interact with the ARFs, which are capable of binding the auxin response element found in the pro-moter of auxin-inducible genes (Kim et al., 1997; Ulmasov et al., 1997a, 1999). Because there is no evidence to suggest that different Aux/IAA proteins have discrete affinities for select sets of Aux/IAAs or ARFs, it appears that the quantity of Aux/IAA protein present in the cell is critical for maintaining equilibrium. Even small changes in the level of a protein can have dramatic effects on the cell's state. For instance, in yeast, a 6-fold change in the intracellular concentration of GAL4 causes a 40-fold change in the expression of GAL1, a gene required for galactose utilization (Griggs and Johnston, 1991).
The gain-of-function phenotype of iaa17/axr3-1 plants might be explained by a dominant negative effect. Because the stabilized iaa17/axr3-1 protein retains its ability to homodimerize and heterodimerize, other Aux/IAA or ARF proteins could be titrated away from their normal partners. The disruption of this web of finely tuned and tightly regulated interactions could lead to the ectopic expression or repression of downstream genes. The altered patterns of gene expression would lead in turn to the pleiotropic phenotypes that are observed.
Mutations in domain III of IAA17/AXR3, which contains a predicted βαα motif, eliminate the capacity of IAA17/AXR3 proteins to homodimerize or heterodimerize. Circular dichroism spectroscopy of recombinant peptides corresponding to domain III has shown that this region contains substantial α-helical secondary structure (Morgan et al., 1999). The location of the reverting amino acid change between the β and α1 segments supports the notion that this protein fold is a prerequisite for stable dimer formation.
The iaa17/axr3-1R3 mutant contains a second site change in domain I that affects the ability of the protein to homodimerize. This suggests that domain I may form an additional site of contact for homodimerization. Alternatively, the second site mutation might alter the secondary structure of the protein in such a way that the ability of the degradation machinery to target the protein is restored and the effects on homodimerization are a secondary consequence of this change.
In addition to auxin resistance, the iaa17/axr3-1 plants exhibit altered responses to other hormones. For instance, several of the iaa17/axr3-1 phenotypes, including ectopic expression from the SAUR-AC1 promoter, can be rescued by the application of exogenous cytokinin (Leyser et al., 1996). This finding implies that the novel or misdirected interactions of a stabilized IAA17/AXR3 protein can be reversed by cytokinin-dependent signals. Furthermore, the iaa17/axr3-1 plants show a resistance to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid, suggesting that improper interactions of IAA17/AXR3 also can affect the output of signaling through the well-characterized ethylene signal transduction pathway. Whether these effects represent the convergence of signaling networks or communication between signaling components remains to be determined.
METHODS
Plant Treatments
Surface-sterilized (0.1 g) wild-type Arabidopsis thaliana seed, ecotype Columbia, and iaa17/axr3-1 seed, ecotype Columbia, were plated on medium containing 0.5 × Murashige and Skoog (1962) salts, 1% sucrose, 1 × B5 vitamins, and 0.4% phytagel. After a 3-day stratification period at 4°C in the dark, seed were grown at 25°C under a 16-hr photoperiod for 7 days. The tissue was collected and was either mock treated or treated with 20 μM indoleacetic acid in 50 mL of 0.5 × Murashige and Skoog (1962) salts for 2 hr in the dark with shaking at 100 rpm. After treatment, the tissue was drained, weighed, and frozen at −80°C until processing.
RNA Gel Blot Analysis
Total RNA was extracted from the treated seedlings as described by Carpenter and Simon (1998). Twenty micrograms of total RNA was glyoxalated, separated on a 1% agarose gel in 10 mM NaPO4, pH 7.0, and transferred onto a GeneScreen membrane using 20 × SSPE (1 × SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4). The blot was washed in 6 × SSPE at room temperature for 5 min and then in 20 mM Tris-HCl, pH 8.0, at 65°C for 5 min. The probes were prepared by labeling the cDNA fragments by random priming (Feinberg and Vogelstein, 1983). Hybridization was performed using Church's buffer (250 mM NaPO4, pH 7.4, 7% SDS, and 1 mM EDTA) at 65°C overnight. The blots were washed in 0.1 × SSPE and 0.5% SDS at 65°C for 1 hr and exposed to X-Omat film (Kodak).
Antibody Production and Purification
A rabbit polyclonal antibody was raised (Genosys, The Woodlands, TX) against an internal peptide of 12 amino acids (NNEPANKEGSTT; amino acids 45 to 56) predicted to be highly specific to IAA17/AXR3. The efficiency and specificity of the antibody was determined by immunoblot analysis of Escherichia coli–expressed wild-type and mutant IAA17/AXR3 proteins. For the immunoprecipitation experiments, preimmune antibodies were purified using protein A–Sepharose as described by the manufacturer (Boehringer Mannheim). Anti-IAA17/AXR3 antibodies were purified from the crude serum by (NH4)2SO4 precipitation, dialysis against 1 × PBS (10 mM NaPO4, pH 6.8, and 130 mM NaCl), and affinity purification using a SulfoLink (Pierce, Rockford, IL) column containing the peptide used for immunization.
Pulse–Chase Analysis
The stability of the IAA17/AXR3 protein in wild-type and iaa17/axr3-1 seedlings was assayed by pulse–chase analysis as described previously (Abel et al., 1994; Oeller and Theologis, 1995). For the labeling, 5 g of wild-type or 4 g of iaa17/axr3-1 etiolated seedlings was incubated in the dark for 2 hr in 20 mL of buffer (1 mM citrate, 1 mM Pipes, pH 6.0, 15 mM sucrose, 1 mM KCl, and 50 μg/mL chloramphenicol) containing 5 mCi of 35S-methionine (Trans label; ICN, Costa Mesa, CA). The chase was performed in buffer containing a 1000-fold excess of unlabeled methionine and cysteine. The incubation medium was changed every 5 min during the chase period. The tissue was rinsed, drained, ground in liquid N2, and lyophilized overnight. The proteins were extracted in boiling 1 × SDS buffer (100 mM NaPO4, pH 7.0, 1% SDS, and 1% β-mercaptoethanol) to inactivate endogenous proteases. After removal of the insoluble material by ultracentrifugation at 100,000g for 20 min, the number of trichloroacetic acid–precipitable counts was determined. The immunoprecipitation was performed using an equal number of trichloroacetic acid–precipitable counts and the protein A–purified preimmune serum or the affinity-purified IAA17/AXR3 antibodies according to published procedures (Oeller and Theologis, 1995). The immunoprecipitates were eluted into boiling 1 × SDS buffer and resolved using SDS-PAGE. The gel was stained with Coomassie Brilliant Blue G 250, destained, and dried. Quantification of the amount of radioactivity present in the IAA17/AXR3 bands was performed using the STORM phosphorimager system (Molecular Dynamics, Sunnyvale, CA).
Plasmid Construction and Mutagenesis
The IAA17/AXR3 coding region (GenBank accession number U49073) that was isolated in a yeast two-hybrid screen (Kim et al., 1997) was subcloned into pBluescript SK+ to create pBSIAA17. The different mutations (iaa17/axr3-1, iaa17/axr3-1R2, and iaa17/axr3-1R3) were introduced in this cDNA using the QuikChange method (Stratagene, La Jolla, CA). The sequence of each plasmid was verified by sequencing.
Subcellular Localization
Translational fusions between β-glucuronidase (GUS) and the different IAA17/AXR3 cDNAs were constructed by subcloning into the pRTL2-GUS/NIaΔBam vector after removing the NIaΔBam fragment (Carrington et al., 1991). The plasmids were coated onto gold particles and transformed in onion epidermis by microprojectile bombardment using a PDS1000He device (Bio-Rad, Hercules, CA) as described by Shieh et al. (1993). After a 20-hr recovery period, tissues were stained with 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid in staining buffer (100 mM KPO4, pH 7.4, 1 mM EDTA, 0.5 mM K-ferricyanide, 0.5 mM K-ferrocyanide, and 0.1% Triton X-100) for 15 to 45 min at 37°C. The reaction was stopped with 5% formaldehyde and 5% acetic acid, and the tissues were rinsed with ethanol (75, 50, and 25%) and finally with water. The epidermis was mounted on microscope slides with mounting buffer (0.1 × PBS, 10 mM NaN3, and 90% glycerol) containing 20 μg/mL 4′-6-diamidino-2-phenylindole (DAPI), a DNA-specific dye (Shieh et al., 1993). The GUS and DAPI staining was visualized using light and UV microscopy, respectively.
Yeast Two-Hybrid Analysis
The Saccharomyces cerevisiae strain Y190 (leu2, trp1, his3, ura3) was used for the two-hybrid assays as described previously (Harper et al., 1993; Kim et al., 1997). The wild-type and mutated IAA17/AXR3 cDNAs were cloned in the pGAD.GH and pGBT9.BS vectors to generate GAL4 activation domain (AD) and binding domain (BD) fusions, respectively. The plasmids were transformed into Y190 using the polyethylene glycol-lithium-acetate method (Gietz et al., 1995) and plated onto the appropriate selective medium. Transformants were grown overnight in selective medium, subcultured into YPD medium (0.5% yeast extract, 1% peptone, and 1% dextrose), and grown until the OD600 was 1 to 1.5 before performing β-galactosidase activity assays (Kim et al., 1997). To assess the activation of the HIS3 reporter gene, 50,000 yeast cells expressing the combinations shown in Figures 4 and 6 were spotted on both synthetic complete medium and synthetic complete medium lacking histidine and supplemented with 25 mM 3-aminotriazole. Proteins extracted from the yeast expressing different constructs were analyzed by immunoblotting using the GAL4-TA (C-10) and GAL4(DBD) (RK5C1) monoclonal antibodies that recognize the AD and BD, respectively, of GAL4 (Santa Cruz Biotechnology, Santa Cruz, CA).
In Vitro Multimerization
Constructs expressing (HIS)6-tagged polypeptides encompassing domain III (βαα motif) of IAA17/AXR3 and iaa17/axr3-1R2 were made. The cDNAs encoding domain III of IAA17/AXR3 (amino acids 101 to 154) and iaa17/axr3-1R2 (amino acids 101 to 158) were amplified by polymerase chain reaction and subcloned using a three-way ligation involving the pQE7 and pQE16 vectors (Qiagen, Valencia, CA). The constructs were transformed into the M15[pREP4] Escherichia coli strain (Qiagen). The bacterial strains were grown in Luria-Bertani medium to 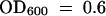 and protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 hr at 30°C. Cells were collected by centrifugation, and the (HIS)6-tagged proteins were purified under denaturing conditions (Qiagen) using a 1-mL column of nickel–nitrilotriacetic acid agarose binding medium. The fractions containing the polypeptides were pooled and dialyzed overnight against 10 mM KPO4, pH 8.0, 100 mM KCl, and 5 mM EDTA using a 3500 MWCO dialysis bag (Spectrum Laboratory, Rancho Dominguez, CA). Polypeptides were concentrated on a Microcon3 spin filter (Millipore, San Jose, CA), and the concentration was determined using the Bio-Rad protein assay kit. The samples were suspended in 1 × SDS sample buffer, heated at 100°C for 5 min, and resolved on a 16.5% Tris–Tricine gel according to Schagger and von Jagow (1987). The gels were fixed, stained with Coomassie Brilliant Blue G 250, and destained as described by Schagger and von Jagow (1987).
and protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 hr at 30°C. Cells were collected by centrifugation, and the (HIS)6-tagged proteins were purified under denaturing conditions (Qiagen) using a 1-mL column of nickel–nitrilotriacetic acid agarose binding medium. The fractions containing the polypeptides were pooled and dialyzed overnight against 10 mM KPO4, pH 8.0, 100 mM KCl, and 5 mM EDTA using a 3500 MWCO dialysis bag (Spectrum Laboratory, Rancho Dominguez, CA). Polypeptides were concentrated on a Microcon3 spin filter (Millipore, San Jose, CA), and the concentration was determined using the Bio-Rad protein assay kit. The samples were suspended in 1 × SDS sample buffer, heated at 100°C for 5 min, and resolved on a 16.5% Tris–Tricine gel according to Schagger and von Jagow (1987). The gels were fixed, stained with Coomassie Brilliant Blue G 250, and destained as described by Schagger and von Jagow (1987).
Acknowledgments
We thank Dr. Ottoline Leyser (York University, UK) for kindly providing the iaa17/axr3-1 seed and Dr. Stanley Fields (University of Washington, Seattle) for the pGAD.GH and pGBT9.BS plasmids. This work was supported by Grant GM-35447 from the National Institutes of Health to A.T.
References
- Abel, S., and Theologis, A. (1995). A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J. 8, 87–96. [DOI] [PubMed] [Google Scholar]
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/ 5- like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Ballas, N., Wong, L.M., and Theologis, A. (1993). Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid–inducible gene, PS-IAA4/5, of pea (Pisum sativum). J. Mol. Biol. 233, 580–596. [DOI] [PubMed] [Google Scholar]
- Ballas, N., Wong, L.M., Ke, M., and Theologis, A. (1995). Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid–inducible gene PS-IAA4/5. Proc. Natl. Acad. Sci. USA 92, 3483–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, C.D., and Simon, A.E. (1998). Preparation of RNA. In Arabidopsis Protocols, J.M. Martínez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 85–89.
- Carrington, J.C., Freed, D.D., and Leinicke, A.J. (1991). Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell 3, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478. [DOI] [PubMed] [Google Scholar]
- Cólon-Carmona, A., Chen, D.L., Yeh, K.-C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- del Pozo, J.C., Timpte, C., Tan, S., Callis, J., and Estelle, M. (1998). The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Schiestl, R.H., Willems, A.R., and Woods, R.A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Griggs, D.W., and Johnston, M. (1991). Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc. Natl. Acad. Sci. USA 88, 8597–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J., Ulmasov, T., and Hagen, G. (1998). The ARF family of transcription factors and their role in plant hormone–responsive transcription. Cell. Mol. Life Sci. 54, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Adami, G.R., Wei, N., Keyomarsi, K., and Elledge, S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin–dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein–protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba, T., Ballas, N., Wong, L.-M., and Theologis, A. (1995). Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum). J. Mol. Biol. 253, 396–413. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Zeng, W., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress–activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M., Lincoln, C.A., Timpte, C., Lammer, D., Turner, J., and Estelle, M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364, 161–164. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. (1999). A ubiquitin ligase complex essential for the NF-B, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 13, 505–510. [DOI] [PubMed] [Google Scholar]
- Mockaitis, K., and Howell, S. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24, 785–796. [DOI] [PubMed] [Google Scholar]
- Morgan, K.E., Zarembinski, T.I., Theologis, A., and Abel, S. (1999). Biochemical characterization of recombinant polypeptides corresponding to the predicted fold in Aux/IAA proteins. FEBS Lett. 454, 283–287. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller, P.W., and Theologis, A. (1995). Induction kinetics of the nuclear proteins encoded by the early indoleacetic acid–inducible genes, PS-IAA4/5 and PS-IAA6, in pea (Pisum sativum L.). Plant J. 7, 37–48. [DOI] [PubMed] [Google Scholar]
- Raumann, B.E., Brown, B.M., and Sauer, R.T. (1994). Major groove DNA recognition by beta-sheets: The ribbon-helix-helix family of gene regulatory proteins. Curr. Opin. Struct. Biol. 4, 36–43. [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Shieh, M.W., Wessler, S.R., and Raikhel, N.V. (1993). Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol. 101, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signaling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]