Abstract
In Arabidopsis, activation of defense responses by flagellin is triggered by the specific recognition of the most conserved domain of flagellin, represented by the peptide flg22, in a process involving the FLS2 gene, which encodes a leucine-rich repeat serine/threonine protein kinase. We show here that the two fls2 mutant alleles, fls2-24 and fls2-17, which were shown previously to confer insensitivity to flg22, also cause impaired flagellin binding. These features are rescued when a functional FLS2 gene is expressed as a transgene in each of the fls2 mutant plants, indicating that FLS2 is necessary for flagellin binding. The point mutation of the fls2-17 allele lies in the kinase domain. A kinase carrying this missense mutation lacked autophosphorylation activity when expressed in Escherichia coli. This indicates that kinase activity is required for binding and probably affects the stability of the flagellin receptor complex. We further show that overexpression of the kinase-associated protein phosphatase (KAPP) in Arabidopsis results in plants that are insensitive to flagellin treatment, and we show reduced flg22 binding in these plants. Furthermore, using the yeast two-hybrid system, we show physical interaction of KAPP with the kinase domain of FLS2. These results suggest that KAPP functions as a negative regulator of the FLS2 signal transduction pathway and that the phosphorylation of FLS2 is necessary for proper binding and signaling of the flagellin receptor complex.
INTRODUCTION
Many biological recognition phenomena involve ligands and receptors. For example, the critical steps of pathogen recognition in plants appear to involve interactions of plant receptors with microbial ligands, the so-called elicitors, and the induction of a phosphorylation cascade that leads to defense responses (Boller, 1995; Yang et al., 1997). One of the most active bacterially derived elicitor molecules in plant cells is flagellin (Felix et al., 1999), which induces rapid defense responses (Gómez-Gómez et al., 1999) and triggers the activation of a phosphorylation cascade (Nühse et al., 2000) in Arabidopsis plants. We recently identified a gene involved in flagellin perception, FLS2, by positional cloning (Gómez-Gómez and Boller, 2000). FLS2 encodes a receptor kinase with an extracellular domain composed of 28 tandem leucine-rich repeats (LRRs) and an intracellular serine/threonine protein kinase domain. Interestingly, the encoded receptor kinase is homologous with the rice resistance gene Xa21, which provides resistance to Xanthomonas oryzae (Song et al., 1995), and the extracellular LRR domain of FLS2 resembles resistance genes from the tomato Cf gene family that provide resistance to various strains of Cladosporium fulvum (Jones et al., 1994; Dixon et al., 1996), suggesting that the recognition pathway for general elicitors such as flagellin may resemble the pathway involved in the highly specific resistance defined by gene-for-gene interactions (Jones and Jones, 1996). In insects and mammals, members of the family of TOLL receptors involved in pathogen recognition also have extracellular LRR domains (Aderem and Ulevitch, 2000). The LRRs form a solvent-exposed parallel β-sheet, which creates a surface that mediates protein–protein interactions (Kobe and Deisenhofer, 1994).
There is precedence from animal systems for the idea that cell membrane serine and threonine kinase–type receptors are composed of several different subunits and that their correct assembly is necessary for the generation of functional receptors (Ebner et al., 1993). From our previous studies, it seems that FLS2 and the product of the FLS1 locus are involved in the formation of the flagellin receptor complex (Gómez-Gómez et al., 1999; Gómez-Gómez and Boller, 2000). The recent characterization of the S-locus receptor kinase (SRK), a receptor-like kinase (RLK) involved in the self-incompatibility response in Brassica spp (Giranton et al., 2000), and the CLAVATA receptor complex involved in meristem determination (Trotochaud et al., 1999) suggests that signaling complexes may be a common feature of receptors of the plant RLK family.
Missense mutations in the kinase domains of CLAVATA1 and other RLKs in plants (reviewed in Torii, 2000) lead to loss of function, confirming the importance of this domain for receptor function. Reversible phosphorylation is a major mechanism for the regulation of intracellular signal transduction pathways. A subset of LRR RLKs is regulated by the kinase-associated protein phosphatase (KAPP) of Arabidopsis (Stone et al., 1994; Braun et al., 1997; Trotochaud et al., 1999). KAPP is considered a downstream regulatory component in RLK signal transduction pathways. It is encoded by a single-copy gene in Arabidopsis and was identified by its binding to the kinase domain of the serine/threonine protein kinase RLK5 (Stone et al., 1994). Given the importance of kinases and phosphatases in pathogen and elicitor signal transduction pathways (Yang et al., 1997), we wanted to determine whether the KAPP protein is associated with FLS2 in the receptor complex and how it affects flagellin binding and action.
Previously, we isolated and identified three mutants representing two independent FLS2 mutant alleles: fls2-24, which has a point mutation in one of the LRR motifs of the extracellular domain; and two mutants, fls2-0 and fls2-17, both of which have the same point mutation in the kinase domain (Gómez-Gómez and Boller, 2000), and we have functionally characterized the flagellin receptor binding site (Z. Bauer, L. Gómez-Gómez, T. Boller, and G. Felix, unpublished data). Here, we show that both alleles fls2-24 and fls2-17 are compromised in flagellin binding and that binding is restored in all transformed fls2 mutant plants carrying the wild-type FLS2 gene. The FLS2 cytoplasmic domain is predicted to contain serine/threonine kinase activity, and we show that the kinase is inactive in the fls2-17 allele. In addition, we show the interaction of KAPP with the kinase domain of FLS2 in a yeast two-hybrid assay. This physical association, together with the unresponsiveness and the reduced binding to flagellin in KAPP-overexpressing plants, supports a model in which FLS2 exists in a dephosphorylated state in these KAPP-overexpressing plants and emphasizes the importance of phosphorylation in the formation of the flagellin receptor complex.
RESULTS
FLS2 Restores flg22 Responsiveness and Flagellin Binding in the fls2-24 Allele
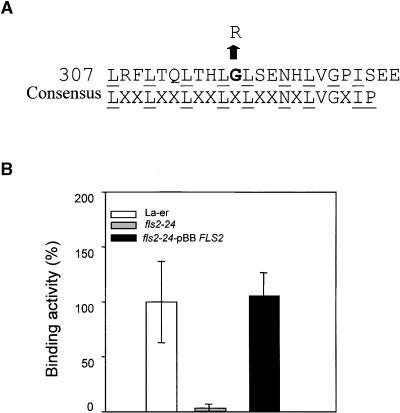
Arabidopsis cells have a high-affinity binding site for flg22 at the plasma membrane (Z. Bauer, L. Gómez-Gómez, T. Boller, and G. Felix, unpublished data). The fls2 mutant alleles were selected by their insensitivity to flg22 (Gómez-Gómez and Boller, 2000). The fls2-24 allele contains a mutation in the 10th LRR domain that does not affect any of the conserved amino acids of the LRR domain (Figure 1A) that are postulated to play a structural role (Kobe and Deisenhofer, 1994). Thus, the point mutation in the fls2-24 allele might disrupt flg22 binding or the interaction with other components of the flagellin receptor complex. In contrast to the Landsberg erecta (Ler) wild-type plants, in all fls2-24 plants tested, the specific binding was below the limit of detection (Figure 1B). To ensure that FLS2 was responsible for the phenotype, we performed complementation experiments. In transgenic fls2-24 plants expressing the wild-type FLS2 gene (designated fls2-24-pBBFLS2), which respond to flagellin in a wild-type manner (Gómez-Gómez and Boller, 2000), flg22 binding was restored (Figure 1B).
Figure 1.
A Point Mutation in the Extracellular Domain of FLS2 Abolishes flg22 Binding.
(A) The derived amino acid sequence of the 10th LRR domain of FLS2 starting with amino acid 307 is shown, along with the point mutation of the fls2-24 allele (G-to-A base change), which results in a change of glycine to arginine, shown in boldface type. The conserved residues of the consensus sequence of the LRR domain are underlined.
(B) Binding activity of 125I-Tyr-flg22 in total extracts of wild-type FLS2 Ler plants, of mutant plants carrying the fls2-24 allele, and in the complemented line fls2-24-pBBFLS2. The mutant and the complemented plants were not measured concomitantly; therefore, the average specific binding of wild-type Ler, which was always measured in parallel, was set as 100%, which corresponded to 4000 and 5100 cpm, respectively. Error bars indicate standard deviation.
The Kinase Catalytic Domain of FLS2 Is Essential for flg22 Binding
The fls2-17 allele has a mutation in one of the 15 invariant residues conserved among all known members of the receptor kinase family in plants (Hanks et al., 1988). The conserved glycine in the IX domain is changed to arginine (Figure 2A). Mutations in this position also have been found in CLV1 alleles (Clark et al., 1997), in the RLK BRI1 involved in brassinosteroid sensitivity (Li and Chory, 1997), and in other serine/threonine protein kinases (Liu et al., 2000). Substitutions of this amino acid result in loss of kinase activity (Hanks et al., 1988) and abolish autophosphorylation activity (Liu et al., 2000; Sessa et al., 2000).
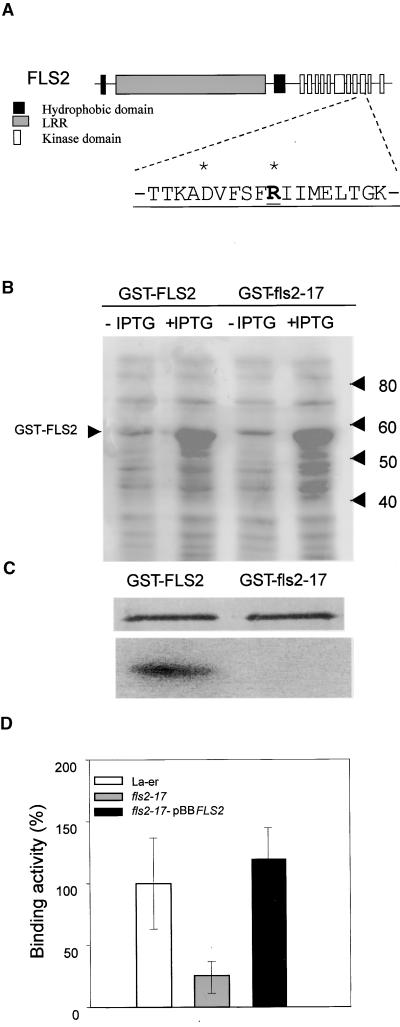
Figure 2.
A Point Mutation in the Kinase Domain of FLS2 Abolishes Kinase Activity and Strongly Reduces Flagellin Binding.
(A) The domain structure of FLS2 is shown schematically with the amino acid sequence of kinase domain IX expanded underneath. Residues conserved in several LRR receptor-like kinases in plants are underlined (Li and Chory, 1997), with asterisks at residues that are present in all kinases. The point mutation in the fls2-17 allele that results in a change of glycine 1064 to arginine (G-to-A base change) is shown in boldface type.
(B) Expression of recombinant proteins consisting of GST fused to the kinase domains of FLS2 and fls2-17 in E. coli. Total proteins were isolated from bacterial cells before and after isopropyl β-d-thiogalactopyranoside (IPTG) induction and resolved by SDS-PAGE. Molecular mass standards in kilodaltons are indicated at right. The position of the FLS2 recombinant protein is indicated at left.
(C) Affinity-purified recombinant fusion proteins (0.5 μg) were subjected to an autophosphorylation reaction. The silver stained SDS–polyacrylamide gel and the corresponding autoradiogram are shown.
(D) Average binding activity of 125I-Tyr-flg22 in homogenates of seven wild-type FLS2 Ler plants, seven fls2-17 plants, and seven plants from the complemented line fls2-17-pBBFLS2. The error bars represent standard deviation values. The mutant and complemented plants were not measured concomitantly; therefore, the average specific binding of wild-type Ler, which was always measured in parallel, was set as 100%, which corresponded to 4000 and 4800 cpm, respectively.
To assess the role of the kinase domain in flg22 binding complex formation, we first determined whether FLS2 encodes a functional protein kinase. The FLS2 kinase domain fused to glutathione S-transferase (GST; GST-FLS2) expressed in Escherichia coli can be autophosphorylated (Figures 2B and 2C). In the same construct, the fls2-17 mutation abolished autophosphorylation (Figure 2C). Because fls2-17 is recessive (Gómez-Gómez and Boller, 2000), the results show that kinase activity is required for FLS2 function.
Although mutations in the intracellular domain of a transmembrane receptor might affect several processes, such as receptor homodimerization and heterodimerization, interaction with other proteins, receptor stability, and enzymatic activity, the insensitivity to flagellin observed in the fls2-17 mutant plants was proposed previously to be due to the lack of activation of downstream components of the signal transduction pathway (Gómez-Gómez and Boller, 2000). However, binding assays in fls2-17 plants revealed a strong reduction in binding activity, although some specific binding (∼20%) was detectable (Figure 2D). In the complemented mutant line fls2-17-pBBFLS2, which reacts normally to flg22 (Gómez-Gómez and Boller, 2000), flagellin binding was restored fully (Figure 2D).
The reduced binding observed in the mutant plants suggests that the cytoplasmic kinase function of FLS2 is required not only for downstream signaling but also for ligand interaction. It is possible that autophosphorylation of FLS2 results in a conformational change of the extracellular LRR domain that allows flagellin binding or is involved in the stability of the flagellin receptor complex. The involvement of receptor phosphorylation in the formation of active receptor complexes has been studied extensively in the epidermal growth factor receptor (Lin et al., 1986) and transforming growth factor receptors (Luo and Lodish, 1997), in which activation requires the phosphorylation of a regulatory segment known as the GS region, which is located upstream of the serine/threonine kinase domain in the cytoplasmic portion of the receptor (Piek et al., 1999). Recently, it has been shown that active kinase is necessary for the assembly of the CLAVATA complex and for CLV3 binding (Trotochaud et al., 2000).
Involvement of KAPP in the Flagellin FLS2 Binding Complex
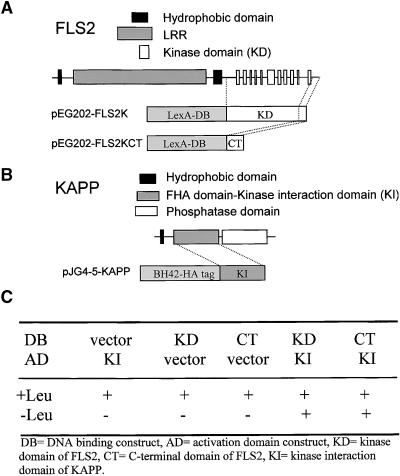
Protein phosphatases play a central role in a variety of signal transduction pathways in plants (Luan, 1998). Several phosphorylated Arabidopsis RLKs have been shown to interact with KAPP. Because of the involvement of phosphorylation in flg22 binding, we decided to determine whether the Arabidopsis FLS2 and KAPP proteins can associate physically by using an LexA version of the yeast two-hybrid assay. Sequence analyses showed that KAPP contains three different functional domains: an N-terminal type I membrane anchor; a kinase interaction (KI) domain containing a 52-residue region that shares sequence identity with FHA domains (Hofmann and Bucher, 1995) and functions as a phosphoprotein binding domain (Li et al., 2000); and a C-terminal protein phosphatase type 2C domain. For the two-hybrid assay, the following protein fusions were expressed simultaneously in the yeast strain EGY-48 containing the reporter plasmid pSH18-34: LexA (DNA binding [DB]) with either the FLS2-KD (pEG202-FLS2K) or the last 60 amino acids from the C-terminal region of FLS2; FLS2-CKD (pEG202-FLS2KCT) in the fusion plasmid pEG202; and B42 (activation domain [AD]) fused to the KI region of KAPP, containing the FHA domain, in the fusion plasmid pJG4-5 (pJG4-5-KAPP). The FLS2 and KAPP protein domains are depicted in Figures 3A and 3B.
Figure 3.
Interaction between the Kinase Domain of FLS2 and the FHA Domain of KAPP in the Yeast Two-Hybrid System.
(A) The diagram depicts the FLS2 protein domains and the fusions with the DB domain used in this two-hybrid assay.
(B) The diagram depicts the KAPP protein domains and the fusion with the AD used in this two-hybrid assay.
(C) Transformed yeast cells were grown at 30°C for 3 days on galactose medium containing (+) or lacking (−) leucine (Leu). Positive interactions between the constructs allowed yeast to grow in a medium lacking leucine.
The detection of protein interactions was based on the expression of the chromosomally integrated reporter gene LEU2, which allows growth in the absence of leucine. We found that yeast transformants containing either of the DB-FLS2 constructs together with the AD-KAPP construct were Leu+ (leucine prototrophs), indicating that the kinase region of FLS2 can interact with the KI domain of KAPP (Figure 3C). The interaction between pEG202-FLS2KCT and pJG4-5-KAPP fusion proteins indicated the presence of critical residues in the C-terminal domain of FLS2 for the interaction with KAPP. The interaction was specific because transformants expressing the DB-FLS2 fusion and AD alone or AD fused to the KI domain and DB alone were Leu− (leucine auxotrophs) (Figure 3C).
The involvement of KAPP in flagellin perception and signaling was assayed in transgenic plants overexpressing the KAPP protein (Williams et al., 1997). Overexpression of KAPP mimicked the fls2 mutant phenotype in terms of lack of growth inhibition and lack of production of reactive oxygen species in response to treatment with 1μM flg22 (Figures 4A and 4B). These results suggest that KAPP functions as a negative regulator of the flagellin signal transduction pathway. We also tested the capacity of these plants to bind flg22. In all of the plants tested, a strong reduction of flagellin binding was observed (Figure 4C). The level of binding was comparable to that obtained for the fls2-17 plants, reinforcing the finding that phosphorylation of FLS2 is required for the proper formation of the flagellin binding core and suggesting that the continuous presence of the KAPP protein maintains the FLS2 kinase domain in a dephosphorylated and inactive state. Examples of this kind of regulation also have been found in several mammal receptors (Suarez-Pestana et al., 1999; Dadke et al., 2000), in which the increase in phosphatase expression antagonizes ligand action by catalyzing the dephosphorylation of the corresponding receptor.
Figure 4.
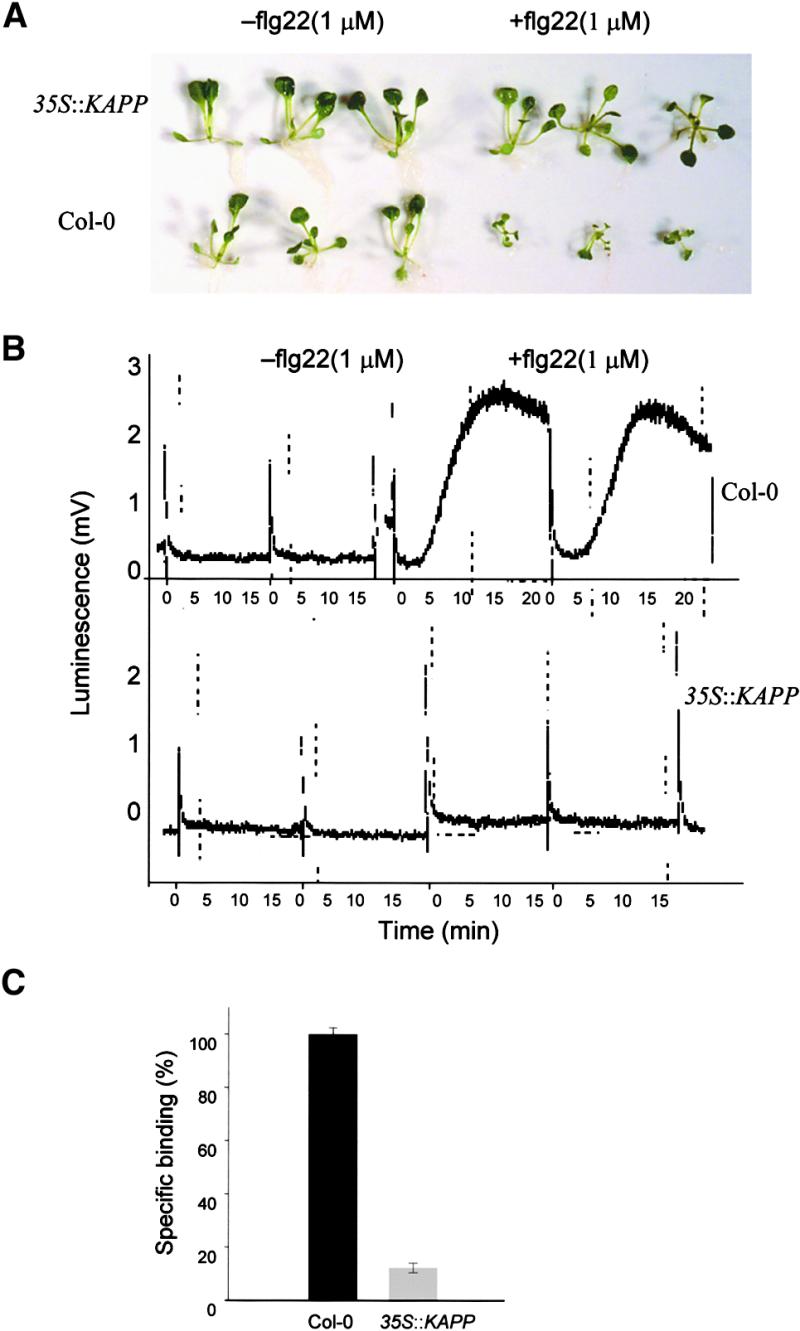
Flagellin Binding and Signaling Are Suppressed in KAPP-Overexpressing Plants.
(A) KAPP-overexpressing seedlings and the corresponding wild-type seedlings were left untreated (−) or treated (+) with 1 μM flg22 for 10 days.
(B) Leaf pieces of wild-type and KAPP-overexpressing plants were examined for the induction of an oxidative burst by 1 μM flg22 in a luminol-based assay.
(C) Average binding activity of 125I-Tyr-flg22 in homogenates of seven wild-type and seven KAPP-overexpressing plants. One hundred percent is equivalent to 8000 cpm. Error bars represent standard deviation.
35S::KAPP, KAPP-overexpressing seedlings; Col-0, wild-type seedlings.
DISCUSSION
Receptor kinases play a key role in important cellular processes. Three functional domains are commonly associated with these proteins: an extracellular domain, a transmembrane domain, and an intracellular catalytic domain. In this study, we demonstrate that FLS2 is localized at the plasma membrane. Both an intact extracellular LRR domain and an intact kinase domain of FLS2 are necessary for high-affinity binding and binding specificity for flagellin. We have shown previously that FLS2 triggered the flg22-dependent responses in stably transformed fls2 mutant plants and that this interaction is very specific; truncated flg22 peptides and the derived Agrobacterium and Rhizobium flagellin peptides were unable to activate flg22-induced responses. Here, we show that the absence of response to flg22 in fls2 mutant plants correlates with an impairment of these mutant alleles in flg22 binding. We also show that both the extracellular and cytoplasmic domains of the FLS2 protein are required for flagellin binding.
The extracellular domains of many receptors are members of the LRR protein superfamily and are responsible for the high-affinity binding of peptides. The predicted site for this interaction is the solvent-exposed parallel β-sheet (Kobe and Deisenhofer, 1994). The LxxLxLxx domain (x is any amino acid) within the LRR lies on the solvent face of the protein with the Leu facing away from the solvent face (Jones and Jones, 1996). The role of variable amino acids (x) is to confer specificity of protein binding to the LRR (Braun et al., 1991; Parniske et al., 1997). The fls2-24 allele has a missense mutation in a variable position in this solvent face domain, indicating that this residue is involved directly in flagellin recognition. On the basis of the results of amino acid substitutions and domain swaps in plant resistance genes encoding LRR-type receptors, it is likely that these LRR domains are responsible for the recognition of the pathogen-derived ligands (Ellis et al., 2000). For instance, experiments at the L locus of flax indicate that important determinants of specificity reside in the LRR domain (Ellis et al., 1999). Similarly, differences in the LRR domains of the Cf gene products are thought to be responsible for ligand binding specificity (Dixon et al., 1996). More recently, direct interaction has been demonstrated between the LRR domains from the resistance gene Pi-ta, which is involved in resistance to strains of the rice blast fungus Magnaporthe grisea (Jia et al., 2000), and the RPS2 gene of Arabidopsis, which is involved in resistance to strains of Pseudomonas syringae that express the avrRpt2 gene (Kunkel et al., 1993), and their corresponding elicitors AvrPi-ta and AvrRpt2, respectively (Jia et al., 2000; Leister and Katagiri, 2000). Thus, there is strong evidence for the involvement of the LRR domains of the defense genes in specific ligand recognition and binding, consistent with their proposed receptor function.
Autophosphorylation has been recognized as an important regulatory mechanism for protein kinases. Autophosphorylation on tyrosine, serine, or threonine residues often induces critical structural changes around the catalytic center of the kinase that result in the activation of kinase activity. This mechanism is used in the activation of the kinase of several receptor kinases and protein kinases and also influences the duration of activation and signaling (Goldsmith and Cobb, 1994; Torii, 2000). The results obtained with the fls2-17 allele provide direct evidence that protein phosphorylation not only is required for receptor activation and signal transduction after ligand binding but also is involved in flagellin recognition and therefore is necessary for the proper assembly of a functional receptor complex. The autophosphorylation experiment confirms the predicted activity of the kinase domain of the FLS2 protein. This kinase domain is not functional in the fls2-17 allele. The kinase domain of FLS2 contains several putative phosphorylation sites, as found in other receptor kinases (Hardie, 1999). By analogy with other protein kinases, phosphorylation at different positions in the kinase domain of FLS2 could control binding and signaling in the flagellin binding complex. The binding data and the KAPP studies indicate that phosphorylation of the FLS2 kinase domain is required for the proper formation of the flagellin binding complex. The phosphorylated residues could induce a conformational change for interaction with other receptor-like molecules or generate docking sites for other components involved in the formation of the flagellin binding core. When flagellin binds, phosphorylation in other residues might upregulate the FLS2 kinase activity, perhaps enhancing the phosphorylation state of subsequent targets in the signal transduction pathway.
Models proposed for plant RLK complexes characterized to date are analogous to that proposed for the transforming growth factor receptors (Torii, 2000). In one model, SRK would function as a type II receptor, with the role of SRK being the recruitment (and perhaps activation) of a second membrane-anchored coreceptor (Giranton et al., 2000). In the case of the CLAVATA complex, several components of the receptor complex have been identified, including the ligand of CLV1, the CLV3 protein (Fletcher et al., 1999), and CLV2, an LRR receptor–like molecule that lacks the kinase domain (Jeong et al., 1999). The Pto receptor complex of tomato, which is involved in resistance to the bacterial pathogen P. syringae pv tomato carrying the avrPto gene (Scofield et al., 1996; Tang et al., 1996), acts at the intracellular level and involves the Pto serine/threonine kinase, the LRR protein Prf, and the bacterially derived ligand avrPto (Bent, 1996). Therefore, signaling through a heteromeric serine/threonine kinase receptor complex might be a universal mechanism present in both the animal and plant kingdoms.
In summary, these studies demonstrate that the FLS2 protein is part of the flagellin binding complex and that phosphorylation plays a key role in the assembly of this complex. Receptors that are composed of multiple protein component complexes permit a tight regulation of cellular activation through a variety of mechanisms. It is not surprising, therefore, that receptors that regulate the activation of defense responses might have similar degrees of complexity. Certainly, additional proteins also are involved in the flagellin receptor complex, a likely candidate being the product of the FLS1 locus (Gómez-Gómez et al., 1999), which is necessary for flagellin signaling (Gómez-Gómez et al., 1999) and binding (Z. Bauer, L. Gómez-Gómez, T. Boller, and G. Felix, unpublished data). Some proteins associated with this complex may be specific for the flg22-signaling pathway, and others may interact with other RLKs as well. Characterization of these other components and their interaction with FLS1 and FLS2 will help to further our understanding of flagellin signaling in the activation of defense responses.
METHODS
Plant Material
Arabidopsis thaliana plants (ecotypes Columbia and Landsberg erecta [Ler]) were germinated and grown as described (Gómez-Gómez et al., 1999). Dr. E. Meyerowitz (Division of Biology, California Institute of Technology, Pasadena) kindly provided kinase-associated protein phosphatase (KAPP)–overexpressing lines in the Columbia background (Williams et al., 1997). The complemented mutant lines fls2-17-pBBFLS2 and fls2-24-pBBFLS2 have been described (Gómez-Gómez and Boller, 2000).
Binding Studies
Binding studies were performed as described (Z. Bauer, L. Gómez-Gómez, T. Boller, and G. Felix, unpublished data). Briefly, homogenates were prepared from individual plants. Aliquots containing 0.2 mg of protein were incubated with 0.6 nM 125I-Tyr-flg22 and with (nonspecific binding) or without (total binding) 10 μM unlabeled flg22. After incubation for 20 min on ice, free label was separated from bound label by vacuum filtration. Radioactivity retained on the filters was determined by γ-counting. Specific binding was calculated by subtracting nonspecific binding from total binding.
Protein Expression
To produce bacterially expressed recombinant proteins, we amplified the kinase region of the FLS2 and fls2-17 cDNAs from amino acid 840 with Pfu polymerase (Stratagene) using the forward primer 5′-GACAAGGGATCGCACTAGCGAAACAGAG-3′, containing a BamHI site, and the reverse primer 5′-TCAACCTTATATAAAACAGAA-ATACA-3′. The polymerase chain reaction (PCR) products were digested with the restriction enzyme BamHI, cloned in frame with glutathione S-transferase (GST) into the BamHI and SmaI sites of pGEX-2TK (Pharmacia Biotech), and transformed into Escherichia coli DH5α. The resulting clones were sequenced to ensure in-frame fusion of KD-FLS2 with GST and to avoid clones that contained PCR-introduced mutations. Following the protocol of the manufacturer (Pharmacia Biotech), E.coli cultures were induced with 1 mM isopropyl β-d-thiogalactopyranoside, pelleted, and resuspended in PBS buffer before lysis by sonication. The cell lysate was cleared by centrifugation at 15,000g, and the fusion proteins were purified from the supernatant using glutatione–Sepharose beads (Pharmacia).
Autophosphorylation
Autophosphorylation experiments were performed essentially as described previously (Stone et al., 1998). A total of 0.5 μg of affinity-purified recombinant fusion proteins was incubated with 1 μCi of γ-32P-ATP in kinase buffer for 15 min at room temperature. The reaction was stopped by adding 5 × SDS sample buffer and analyzed by 10% SDS-PAGE and autoradiography.
Flagellin Treatments and Oxidative Burst Measurements
The treatments with the flg22 peptide and the oxidative burst experiments were performed as described (Gómez- Gómez et al., 1999).
DNA Manipulations and PCR Reactions
Standard protocols were used for PCR reactions, DNA cloning, plasmid isolation, restriction enzyme digestion, and DNA analysis (Ausubel et al., 1994).
Two-Hybrid Assay
Protein fusions to the bacterial repressor LexA DNA binding (DB) domain were made using the plasmid pEG202-NLS, which contains the SV40 nuclear localization sequence between LexA and the polylinker, and protein fusions to the activation domain (AD) B42 were made using the plasmid pJG4-5. DNA fragments were amplified by PCR from cDNA clones using primers designed to incorporate restriction sites in the correct reading frame. All subcloned PCR products were verified by sequencing. The kinase domain of FLS2, corresponding to amino acids 870 to 1150 (GenBank accession number AB010698) was used to make the kinase domain and C-terminal domain constructs. The primers used to generate the kinase domain sequence of FLS2 were as follows: forward primer, 5′-TCTTGTCCTGCTTCTTGTTCTG-3′, and reverse primer, 5′-TAGGTCGACAACTTCTCGATCCTCGTTAC-3′. For the C-terminal domain construct, the following primers were used: forward primer, 5′-TAGGGA-TCCTCTATTGTTTCTCTGAAACA-3′, and reverse primer, 5′-CAGGTG-CACGAACTTCTCGATCCTCGTTACG-3′. The kinase interaction (KI) domain (FHA; amino acids 50 to 337) of the KAPP cDNA (Li et al., 1999) was used to determine the interaction with the FLS2 constructs. The primers used to generate the FHA fragment were as follows: forward primer, 5′-CGGAATTCGGGATTTGCAGAGACCA-3′, and reverse primer, 5′-TCGGATCCGGAAGTATCGAAATCTA-3′.
The yeast strain EGY48 and plasmids pEG202-NLS, pSH18-34, and pJG4-5 were used in these studies. The partial genotype of EGY48 is MATa trp1 his3 ura3 leu2::4 LexAop-LEU2. The yeast strain was transformed serially with the reporter plasmid pSH18-34 and then with the LexA fusion and B42 fusion using a lithium acetate–based protocol (Gietz et al., 1992). Standard media were used for growth, glucose was used as the carbon source for transformation, and 1% raffinose and 2% galactose were used as the carbon sources for the two-hybrid assay. The ability to drive the expression of the yeast LEU2 reporter gene was tested by growing transformants on selective medium lacking tryptophan, leucine, uracil, and histidine.
Acknowledgments
We thank F. Fischer, P. Müller, and H. Angliker (Friedrich Miescher-Institute) for the synthesis of various peptides and oligonucleotides and for DNA sequencing. We thank Drs. M. Collinge and H. Rothnie (Friedrich Miescher-Institute) for critical reading of the manuscript. This work was supported in part by Swiss National Science Foundation Grant No. 31-056815.99.
References
- Aderem, A., and Ulevitch, R.J. (2000). Toll-like receptors in the induction of the innate immune response. Nature 406 782–787. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates/Wiley Interscience).
- Bent, A.F. (1996). Plant disease resistance genes—Function meets structure. Plant Cell 8 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. (1995). Chemoperception of microbial signals in plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 189–214. [Google Scholar]
- Braun, D.M., Stone, J.M., and Walker, J.C. (1997). Interaction of the maize and Arabidopsis kinase interaction domains with a subset of receptor-like protein kinases: Implications for transmembrane signalling in plants. Plant J. 12 83–95. [DOI] [PubMed] [Google Scholar]
- Braun, T., Schofield, P.R., and Sprengel, R. (1991). Amino-terminal leucine-rich repeats in gonadotropin receptors determine hormone selectivity. EMBO J. 10 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585. [DOI] [PubMed] [Google Scholar]
- Dadke, S.S., Li, H.C., Kusari, A.B., Begum, N., and Kusari, J. (2000). Elevated expression and activity of protein-tyrosine phosphatase 1B in skeletal muscle of insulin-resistant type II diabetic Goto-Kakizaki rats. Biochem. Biophys. Res. Commun. 274 583–589. [DOI] [PubMed] [Google Scholar]
- Dixon, M.S., Jones, D.A., Keddie, J.S., Thomas, C.M., Harrison, K., and Jones, J.D.G. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84 451–459. [DOI] [PubMed] [Google Scholar]
- Ebner, R., Chen, R.H., Shum, L., Lawle, S., Zioncheck, T.F., Lee, T.F. Lopez, A.R., and Derynck, R. (1993). Cloning of the type I TGF-β receptor and its effect on TGF-β binding to the type II receptor. Science 260 1344–1348. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3 278–284. [DOI] [PubMed] [Google Scholar]
- Felix, G., Duran, J.D., Volko, S., and Boller, T. (1999). Plants recognise bacteria through the most conserved domain of flagellin. Plant J. 18 265–276. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20 1425–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giranton, J.-L., Dumas, C., Cock, J.M., and Gaude, T. (2000). The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc. Natl. Acad. Sci. USA 97 3759–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith, E.J., and Cobb, M.H. (1994). Protein kinases. Curr. Opin. Struct. Biol. 4 833–840. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez, L., and Boller, T. (2000). FLS2: A LRR receptor-like kinase involved in recognition of the flagellin elicitor in Arabidopsis. Mol. Cell 5 1–20. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez, L., Felix, G., and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18 277–284. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., Quinn, A.M., and Hunter, T. (1988). The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 241 42–52. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G. (1999). Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 97–131. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Bucher, P. (1995). The FHA domain: A putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. 20 347–349. [DOI] [PubMed] [Google Scholar]
- Jeong, S., Trotochaud, A.E., and. Clark S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., and Jones, J.D.G. (1996). The roles of leucine rich repeats in plant defences. Adv. Bot. Res. 24 89–167. [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266 789–793. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 19 415–421. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., Bent, A.F., Dahlbeck, D., Innes, R.W., and Staskawicz, B.J. (1993). RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, R.T., and Katagiri, F. (2000). A resistance gene product of the nucleotide binding site leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 22 345–354. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in Brassinosteroid signal transduction. Cell 90 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., Smith, G.P., and Walker, J.C. (1999). Kinase interaction domain of kinase-associated protein phosphatase, a phosphoprotein-binding domain. Proc. Natl. Acad. Sci. USA 96 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Lee, G., Van Doren, S.R., and Walker, J.C. (2000). The FHA domain mediates phosphoprotein interactions. J. Cell Sci. 113 4143–4149. [DOI] [PubMed] [Google Scholar]
- Lin, C.R., Chen, W.S., Lazar, C.S., Carpenter, C.D., Gill, G.N., Evans, R.M., and Rosenfeld, M.G. (1986). Protein kinase phosphorylation at Thr 654 of the unoccupied EGF receptor and EGF binding regulate functional receptor loss by independent mechanisms. Cell 44 839–848. [DOI] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, S. (1998). Protein phosphatases and signaling cascades in higher plants. Trends Plant Sci. 3 271–275. [Google Scholar]
- Luo, K., and Lodish, H.F. (1997). Positive and negative regulation of type II TGF-β receptor signal transduction by autophosphorylation on multiple serine residues. EMBO J. 16 1970–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse, T.S., Peck, S.C., Hirt, H., and Boller, T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275 7521–7526. [DOI] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B., and Jones, J.D. (1997). Novel disease resistance specificities result from se-quence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91 821–832. [DOI] [PubMed] [Google Scholar]
- Piek, E., Heldin, C.-H., and Ten Dijke, P. (1999). Specificity, diversity, and regulation in TGF-β superfamily signaling. FASEB J. 13 2105–2124. [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274 2063–2065. [DOI] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., and Martin, G.B. (2000). Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto–Pto-mediated hypersensitive response. EMBO J. 19 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase–like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., Collinge, M.A., Smith, R.D., Horn, M.A., and Walker, J.C. (1994). Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 266 793–795. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., Trotochaud, A.E., Walker, J.C., and Clark, E. (1998). Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 117 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Pestana, E., Tenev, T., Gross, S., Stoyanov, B., Ogata, M., and Bohmer, F.D. (1999). The transmembrane protein tyrosine phosphatase RPTPsigma modulates signaling of the epidermal growth factor receptor in A431 cells. Oncogene 18 4069–4079. [DOI] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274 2060–2063. [DOI] [PubMed] [Google Scholar]
- Torii, K.U. (2000). Receptor kinase activation and signal transduction in plants: An emerging picture. Curr. Opin. Plant Biol. 3 361–367. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Jeong, S., and Clark, S.E. (2000). CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289 613–617. [DOI] [PubMed] [Google Scholar]
- Williams, R.W., Wilson, J.M., and Meyerowitz, E.M. (1997). A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 94 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11 1621–1639. [DOI] [PubMed] [Google Scholar]